The MALAT1 long noncoding RNA is a Pol II transcript lacking a poly(A) tail that is nonetheless abundantly expressed at a level comparable with many protein-coding housekeeping genes. The Sharp laboratory now identifies a highly conserved AU-rich triple-helical structure that protects the 3′ ends of these long noncoding RNAs from exonucleases. This structure supports the transport, stability, and translation of an RNA, while also allowing efficient repression by microRNAs. The work provides new insight into how transcripts lacking poly(A) tails are stabilized and regulated.
Keywords: MALAT1, MEN β, NEAT1, translation, uridylation, RNA decay, RNA stability
Abstract
The MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) locus is misregulated in many human cancers and produces an abundant long nuclear-retained noncoding RNA. Despite being transcribed by RNA polymerase II, the 3′ end of MALAT1 is produced not by canonical cleavage/polyadenylation but instead by recognition and cleavage of a tRNA-like structure by RNase P. Mature MALAT1 thus lacks a poly(A) tail yet is expressed at a level higher than many protein-coding genes in vivo. Here we show that the 3′ ends of MALAT1 and the MEN β long noncoding RNAs are protected from 3′–5′ exonucleases by highly conserved triple helical structures. Surprisingly, when these structures are placed downstream from an ORF, the transcript is efficiently translated in vivo despite the lack of a poly(A) tail. The triple helix therefore also functions as a translational enhancer, and mutations in this region separate this translation activity from simple effects on RNA stability or transport. We further found that a transcript ending in a triple helix is efficiently repressed by microRNAs in vivo, arguing against a major role for the poly(A) tail in microRNA-mediated silencing. These results provide new insights into how transcripts that lack poly(A) tails are stabilized and regulated and suggest that RNA triple-helical structures likely have key regulatory functions in vivo.
Processing the 3′ end of a nascent transcript is critical for termination of RNA polymerase and for ensuring the proper functionality of the mature RNA. During normal development and in the progression of diseases such as cancer, 3′ end cleavage site usage frequently changes, resulting in additional sequence motifs being included (or excluded) at the 3′ ends of mature RNAs that can affect the transcripts' stability, subcellular localization, or function (for review, see Lutz and Moreira 2011). Virtually all long RNA polymerase II (Pol II) transcripts terminate in a poly(A) tail that is generated by endonucleolytic cleavage followed by the addition of adenosine (A) residues in a nontemplated fashion (Moore and Sharp 1985; for review, see Colgan and Manley 1997; Zhao et al. 1999; Proudfoot 2004). However, recent large-scale studies of the human transcriptome indicate that transcription is pervasive throughout the genome (for review, see Wilusz et al. 2009) and suggest that a significant fraction (possibly >25%) of long Pol II transcripts present in cells may lack a canonical poly(A) tail (Cheng et al. 2005; Wu et al. 2008; Yang et al. 2011a). Although some of these transcripts are likely degradation intermediates, there are well-characterized stable Pol II transcripts that lack a poly(A) tail, such as replication-dependent histone mRNAs. Following U7 small nuclear RNA (snRNA)-guided endonucleolytic cleavage at their 3′ ends, histone mRNAs have a highly conserved stem–loop structure in their 3′ untranslated regions (UTRs) that is functionally analogous to a poly(A) tail, as it ensures RNA stability and enhances translational efficiency (for review, see Marzluff et al. 2008).
