The zinc finger protein ZFP57 is expressed early in embryogenesis and down-regulated when embryonic stem cells differentiate. Liu et al. present the crystal structure of the ZFP57 DNA-binding domain in complex with fully methylated DNA. The work provides valuable insight into 5mC recognition as well as the effect of ZFP57 point mutations on DNA binding in transient neonatal diabetes.
Keywords: epigenetics, DNA methylation, imprinting, CpG islands, TGCCGC element, zinc finger protein Zfp57
Abstract
Zinc finger transcription factor Zfp57 recognizes the methylated CpG within the TGCCGC element. We determined the structure of the DNA-binding domain of Zfp57, consisting of two adjacent zinc fingers, in complex with fully methylated DNA at 1.0 Å resolution. The first zinc finger contacts the 5′ half (TGC), and the second recognizes the 3′ half (CGC) of the recognition sequence. Zfp57 recognizes the two 5-methylcytosines (5mCs) asymmetrically: One involves hydrophobic interactions with Arg178, which also interacts with the neighboring 3′ guanine and forms a 5mC–Arg-G interaction, while the other involves a layer of ordered water molecules. Two point mutations in patients with transient neonatal diabetes abolish DNA-binding activity. Zfp57 has reduced binding affinity for unmodified DNA and the oxidative products of 5mC.
Genomic imprinting is an epigenetic regulation phenomenon common in animals and plants (Feng et al. 2010). Different from regular genes, which are expressed from both paternal and maternal alleles, imprinted genes are expressed from only one of the two parental alleles in a parent-of-origin-specific manner (Feil and Berger 2007). The monoallelic expression of most known imprinted genes, localized in clusters, are regulated by DNA methylation of CpG-rich sequences known as imprinting control regions (ICRs) (Williamson et al. 2006) that originate from the oocyte or sperm. Thus, protein domains involved in discrimination of methylated versus nonmethylated cytosine in DNA are important in targeting different chromatin-modifying and chromatin remodeling activities to ICRs with or without the modification. Human diseases can result from defects in DNA methylation, including known imprinting-associated disorders (Amor and Halliday 2008; Mackay and Temple 2010; Wilkins and Ubeda 2011).
The best-known modified DNA recognition domains are those that recognize methylated cytosine in either fully methylated CpG dinucleotides by methyl-binding domains (MBDs) (Dhasarathy and Wade 2008; Guy et al. 2011) or hemimethylated CpG sites—transiently generated during DNA replication—by SET and RING finger-associated (SRA) domains (Hashimoto et al. 2009). A third class of mammalian proteins that recognize methylated DNA is the C2H2 zinc finger proteins, which preferentially bind to methylated CpG within a specific sequence (Sasai et al. 2010).
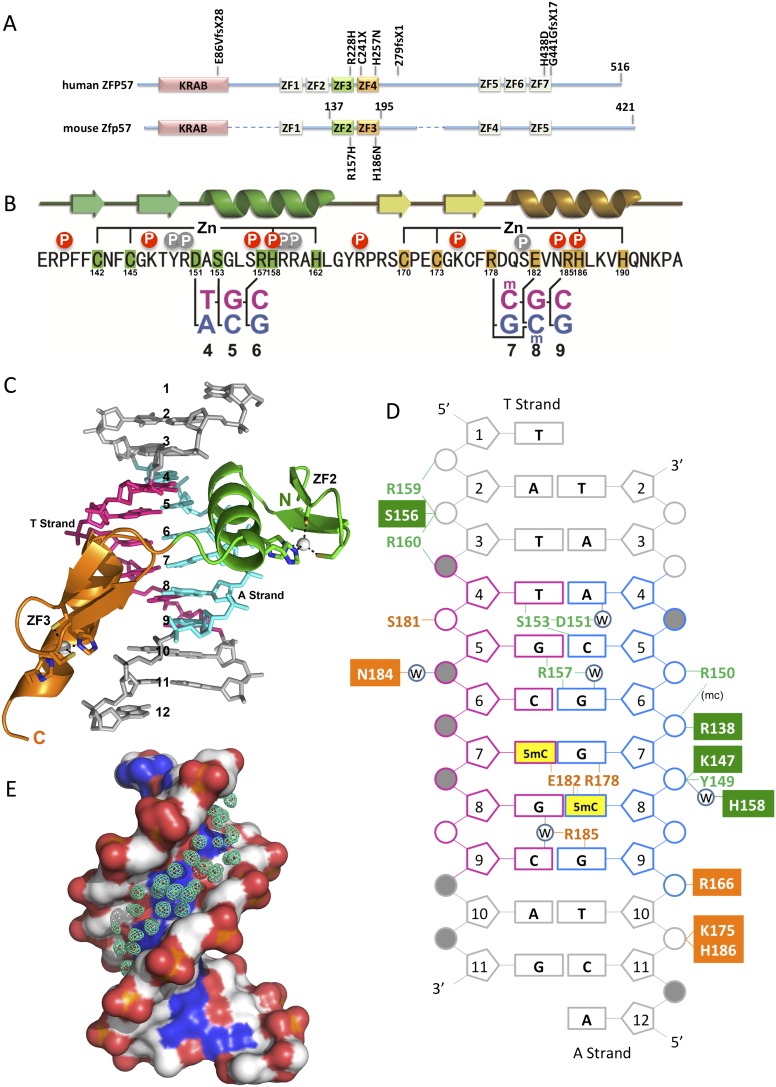
Zinc finger protein Zfp57 is one of a group of genes expressed in very early embryogenesis and down-regulated upon differentiation of embryonic stem cells (Loh et al. 2007). Loss of Zfp57 function in the developing mouse zygote causes partial neonatal lethality, whereas eliminating both the maternal and zygotic function of Zfp57 results in embryonic lethality (Li et al. 2008). Zfp57 contributes to the maintenance of both maternally and paternally imprinted loci, and at one locus, Zfp57 is also involved in imprint establishment (Li et al. 2008). ZFP57 on chromosome 6p22 encodes a protein of 516 amino acids in humans and 421 amino acids in mice, including a predicted Krüppel-associated box (KRAB) followed by seven (in humans) or five (in mice) putative C2H2 zinc fingers (Fig. 1A; Supplemental Fig. S1). ZFP57 mutations—either deletions or missense mutations—have been found in patients with transient neonatal diabetes (Fig. 1A; Mackay et al. 2008), suggesting that ZFP57 plays a comparable role in human development. Recently, a fragment of mouse Zfp57 comprising the two highly conserved and adjacent zinc fingers ZF2 and ZF3 (Fig. 1A) has been shown to recognize the TGCCGC element (with the underlined CpG methylated) found in all known murine ICRs as well as in several tens of additional loci (Quenneville et al. 2011). Here we report the atomic resolution crystal structure of the ZF2–3 fingers of mouse Zfp57 in a complex with the methylated TGCCGC element and present the mechanism of sequence- and methylation-specific recognition of DNA.
Figure 1.
Overall structure of the Zfp57 DNA-binding domain. (A) Schematic representation of human ZFP57 and mouse Zfp57. The mouse Zfp57 regions, corresponding to human ZF2 and ZF6, have mutations/deletions of the zinc ligands (see Supplemental Fig. S1). In addition, the corresponding ZF1 in the mouse protein has a point mutation in one of the histidine zinc ligands. The human mutations are listed above the sequence (Mackay et al. 2008). (B) The sequence of DNA-binding domain (ZF2–3) of mouse Zfp57 and the secondary structure are shown. (Arrows) β strands; (ribbons) α helices. The positions highlighted are responsible for Zn ligand binding and DNA base-specific interactions, and the circles with a “P” are amino acids that interact with the DNA phosphate backbone. The red circles indicate the phosphates contacted by the conserved residues at the corresponding positions between ZF2 and ZF3, and the gray circles indicate the contact specific for each zinc finger. (C) The zinc fingers bind in the major groove of DNA with ZF2 (green) and ZF3 (orange). (D) Summary of the Zfp57–DNA interactions. The T strand (in magenta) is the strand containing the 5′ T of TGCCGC, and the A strand (in blue) is the strand containing the 3′ A of the opposite strand. ZF2 residues are labeled in green, and ZF3 residues are labeled in orange. Yellow boxes represent 5mC. Filled square boxes indicate the conserved phosphate interactions by residues at the corresponding positions between ZF2 and ZF3. Filled circles indicate that the phosphate groups have alternative conformations (Supplemental Fig. S2A). Large numbers of water (w)-mediated hydrogen bonds are not shown. (E) A stream of water molecules occupies the DNA minor groove.
Results and Discussion
Overall structure
A mouse Zfp57 fragment (residues 137–195) consisting of a tandem array of two fingers ZF2–3 (Fig. 1B) was used for cocrystallization with a 10-base-pair (bp) oligonucleotide containing the fully methylated CpG within the TGCCGC element, plus a 5′-overhanging thymine on the T strand and a 5′-overhanging adenine on the A strand (Fig. 1C,D). We crystallized the protein–DNA complex in space group C2221, containing one complex per crystallographic asymmetric unit, and determined the structure to the resolution of 1.0 Å (Supplemental Table 1). The DNA molecules are coaxially stacked, with the overhanging A and T forming a Hoogsteen base pair with neighboring DNA molecules, thus forming a pseudocontinuous duplex (Supplemental Fig. S2A). The atomic resolution structure, with an overall crystallographic thermal B factor of ∼12.3 Å2, allowed us to position every atom of the 22 DNA nucleotides, every atom of protein residues of 137–192 (except the side chain of Lys193 and the last two residues at the C terminus), and 257 water molecules. Almost all polar atoms are involved in intermolecular and intramolecular as well as water-mediated interactions.
