Abstract
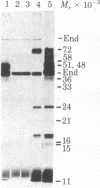
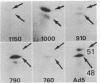
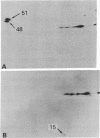
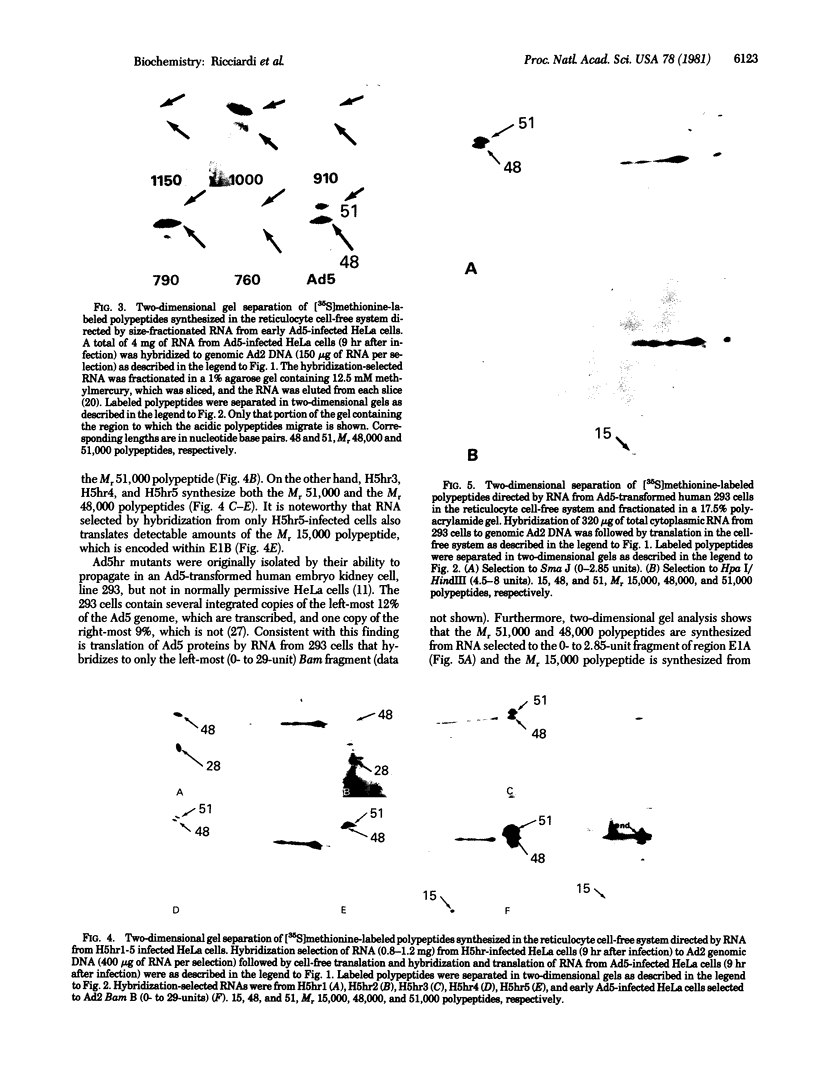
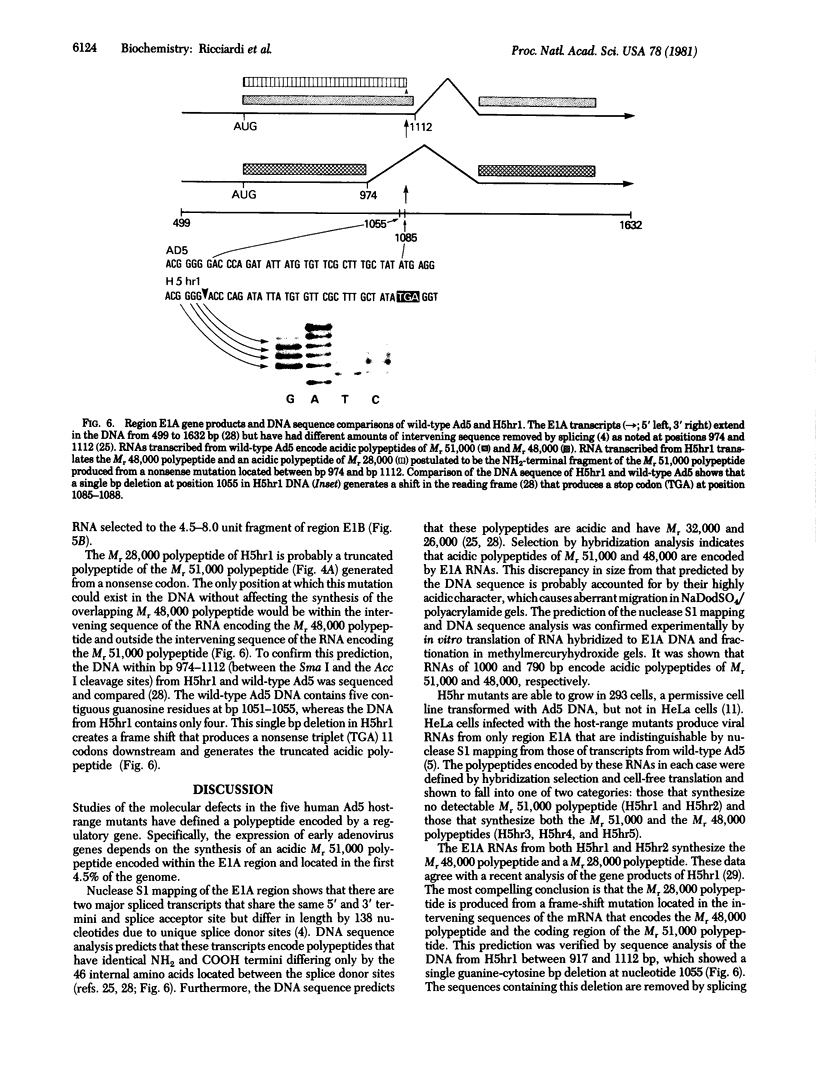
Host-range mutants of adenovirus 5 that contain a defect in region E1A (0-4.5 units) fail to replicate in HeLa cells and to transform rodent cells. In HeLa cells, these mutants synthesize only the two RNAs from E1A that share the same 5' and 3' termini but differ in length by the amount of internal sequence removed by splicing. RNA from wild-type virus, selected by hybridization to DNA from region E1A, translates into polypeptides of Mr 51,000 and 48,000 that are highly acidic in isoelectric focusing gels. These acidic Mr 51,000 and Mr 48,000 polypeptides are encoded by the longer and shorter E1A RNAs, respectively. Two of the host-range mutants, H5hr1 and H5hr2, fail to synthesize the Mr 51,000 polypeptide but do produce the Mr 48,000 polypeptide and a novel polypeptide thought to be a truncated portion of the Mr 51,000 polypeptide. H5hr1 and H5hr2 are hypothesized to have termination codons in sequences found only in RNA encoding the Mr 51,000 polypeptide. This prediction is verified for H5hr1 by DNA sequence analysis. The other three host-range mutants (H5hr3-5) synthesize both acidic polypeptides and are predicted to be missense. These results strongly imply that the Mr 51,000 polypeptide, alone or in combination with the Mr 48,000 polypeptide, is needed to regulate expression of adjacent viral genes during the early phase of adenovirus infection.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiello L., Guilfoyle R., Huebner K., Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293). Virology. 1979 Apr 30;94(2):460–469. doi: 10.1016/0042-6822(79)90476-8. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Birkenmeier C. S. Inhibition of intractable nucleases with ribonucleoside--vanadyl complexes: isolation of messenger ribonucleic acid from resting lymphocytes. Biochemistry. 1979 Nov 13;18(23):5143–5149. doi: 10.1021/bi00590a018. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Lee F., Harrison T., Williams J., Sharp P. A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell. 1979 Aug;17(4):935–944. doi: 10.1016/0092-8674(79)90333-7. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Ultraviolet mapping of the adenovirus 2 early promoters. Cell. 1977 Sep;12(1):45–55. doi: 10.1016/0092-8674(77)90184-2. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Esche H., Mathews M. B., Lewis J. B. Proteins and messenger RNAs of the transforming region of wild-type and mutant adenoviruses. J Mol Biol. 1980 Sep 25;142(3):399–417. doi: 10.1016/0022-2836(80)90279-x. