Abstract
A new generation of genetic studies of diabetes is underway. Following from initial genome-wide association (GWA) studies, more recent approaches have used genotyping arrays of more densely spaced markers, imputation of ungenotyped variants based on improved reference haplotype panels, and sequencing of protein-coding exomes and whole genomes. Experimental and statistical advances make possible the identification of novel variants and loci contributing to trait variation and disease risk. Integration of sequence variants with functional analysis is critical to interpreting the consequences of identified variants. We briefly review these methods and technologies and describe how they will continue to expand our understanding of the genetic risk factors and underlying biology of diabetes.
Keywords: genotyping, genome-wide association, sequencing, imputation, exome, genome, fine-mapping, diabetes, quantitative traits, metabochip, single nucleotide polymorphism
Introduction
GWA studies have made progress toward understanding the inherited basis of type 1 and type 2 diabetes by detecting disease-associated DNA variants, usually with allele frequencies greater than 5%. More than 50 genome-wide-significant (P < 5 × 10−8) loci have been described in European, East Asian and South Asian populations [1–5]. The next generation of genome-wide studies is underway, identifying additional variants at both known and novel loci. These newer studies consist of more detailed studies based on high-density genotyping arrays, fine-mapping of known loci, sequencing of whole genomes and whole exomes, and integrating sequence results with functional studies.
The decreasing costs of sequencing both exomes and genomes are enabling lower frequency DNA variants to be identified and tested for association with diabetes and related traits such as fasting glucose. Typically, low frequency refers to minor allele frequencies equal to or greater than 0.5% and less than 5%, and rare refers to allele frequencies less than 0.5%. As more individuals are sequenced, more of these lower frequency and rare variants can be detected. The least frequent variants are private to a family or an individual, and sequencing all individuals in large population-based studies to find more of the rarest variants remains cost-prohibitive. The power to detect the association of a lower frequency variant with a disease or trait depends on the magnitude of its effect. Relative to more common variants in a fixed sample size, low frequency variants need to have stronger effects to be detected.
The 1000 Genomes Project demonstrated the characteristics of variants that will be identified by sequencing genomes [6]. This project was the first to provide a comprehensive resource of human genetic variation based on the genomes of hundreds of people. The data generated include the genome position, allele frequency and local haplotype structure of more than 15 million single nucleotide polymorphisms (SNPs), more than 1 million short insertions and deletions, and more than 20,000 structural variants, most of which had not been described previously. A pilot project focused on exons of 1,000 genes in ~700 samples identified >12,000 SNPs, 70% of which were newly discovered. Of these coding SNPs, 74% had a frequency <1%; this work confirmed that many rare coding variants are population-specific and enriched for functional variants [7]. Sequencing also finds copy number variants that are largely missing from early genome-wide genotyping arrays. Sequencing of 185 genomes identified 22,025 deletions and 6,000 additional structural variants, including insertions and tandem duplications [8]. Based on sequence data, human genomes typically contain approximately 100 genuine loss-of-function variants and ~20 genes that are completely inactivated [9].
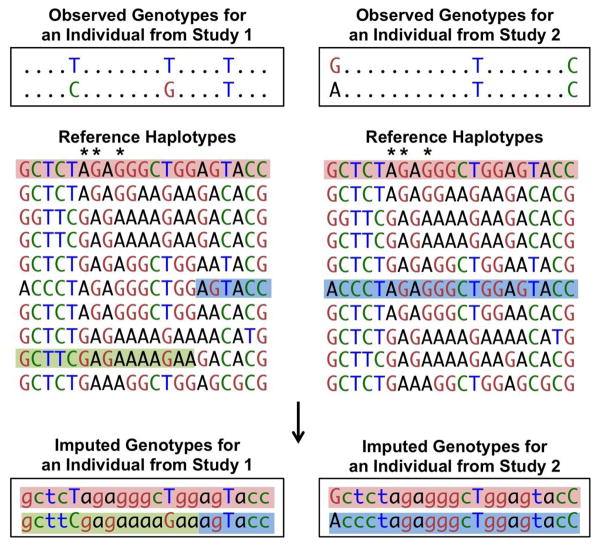
Extending work from the HapMap consortium [10], the 1000 Genomes Project provides the best resource to date for reference information on the patterns of linkage disequilibrium in the human genome [6]. Two variants exhibit linkage disequilibrium when genotypes of one variant are partially or fully correlated with the genotypes of a second variant, usually because the variants are located close enough to each other on a chromosome that no or limited recombination has taken place between them. Two variants exhibit perfect linkage disequilibrium when the genotypes of one variant perfectly predict the genotypes of the second variant, as shown by SNPs with asterisks in Figure 1. Due to linkage disequilibrium, contiguous sets of SNPs exist in many fewer unique haplotypes than randomly assorted alleles. The presence of limited numbers of haplotypes enables ungenotyped alleles to be predicted using imputation methods. Genotype array-based association studies of approximately 300,000 to 1 million variants can represent 61–89% of the common variation in the genome based on a correlation of at least .8 with European ancestry HapMap SNPs. Using imputation, coverage can be extended to 82–95% of the common variation [11].