Each of the two classic C2H2 zinc fingers contain two β strands and one helix, coordinating one zinc ion tetrahedrally via two cysteines from the β strands and two histidines from the helix (Fig. 1B,C). The zinc fingers bind in the major groove of the DNA, which shows an ordered B-DNA conformation (Supplemental Table 2). Although there is no specific protein interaction in the DNA minor groove, a monolayer of ordered solvent sites (modeled as water molecules) is clearly visible (Fig. 1E). Each zinc finger recognizes 3 bp, with ZF2 interacting with the 5′ half (TGC) and ZF3 interacting with the 3′ half (CGC) of the TGCCGC element (Fig. 1C,D). The protein makes phosphate contacts spanning 9 bp, with each zinc finger making conserved contacts with the two phosphates immediately 5′ to its respective 3-bp recognition sequence (Fig. 1D). Arg138, Lys147, and His158 of ZF2 interact with the two phosphates 5′ to guanine 6 (G6), while the corresponding Arg166, Lys175, and His186 of ZF3 interact with the two phosphates 5′ to the G9 on the A strand (Fig. 1D).
DNA base-specific recognition in crystal
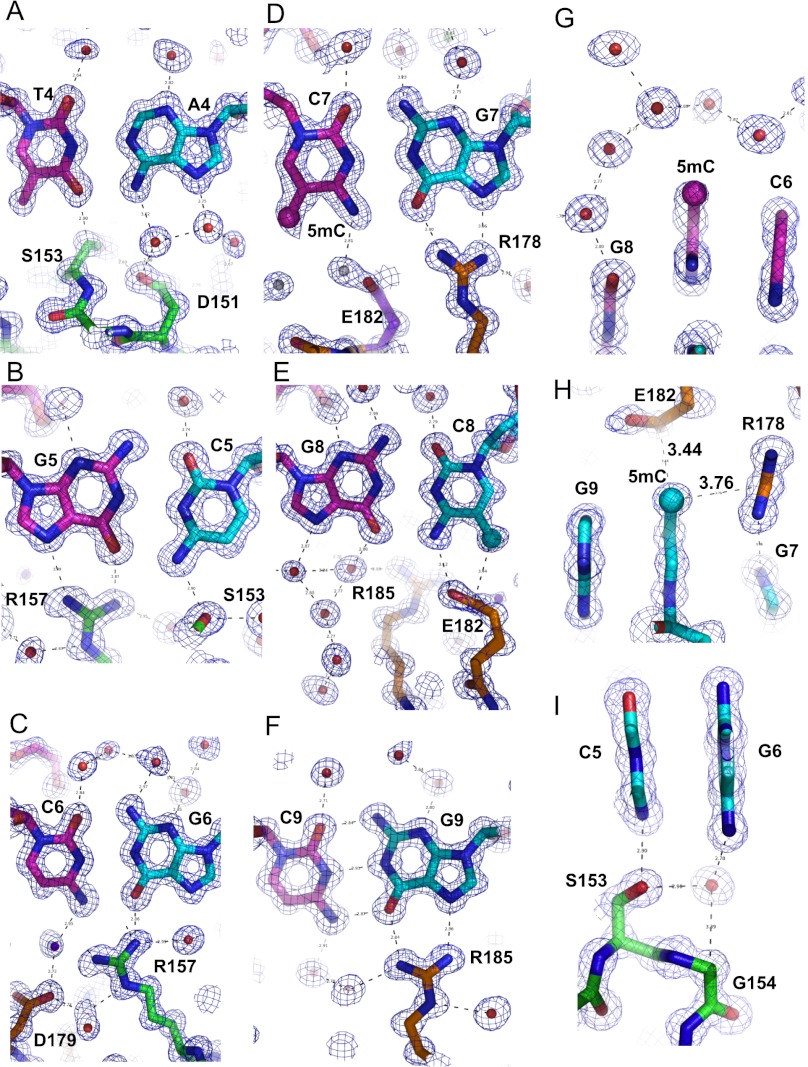
All 6 bp of the recognition sequence TGCCGC (base pairs 4–9 in Fig. 1D) have direct interactions with Zfp57 residues. The O4 atom of thymine 4 (T4) is 2.98 Å away from the Cβ atom of Ser153, forming a C=OH-C type of hydrogen bond, while the exocyclic amino group N6 (NH2) of adenine 4 (A4) forms a water-mediated hydrogen bond with Asp151 (Fig. 2A). Direct interaction with the carbonyl oxygen O4 atom of thymine distinguishes it from that of the N4 amino group of cytosine. Mutation of the T:A base pair to C:G resulted in loss of binding affinity by a factor of ∼110 (Fig. 3A).
Figure 2.
