This review focuses on piRNAs, the small noncoding RNAs that protect the germline genome from invasive transposable elements. Siomi and colleagues highlight newly identified piRNA factors and novel insights into the piRNA world, providing a timely overview of the current status of piRNA research that will be especially valuable given the flurry of recent studies on the topic.
Keywords: PIWI, germline, piRNA
Abstract
PIWI-interacting RNAs (piRNAs) are endogenous small noncoding RNAs that act as guardians of the genome, protecting it from invasive transposable elements in the germline. Animals lacking piRNA functions show defects in gametogenesis and exhibit sterility. Their descendants are also predisposed to inheriting mutations. Thus, the piRNA pathway has evolved to repress transposons post-transcriptionally and/or transcriptionally. A growing number of studies on piRNAs have investigated piRNA-mediated gene silencing, including piRNA biogenesis. However, piRNAs remain the most enigmatic among all of the silencing-inducing small RNAs because of their complexity and uniqueness. Although piRNAs have been previously suggested to be germline-specific, recent studies have shown that piRNAs also play crucial roles in nongonadal cells. Furthermore, piRNAs have also recently been shown to have roles in multigenerational epigenetic phenomena in worms. The purpose of this review is to highlight new piRNA factors and novel insights in the piRNA world.
RNA silencing controls gene expression in a spatiotemporal-specific manner to tightly regulate the development and homeostasis of living bodies (Bartel 2009; Malone and Hannon 2009). The key components of RNA silencing—small noncoding RNAs and Argonaute proteins—associate stoichiometrically to form RNA-induced silencing complexes (RISCs), the effector complexes in RNA silencing (Siomi and Siomi 2009). RISCs are directed to the target genes based on the complementarities between small RNAs and target gene transcripts and inhibit their expression by cleaving the transcripts or inducing translational inhibition, RNA instability, and/or heterochromatinization (Moazed 2009; Siomi et al. 2011).
Animal species express three types of endogenous silencing-inducing small RNAs: microRNAs (miRNAs), endogenous siRNAs (endo-siRNAs), and PIWI-interacting RNAs (piRNAs). These small RNAs can be classified on the basis of their origins, processing factors, and Argonaute-binding partners (Kim et al. 2009). piRNAs are generated from single-stranded precursors in a manner independent of RNase III enzymes (Vagin et al. 2006; Brennecke et al. 2007; Houwing et al. 2007), which are, in contrast, necessary for endo-siRNA and miRNA biogenesis. piRNAs associate with PIWI subfamily members of the Argonaute family of proteins, while endo-siRNAs and miRNAs associate with AGO subfamily members. piRNAs are normally 24–32 nucleotides (nt) long, but endo-siRNAs and miRNAs are 20–23 nt in most cases. Thus, the uniqueness of piRNAs is obvious.
piRNAs arise from intergenic repetitive elements in the genome called piRNA clusters (Brennecke et al. 2007). piRNA clusters span a wide region of the genome, occasionally consisting of >100,000 bases, and are mostly comprised of various transposable DNA elements and their remnants. Thus, piRNAs, especially those in Drosophila gonads and mouse prepachytene piRNAs (piRNAs expressed before the pachytene stage of meiosis in spermatogenesis), are enriched in transposon sequences. Most of these piRNAs have an antisense orientation to transposon transcripts and hence can induce silencing by hybridizing with them (Saito and Siomi 2010). Loss-of-function mutations in piRNAs and their cofactors, PIWI proteins, derepress transposons, allowing them to insert copies of themselves or relocate within the genome in a random fashion (Kalmykova et al. 2005). This selfish event activates the Chk2 DNA damage checkpoint, resulting in defects of, for example, microtubule organization and axis specification during gonadal development, which often lead to infertility (Khurana and Theurkauf 2010).
The unique characteristics of piRNAs have attracted many researchers, encouraging them to unveil the mystery of piRNAs. Remarkable progress has been made, especially in the area of biogenesis. A comprehensive computational analysis of piRNA populations in the gonads of various animals—both wild type and mutants showing defects in oogenesis and/or spermatogenesis—led to two models for piRNA biogenesis: the primary processing pathway and the amplification loop (see below, “Outline of the piRNA Biogenesis Pathway”).
However, the mechanisms underlying piRNA biogenesis and functions remain largely unknown, mainly because the piRNA pathway has little in common with the endo-siRNA and miRNA pathways as well as the restriction of piRNA territories to the reproductive tissues. Nonetheless, recent studies have suggested a possible function of piRNAs in nongonadal cells such as neuronal cells in Aplysia and mammals (Lee et al. 2011; Rajasethupathy et al. 2012). Other studies have characterized new piRNA factors, Vreteno (Vret) and Shutdown (Shu) (Handler et al. 2011; Zamparini et al. 2011; Olivieri et al. 2012; Preall et al. 2012; Xiol et al. 2012), and revealed Zucchini (Zuc) as an endonuclease necessary for primary piRNA biogenesis (Ipsaro et al. 2012; Nishimasu et al. 2012). Multigenerational epigenetic phenomena in nematodes are now known to involve piRNAs (Ashe et al. 2012; Shirayama et al. 2012). Thus, the visibility of the piRNA field has rapidly broadened.
Outline of the piRNA biogenesis pathway
The primary processing pathway
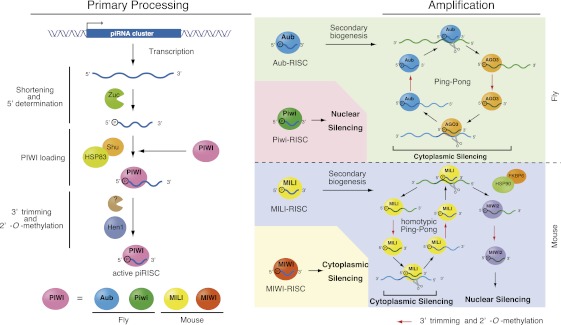
Nascent transcripts arising from piRNA clusters are processed into piRNA-like molecules, which are then loaded onto PIWI proteins (Fig. 1). The factors involved in the transcription of piRNA clusters and its regulation remain elusive. The current model proposes that the 5′ ends of piRNAs are determined prior to loading onto PIWI proteins. However, the 3′ ends of piRNAs harbor extra bases, which need to be trimmed upon association with PIWI proteins (Kawaoka et al. 2011; Vourekas et al. 2012). The lengths of mature piRNAs are determined during this step, largely depending on the sizes of the PIWI proteins. Thus, piRNAs associated with individual PIWIs show a similar but distinct size distribution (Siomi et al. 2011). The factors responsible for the 3′ trimming remain elusive. Upon maturation, the 3′ ends of piRNAs are 2′-O-methylated by Hen1/Pimet, which is associated with PIWI proteins (Horwich et al. 2007; Saito et al. 2007). This modification maintains the stability of piRNAs in vivo (Horwich et al. 2007; Kamminga et al. 2010, 2012; Billi et al. 2012). Hen1 was first discovered in Arabidopsis as the enzyme responsible for 2′-O-methylation of miRNAs (Chen 2005). However, in animals, Hen1 does not 2′-O-methylate miRNAs. Although there is no direct evidence for this, the high turnover rate for animals' miRNAs may serve to spatiotemporally regulate their function. Only selected PIWI proteins are loaded with primary piRNAs. For instance, Drosophila expresses three PIWI proteins: Of these, Aubergine (Aub) and Piwi, but not Ago3, associate with primary piRNAs (Li et al. 2009; Malone et al. 2009; Saito et al. 2009). In mice, among three PIWI proteins, MIWI and MILI associate with primary piRNAs, whereas MIWI2 mostly associates with secondary piRNAs, as does Ago3 in flies (see below, “The Amplification Loop”; Siomi et al. 2011). The mechanism underlying this PIWI selection remains undetermined.