Figure 1.
The top sequences represent the observed genotypes of two individuals from different studies that used different genome-wide arrays. Different markers are available on the arrays, and in this example, only one shared marker is available for the two individuals. The reference haplotypes contain many more markers (SNPs, and, more recently, insertions or deletions) than the genotyped samples. Reference haplotypes may be from the HapMap Project, the 1000 Genomes Project, or other sequenced or densely genotyped samples. Haplotypes from the reference samples that are consistent with the observed individual haplotypes are highlighted. These reference haplotypes are used to fill in (impute) the unobserved genotypes in the study individuals (bottom). More than one reference haplotype can be consistent with an individual’s phased genotypes. To account for this, imputation programs provide the probability of each genotype. Asterisks indicate markers that are in perfect linkage disequilibrium within the reference panel; an ‘A’ at the first asterisk marker is always observed with ‘G’ at the second asterisk marker and ‘G’ at the third asterisk marker.
For any given cohort, practical considerations influence the choice to genotype and/or sequence the available samples (Table 1). As genotyping and sequencing reagents and technologies become less expensive and more widely available, samples may be subjected to multiple genotyping and sequencing analyses, yielding ever more complete data. Genotyping usually costs less per sample and currently involves easier data processing than sequencing, although data is generated only for variants that were known and included on the genotyping array. The data generated by genome-wide genotyping arrays can be extended to include a much larger set of variants by performing genotype imputation (Fig. 1) [12]. Imputation requires no reagents and thus relatively little cost other than computational time. Sequencing of sample genomes or exomes allows discovery of increasingly less frequent SNPs, insertions and deletions, but is currently expensive both in terms of producing and processing the data. Examples of the next generation of genome-wide studies are described in the following sections.
Table 1.
Characteristics of genotyping and sequencing technologies
| Relative cost | Coding variant coverage | Regulatory variant coverage | |
|---|---|---|---|
| Original GWA array | + | + | + |
| Denser GWA arrays | + | ++ | ++ |
| IBC array | − | selected | selected |
| Metabochip/Immunochip | − | selected | selected |
| Exome chip | − | ++ | − |
| Imputation of untyped variants | − | ++ | ++ |
| Exome sequencing | ++ | +++ | − |
| Low read depth genome sequencing | ++ | ++ | ++ |
| High read depth genome sequencing | +++ | +++ | +++ |
Symbols indicate lower to higher cost and lower to better coverage: −, +, ++, +++.
Dense genotyping arrays
New results are being generated through use of several types of dense genotyping arrays. Variants were selected for these arrays with different strategies or specific goals: more complete coverage of variants across the genome, content focused on candidate genes, follow up of metabolic or autoimmune and inflammatory trait GWA studies, or known variants in the exome. The more focused arrays allow genotyping of 10’s to 100’s of thousands of samples at relatively low cost per sample. They can provide data on known low frequency and rare variants that were not genotyped or able to be imputed with confidence from the original GWA genotype arrays.
One type of higher density genotyping array includes those designed to provide broad coverage of the common and less frequent variation in the genome. Compared to previously available genome-wide arrays, these arrays include more SNPs (>1 million) and greatly increased coverage of low frequency variants. The HumanOmni2.5 BeadChip, for example, includes common and low frequency variants discovered by the sequencing of the 1000 Genomes Project [13].