Details of Zfp57–DNA base-specific interactions. (A) The T:A base pair at position 4 interacts with Ser153 and Asp151. Electron densities (2Fo–Fc) contoured at 1σ above the mean are shown. The T strand and A strand are colored in magenta and blue, respectively. (B) The interaction of the G:C base pair at position 5 with Arg157 and Ser153. (C) The interaction of the C:G base pair at position 6 with Arg157. (D) The interaction of the 5mC:G base pair at position 7 with Arg178. (E) The interaction of the G:5mC base pair at position 8 with Glu182. (F) The interaction of the C:G base pair at position 9 with Arg185. (G) A layer of water molecules shields the methyl group of 5mC at position 7. (H) Arg178 and Glu182 are in hydrophobic contact with the methyl group of 5mC at position 8. (I) Gly154 is near the unmodified cytosine at position 5 and guanine at position 6.
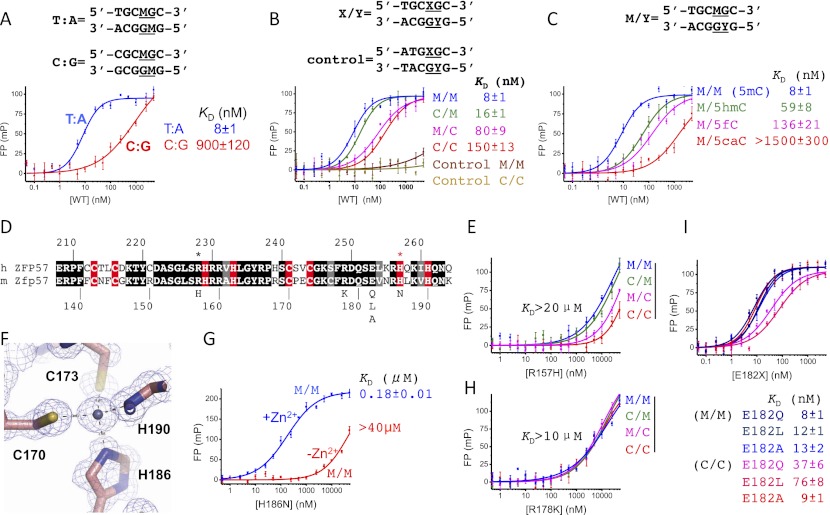
Figure 3.
The effects of DNA sequence, 5mC oxidation, and Zfp57 mutants on DNA binding. (A) The first T:A base pair of the TGCCGC element is important for recognition by Zfp57. (B) Binding affinities between ZF2–3 and TGCCGC with four different modification states (M/M, C/M, M/C, and C/C, where M is 5mC and C is Cyt). A 5′ half-mutated sequence (ATGCGC) was used for negative control. (C) The effect of 5mC oxidation ([5hmC] 5-hydroxymethylation; [5fC] 5-formylation; [5caC] 5-carboxylation) on DNA binding. (D) The sequence alignment between human ZFP57 and mouse Zfp57, with white-on-black residues being invariant among the corresponding regions of sequences examined, gray-highlighted positions conserved, and white-on-red being zinc ligands. (E) The effect of the R157H mutant on DNA binding. Although there is significantly reduced affinity, fully methylated DNA (M/M) is still preferred over hemimethylated DNA (C/M and M/C) and unmodified DNA (C/C) by this mutant. (F) His186 is one of the zinc ligands. (G) The effect of Zn2+ (1 mM ZnCl2 added in the reaction buffer) on DNA binding by the H186N mutant. (H) R178K loses the discrimination of methylation status. (I) E182 is dispensable for binding methylated DNA.
The next 5 bp are all G:C pairs. Three out of five guanines (G5, G7, and G9) make bifurcated hydrogen-bonding interactions (via the O6 and N7 atoms) with Arg157, Arg178, and Arg185, respectively (Fig. 2B,D,F). Arg157 also makes a direct hydrogen bond to the O6 atom of G6 (Fig. 2C), while Arg185 makes a water-mediated interaction with the O6 atom of G8 (Fig. 2E). Direct interactions with the O6 atom of guanine distinguish it from that of the N6 atom of adenine. Mutating each base pair to A:T at positions 4, 5, 6, and 9 (outside of the CpG dinucleotide) resulted in loss of binding (Quenneville et al. 2011).
The N4 atom of cytosine 5 (C5) forms a direct hydrogen bond with the side chain hydroxyl oxygen of Ser153 (Fig. 2B), while the two other unmodified cytosine residues (C6 and C9) are involved in water-mediated interactions (Fig. 2C,F). The two 5-methylcytosines (5mCs; C7 and C8) exhibit very different patterns of interactions. A layer of ordered water molecules encloses the methyl group of 5mC at position 7 (Fig. 2G). The methyl group of 5mC at position 8 has van der Waals contacts with the guanidino group of Arg178 and the carboxyl group of Glu182 (Fig. 2H), while one of its carboxylate oxygen atoms interacts with the N4 atom of the same 5mC residue (Fig. 2E). The residues involved in the base-specific interactions are invariant between mouse and human Zfp57 proteins (Supplemental Fig. S1) and are located, from each zinc finger, in the N-terminal portion of the helix and the preceding loop (Supplemental Fig. S2B).