Figure 1.
piRNA biogenesis in Drosophila and mice consists of the primary piRNA processing pathway and the amplification loop. The left side shows a model of the primary processing pathway in flies. The primary transcripts of piRNA clusters are shortened into piRNA-like small RNAs. The factors necessary for this process largely remain to be determined. Recent studies have suggested that Zucchini might be an endonuclease forming the 5′ ends of piRNAs. The mouse Zucchini homolog (MitoPLD) might also function as a nuclease to produce primary piRNAs. piRNA-like small RNAs are loaded onto PIWI proteins (shown in pink) and then trimmed from the 3′ end to the size of mature piRNAs by an unknown nuclease. Hsp83/Shu may play a role in the Piwi loading step. piRNAs are 2′-O-methylated by HEN1/Pimet. In flies, the PIWI proteins that associate with primary piRNAs are Piwi (green) and Aub (blue). Piwi associated with piRNAs is translocated to the nucleus and thus likely does not contribute to the amplification loop. Aub associated with piRNAs now triggers the Ping-Pong cycle by cleaving transposon transcripts. Ago3 (orange) loaded with secondary piRNAs in turn produces piRNAs that associate with Aub. Transposon transcripts are cleaved through this amplification cycle, resulting in cytoplasmic transposon silencing. In mice, primary piRNAs are loaded onto MILI (yellow) and MIWI (red). MIWI associated with pachytene piRNAs functions in cytoplasmic silencing. The targets remain largely unknown. MILI associated with primary piRNAs contributes to the Ping-Pong cycle to produce piRNAs that associate with MIWI2 (purple), upon which MIWI2 is localized to the nucleus to accomplish nuclear silencing. The contribution of MIWI2 to the Ping-Pong cycle may be negligible. MILI might operate a homotypic Ping-Pong cycle as indicated. HSP90/FKBP6 plays a role in producing secondary piRNAs that associate with MIWI2. HEN1/Pimet 2′-O-methylates secondary piRNAs, the products of the Ping-Pong cycle.
The amplification loop
Primary piRNAs are subjected to an amplification system to enforce the high expression of piRNAs in the germline: This system is called the amplification loop or the Ping-Pong cycle (Fig. 1; Brennecke et al. 2007; Gunawardane et al. 2007; Houwing et al. 2008). In this system, Aub in flies and MILI in mice associated with primary piRNAs cleave the target RNAs via their Slicer (endonuclease) activity (Saito et al. 2006; Gunawardane et al. 2007; Nishida et al. 2007). This process forms the 5′ ends of secondary piRNAs. In Drosophila, maternally deposited piRNAs can also trigger the Ping-Pong cycle, as do primary piRNAs (Brennecke et al. 2008). The cleavage products are then transferred onto other PIWI members, Ago3 in flies and MIWI2 in mice, and are trimmed from the 3′ end to give rise to mature piRNAs. This step is mechanistically equivalent to that observed in the primary processing pathway (Fig. 1), although the factors required might differ. In turn, in flies, Ago3 associated with secondary piRNAs cleaves its targets, giving rise to secondary piRNAs, which associate with Aub (Brennecke et al. 2007; Gunawardane et al. 2007; Li et al. 2009; Malone et al. 2009). Through these reciprocal Slicer-mediated cleavage reactions between Aub and Ago3, the heterotypic Ping-Pong cycle persists in Drosophila (Fig. 1). Piwi is translocated to the nucleus upon primary piRNA loading (Haase et al. 2010; Olivieri et al. 2010; Saito et al. 2010; Ishizu et al. 2011); therefore, Piwi barely contributes to secondary piRNA production via the amplification loop (Malone et al. 2009; Olivieri et al. 2012). In the Drosophila gonadal soma, Piwi is expressed and associates with primary piRNAs. However, the expression levels of Aub and Ago3 are too low for them to operate in the amplification loop. Thus, the situation in ovarian somatic cells, such as follicle cells, is different from that in germ cells.
Like Piwi in Drosophila, in mice, MIWI2 loaded with secondary piRNAs is localized to the nucleus upon piRNA loading (Fig. 1; Carmell et al. 2007). Thus, MIWI2 likely does not contribute to the bearing of secondary piRNAs via the Ping-Pong cycle. The pathway for producing MIWI2–piRNAs is thus considered to be the “one-way” secondary piRNA biogenesis pathway (Aravin et al. 2008; De Fazio et al. 2011; Reuter et al. 2011), although there is evidence supporting the notion that the MILI–piRNA complex may operate a homotypic Ping-Pong cycle in mouse testes (De Fazio et al. 2011). MIWI is associated with pachytene piRNAs (piRNAs expressed starting at the pachytene stage of meiosis in mouse spermatogenesis), which are also barely involved in the amplification loop (Beyret et al. 2012). Based on these observations, it is highly likely that a heterotypic Ping-Pong cycle does not operate in mice, unlike in Drosophila (Fig. 1). Although the details of the Ping-Pong cycle differ slightly between flies and mice, secondary piRNAs are 2′-O-methylated by Hen1/Pimet in both species (Fig. 1).
Because the amplification loop depends on the Slicer activity of PIWI proteins, secondary piRNAs on Aub in flies and MILI in mice show complete complementarity to the piRNAs on Ago3 (in flies) and MIWI2 (in mice), respectively, through the first 10 bases from their 5′ ends (Brennecke et al. 2007; Gunawardane et al. 2007). In addition, Aub-piRNAs show a strong bias for uracil at the 5′ end (1-U), and, accordingly, Ago3-piRNAs tend to have adenosine at the 10th nucleotide from the 5′ end (10-A). These are typical signatures of piRNAs made via the Ping-Pong cycle or one-way secondary piRNA biogenesis, which is highly conserved in animal species (Grimson et al. 2008).