Other genotyping arrays focus on specific genes or regions of interest. The IBC array contains ~50,000 SNPs focused on ~2,000 cardiovascular, inflammatory and metabolic genes and was designed to capture genetic diversity across populations [14]. A recent report described IBC array variants associated with type 2 diabetes in ~87,000 multi-ethnic population-based samples [15] and meta-analysis with existing GWA data from additional samples. By analyzing larger sample sizes and including individuals with non-European ancestry, two novel loci, GATAD2A/CILP2 and BCL2, were found to be associated with type 2 diabetes at genome-wide significance, and three loci achieved a less stringent threshold of study-wide significance. A low frequency variant (allele frequency 3%) in HNF1A was confirmed to be associated with type 2 diabetes.
Two high-density arrays of ~200,000 SNPs each, termed the Metabochip (formally the CardioMetabochip) and the Immunochip, were designed to cost-effectively test thousands of suggestive GWA signals in additional samples and to enable detailed fine-mapping of selected GWA loci [16–18]. The Metabochip focuses on cardiovascular and metabolic traits and diseases, while the Immunochip focuses on major autoimmune and inflammatory traits and diseases. A large-scale meta-analysis of Metabochip variants in primarily European subjects were recently reported for type 2 diabetes [19]. New loci were identified in or near six loci not previously reported for another metabolic trait (Table 2). Of these, the BCAR1 signal also is genome-wide significant for type 1 diabetes, although risk is conferred by the opposite allele to type 2 diabetes [5]. The type 2 diabetes Metabochip analysis also identified four loci harboring evidence of more than one independently associated variant, near KCNQ1, CDKN2A, DGKB, and MC4R. Among 36 of the previously known GWA loci, low-frequency alleles were found to be associated with a stronger risk of type 2 diabetes only at two loci, PROX1 and KLF14. These variants with frequencies of 2–3% may be responsible for or contribute to the GWA signals at these loci. Although the Metabochip does not contain all low-frequency and rare alleles, the data suggest that the underlying functional variants at most previously discovered GWA loci are common (also see [20]).
Table 2.
Loci identified using Metabochip to be associated with type 2 diabetes, fasting glucose, fasting insulin or 2-hour glucose
| Trait | SNP | Chr | Mb | Gene | Alleles | Freq | Odds ratio (CI)or Effect (SE) | P value |
|---|---|---|---|---|---|---|---|---|
| Type 2 diabetes | rs12571751 | 10 | 80.6 | ZMIZ1 | A/G | .52 | 1.08 (1.05–1.10) | 1.0 × 10−10 |
| Type 2 diabetes | rs516946 | 8 | 41.6 | ANK1 | C/T | .76 | 1.09 (1.06–1.12) | 2.5 × 10−10 |
| Type 2 diabetes | rs10842994 | 12 | 27.9 | KLHDC5 | C/T | .80 | 1.10 (1.06–1.13) | 6.1 × 10−10 |
| Type 2 diabetes | rs2796441 | 9 | 83.5 | TLE1 | G/A | .57 | 1.07 (1.05–1.10) | 5.4 × 10−9 |
| Type 2 diabetes | rs459193 | 5 | 55.8 | ANKRD55 | G/A | .70 | 1.08 (1.05–1.11) | 6.0 × 10−9 |
| Type 2 diabetes | rs7202877 | 16 | 73.8 | BCAR1 | T/G | .89 | 1.12 (1.07–1.16) | 3.5 × 10−8 |
| Fasting glucose | rs16913693 | 9 | 110.7 | IKBKAP | T/G | .97 | .0434 (.007) | 3.5 × 10−11 |
| Fasting glucose | rs3829109 | 9 | 138.4 | DNLZ | G/A | .71 | .0172 (.003) | 1.1 × 10−10 |
| Fasting glucose | rs3783347 | 14 | 99.9 | WARS | G/T | .79 | .0168 (.003) | 1.3 × 10−10 |
| Fasting glucose | rs10747083 | 12 | 131.6 | P2RX2 | A/G | .66 | .0133 (.002) | 7.6 × 10−9 |
| Fasting glucose | rs6072275 | 20 | 39.2 | TOP1 | A/G | .16 | .0159 (.003) | 1.7 × 10−8 |
| Fasting glucose | rs576674 | 13 | 32.5 | KL | G/A | .15 | .0167 (.003) | 2.3 × 10−8 |
| Fasting glucose | rs11715915 | 3 | 49.4 | AMT | C/T | .68 | .0120 (.002) | 4.9 × 10−8 |
| Fasting glucose adjusted for BMI | rs17762454 | 6 | 7.2 | RREB1 | T/C | .26 | .0140 (.002) | 9.6 × 10−9 |
| Fasting glucose adjusted for BMI | rs2657879 | 12 | 55.2 | GLS2 | G/A | .18 | .0157 (.003) | 3.9 × 10−8 |
| Fasting insulin | rs9884482 | 4 | 106.3 | TET2 | C/T | .39 | .0165 (.002) | 1.4 × 10−11 |