Sequence and methyl-specific binding in solution
To explore the effect of DNA methylation and sequence variation on Zfp57 binding, we measured the dissociation constants (KD) between ZF2–3 fingers and double-stranded oligonucleotides containing a single CpG dinucleotide using fluorescence polarization analysis. ZF2–3 fingers bind the fully methylated TGCCGC sequence (M/M) with a KD of 8 nM (Fig. 3B). Deleting the methyl group on the T strand (C/M) increased the KD to 16 nM (a twofold weaker binding), whereas deleting the methyl group on the A strand weakened the binding by a factor of 10 with a KD of 80 nM (Fig. 3B). This difference in the contribution of the two strand-specific methylations to the binding affinity fits perfectly well with the asymmetric interactions observed in the crystal structure: 5mC at position 7 of the T strand interacts with a layer of ordered water molecules (Fig. 2G), and 5mC at position 8 of the A strand is directly involved in hydrophobic interactions (Fig. 2H). The reduction of binding affinity to the unmodified TGCCGC sequence (C/C) is the accumulative effect of removing both methyl groups (∼20-fold). Mutating 5′ TGC to ATG abolished the binding of ZF2–3, regardless of the methylation status of the 3′ CpG site (Fig. 3B). Together with Figure 3A, these data indicate that the interaction between ZF2–3 and the DNA depends on both the specific sequence and the methylation state of TGCCGC element.
Mammalian Tet (ten eleven translocation) proteins convert 5mC to 5-hydroxymethylcytosine (5hmC) (Tahiliani et al. 2009), which can be further oxidized by Tet proteins to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (He et al. 2011; Ito et al. 2011). 5hmC is a constituent of nuclear DNA, present in many tissues and cell types (Globisch et al. 2010) but relatively enriched in embryonic stem cells (Tahiliani et al. 2009; Booth et al. 2012; Yu et al. 2012) and Purkinje neurons (Kriaucionis and Heintz 2009). The effects of human ZFP57 and mouse Zfp57 on DNA methylation of ICRs were analyzed by bisulfite sequencing (Li et al. 2008; Mackay et al. 2008; Quenneville et al. 2011). Unfortunately, this method cannot discriminate 5mC from 5hmC (Huang et al. 2010; Jin et al. 2010) or 5caC from normal cytosine (He et al. 2011). We thus examined the effects of 5mC oxidation on DNA binding by ZF2–3. Because the methylation on the A strand is more important in binding, we replaced 5mC at position 8 with three different oxidation states. Each hydroxylation event from M/M to M/5hmC to M/5fC to M/5caC resulted in weaker binding (by a factor of 7, 2, and 11, respectively) (Fig. 3C). These results indicate that the effect of 5hmC on binding is similar to that of A-strand demethylation, the effect of 5fC is similar to loss of methyl groups on both strands, and the effect of 5caC is similar to nonspecific binding.
Mutations in human ZFP57 have been found in patients of transient neonatal diabetes, causing hypomethylation at multiple imprinted loci (Mackay et al. 2008). Two missense mutations occur in the region of human ZFP57, corresponding to the ZF2–3 of mouse Zfp57 (Fig. 1A). Because the sequences in this region are highly conserved between humans and mice (Fig. 3D), we examined the effects of these mutations. The R228H mutation in human ZFP57 corresponds to R157H in mouse Zfp57, which abolished ZF2–3 binding of DNA (Fig. 3E), probably due to the disruption of the interactions between Arg157 and G5 and possibly G6 (Fig. 2B,C). H257N in human ZFP57 corresponds to H186N in the mouse protein. His186 is one of the four zinc ligands in ZF3 (Fig. 3F) and the H186N mutant decreased DNA binding by a factor of at least 5000 (Fig. 3G). Interestingly, adding exogenous Zn2+ rescued the binding of methylated DNA by the H186N mutant considerably (KD of 180 nM) (Fig. 3G), to the level of wild-type protein binding of the unmodified TGCCGC element (KD of 150 nM) (Fig. 3B). Apparently, the additional zinc ions allowed the mutant protein to bind and be stabilized. The weakened DNA binding by the mutants might provide an opportunity for other enzymes (such as Tet proteins, APOBEC deaminases, and DNA repair enzymes TDG and MBD4) to compete and further modify 5mC, resulting in the hypomethylation phenomenon associated with human mutations (Mackay et al. 2008).