In the Ping-Pong cycle, transposon transcripts are cleaved by piRISCs. This means that the Ping-Pong cycle accomplishes dual tasks simultaneously, producing secondary piRNAs and silencing transposons by cleaving their transcripts. In mice, primary piRNAs on MILI are predominantly “sense” to transposon transcripts and are unable to target transposon transcripts. Nuclear MIWI2 associates with antisense piRNAs and thus is capable of inducing transposon silencing, regardless of whether nuclear silencing targets transposon loci directly or via nascent RNAs transcribed from transposon loci. MIWI2 (and also MILI) has been linked to DNA methylation of target gene loci in the genome (Aravin et al. 2008). However, the molecular mechanisms underlying this action remain unclear. The functions of pachytene piRNAs loaded onto MIWI are unclear.
piRNA biogenesis factors: Tudor domain-containing (TDRD) proteins
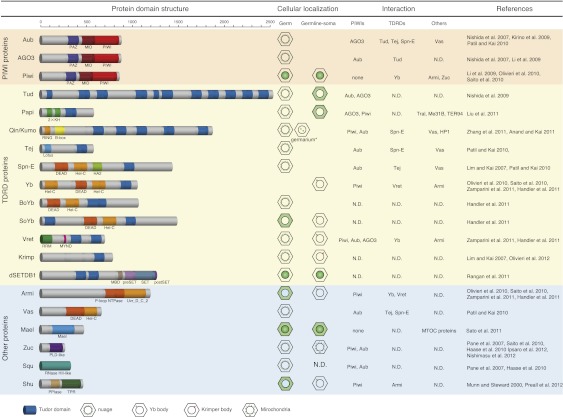
piRNA biogenesis requires several factors other than PIWI proteins, involving Tudor superfamily members or TDRD proteins (Fig. 2; Siomi et al. 2010). TDRD proteins specifically associate with particular protein substrates through symmetrical dimethyl arginines (sDMAs) or asymmetrical dimethyl arginines (aDMAs) in the substrates (Chen et al. 2011). This protein–protein interaction has been shown to play a role in aspects of RNA metabolism such as splicing (Neuenkirchen et al. 2008). RNA silencing is no exception. PIWI proteins contain sDMAs toward their N-terminal ends (Chen et al. 2009; Kirino et al. 2009; Nishida et al. 2009; Reuter et al. 2009; Vagin et al. 2009). An enzyme responsible for the sDMA modification is PRMT5/DART5/Capsleen (Kirino et al. 2009). To date, 11 TDRD proteins in flies (Tudor [Tud], Partner of piwis [Papi], Qin/Kumo, Tejas, Spindle-E [Spn-E], Yb, Brother of Yb [BoYb], Sister of Yb [SoYb], Krimper [Krimp], dSETDB1, and Vret) (Fig. 2; Anand and Kai 2011; Handler et al. 2011; Liu et al. 2011; Rangan et al. 2011; Zamparini et al. 2011; Zhang et al. 2011) and seven in mice (TDRD1, TDRD2, TDRD4, TDRD5, TDRD6, TDRD7, and TDRD9) (Siomi et al. 2010) have been known to be involved in the piRNA pathway.
Figure 2.
piRNA biogenesis factors in Drosophila. Drosophila piRNA factors can be subgrouped into PIWI proteins, TDRD proteins, and others. The cellular localizations in ovaries and protein–protein interaction partners of individual factors are summarized on the right. (N.D.) Not determined. Interaction of Tral, Me31B, and TER94 with Papi was reported by Liu et al. (2011). HP1 interaction with Qin/Kumo was reported by Anand and Kai (2011). In addition to perinuclear localization in the nurse cells, Kumo also appears as foci in the nuclei of the germ cells in the germarium (indicated by an asterisk). Interaction of MTOC proteins with Mael was reported by Sato et al. (2011).
Earlier genetic studies in flies showed that Tud mutants phenocopy Aub mutants (Arkov et al. 2006). More recent studies have revealed that Tud, consisting of 11 Tud domains (Fig. 2), associates with both Aub and Ago3 in an sDMA-dependent manner to act as a “platform” for the Ping-Pong cycle (Nishida et al. 2009). In tud mutants, Aub and Ago3 associate with piRNAs more abundantly than in wild-type flies; however, these associations are likely predominantly aberrant. Thus, Tud is necessary for the quality control of piRNAs in the germline (Nishida et al. 2009). TDRD6 in mice, containing seven Tud domains, is thought to be the nearest homolog of Drosophila Tud. TDRD6 binds mouse Vasa homolog (MVH) (see below), MIWI, MILI, and TDRD1 (Kirino et al. 2010), although the function of TDRD6 in piRNA biogenesis is not yet clear.
Qin/Kumo (CG14303) (Anand and Kai 2011; Zhang et al. 2011) and Vret (CG4771) (Handler et al. 2011; Zamparini et al. 2011) are newly characterized Drosophila TDRD genes (Fig. 2). Qin/Kumo encodes a protein containing one RING domain and two B-box domains in the N-terminal region, followed by five Tud domains. The nearest homolog of Qin/Kumo in mice is TDRD4/RNF17 (Pan et al. 2005). Qin/Kumo localizes to the nuage, a nonmembranous perinuclear structure found in animal germ cells, which is implicated as the site of piRNA biogenesis (Anand and Kai 2011). Qin was named after the famous ancient Chinese dynasty (Zhang et al. 2011), while Kumo in Japanese means “cloud,” the French for which is nuage (Anand and Kai 2011). Mutations in Qin/Kumo cause most nuage components, including Aub and Ago3, not to accumulate to the nuage; thus, Qin/Kumo is considered to be the core of the nuage (Anand and Kai 2011). Loss of function of Qin/Kumo induces homotypic Ping-Pong among Aub proteins, instead of natural heterotopic Ping-Pong between Aub and Ago3, although the expression level of Ago3 in qin/kumo mutants is greater than that in ago3 heterozygotes (Zhang et al. 2011). Mutations in Qin/Kumo weaken the association between Aub and Ago3; this might be the cause of the weakening of heterotopic Ping-Pong between Aub and Ago3. A comprehensive bioinformatic analysis of piRNAs revealed that the homotypic Ping-Pong in qin/kumo mutants is, however, futile, resulting in derepression of transposons and DNA damage in the ovaries. Thus, Qin/Kumo enforces the heterotypic amplification loop between Aub and Ago3 to quality-control piRNAs to ensure transposon silencing. It is of interest to note that the functions of Tud and Qin/Kumo are identical to each other; namely, both proteins function in the quality control of piRNAs. However, tud and qin/kumo mutants show different phenotypes. One could infer that Tud and Qin/Kumo have additional functionalities and/or specificities that contribute to successful Ping-Pong in the germ cells.