| Fasting insulin | rs1167800 | 7 | 75.0 | HIP1 | A/G | .54 | .0156 (.003) | 2.6 × 10−9 |
| Fasting insulin | rs731839 | 19 | 38.6 | PEPD | G/A | .34 | .0145 (.003) | 1.7 × 10−8 |
| Fasting insulin | rs1530559 | 2 | 135.5 | YSK4 | A/G | .52 | .0145 (.003) | 3.4 × 10−8 |
| Fasting insulin adjusted for BMI | rs3822072 | 4 | 90.0 | FAM13A | A/G | .48 | .0116 (.002) | 1.8 × 10−8 |
| 2-hour glucose | rs1019503 | 5 | 96.3 | ERAP2 | A/G | .48 | .0628 (.011) | 8.9 × 10−9 |
The subset of loci shown have not previously been reported to be associated with another metabolic trait at genome-wide significance in Europeans [19, 21].
Chr, chromosome; Mb, megabase position; Gene, a nearby gene; Freq, frequency of the type 2 diabetes risk-increasing or quantitative trait-raising allele, listed first; CI, 95% confidence interval; SE, standard error.
Odds ratios are shown for type 2 diabetes loci. For quantitative traits, effects are shown in units of untransformed fasting glucose level, natural log-transformed fasting insulin level, or untransformed 2-hour glucose level.
A large-scale meta-analysis of Metabochip SNPs in European subjects was also recently reported for glycemic traits [21]. This study identified 19 novel loci for fasting glucose, 15 for fasting insulin and four for glucose levels after an oral glucose load. Of these, 15 were not previously reported for another metabolic trait (Table 2). The known GWA loci were evaluated for more strongly associated variants, an analysis that identified a common promoter SNP at GCK that may be driving the association signal. Further analysis of the high density SNPs is warranted to determine whether additional low-frequency variants can be identified that either contribute to or are independent of the GWA signals.
A fourth type of newly available genotyping array has been designed to facilitate analysis of coding variants in large numbers of subjects [22]. These ‘exome chips’ contain >200,000 SNPs, focused on variants that change the protein sequence. Many variants included on such arrays were only recently identified by exome sequencing studies and have not yet been analyzed for association with diabetes or related traits in very large samples. Analysis of exome chip data from thousands of individuals may identify both novel variants at known loci and novel loci. Compared to GWA analysis, identification of a locus via a low frequency variant that changes the protein sequence offers the potential to jump directly to a candidate variant for functional study.
Imputation
In genotype imputation, the genotypes at untested markers are predicted by inferring haplotypes in the genotyped samples, matching those haplotypes to the most similar ones from reference samples (e.g. from the HapMap [10] or 1000 Genomes Projects [6]), and then recording the allele(s) present in the matching reference sample haplotypes (Fig. 1) [11, 23, 24]. Imputation was developed, in part, to enable combination of results from studies genotyped on different platforms, and thus with different SNP sets (e.g. [25] and [1]). Imputation is now performed using reference panels with ever increasing numbers of variants and is currently being implemented in cross-ancestry meta-analyses.
Recently, imputation has been performed based on reference haplotypes from the 1000 Genomes Project. Using data from the Wellcome Trust Case Control Consortium phase I study of seven diseases including type 1 and type 2 diabetes, 1000 Genomes imputation identified association signals that were not found using the original genotype data or HapMap-based imputation data, one signal within IL2RA for type 1 diabetes and one near CDKN2A for type 2 diabetes [26]. Both signals had been identified by GWA before this proof-of-principle study. This analysis also refined association signals near CUX2 for type 1 diabetes and in IL23R for Crohn’s disease. Subsequent larger type 2 diabetes meta-analyses of data imputed using the 1000 Genomes reference panel [27] and larger reference panels are underway.