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Sambrook J., Williams J. F., Sharp P. A. Viral nucleic acid sequences in transformed cells. IV. A study of the sequences of adenovirus 5 DNA and RNA in four lines of adenovirus 5-transformed rodent cells using specific fragments of the viral genome. Virology. 1976 Jul 15;72(2):456–470. doi: 10.1016/0042-6822(76)90174-4. [DOI] [PubMed] [Google Scholar]
- Frost E., Williams J. Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology. 1978 Nov;91(1):39–50. doi: 10.1016/0042-6822(78)90353-7. [DOI] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert D. N., Spector D. J., Raskas H. J. In vitro translation products specified by the transforming region of adenovirus type 2. J Virol. 1979 Sep;31(3):621–629. doi: 10.1128/jvi.31.3.621-629.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison T., Graham F., Williams J. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology. 1977 Mar;77(1):319–329. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- Hopper J. E., Broach J. R., Rowe L. B. Regulation of the galactose pathway in Saccharomyces cerevisiae: induction of uridyl transferase mRNA and dependency on GAL4 gene function. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2878–2882. doi: 10.1073/pnas.75.6.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. B., Mathews M. B. Control of adenovirus early gene expression: a class of immediate early products. Cell. 1980 Aug;21(1):303–313. doi: 10.1016/0092-8674(80)90138-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGrogan M., Spector D. J., Goldenberg C. J., Halbert D., Raskas H. J. Purification of specific adenovirus 2 RNAs by preparative hybridization and selective thermal elution. Nucleic Acids Res. 1979 Feb;6(2):593–607. doi: 10.1093/nar/6.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. S., Ricciardi R. P., Roberts B. E., Paterson B. M., Mathews M. B. Arrangement of messenger RNAs and protein coding sequences in the major late transcription unit of adenovirus 2. J Mol Biol. 1980 Oct 5;142(4):455–488. doi: 10.1016/0022-2836(80)90258-2. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Ginsberg H. S., Blanchard J. M., Wilson M. C., Darnell J. E., Jr Regulation of the primary expression of the early adenovirus transcription units. J Virol. 1979 Dec;32(3):727–733. doi: 10.1128/jvi.32.3.727-733.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Perricaudet M., Akusjärvi G., Virtanen A., Pettersson U. Structure of two spliced mRNAs from the transforming region of human subgroup C adenoviruses. Nature. 1979 Oct 25;281(5733):694–696. doi: 10.1038/281694a0. [DOI] [PubMed] [Google Scholar]