Qin/Kumo is localized to the nucleus in the germarium. A potential nuclear function of Qin/Kumo was suggested; namely, maintaining the transcription level of the bidirectional piRNA cluster 42AB, as does the HP1 homolog Rhino (Klattenhoff et al. 2009; Anand and Kai 2011). However, Qin/Kumo associates with HP1 but not with Rhino. Findings to date suggest that Qin/Kumo regulates the level of bidirectional piRNA clusters by restricting HP1 binding to the clusters by physically associating with HP1. However, the important question of how Qin/Kumo/HP1 selectively regulates piRNA clusters remains to be addressed. The roles of Qin/Kumo in transcription of piRNA clusters and piRNA biogenesis are not yet known. However, the Qin/Kumo studies by Anand and Kai (2011) and Zhang et al. (2011) raised the interesting idea that ubiquitination might be involved in the piRNA pathway because the RING domain is known to specify the activity of E3 ligases, which specifically modify their substrates with ubiquitins. It is of great interest to address how ubiquitination is molecularly linked to piRNA biogenesis, if ubiquitination is indeed involved in the pathway.
Drosophila Vret consists of one RNA recognition motif (RRM), one MYND domain, and two Tud domains and is necessary for piRNA-based transposon regulation in both germ cells and gonadal somas (Fig. 2; Handler et al. 2011; Zamparini et al. 2011). Loss of function of Vret abolishes the primary piRNA processing activity, whereas the Ping-Pong pathway is not greatly affected by vret mutations. Thus, Vret plays a role exclusively in primary piRNA production, although the expression of Vret can also be observed in germ cells, where the Ping-Pong amplification takes place. Vret interacts with the somatic primary piRNA factors Armitage (Armi), Yb, and Zuc (Fig. 3; see below). The transcriptional levels of piRNA clusters are not changed in vret mutants. Thus, the requirement for Vret in the piRNA pathway is distinct from that for Qin/Kumo, although both Vret and Qin/Kumo are TDRD members and bind PIWI proteins. Vret contains a RRM, suggesting a direct association of Vret with RNA molecules. Investigations to identify the RNA-binding substrates are awaited. It is noted that no obvious homolog of Vret has been found in mice.
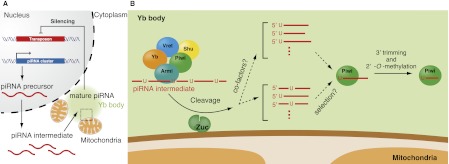
Figure 3.
Zucchini in primary piRNA biogenesis in Drosophila ovarian somas. (A) Long, single-stranded piRNA precursors are transcribed from piRNA clusters in the genome and are processed into piRNA intermediates through an unknown mechanism. (B) An enlarged cartoon of the area shown by a dotted square in A: The RNA helicase Armi and the Tud domain-containing RNA helicase Yb localize to Yb bodies, which are often located adjacent to mitochondria. Armi and Yb form a complex that contains piRNA intermediates with 5′-hydroxyl and 3′-cyclic phosphate ends. Nascent, piRNA-free Piwi transiently localizes to Yb bodies to interact with the Armi–Yb complex. Zuc is anchored on the outer surfaces of mitochondria with the catalytic site facing the cytosol and processes piRNA intermediates into piRNA fragments, which are loaded onto Piwi. Primary piRNAs show a strong bias for 1-U, although Zuc endonuclease showed little sequence specificity in vitro. These findings lead to the possibility that cofactors may be involved in 1-U determination in vivo (“cofactors?”). Alternatively, Piwi may preferentially bind 1-U piRNAs among all Zuc cleavage products (“selection?”). Vret and Shu are newly characterized piRNA factors that localize to Yb-bodies. The functions of Vret and Shu have not been fully elucidated. The enzymes participating in 3′ end formation of piRNAs remain to be identified.
piRNA biogenesis factors: other than PIWI and TDRD proteins
Several non-PIWI, non-TDRD proteins are also involved in piRNA biogenesis: These include Vasa (Vas), Maelstrom (Mael), Armi, Zuc, Squash (Squ), and Shu in Drosophila (Fig. 2). All of these factors except Squ are conserved in mice: The mouse homologs are MVH, Mael, MOV10/MOV10l1, MitoPLD/PLD6, and FKBP6, respectively (Soper et al. 2008; Frost et al. 2010; Huang et al. 2011; Watanabe et al. 2011; Xiol et al. 2012). Most of these factors play a role in the amplification loop. The representative is Vas, a germ cell-specific DEAD-box-type RNA helicase required for pole cell development in Drosophila (Lasko and Ashburner 1990).
Vas was implied to regulate translation of target genes, such as Gurken, because it associates with the translation initiation factor eIF5B, and Gurken does not accumulate in oocytes in vas mutants (Carrera et al. 2000). Translation of oskar and nanos may also be activated by Vas in the pole plasm. MVH also has pivotal roles specifically in male germ cells in mice (Tanaka et al. 2000). MVH-deficient mouse testes phenocopy MILI- and MIWI2-deficient testes; IAP and Line-1 retrotransposons are derepressed, and de novo DNA methylation of their regulatory regions is impaired (Kuramochi-Miyagawa et al. 2010). Without MVH function, MIWI2 becomes almost free from secondary piRNAs, although MILI's association with primary piRNAs is maintained. Thus, MVH is considered to be necessary for the Ping-Pong cycle. Mutations in Vas in Drosophila also impair piRNA accumulation in the gonads and derepress transposons (Lim and Kai 2007; Nagao et al. 2010), although the underlying mechanism remains to be elucidated. Ovarian somas in Drosophila are devoid of Vas expression, suggesting that the somatic primary processing does not require Vas.
Many non-PIWI, non-TDRD piRNA factors also play a role in primary processing. Drosophila ovarian somatic cells use the primary processing pathway but not the Ping-Pong cycle (Lau et al. 2009; Li et al. 2009; Malone et al. 2009; Saito et al. 2009) simply because they do not express Aub and Ago3. In these cells, primary processing occurs in perinuclear Yb bodies. The core component Yb and other piRNA factors—Armi, Shu, and Vret—accumulate in Yb bodies, where they exert their functions (Fig. 3). Upon processing, mature primary piRNAs are loaded onto Piwi to assemble active piRISCs. This assembly allows piRISCs to be transported to the nucleus, where they function in transposon silencing (Saito et al. 2009; Ishizu et al. 2011). Piwi Slicer activity is dispensable for nuclear silencing (Saito et al. 2009), although it is still unclear whether the silencing occurs transcriptionally or post-transcriptionally.