We also made conservative substitutions for the residues (R178K and E182Q) surrounding the methyl group of 5mC at position 8 of the A strand (Fig. 2H). The R178K mutant abolished DNA binding to the same degree as that of R157H (Fig. 3, cf. H and E). Both arginines are involved in the base-specific interactions with G5 (Arg157) and G7 (Arg178), respectively (Fig. 2B,D). However, while R178K is insensitive to the states of DNA methylation, R157H retains discriminative, albeit much less so, of fully, hemimethylated, and nonmethylated sequences (Fig. 3, cf. H and E), suggesting that Arg178 is a fundamental element for methyl group recognition.
As expected, E182Q had no effect on methylated DNA binding (Fig. 3I) because the mutant could maintain the same interactions with the 5mC at position 8 (Fig. 2E). Similarly, E182L (which might preserve the hydrophobic interaction with the methyl group) and E182A have a minimal effect on methylated DNA binding (decreasing the binding by a factor of 1.5), suggesting that the side chain of Glu182 is dispensable for methyl group recognition. Unexpectedly, E182Q and E182L have enhanced binding affinity to unmodified DNA (C/C) by a factor of 4 and 2, respectively, in comparison with that of the wild-type protein (Fig. 3I). More dramatically, the E182A mutant binds the unmodified and methylated DNA equally well. Apparently, the side chain of Glu182 (the size and the charge) negatively impacts the binding of unmodified C8. We note that the residue corresponding to Glu182 is Gly154 in ZF2 (Supplemental Fig. S2B), which is near the unmodified C5 (Fig. 2I).
Zfp57 and MBD proteins share a common mode of 5mC recognition
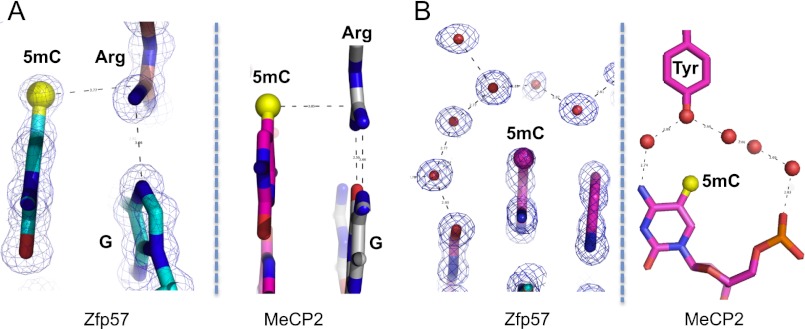
Two other protein domains—namely, SET and RING-associated (SRA) domain (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008; Rajakumara et al. 2011) and MBD (Ohki et al. 2001; Ho et al. 2008; Scarsdale et al. 2011)—have been structurally characterized in their recognition of 5mC. The mammalian SRA domain flips the 5mC of a hemimethylated CpG into a binding cage with the methyl group in hydrophobic interactions with the side chains of two tyrosines and one serine (Supplemental Fig. S3A). MBDs bind the symmetrical, fully methylated CpG site with two arginines, each interacting with one guanine in a bifurcated hydrogen-bonding pattern (Supplemental Fig. S3B–D). Like Arg178 of Zfp57, the two MBD arginines are also engaged in van der Waals contacts with the methyl group of the neighboring 5′ 5mC of the same strand (namely, a 5mC–Arg-G interaction). Unlike Zfp57, which uses one 5mC–Arg-G to interact with the fully methylated DNA within a nonpalindromic recognition sequence in an asymmetric way, the MBD uses two pairs of 5mC–Arg-G interactions for the palindromic fully methylated CpG site (Fig. 4A). In addition, Zfp57 uses a layer of hydration for one of the two methyl groups to increase the binding by a factor of 2, whereas MBD uses a tyrosine-associated water-mediated interaction with one of the two methyl groups to further enhance the methyl DNA binding (Fig. 4B; Ho et al. 2008; Scarsdale et al. 2011). In the latter case, the hydroxyl oxygen atom of the tyrosine functions as one of the water molecules in a water network surrounding the methyl group. Mutating the corresponding tyrosine to phenylalanine (Y123F) in MeCP2 reduced the affinity for methylated DNA only by a factor of ∼1.5 (Ho et al. 2008), the same effect as we observed for the E182L and E182A mutations in Zfp57 (Fig. 3I) or as lost the methyl–water interaction in the T strand (twofold). In summary, we discovered that Zfp57 and MBD proteins use an evolutionally conserved mechanism (5mC–Arg-G) for recognition of 5mC, despite their unrelated protein sequences and structures.
Figure 4.
A common mode of the 5mC–Arg-G interaction between Zfp57 and MBDs. (A) The 5mC–Arg-G interaction in Zfp57 (left panel) and MeCP2 (right panel). MBD proteins use two pairs of 5mC–Arg-G interactions for recognition of symmetric 5mCpG dinucleotides. (B) A layer of hydration surrounds one of the 5mC methyl group in Zfp57 (left panel) and MeCP2 (right panel).