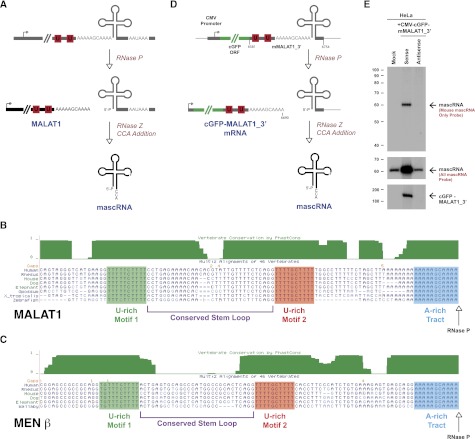
Recent work has identified additional Pol II transcripts that are subjected to noncanonical 3′ end processing mechanisms (for review, see Wilusz and Spector 2010). In particular, enzymes with well-known roles in other RNA processing events, such as pre-mRNA splicing (Box et al. 2008) and tRNA biogenesis, have been shown to cleave certain nascent transcripts to generate mature 3′ ends. In its well-characterized role, RNase P endonucleolytically cleaves tRNA precursors to produce the mature 5′ termini of functional tRNAs (for review, see Kirsebom 2007). One of us (Wilusz et al. 2008) previously showed that RNase P also generates the mature 3′ end of the long noncoding RNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), also known as NEAT2, despite the presence of a nearby polyadenylation signal. Cleavage by RNase P simultaneously generates the mature 3′ end of the ∼6.7-kb MALAT1 noncoding RNA and the 5′ end of a small tRNA-like transcript (Fig. 1A). Additional enzymes involved in tRNA biogenesis, including RNase Z and the CCA-adding enzyme, further process the small RNA to generate the mature 61-nucleotide (nt) transcript known as mascRNA (MALAT1-associated small cytoplasmic RNA) (Wilusz et al. 2008).
Figure 1.
The 3′ end of MALAT1 is highly conserved and cleaved by RNase P. (A) Although there is a polyadenylation signal at the 3′ end of the MALAT1 locus, MALAT1 is primarily processed via an upstream cleavage mechanism that is mediated by the tRNA biogenesis machinery. RNase P cleavage simultaneously generates the mature 3′ end of MALAT1 and the 5′ end of mascRNA. The tRNA-like small RNA is subsequently cleaved by RNase Z and subjected to CCA addition. (B) Immediately upstream of the MALAT1 RNase P cleavage site (denoted by an arrow) is a highly evolutionarily conserved A-rich tract. Further upstream are two nearly perfectly conserved U-rich motifs separated by a predicted stem–loop structure. (C) Similar motifs are present upstream of the MEN β RNase P cleavage site. (D) The CMV-cGFP-mMALAT1_3′ sense expression plasmid was generated by placing nucleotides 6581–6754 of mouse MALAT1 downstream from the cGFP ORF. No polyadenylation signal is present at the 3′ end. (E) After transfecting the plasmids into HeLa cells, Northern blots were performed to detect expression of mascRNA and cGFP-MALAT1_3′ RNA. To verify that the 3′ end of cGFP-MALAT1_3′ RNA was accurately generated and that no additional nucleotides were added post-transcriptionally, RNase H digestion was performed prior to Northern blot analysis.
The long MALAT1 transcript is retained in the nucleus in nuclear speckles (Hutchinson et al. 2007), where it has been proposed to regulate alternative splicing (Tripathi et al. 2010), transcriptional activation (Yang et al. 2011b), and the expression of nearby genes in cis (Nakagawa et al. 2012; Zhang et al. 2012). Although the MALAT1 locus appears to be dispensable for mouse development (Eissmann et al. 2012; Nakagawa et al. 2012; Zhang et al. 2012), MALAT1 is overexpressed in many human cancers (Ji et al. 2003; Lin et al. 2007; Lai et al. 2011), suggesting that it may have an important function during cancer progression. Furthermore, chromosomal translocation breakpoints (Davis et al. 2003; Kuiper et al. 2003; Rajaram et al. 2007) as well as point mutations and short deletions (Ellis et al. 2012) associated with cancer have been identified within MALAT1.
Despite lacking a canonical poly(A) tail, MALAT1 is among the most abundant long noncoding RNAs in mouse and human cells. In fact, MALAT1 is expressed at a level comparable with or higher than many protein-coding genes, including β-actin or GAPDH (Zhang et al. 2012). How, then, is the 3′ end of MALAT1 protected from degradation? As previously noted, although the 3′ end of MALAT1 is generated via a mechanism distinct from canonical cleavage/polyadenylation, the mature MALAT1 transcript has a short A-rich tract on its 3′ end (Wilusz et al. 2008; Wilusz and Spector 2010). Rather than being added on post-transcriptionally, as occurs during polyadenylation, the MALAT1 poly(A) tail-like moiety is encoded in the genome and thus is part of the nascent transcript (Fig. 1A). From humans to fish, this A-rich motif, along with two upstream U-rich motifs and a stem–loop structure, is highly evolutionarily conserved (Fig. 1B), suggesting the functional relevance of these sequences. Similar highly conserved A- and U-rich motifs are present at the 3′ end of the MEN β long nuclear-retained noncoding RNA, also known as NEAT1_2, which is also processed at its 3′ end by RNase P (Fig. 1C; Sunwoo et al. 2009). However, the function of these motifs as well as the molecular mechanism by which the 3′ ends of MALAT1 and MEN β are protected to allow the transcripts to accumulate to high levels have not been investigated.