Zuc, a member of the phospholipase D (PLD) superfamily, is required for primary piRNA biogenesis in Drosophila ovarian somatic cells. Zuc has been considered a candidate ribonuclease for piRNA maturation (Pane et al. 2007; Saito et al. 2009, 2010; Haase et al. 2010; Olivieri et al. 2010). This is because a bacterial protein, Nuc, another member of the PLD superfamily, has endonuclease activity (Pohlman et al. 1993). MitoPLD, the mammalian homolog of Zuc, also plays a crucial role in piRNA biogenesis (Huang et al. 2011; Watanabe et al. 2011). Previous reports showed that MitoPLD generates a lipid signal molecule, phosphatidic acid, using mitochondrial lipid cardiolipin as a substrate (Choi et al. 2006; Huang et al. 2011), although MitoPLD showed no nuclease activity toward DNA and RNA substrates in in vitro assays (Watanabe et al. 2011). These observations suggested that mitochondrial morphology regulated by the mitochondrial lipid signaling pathway might be related to piRNA production. Like MitoPLD, Zuc localizes to mitochondria in Drosophila cells (Saito et al. 2010). However, it was unclear whether Drosophila and mammalian Zuc have conserved enzymatic functions. Furthermore, these studies could not rule out the possibility that Zuc/MitoPLD might exhibit a nuclease activity in intracellular environments.
Recent crystal structural analyses revealed that, similar to other PLD enzymes, Zuc/MitoPLD forms dimers with a single active site at the subunit interface (Ipsaro et al. 2012; Nishimasu et al. 2012). The active site contains two conserved catalytic HKD (His–Lys–Asp) motifs, one from each subunit. Zuc was found by in vitro nuclease assays to have single-strand-specific endonuclease activity. Mutational analyses showed that Zuc cleaves the target phosphodiester bond in a manner similar to other PLD family nucleases, by leaving products with a 5′-phosphate terminus. Since the 5′-phosphate terminus is one of the hallmarks of mature piRNAs, it was postulated that Zuc cleaves piRNA intermediates to generate the 5′ ends of piRNAs (Fig. 3). Zuc showed little sequence specificity in vitro. These findings appear to contradict the finding that primary piRNAs show a strong bias for 1-U. One possibility might be that cofactors—for example, TDRD proteins—are involved in 1-U determination in vivo. An alternative possibility is that Piwi selects 1-U piRNAs among all Zuc cleavage products to bind. In any case, these findings suggest that Zuc plays a role equivalent to that of Aub/Ago3 in the Ping-Pong cycle; that is, to form the 5′ end of piRNAs.
Shu is a female sterile gene in Drosophila (Schüpbach and Wieschaus 1991) encoding an evolutionarily conserved cochaperone of the immunophilin class that harbors a peptidyl-propyl-cis/trans-isomerase domain and a tetratricopeptide repeat (TPR) domain (Munn and Steward 2000). Recent studies identified Shu as a new piRNA factor necessary for both primary processing and the amplification loop (Olivieri et al. 2012; Preall et al. 2012). Indeed, Shu was found to be a component of both the nuage and Yb bodies in germ cells and ovarian somas, respectively, both of which are considered to be centers of piRNA biogenesis. Mutations in Shu in ovarian somas do not affect the localization of Yb, Armi, and Vret, meaning that Shu is located downstream from these molecules. However, the Yb body localization of Shu depends on Piwi. Shu may be unnecessary for somatic primary piRNA biogenesis but rather may be required for Piwi loading. In mice, FKBP6 (a Shu homolog in mice), which is essential for male fertility (Crackower et al. 2003), was determined to be unnecessary for primary processing, unlike Shu in flies, but required for the amplification as a remover of 16-nt slicing products (Xiol et al. 2012). FKBP6 associates with Hsp90 through its TPR domain and plays a role in producing secondary piRNAs that associate with MIWI2. Hsp90 has been implicated in piRNA biogenesis (Specchia et al. 2010). FKBP6 also binds with PIWI proteins (Vagin et al. 2009). Inhibition of HSP90 results in the accumulation of ∼16-nt by-products of piRNA amplification specifically on BmAgo3, one of two PIWI proteins expressed in cultured Bombyx germ cells. One proposal is that the chaperone machinery consisting of HSP90 and FKBP6 removes the 16-nt by-products of piRNA amplification from BmAgo3, facilitating its turnover in the Ping-Pong pathway to enforce the high expression of piRNAs in germ cells. How, then, are the by-products on SIWI, another PIWI in Bombyx, removed from SIWI? One suggestion is that other chaperone machinery similar to HSP90/FKBP6 might exist for SIWI, although currently this remains unclear.
piRNA functions outside of the germline cells
A unique, fascinating study using the Aplysia CNS revealed that piRNAs can play roles in regulating memory storage in the brain by silencing CREB2, the major inhibitory constraint of memory formation (Rajasethupathy et al. 2012).
piRNAs in the Aplysia CNS cluster in the genome (372 clusters were found in the study), similar to the piRNAs in germline cells. Neuronal piRNAs preferably contain 1-U, are 2′-O-methylated, and induce DNA methylation of target genes, including CREB2, a transcriptional repressor of memory. Notably, all of these features are hallmarks of piRNAs in germline cells. The association of acaPiwi (the single PIWI in Aplysia) with piRNAs was also confirmed experimentally.
One or a few individual piRNAs were frequently cloned from the same cluster, suggesting that some piRNAs are selectively enriched in the CNS. In this regard, Aplysia neuronal piRNAs may be similar to Suppressor of Stellate [Su(Ste)]-derived piRNAs in the Drosophila testis (Nishida et al. 2007). Su(Ste) piRNAs were suggested to be the products of primary processing (Nagao et al. 2010), and neuronal piRNAs in Aplysia may be produced by an equivalent pathway. Aplysia expresses only PIWI (acaPiwi); thus, the animal lacks the amplification loop. As a result, all Aplysia piRNAs are considered to be primary piRNAs. It remains unclear whether primary piRNA biogenesis factors are conserved in Aplysia. acaPiwi is localized to the nucleus, as is MIWI2 in mouse testis and Piwi in fly gonads. Nuclear localization of MIWI2 and Piwi is achieved by piRNA loading onto the proteins. It would be of interest to determine whether piRNA loading is necessary for the nuclear localization of acaPiwi. Whether nuclear localization of acaPiwi can also be observed in the ovotestes remains to be determined.