Sequence genotype calling methods based on the principles of imputation can also be used to increase accuracy and density of genotypes in individuals with low-depth sequence data [11]. This approach uses genotype information from other sequenced individuals to increase the accuracy of the genotype calls. More accurate calls can be obtained as the number of sequenced individuals increases.
Sequencing of exomes and genomes
While high-density arrays and imputation will allow many low-frequency and rare variants to be studied, the fixed-content chips and imputation reference panels are limited to sequence variants that are already known. The goal of sequencing is to identify further variants that may contribute to disease. Next-generation sequencing using highly parallel technologies has been available for several years, and costs continue to decrease. Exome sequencing currently costs about five-fold less than genome sequencing. However, sequencing a sample still costs 10- to 100-fold more than genotyping a fixed-content array, and substantially more data management is needed.
In recent years, exome sequencing has been used to discover the genes responsible for many monogenic disorders [28]. The exome is enriched for variants with functional consequences that are frequently observed to be responsible for monogenic disorders. Exome sequencing has been used to diagnose MODY in patients in whom disease variants have not yet been identified [29–31]. In one study, three affected relatives and one unaffected relative from a large family were sequenced. Of the 324 variants that were subsequently genotyped in additional family members and controls, only one segregated with disease, Glu227Lys in KCNJ11. These data implicated KCNJ11 as the 13th MODY gene [30].
Exome sequencing also can be used to discover coding variants contributing to complex traits. Given the mutation rate in protein-coding genes, almost every gene is expected to contain variants that affect function, even if the variants are rare [32]. Exome sequencing can be expected to provide insights into complex traits because less comprehensive candidate gene sequencing studies have successfully identified coding variants [33, 34]. Exon sequencing at a type 1 diabetes GWA locus identified four rare variants in IFIH1 that independently lowered diabetes risk and are predicted to alter the expression or structure of the protein [35]. At known MODY gene and GWA locus HNF4A, the low-frequency coding variant HNF4A Thr130Ile shows suggestive evidence (P = 2.1 × 10−5) of influencing type 2 diabetes risk [36]. This variant has been shown to decrease the function of HNF4A in cultured cells [37, 38].
Analysis of rare variants requires statistical methods that differ from methods used to test association with common variants [32]. Typically, to obtain reasonable power, rare variants need to be tested in groups aggregated by gene or other functional units. Several new statistical tests have been designed for rare variants [39]; the tests differ in their power to detect evidence of association based on the number of variants, number of causal variants, allele frequency, effect sizes, and consistency of direction of effect relative to the less common allele.
Whole genome sequencing offers the ultimate opportunity to identify genetic variants for diabetes, including both coding and regulatory variants. High-depth coverage is needed to identify the rarest variants accurately, but remains expensive for large sample sizes. Low-depth coverage sequencing offers a less expensive strategy for identifying genetic variants in larger numbers of individuals [40]. As more individuals are sequenced, more accurate genotype calls can be generated for a given sequence depth. Sequencing 400 individuals at 30× (high) average read depth each requires similar sequencing capacity as sequencing 3000 individuals at 4× (low) average read depth. Both designs detect variants with frequency > 0.5%; the low-depth coverage design detects more variants with frequency 0.2%–0.5%, but fewer of the variants with frequency < 0.2%. At the lower read-depths, genotype accuracy is lower but still very good [40]. In the example of 4× coverage, genotype accuracy of variants with frequencies > 0.1% remains >99.6%.
At least three large exome and genome sequencing projects are ongoing to discover variants influencing type 2 diabetes and related traits. The Go-T2D study is performing low-coverage whole-genome sequencing, deep exome sequencing, and 2.5M SNP array genotyping of 1,425 type 2 diabetes cases and 1,425 controls from Northern Europe [41]. The T2D-GENES Project 1 study is performing exome sequencing of 5,000 type 2 diabetes cases and 5,000 controls from five ancestral groups, and the T2D-GENES Project 2 study is performing deep whole genome sequencing of >500 individuals from 20 large Mexican American pedigrees [42]. These projects will detect many novel low-frequency and rare variants that, when analyzed in sufficiently large numbers of subjects, can be expected to identify new insights into the genetic basis for disease.