Unlike MBD proteins, which use a pair of 5mC–Arg-G interactions to recognize the symmetric 5mCpG dinucleotide, Zfp57 binds 5mCpG asymmetrically within an asymmetric 6-bp sequence (with the methylation of the A strand being more important for binding than that of the T strand). Interestingly, the asymmetric recognition has also been observed for the binding of an unmodified CpG by CTCF (Hark et al. 2000; Renda et al. 2007). The TGCCGC element is part of the sequence recognized by CTCF at the mouse H19/Igf2 ICR locus (Hark et al. 2000; Quenneville et al. 2011). The methylated CpG site recognized by Zfp57 corresponds to the single conserved CpG of the five CTCF-binding sites of mouse H19/Igf2 ICRs. CTCF binding was inhibited by T-strand methylation, whereas hemimethylation of the A strand had a minimal effect (Hark et al. 2000; Renda et al. 2007). CTCF binding of hemimethylated ICR, transiently generated during semiconservative DNA replication, had been suggested as a possible mechanism for passively demethylating the paternal chromosome in the female germline (Hark et al. 2000). Intriguingly, both Zfp57 and CTCF are capable of binding a hemimethylated CpG site with the T strand unmodified and the A strand methylated, but with much reduced affinities for the T-strand-methylated and A-strand-unmodified hemimethylated CpG site.
Another possible way to generate an asymmetric modification is via Tet-mediated 5mC oxidation, generating asymmetric hydroxymethylation at CpG sequences (Yu et al. 2012), with one strand methylated and the opposite strand hydroxymethylated (M/H). Recent analysis revealed that 5hmC is strand-biased toward G-rich sequences (Stroud et al. 2011; Yu et al. 2012). The functional significance of asymmetric hydroxymethylation at the CpG site within an asymmetric sequence recognized by Zfp57 (as well as CTCF) has yet to be studied.
Despite tremendous advances in understanding the molecular machinery of DNA methylation, we do not have an adequate explanation for why specific sequences at ICRs and other targeted loci become methylated or demethylated via either an enzyme-catalyzed active pathway (He et al. 2011) or a DNA replication-dependent passive process (Inoue and Zhang 2011; Inoue et al. 2011). Thus, the discovery of Zfp57 recognition of the TGCCGC element, which is present at all known murine and some human ICRs (Supplemental Material) and CpG islands (Quenneville et al. 2011), provided the first example of sequence-specific as well as methylation status recognition. We further analyzed quantitatively the 15 experimentally determined ICRs in mouse embryos (Kobayashi et al. 2006) and the corresponding regions in sperm and oocytes (Tomizawa et al. 2011). We found that the frequencies of occurrence of the TGCCGC element in the embryonic ICRs (∼1.65 × 10−3), the gametic ICRs (∼1.57 × 10−3), and CpG islands (∼1.29 × 10−3) are >20-fold higher in comparison with that of the whole genome (0.06 × 10−3) (Supplemental Table 3). Our atomic model of Zfp57 recognition of the TGCCGC element defines one of the initial steps along the Zfp57-mediated molecular mechanism in the maintenance of DNA methylation within a specific sequence.
Materials and methods
Expression, purification, mutagenesis, and crystallography
The DNA fragment encoding mouse Zfp57 residues 137–195 (pXC1127) and its mutants (R157H, H186N, R178K, E182Q, E182L, and E182A) were cloned into pGEX6P-1 vector and expressed in Escherichia coli BL21 (DE3) Codon-plus RIL. Cells were cultured in Luria-Bertani medium at 37°C until OD600 reached 1.0 before inducing protein expression with 0.2 mM isopropyl β-D-1-thiogalactopyranoside overnight at 16°C. Cells were harvested and lysed in 20 mM Tris-HCl (pH 7.5), 250 mM NaCl, and 5% (v/v) glycerol by sonication, followed by centrifugation at 18,000 rpm for 60 min. The recombinant GST-tagged protein was purified with Glutathione Sepharose 4B, and the GST tag was removed by incubating with PreScission protease overnight at 4°C. Protein was further purified to homogeneity by three columns of HiTrap-Q, HiTrap-SP, and Superdex-200 (16/60) and concentrated to 25 mg mL−1 in 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5% (v/v) glycerol, and 1 mM tris(2-carboxyethyl)phosphine (TCEP). The yields of the mutant proteins were lower than that of the wild-type protein: ∼90% (R178K), 85% (E182Q), 73% (E182A), 65% (H186N), 60% (R157H), and 30% (E182L).