Knockdown of acaPiwi induces up-regulation of CREB2 but does not affect the related genes C/EBP and CPEB. Rajasethupathy et al. (2012) suggested that acaPiwi has target genes other than CREB2, although these are currently unknown. Inhibition of acaPiwi completely abolished the serotonin-dependent increase in methylation at the promoter of CREB2 (Rajasethupathy et al. 2012); however, the details of the mechanism underlying this effect are also unknown.
The following questions remain to be addressed: (1) What is the benefit for Aplysia in using miRNA and piRNA to silence CREB1 and CREB2, respectively? (2) Do mammals also use piRNAs in brains to establish long-term memory storage? The answers to these questions are eagerly awaited.
A recent study found piRNAs in the mouse hippocampus (Lee et al. 2011). These piRNAs associate with MIWI to form an effector piRISC, which is localized to the cytoplasm in mouse hippocampal neurons. piRNA function seems to be required for dendritic spine development because inhibition of piRNA function led to a decrease in dendrite spine area. Computational analyses have suggested target genes of these hippocampal piRNAs. However, it remains to be determined whether expression of these genes is regulated by piRNAs.
piRNAs in Caenorhabditis elegans
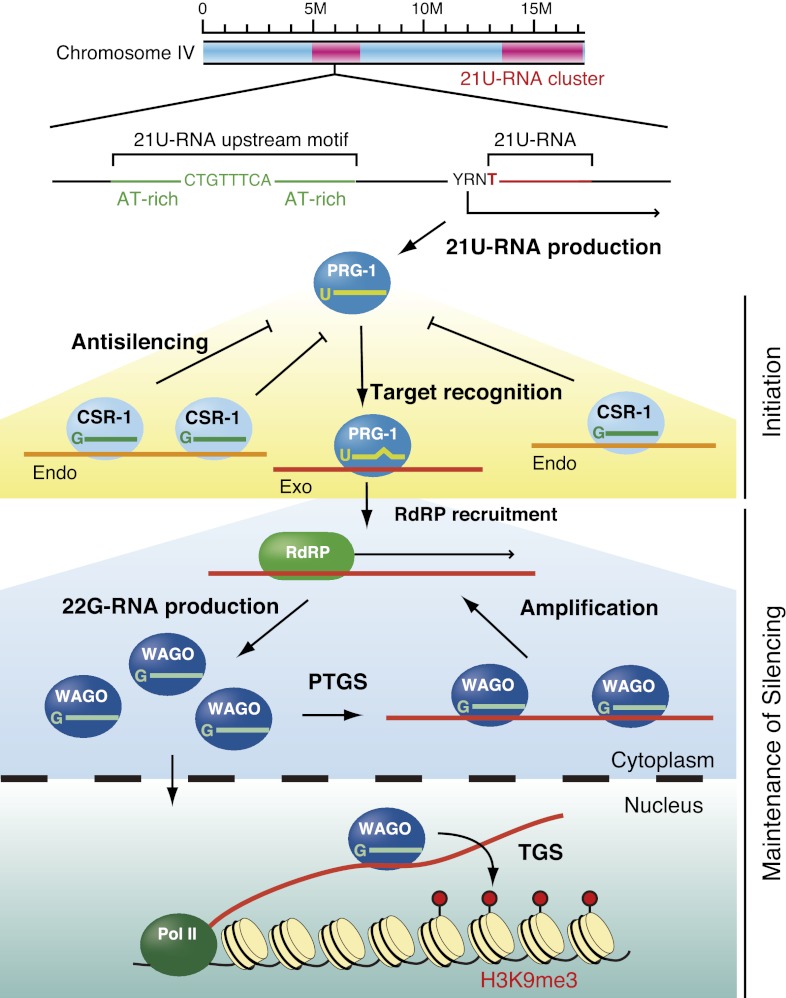
One of the outstanding questions in piRNA research is how the piRNA system selects transposable elements—and not other cellular genes—as targets to silence. In other words, how does the system discriminate between foreign nucleic acid elements and cellular mRNAs? This question has conceptual similarities to questions about immune systems, which achieve “self” versus “nonself” recognition. Recent studies in C. elegans have provided important insights into piRNA-mediated genome-wide surveillance of germline transcripts, leading to long-term silencing of transcripts recognized as nonself and to licensing to protect cellular protein-coding genes as self from piRNA-mediated silencing (Fig. 4; Ashe et al. 2012; Bagijn et al. 2012; Lee et al. 2012; Luteijn et al. 2012; Shirayama et al. 2012).
Figure 4.
piRNAs in C. elegans. C. elegans piRNAs are 21 nt in length with 1-U (21U-RNAs) and are loaded onto the PIWI protein PRG-1. piRNAs are genomically encoded in two large clusters on chromosome IV. These clusters are depleted of protein-coding genes. piRNAs have a characteristic sequence motif ∼42 nt upstream of the start of the small RNA, which may be required for RNA polymerase II (Pol II) transcription. piRNAs scan virtually all transcripts expressed in the germline and guide PRG-1 to targets by means of relaxed base-pairing with up to four mismatches. PRG-1–piRNA complexes then recruit RNA-dependent RNA polymerases (RdRPs) to the target site to produce 22G-RNAs (which are loaded onto WAGOs) that silence foreign genetic elements (Exo). WAGOs that localize to the cytoplasm mediate mRNA turnover (PTGS), whereas WAGOs that localize to the nucleus mediate transcriptional gene silencing (TGS). The CSR-1 22G RNA pathway may provide a memory of self (Endo) and act as an anti-silencing signal.
The piRNAs in C. elegans are 21-U RNAs, a population of 21-nt small RNAs characterized by a 1-U bias and a characteristic sequence motif ∼42 nt upstream of the start of the small RNA (Ruby et al. 2006; Batista et al. 2008). 21-U RNAs appear to be derived from thousands of individual, autonomously expressed loci broadly scattered in two large clusters on chromosome IV (Ruby et al. 2006; Batista et al. 2008). The most conserved 8-mer sequence (CTGTTTCA) in the upstream region is required for their individual expression and is specifically recognized by Forkhead family transcription factors (Cecere et al. 2012). piRNAs are expressed in germline cells, where they interact with the PIWI protein PRG-1. Mutations in prg-1 result in a reduced brood size and a temperature-sensitive sterile phenotype, consistent with the notion that PIWI proteins are linked to germline maintenance. Like the abundant pachytene piRNAs in mammals (Aravin et al. 2006; Lau et al. 2006), C. elegans piRNAs encode remarkable sequence diversity and yet lack obvious targets. Although the piRNA pathway is implicated in the silencing of only a single transposon, Tc3, by acting upstream of secondary siRNA production via RNA-dependent RNA polymerase (RdRP) activity (Das et al. 2008), the lack of perfect complementarity to Tc3 and other transposable elements displayed by ∼16,000 different piRNAs encoded in the C. elegans genome has been a long-standing conundrum.