Integrating sequence variants with functional studies
Although thousands of variants are being identified by sequencing, many of them novel, determining which variants alter protein or regulatory function remains challenging. Sequencing an individual to understand disease pathophysiology may lead to multiple apparently equivalent putative disease variants. Gene-based tests of association that simultaneously evaluate multiple variants likely will provide sets of variants but not specifically identify the functional variants. Computational approaches to assess deleteriousness are improving, but remain imperfect [43]. Two recent studies functionally tested variants identified by exon sequencing and identified rare variants with strong biological effects and potential clinical significance [44, 45].
In one study, sequencing the exons of melatonin receptor 1B (MTNR1B) in >7,600 individuals identified 40 nonsynonymous variants that change the protein sequence [44]. GWA studies had identified common variants near MTNR1B associated with fasting glucose levels and type 2 diabetes [46, 47]. The 40 variants identified by sequencing included 36 that were very rare, with minor allele frequency less than 0.1%, and not yet present in SNP databases. A pooled analysis of these 36 rare variants showed evidence of association with type 2 diabetes (P = 1.6 × 10−4). Proteins containing each of these variants were individually expressed in human HEK293 cells and evaluated for melatonin binding and three measures of signaling activity. Fourteen of the variants showed a partial or total loss of function, and subsequent tests of association demonstrated that compared to the variants with neutral effects on function, the loss-of-function variants were much more strongly associated with type 2 diabetes. Comparison of experimental results to a bioinformatics prediction of functional consequence showed only 60% concordance, confirming the need for such functional studies of individual variants.
In another recent study, sequencing the exons of glucokinase regulatory protein (GCKR) in 800 individuals identified 19 nonsynonymous variants [45]. GCKR variants have been associated with several metabolic traits, and GCKR is known to play a role in glucose homeostasis [48]. The 19 variants identified include the common Pro446Leu substitution, which has been shown to increase active cytosolic glucokinase [49]. The proteins containing each of the variants were individually evaluated for cellular localization, ability to interact with glucokinase, and kinetic activity. Most of the variants had functional effects consistent with loss-of-function, although two exhibited a potential gain-of-function. Experimental analysis of function will continue to be critical to interpretation of the role of variants identified by sequencing.
Conclusions
How will sequencing genomes influence the health of people at risk for or affected with diabetes? The more complete understanding of the biological mechanisms underlying diabetes derived from these studies may lead to identification of novel drug targets. Individuals with variants in genes responsible for MODY or neonatal diabetes respond better to specific drugs [50, 51], and sequencing may identify small numbers of individuals with combinations of rarer, more highly penetrant variants that respond better to specific therapeutic options. Although sets of known variants for type 2 diabetes do not add substantially to prediction of type 2 diabetes development in the overall population [52, 53], identification of individuals at greater or lower genetic risk for diabetes within the overall population or in specific subgroups, such as younger onset or leaner individuals [54, 55] could lead to better targeted health information and also allow identification of higher risk individuals for more efficient design of clinical trials for disease prevention.
Hundreds if not thousands of additional diabetes loci likely will be identified in the future. These loci will almost certainly be a combination of common variants with modest effects and low frequency to rare variants with a range from very modest to strong effects, although their contribution by frequency currently is not clear [56]. A variety of strategies will lead to their discovery: analysis of increasingly larger numbers of subjects, identification of successively rarer variants via sequencing and imputation, investigation of specialized phenotypes that may provide new insights into diabetes, and improved analysis methods that incorporate functional information. Identification of the underlying causal variants at these loci remains challenging. Identification of new traits or sets of known traits that are influenced by the same genetic variants may provide hints into the biology of specific variants or loci [1, 2, 57]. Identification of overlap between diabetes-associated variants and regulatory elements that influence gene expression, transcription factor binding and methylation in relevant tissues may help narrow the possible sets of causal variants [58–61]. Likewise, tests for association that can directly incorporate functional information by upweighting or advantaging the contribution of specified variants may help identify loci [62–64]. Informed by association results, experimental work in vitro, in cells and with model organisms will be necessary to understand the underlying biological mechanisms and pathophysiology.