Here, we use a newly developed expression plasmid that accurately recapitulates MALAT1 3′ end processing in vivo to show that these highly conserved A- and U-rich motifs form a triple-helical structure. Formation of the triple helix does not affect RNase P processing or mascRNA biogenesis but is instead critical for protecting the 3′ end of MALAT1 from 3′–5′ exonucleases. Surprisingly, when the 3′ end of MALAT1 or MEN β was placed downstream from an ORF, the transcript was efficiently translated in vivo despite the absence of a poly(A) tail. The triple helix structure thus strongly promotes both RNA stability and translation, suggesting that these long noncoding RNAs may interact with the protein synthesis machinery or even be translated under certain conditions. In addition, mutational analysis was used to show that the RNA stability and translational control functions can be separated. Finally, as this expression system provides a unique way to generate a stable transcript lacking a poly(A) tail in vivo, we explored the role of the poly(A) tail in microRNA-mediated repression. These results provide important new insights into how MALAT1, MEN β, and likely other transcripts that lack a poly(A) tail are stabilized, regulated, and thus able to perform important cellular functions.
Results
Generation of an expression plasmid that accurately recapitulates MALAT1 3′ end processing
Although it is clear that MALAT1 is cleaved to generate mascRNA (Wilusz et al. 2008), a plasmid expression system that recapitulates this processing event in vivo has not been reported. We thus set out to generate such a plasmid by inserting downstream from a CMV promoter the coral green fluorescent protein (cGFP) ORF followed by a 174-nt fragment of the 3′ end of the mouse MALAT1 locus (nucleotides 6581–6754 of mMALAT1) (Fig. 1D). This region, denoted as mMALAT1_3′, is highly evolutionarily conserved from humans to zebrafish (Fig. 1B) and includes the well-conserved U- and A-rich motifs, the RNase P cleavage site (after nucleotide 6690), and mascRNA (nucleotides 6691–6748). As a control, a plasmid with the mMALAT1_3′ region cloned downstream from cGFP in the antisense direction was generated to verify that mascRNA expression is dependent on processing from the CMV-driven transcript.
The CMV-cGFP-mMALAT1_3′ sense and antisense plasmids were transiently transfected into human HeLa cells, and total RNA was isolated 24 h later. For this system to accurately recapitulate MALAT1 3′ end processing, it must generate two transcripts: the ∼850-nt cGFP-MALAT1_3′ RNA and mature (61-nt) mouse mascRNA that has been processed by RNases P and Z as well as by the CCA-adding enzyme (Fig. 1D). There are four sequence changes between mouse and human mascRNA (Wilusz et al. 2008), allowing us to design oligo probes that either distinguish between the homologs (probe denoted as “mouse mascRNA only” in Fig. 1E) or detect both mouse and human mascRNA (probe denoted as “all mascRNA” in Fig. 1E) by Northern blot analysis. In cells transfected with the sense, but not antisense, expression plasmid, mature mascRNA was generated and expressed ∼16-fold over the level observed in mock-treated cells (Fig. 1E). 3′ RACE PCR was used to confirm that mascRNA generated from the plasmid was properly processed and had CCA post-transcriptionally added to its 3′ end (data not shown). In parallel, mutant mascRNA transcripts expressed using this plasmid were subjected to CCACCA addition and rapidly degraded in vivo (Supplemental Fig. 1), confirming our previous finding that the CCA-adding enzyme plays a key role in tRNA quality control (Wilusz et al. 2011). We thus conclude that our plasmid generates bona fide mascRNA.