A combination of C. elegans genetics together with bioinformatics for small RNAs answered a very important question about how 21-U RNAs recognize and silence target genes (Ashe et al. 2012; Bagijn et al. 2012; Lee et al. 2012; Shirayama et al. 2012). Compelling evidence supported the concept that PRG-1–piRNA complexes scan foreign RNA sequences and drive the production and initial amplification of RdRP-dependent siRNAs known as 22G-RNAs, which are loaded onto worm-specific Argonaute members, WAGO proteins. An important recurring theme that emerged is that RNA silencing in C. elegans occurs in a two-step pathway, and multiple C. elegans small RNA pathways converge on WAGO proteins, including the RDE-1 exogenous siRNA pathway (RNAi) and the ERGO-1 endogenous siRNA pathway (Yigit et al. 2006; Pak and Fire 2007; Gu et al. 2009). Amplification of the silencing signal in the two-step system occurs via the production of secondary siRNAs (22G-RNAs) by RdRPs that are then discriminately bound to WAGO proteins, which mediate downstream silencing; WAGOs that localize to the cytoplasm mediate mRNA turnover, whereas WAGOs that localize to the nucleus mediate transcriptional silencing (Guang et al. 2008; Gu et al. 2009). Once a piRNA initiates the two-step pathway against nonself elements—a process that is independent of PRG-1 Slicer activity (Bagijn et al. 2012)—the silencing is permanently heritable and no longer requires piRNAs and PRG-1 (Ashe et al. 2012; Luteijn et al. 2012; Shirayama et al. 2012). This phenomenon (which involves nuclear WAGO proteins and canonical chromatic modifications, including H3K9me3) is referred to as RNA-induced epigenetic silencing or RNAe (Shirayama et al. 2012). Thus, piRNAs represent the initial triggers of siRNA amplification and heterochromatin formation, revealing intriguing parallels with the roles of fission yeast primal RNAs (priRNAs) that nucleate a positive feedback loop of siRNA production, which then promotes heterochromatin formation (Halic and Moazed 2010).
Mutagenesis studies and genome-wide computational predictions revealed that piRNAs guide PRG-1 to target transcripts by base-pairing with up to four mismatches, initiating localized generation of 22G-RNAs within a ±50-nt window around the site of piRNA complementarity on the target RNA (Ashe et al. 2012; Lee et al. 2012). However, just like the target recognition of miRNAs (Bartel 2009) and similar to the target recognition of mouse piRNAs (Reuter et al. 2011), the 5′ end of the C. elegans piRNA, particularly the seed sequence (nucleotides 2–8), appears critical for targeting. Thus, C. elegans piRNAs have the potential to silence transcripts in trans through imperfectly complementary sites. However, such relaxed targeting requirements imply that piRNAs with ∼16,000 distinct sequences target not only multiple transposable elements and exogenously introduced elements (transgenes), but also most cellular protein-coding transcripts. How do piRNAs selectively target foreign elements and avoid targeting endogenous protein-coding genes? Perhaps piRNAs that silence protein-coding genes are negatively selected, as depletion of mismatch-tolerant piRNAs that base-pair with germline protein-coding transcripts is observed (Bagijn et al. 2012; Lee et al. 2012). Instead, piRNA clusters tend to overlap transposon ends, suggesting the occurrence of recent transposon integrations downstream from the sequence motif thought to be required for piRNA biogenesis. Thus, the C. elegans piRNA system bears many conceptual similarities to Drosophila piRNA clusters. These systems must cope with a diverse range of foreign genetic elements that show little similarity at the primary sequence level. The strategies used to build a piRNA repertoire seem to rely on transposition by chance into piRNA clusters, which can then become fixed by evolutionary selection. In both cases, piRNA clusters act as traps for transposable elements.
In addition, germline mRNAs seem to be protected from piRNA-induced silencing, and this is potentially mediated by Argonaute CSR-1. CSR-1 lies in one of two distinct germline 22G-RNA pathways, and the interacting 22G-RNAs are antisense to virtually all germline cell-expressed protein-coding genes (Claycomb et al. 2009). However, endogenous CSR-1 22G-RNA targets do not appear to be down-regulated by CSR-1. CSR-1 22G-RNAs are expressed at low levels relative to WAGO 22G-RNAs, perhaps below the threshold required to trigger mRNA turnover. piRNAs that map to CSR-1 22G-RNA targets trigger a less robust secondary siRNA response compared with those mapping to WAGO 22G-RNA targets. Perhaps CSR-1 22G-RNA complexes can compete for binding sites with PRG-1–piRNA and/or WAGO 22G-RNA complexes. This finding led to the hypothesis that CSR-1 22G-RNA complexes function as a memory of previous gene expression in germline cells and that the CSR-1-dependent licensing (“anti-silencing”) protects endogenous protein-coding genes from piRNA-mediated silencing (Shirayama et al. 2012). Thus, the balance between nonself recognition by the PRG-1–piRNA pathway and self recognition by the CSR-1–22G-RNA pathway might determine the outcome of gene expression in germline cells.
Although recent biochemical analyses revealed no obvious targets of mouse pachytene piRNAs (Vourekas et al. 2012), the findings in C. elegans may spur re-examination of the idea that pachytene piRNAs are the end products of RNA processing in mice using extensive genome-wide computational analysis to identify targets with relaxed base-pairing.
Perspective
The molecular mechanisms underlying piRNA biogenesis and functions are complex and diverse; therefore, many questions still need to be answered. The first question is related to the discrimination of piRNA precursors from non-piRNA precursors. As noted above, the transcripts arising from piRNA clusters and a few specific protein-coding mRNAs, including traffic jam, are recognized as piRNA precursors and give rise to piRNAs (Robine et al. 2009; Saito et al. 2009). piRNA clusters normally span a wide region in the genome, and thus it is feasible to think that they are transcribed by RNA polymerase II and later are capped and poly(A)-tailed at the 5′ and 3′ ends like normal mRNAs. Why and how are regular mRNAs able to escape being victims of the piRNA production system in vivo? Answering this question is crucial because the answer will enhance our understanding of the mechanism of piRNA biogenesis at the molecular level. The second question is related to the identity and nature of piRNA biogenesis factors required upstream of Zuc. Mutations in Zuc lead to the accumulation of piRNA intermediates but impair piRNA maturation. The intermediates are approximately several hundred bases long, although piRNA precursors are much larger, meaning that other factors downsize the piRNA primary precursors into their intermediates. Establishing an in vitro system to recapitulate piRNA processing from piRNA cluster transcripts in combination with cellular fractionation would be helpful for dissecting the piRNA biology. Cultured cell lines established from ovaries and/or testes of various species or induced pluripotent stem cells would also be useful for biochemical studies. Ultimately, we strive to understand the piRNA system in humans.