Acknowledgments
We acknowledge support from National Institutes of Health grants DK62370, DK72193, DK88389, HG000376, and DK93757.
Footnotes
Disclosure
No potential conflicts of interest relevant to this article were reported.
Contributor Information
Karen L. Mohlke, Email: mohlke@med.unc.edu, 5096 Genetic Medicine, 120 Mason Farm Drive, University of North Carolina, Chapel Hill, NC 27599-7264, USA, Tel: 919-966-2913, Fax: 919-843-0291.
Laura J. Scott, Email: ljst@umich.edu, M4134 SPH II, 1415 Washington Heights, University of Michigan, Ann Arbor, MI 48109-2029, USA, Tel: 734-763-0006, Fax: 734-763-2215.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kooner JS, Saleheen D, Sim X, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.1000 Genomes Project Consortium: A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. Provided a comprehensive resource on human genetic variation based on the genomes of hundreds of people. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marth GT, Yu F, Indap AR, et al. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12:R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills RE, Walter K, Stewart C, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.MacArthur DG, Balasubramanian S, Frankish A, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. Evaluated loss-of-function variants from 185 human genomes and determined their prevalence and properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Willer CJ, Ding J, et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 13. [Accessed July 2012.];HumanOmni2.5S Data Sheet. Available at http://www.illumina.com/documents/products/datasheets/datasheetomni25S.pdf.
- 14.Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saxena R, Elbers CC, Guo Y, et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90:410–425. doi: 10.1016/j.ajhg.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthr Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buyske S, Wu Y, Carty CL, et al. Evaluation of the metabochip genotyping array in African Americans and implications for fine mapping of GWAS-identified loci: the PAGE study. PLoS One. 2012;7:e35651. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voight BF, Kang HM, Ding J, et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012 doi: 10.1038/ng.2383. In press Used the metabochip to perform the largest yet meta-analysis of type 2 diabetes and identified novel loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan W, Boehnke M, Pluzhnikov A, et al. Identifying plausible genetic models based on association and linkage results: Application to type 2 diabetes. Genet Epidemiol. 2012 doi: 10.1002/gepi.21668. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Scott RA, Lagou V, Welch RP, et al. Large-scale association study using the Metabochip array reveals new loci influencing glycemic traits and provides insight into the underlying biological pathways. Nat Genet. 2012 doi: 10.1038/ng.2385. In press. Used the metabochip to perform the largest yet meta-analysis of glycemic traits and identified 35 loci not previously described in European genome-wide approaches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. [Accessed July 2012.];Exome Chip Design. Available at http://genome.sph.umich.edu/wiki/Exome Chip Design.
- 23.Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 24.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Ellinghaus D, Franke A, et al. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur J Hum Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prokopenko I, Ma C, Magi R, et al. Search for novel type 2 diabetes susceptibility loci using genome-wide association studies imputed from a 1000 Genomes reference panel [abstract 139]. Presented at American Diabetes Association 72nd Scientific Sessions; Philadelphia, PA. June 8–12, 2012. [Google Scholar]
- 28.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 29.Johansson S, Irgens H, Chudasama KK, et al. Exome sequencing and genetic testing for MODY. PLoS One. 2012;7:e38050. doi: 10.1371/journal.pone.0038050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Bonnefond A, Philippe J, Durand E, et al. Whole-exome sequencing and high throughput genotyping identified KCNJ11 as the thirteenth MODY gene. PLoS One. 2012;7:e37423. doi: 10.1371/journal.pone.0037423. Used exome sequencing to identify KCNJ11 as a MODY gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thanabalasingham G, Pal A, Selwood MP, et al. Systematic assessment of etiology in adults with a clinical diagnosis of young-onset type 2 diabetes is a successful strategy for identifying maturity-onset diabetes of the young. Diabetes Care. 2012;35:1206–1212. doi: 10.2337/dc11-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiezun A, Garimella K, Do R, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen JC, Kiss RS, Pertsemlidis A, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 34.Ahituv N, Kavaslar N, Schackwitz W, et al. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80:779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nejentsev S, Walker N, Riches D, et al. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jafar-Mohammadi B, Groves CJ, Gjesing AP, et al. A role for coding functional variants in HNF4A in type 2 diabetes susceptibility. Diabetologia. 2011;54:111–119. doi: 10.1007/s00125-010-1916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Q, Yamagata K, Miura A, et al. T130I mutation in HNF-4 alpha gene is a loss-of-function mutation in hepatocytes and is associated with late-onset Type 2 diabetes mellitus in Japanese subjects. Diabetologia. 2003;46:567–573. doi: 10.1007/s00125-003-1067-y. [DOI] [PubMed] [Google Scholar]