The purified ZF2-3 protein was incubated with annealed oligonucleotides in a molar ratio of ∼1:1 for 1 h on ice before crystallization. The final solution contained ∼1.5 mM protein/DNA complex in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5% glycerol, and 0.5 mM TCEP. Crystals were obtained by the sitting-drop method; the mother liquor contained 25%–30% 2-methyl-2,4-pentanediol, 12.5%–17.5% polyethylene glycol 8000, 100 mM CaCl2, and 100 mM acetate (pH 4.6). Rectangular crystals with a maximum dimension of 0.5 mm grew within 3 d at 16°C. X-ray diffraction data are summarized in Supplemental Table 1.
Fluorescence-based DNA-binding assay
Fluorescence polarization assays were performed in 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5% (v/v) glycerol, and 1 mM TCEP at room temperature (∼22°C) using a Synergy 4 Microplate Reader (BioTek). The wavelengths of fluorescence excitation and emission were 490 and 524 nm, respectively. Fluorescent-labeled DNA probe (1 nM) and various amounts of ZF2-3 protein (wild type or mutants) with a final volume of 50 μL were incubated in a 384-well plate for 0.5 h before measurement. The sequences of 6-carboxy-fluorescein (FAM)-labeled double-stranded oligonucleotides were FAM-5′-TATTGCXGCAG-3′and 3′-TAACGGYGTCA-5′ (where X and Y were C, 5mC, 5hmC, 5fC, or 5caC). The control DNA sequences were FAM-5′-CCATGXGCTGAC-3′ and 3′-GGTACGYGACTG-5′ (where X and Y were C or 5mC). Curves were fit individually using Origin 7.5 software (OriginLab). KD were calculated as [mP] = [maximum mP] × [C]/(KD + [C]) + [baseline mP], where [mP] is millipolarization and [C] is protein concentration. Averaged KD and its standard error were reported.
Accession numbers
The coordinates and structure factors of the mouse Zfp57 DNA-binding domain–DNA complexes have been deposited in Protein Data Bank with accession numbers 4GZN.
Acknowledgments
We sincerely thank Brenda Baker of New England Biolabs for DNA oligonucleotide synthesis; Taiping Chen of the M.D. Anderson Cancer Center for the assembly of human DMR sequences; and John R. Horton for maintaining our local X-ray facility, help with X-ray data collection in the Advanced Photon Source, and comments on the manuscript. U.S. National Institutes of Health (grant GM049245-18) supported this work. Y.L. initiated this project and performed all experiments. H.T. and H.S. analyzed mouse ICR sequences. X. Z. and X.C. organized and designed the scope of the study, and all were involved in analyzing data and preparing the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.202200.112.
References
- Amor DJ, Halliday J 2008. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod 23: 2826–2834 [DOI] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M 2008. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455: 818–821 [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S 2008. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455: 822–825 [DOI] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937 [DOI] [PubMed] [Google Scholar]
- Dhasarathy A, Wade PA 2008. The MBD protein family-reading an epigenetic mark? Mutat Res 647: 39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Berger F 2007. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23: 192–199 [DOI] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W 2010. Epigenetic reprogramming in plant and animal development. Science 330: 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T 2010. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5: e15367 doi: 10.1371/journal.pone.0015367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Cheval H, Selfridge J, Bird A 2011. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol 27: 631–652 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X 2008. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455: 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Cheng X 2009. UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics 4: 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD 2008. MeCP2 binding to DNA depends upon hydration at Methyl-CpG. Mol Cell 29: 525–531 [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A 2010. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 5: e8888 doi: 10.1371/journal.pone.0008888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y 2011. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y 2011. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21: 1670–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP 2010. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38: e125 doi: 10.1093/nar/gkg223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H 2006. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): Paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res 113: 130–137 [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC 2008. A maternal–zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH 2007. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21: 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Temple IK 2010. Transient neonatal diabetes mellitus type 1. Am J Med Genet C Semin Med Genet 154C: 335–342 [DOI] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 40: 949–951 [DOI] [PubMed] [Google Scholar]
- Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M 2001. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell 105: 487–497 [DOI] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE 2011. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev 25: 137–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV 2007. Critical DNA binding interactions of the insulator protein CTCF: A small number of zinc fingers mediate strong binding, and a single finger–DNA interaction controls binding at imprinted loci. J Biol Chem 282: 33336–33345 [DOI] [PubMed] [Google Scholar]
- Sasai N, Nakao M, Defossez PA 2010. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res 38: 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsdale JN, Webb HD, Ginder GD, Williams DC Jr 2011. Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence. Nucleic Acids Res 39: 6741–6752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE 2011. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 12: R54 doi: 10.1186/gb-2011-12-6-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H 2011. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 138: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Ubeda F 2011. Diseases associated with genomic imprinting. Prog Mol Biol Transl Sci 101: 401–445 [DOI] [PubMed] [Google Scholar]
- Williamson CM, Turner MD, Ball ST, Nottingham WT, Glenister P, Fray M, Tymowska-Lalanne Z, Plagge A, Powles-Glover N, Kelsey G, et al. 2006. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet 38: 350–355 [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149: 1368–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]