To determine whether the sense plasmid expresses cGFP-MALAT1_3′ RNA that is stable and properly processed by RNase P at its 3′ end in vivo, total RNA from the transfections was first hybridized to an oligo complementary to near the 3′ end of the cGFP ORF and subjected to RNase H digestion. Cleavage of the transcript to a smaller size allowed Northern blots with a high resolution to be performed to verify the accuracy of RNase P cleavage. A single band of the expected size (190 nt) was observed with the sense, but not antisense, plasmid (Fig. 1E). These results indicate that the cGFP-MALAT1_3′ sense primary transcript is efficiently cleaved by RNase P to generate both expected mature transcripts (Fig. 1D) and thus accurately recapitulates 3′ end processing of MALAT1 in vivo. The antisense plasmid likely failed to produce a stable cGFP mRNA, as the transcript contained no functional polyadenylation signals, causing the transcript to be rapidly degraded by nuclear surveillance pathways.
As mascRNA is efficiently produced from the CMV promoter-driven transcript (Fig. 1E), we conclude that the MALAT1 promoter is not required for the recruitment of RNase P or any of the other tRNA processing factors to the nascent RNA. Furthermore, we found that the only region of the MALAT1 primary transcript that is required for mascRNA generation in vivo is the tRNA-like structure itself (Supplemental Fig. 2). Consistent with current models of substrate recognition by RNase P (Kirsebom 2007), the enzyme will probably recognize and cleave any tRNA-like structure, regardless of the promoter used to generate the transcript. Indeed, placing the MEN β tRNA-like structure downstream from cGFP in our expression system similarly resulted in efficient RNase P cleavage (Supplemental Fig. 3).
The conserved U-rich motifs protect the 3′ end of MALAT1 from degradation
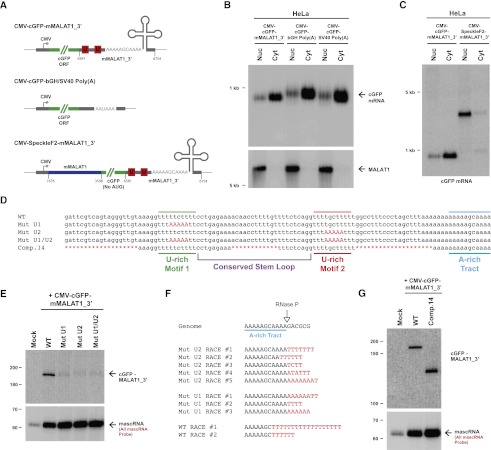
As the highly conserved U- and A-rich motifs present immediately upstream of the MALAT1 RNase P cleavage site were not required for mascRNA biogenesis (Fig. 1B; Supplemental Fig. 2), we hypothesized that they may instead function to prevent nuclear export of MALAT1 and/or stabilize the long noncoding RNA post-RNase P cleavage. Using biochemical fractionation to separate nuclear and cytoplasmic total RNA from transfected HeLa cells, we found that the cGFP-MALAT1_3′ reporter RNA was efficiently exported to the cytoplasm (Fig. 2B). In fact, the transcript was exported as efficiently as a cGFP transcript ending in a canonical poly(A) tail (Fig. 2A,B). Therefore, the 3′ end of MALAT1 does not function in nuclear retention. We instead identified a region within the body of mouse MALAT1 (nucleotides 1676–3598) that, when inserted into our expression construct (to generate the CMV-SpeckleF2-mMALAT1_3′ plasmid) (Fig. 2A), was sufficient to cause nuclear retention (Fig. 2C). This is consistent with previous reports that indicated that this region is important for targeting endogenous MALAT1 to nuclear speckles (Tripathi et al. 2010; Miyagawa et al. 2012).
Figure 2.