- 38.Ek J, Rose CS, Jensen DP, et al. The functional Thr130Ile and Val255Met polymorphisms of the hepatocyte nuclear factor-4 alpha (HNF4A): gene associations with type 2 diabetes or altered beta-cell function among Danes. J Clin Endocrinol Metab. 2005;90:3054–3059. doi: 10.1210/jc.2004-2159. [DOI] [PubMed] [Google Scholar]
- 39.Stitziel NO, Kiezun A, Sunyaev S. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Sidore C, Kang HM, et al. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang HM, Gaulton K, Voight BF, et al. Sequencing and genotyping thousands of European genomes and exomes to better understand the genetic architecture of type 2 diabetes: the GoT2D Study [abstract 190]. Presented at International Congress of Human Genetics; Montreal, Canada. October 11–15, 2011. [Google Scholar]
- 42.Almeida M, Jun G, Teslovich TM, et al. Whole genome sequencing to discover type 2 diabetes risk genes in Mexican American pedigrees: T2D-GENES consortium project 2 [abstract 141]. Presented at American Diabetes Association 72nd Scientific Sessions; Philadelphia, PA. June 8–12, 2012. [Google Scholar]
- 43.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 44••.Bonnefond A, Clement N, Fawcett K, et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44:297–301. doi: 10.1038/ng.1053. Used exon sequencing to identify 40 nonsynonymous variants in MTNR1B and functionally characterized the variant proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Rees MG, Ng D, Ruppert S, et al. Correlation of rare coding variants in the gene encoding human glucokinase regulatory protein with phenotypic, cellular, and kinetic outcomes. J Clin Invest. 2012;122:205–217. doi: 10.1172/JCI46425. Used exon sequencing to identify 19 nonsynonymous variants in GCKR and functionally characterized the variant proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 47.Prokopenko I, Langenberg C, Florez JC, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Bunt M, Gloyn AL. From genetic association to molecular mechanism. Curr Diab Rep. 2010;10:452–466. doi: 10.1007/s11892-010-0150-2. [DOI] [PubMed] [Google Scholar]
- 49.Rees MG, Wincovitch S, Schultz J, et al. Cellular characterisation of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia. 2012;55:114–122. doi: 10.1007/s00125-011-2348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearson ER, Starkey BJ, Powell RJ, et al. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet. 2003;362:1275–1281. doi: 10.1016/S0140-6736(03)14571-0. [DOI] [PubMed] [Google Scholar]
- 51.Slingerland AS, Hattersley AT. Mutations in the Kir6. 2 subunit of the KATP channel and permanent neonatal diabetes: new insights and new treatment. Annals of Medicine. 2005;37:186–195. doi: 10.1080/07853890510007287. [DOI] [PubMed] [Google Scholar]
- 52.Talmud PJ, Hingorani AD, Cooper JA, et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 2010;340:b4838. doi: 10.1136/bmj.b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 54.de Miguel-Yanes JM, Shrader P, Pencina MJ, et al. Genetic risk reclassification for type 2 diabetes by age below or above 50 years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care. 2011;34:121–125. doi: 10.2337/dc10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perry JR, Voight BF, Yengo L, et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS Genet. 2012;8:e1002741. doi: 10.1371/journal.pgen.1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingelsson E, Langenberg C, Hivert MF, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59:1266–1275. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stitzel ML, Sethupathy P, Pearson DS, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gamazon ER, Badner JA, Cheng L, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiat. 2012 doi: 10.1038/mp.2011.174. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cookson W, Liang L, Abecasis G, et al. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darnell G, Duong D, Han B, Eskin E. Incorporating prior information into association studies. Bioinformatics. 2012;28:i147–i153. doi: 10.1093/bioinformatics/bts235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012 doi: 10.1093/biostatistics/kxs014. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]