- Persson H., Monstein H. J., Akusjärvi G., Philipson L. Adenovirus early gene products may control viral mRNA accumulation and translation in vivo. Cell. 1981 Feb;23(2):485–496. doi: 10.1016/0092-8674(81)90144-6. [DOI] [PubMed] [Google Scholar]
- Pettersson U., Sambrook J. Amount of viral DNA in the genome of cells transformed by adenovirus type 2. J Mol Biol. 1973 Jan;73(1):125–130. doi: 10.1016/0022-2836(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Ricciardi R. P., Miller J. S., Roberts B. E. Purification and mapping of specific mRNAs by hybridization-selection and cell-free translation. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4927–4931. doi: 10.1073/pnas.76.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S. R., Flint S. J., Levine A. J. Identification of the adenovirus early proteins and their genomic map positions. Virology. 1980 Jan 30;100(2):419–432. doi: 10.1016/0042-6822(80)90533-4. [DOI] [PubMed] [Google Scholar]
- Ross S. R., Levine A. J., Galos R. S., Williams J., Shenk T. Early viral proteins in HeLa cells infected with adenovirus type 5 host range mutants. Virology. 1980 Jun;103(2):475–492. doi: 10.1016/0042-6822(80)90205-6. [DOI] [PubMed] [Google Scholar]
- Schrier P. I., Van Den Elsen P. J., Hertoghs J. J., Van Der Eb A. J. Characterization of tumor antigens in cells transformed by fragments of adenovirus type 5 DNA. Virology. 1979 Dec;99(2):372–385. doi: 10.1016/0042-6822(79)90016-3. [DOI] [PubMed] [Google Scholar]
- Shenk T., Jones N., Colby W., Fowlkes D. Functional analysis of adenovirus-5 host-range deletion mutants defective for transformation of rat embryo cells. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):367–375. doi: 10.1101/sqb.1980.044.01.041. [DOI] [PubMed] [Google Scholar]
- Spector D. J., McGrogan M., Raskas H. J. Regulation of the appearance of cytoplasmic RNAs from region 1 of the adenovirus 2 genome. J Mol Biol. 1978 Dec 15;126(3):395–414. doi: 10.1016/0022-2836(78)90048-7. [DOI] [PubMed] [Google Scholar]
- Van Ormondt H., Maat J., De Waard A., Van der Eb A. J. The nucleotide sequence of the transforming HpaI-E fragment of adenovirus type 5 DNA. Gene. 1978 Dec;4(4):309–328. doi: 10.1016/0378-1119(78)90048-3. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. F. Enhancement of adenovirus plaque formation on HeLa cells by magnesium chloride. J Gen Virol. 1970 Dec;9(3):251–255. doi: 10.1099/0022-1317-9-3-251. [DOI] [PubMed] [Google Scholar]