Impairment of piRNA expression and mutations in PIWI proteins result in sterility in animals. Are there any human reproductive diseases, such as azoospermia, that involve piRNA clusters and PIWI loci? Whole-genome sequencing of patients may provide the answer to this question. Interestingly, the human genome contains one additional PIWI gene, PIWIL3 (HIWI3 in humans), compared with mouse and fly genomes. Since the current piRNA biogenesis model was based on insights gained from investigations mainly involving mice and flies, it obviously excludes PIWIL3. Thus, to understand the piRNA pathway correctly, new insights into the molecular function of PIWIL3 should be incorporated. The role of piRNAs in cancer is another area that warrants investigation, as there is evidence that PIWI proteins and piRNAs may have important roles in epigenetic regulation in tumorigenesis (Siddiqi and Matushansky 2012), as has been shown in gametogenesis. It will be of great interest to examine whether impairment of the functions of piRNAs and/or PIWI proteins in cancer cells would inhibit tumorigenesis.
Acknowledgments
We thank Kaoru Sato and other members of the Siomi laboratories at Keio University School of Medicine and at the Graduate School of Science, The University of Tokyo, for useful discussion and comments. H.I. is supported by JSPS (Japan Society for the promotion of Science). H.S. and M.C.S. are supported by MEXT (Ministry of Education, Culture, Sports, Science, and Technology). M.C.S. is supported by CREST (Core Research for Evolutional Science and Technology) from JST (Japanese Science and Technology Agency).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.203786.112.
References
- Anand A, Kai T 2011. The tudor domain protein Kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J 31: 870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov AL, Wang J-YS, Ramos A, Lehmann R 2006. The role of Tudor domains in germline development and polar granule architecture. Development 133: 4053–4062 [DOI] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, Cording AC, Doebley A-L, Goldstein LD, Lehrbach NJ, Le Pen J, et al. 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyret E, Liu N, Lin H 2012. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res 22: 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK 2012. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet 8: e1002617 doi: 10.1371/journal.pgen.1002617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Carrera P, Johnstone O, Nakamura A, Casanova J, Jäckle H, Lasko P 2000. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell 5: 181–187 [DOI] [PubMed] [Google Scholar]
- Cecere G, Zheng GXY, Mansisidor AR, Klymko KE, Grishok A 2012. Promoters recognized by forkhead proteins exist for individual 21U-RNAs. Mol Cell 47: 734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X 2005. MicroRNA biogenesis and function in plants. FEBS Lett 579: 5923–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, et al. 2009. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci 106: 20336–20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, Pawson T 2011. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol 12: 629–642 [DOI] [PubMed] [Google Scholar]
- Choi S-Y, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA 2006. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol 8: 1255–1262 [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. 2009. The argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower MA, Kolas NK, Noguchi J, Sarao R, Kikuchi K, Kaneko H, Kobayashi E, Kawai Y, Kozieradzki I, Landers R, et al. 2003. Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science 300: 1291–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263 [DOI] [PubMed] [Google Scholar]
- Frost RJA, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN 2010. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci 107: 11847–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S 2008. An argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D 2010. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140: 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J 2011. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J 30: 3977–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi S-Y, Sarma K, Ren H, Morris AJ, Frohman MA 2011. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase A, Knott SR, Joshua-Tor L, Hannon GJ 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature doi: 10.1038/nature11502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Nagao A, Siomi H 2011. Gatekeepers for Piwi–piRNA complexes to enter the nucleus. Curr Opin Genet Dev 21: 484–490 [DOI] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA 2005. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res 33: 2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJT, Roovers EF, Ladurner P, Berezikov E, Ketting RF 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 29: 3688–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJT, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF 2012. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet 8: e1002702 doi: 10.1371/journal.pgen.1002702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y 2011. 3′ End formation of PIWI-interacting RNAs in vitro. Mol Cell 43: 1015–1022 [DOI] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W 2010. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol 191: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z 2010. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem 285: 8148–8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. 2010. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev 24: 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M 1990. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev 4: 905–921 [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC 2009. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS 2011. Identification of piRNAs in the central nervous system. RNA 17: 1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Gu W, Shirayama M, Youngman E, Conte D, Mello CC 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AKA, Kai TT 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H 2011. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 138: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJT, Almeida MV, Roovers EF, Berezikov E, Ketting RF 2012. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J 31: 3422–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn K, Steward R 2000. The shut-down gene of Drosophila melanogaster encodes a novel FK506-binding protein essential for the formation of germline cysts during oogenesis. Genetics 156: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H 2010. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 16: 2503–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U 2008. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett 582: 1997–2003 [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC 2007. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al. 2009. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 28: 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Matsumoto N, Nishizawa T, Bonnefond L, Nakanaga K, Aoki J, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature doi: 10.1038/nature11509 [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J 2012. The cochaperone Shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell 47: 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ 2005. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development 132: 4029–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T 2010. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol 20: 724–730 [DOI] [PubMed] [Google Scholar]
- Pohlman RF, Liu F, Wang L, Moré MI, Winans SC 1993. Genetic and biochemical analysis of an endonuclease encoded by the IncN plasmid pKM101. Nucleic Acids Res 21: 4867–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ 2012. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 18: 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER 2012. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 149: 693–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R 2011. piRNA production requires heterochromatin formation in Drosophila. Curr Biol 21: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267 [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC 2009. A broadly conserved pathway generates 3′ UTR-directed primary piRNAs. Curr Biol 19: 2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Saito K, Siomi MC 2010. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell 19: 687–697 [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nishida KM, Shibuya A, Siomi MC, Siomi H 2011. Maelstrom coordinates microtubule organization during Drosophila oogenesis through interaction with components of the MTOC. Genes Dev 25: 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, Mello CC 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi S, Matushansky I 2012. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem 113: 373–380 [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2009. On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Mannen T, Siomi H 2010. How does the Royal Family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev 24: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA 2011. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Soper SFC, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A 2008. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15: 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specchia V, Piacentini L, Tritto P, Fanti L, D'Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP 2010. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463: 662–665 [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T 2000. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 14: 841–853 [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA 2009. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosakawa M, Reuter M, Yang Z, Berninger P, Oalencia A, et al. 2012. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47: 970–979 [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen C-CG, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R 2011. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 138: 4039–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD 2011. Heterotypic piRNA ping-pong requires Qin, a protein with both E3 ligase and tudor domains. Mol Cell 44: 572–584 [DOI] [PMC free article] [PubMed] [Google Scholar]