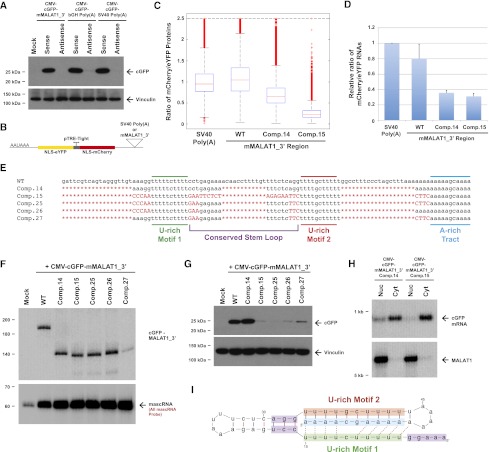
The U-rich motifs inhibit uridylation and degradation of the 3′ end of MALAT1. (A) Schematics of cGFP expression plasmids used in this study. (Middle) To generate a cGFP transcript ending in a canonical poly(A) tail, the mMALAT1_3′ region was replaced with either the bovine growth hormone (bGH) or the SV40 polyadenylation signal. (Bottom) To generate a nuclear-retained cGFP transcript, nucleotides 1676–3598 of mMALAT1 was placed upstream of cGFP. (B) Transfected HeLa cells were fractionated to isolate nuclear and cytoplasmic total RNA, which was then subjected to Northern blot analysis with a probe to the cGFP ORF. A probe to endogenous MALAT1 was used as a control for fractionation efficiency. (C) The SpeckleF2-MALAT1_3′ transcript was efficiently retained in the nucleus. (D) Mutations or deletions (denoted in red) were introduced into the mMALAT1_3′ region of the CMV-cGFP-mMALAT1_3′ expression plasmid. (E) The wild-type (WT) or mutant plasmids were transfected into HeLa cells, and Northern blots were performed. RNase H treatment was performed prior to the Northern blot that detected cGFP-MALAT1_3′ RNA. (F) A ligation-mediated 3′ RACE approach was used to examine the 3′ ends of cGFP-MALAT1_3′ transcripts undergoing degradation. Nucleotides added post-transcriptionally are in red. (G) RNase H treatment followed by Northern blotting was used to show that the cGFP-MALAT1_3′ Comp.14 transcript is stable. As 51 nt were deleted to generate the Comp.14 transcript, a band of only 139 nt is expected.
To instead explore a possible role for the highly conserved U-rich motifs in MALAT1 RNA stability, we generated and transfected cGFP-mMALAT1_3′ expression plasmids containing 5-nt mutations in U-rich motif 1, U-rich motif 2, or both motifs (Fig. 2D). These mutations had no effect on RNase P cleavage or mascRNA biogenesis (Fig. 2E, bottom) but caused the mature cGFP-MALAT1_3′ RNA to be efficiently degraded (Fig. 2E, top). Introducing similar mutations into the nuclear-retained reporter transcript also caused the RNA to be undetectable by Northern blot analysis (Supplemental Fig. 4B), indicating that U-rich motifs 1 and 2 are both required for stabilizing the 3′ end of MALAT1 in the nucleus and cytoplasm.
A ligation-based 3′ RACE PCR approach was used to gain insight into the mechanism by which the mutant transcripts are degraded. In addition to detecting transcripts simply degraded from their 3′ ends to various extents, we surprisingly detected numerous cGFP-MALAT1_3′ transcripts ending in short post-transcriptionally added U-rich tails (10 out of 56 sequenced RACE clones), implicating uridylation in the degradation of both the wild-type and mutant MALAT1 3′ ends (Fig. 2F). Several degradation patterns were observed: (1) untemplated adenylation of the MALAT1 3′ end prior to uridylation (e.g., Mut U1 RACE #1), (2) addition of a U-rich tail to the full-length transcript (e.g., Mut U2 RACE #1), and (3) partial degradation of the 3′ end prior to uridylation (e.g., wild-type RACE #1) (Fig. 2F). This last pattern is particularly interesting, as it suggests that a 3′–5′ exonuclease stalled as it was degrading the MALAT1 3′ end. The U-tail was then likely added to provide a new single-stranded tail for an exonuclease to recognize and restart the decay process (Houseley et al. 2006). We also detected uridylated decay intermediates using the nuclear-retained reporter transcript (Supplemental Fig. 4C), indicating that uridylation likely occurs in both the nucleus and the cytoplasm. These results indicate that U-rich motifs 1 and 2 are likely critical for stabilizing the 3′ end of MALAT1 by preventing uridylation and degradation by 3′–5′ exonucleases.
A triple helix forms at the 3′ ends of MALAT1 and MEN β
Having identified U-rich motifs 1 and 2 as being critical for MALAT1 3′ end stability, we investigated the minimal sequence elements required to stabilize the 3′ end of the cGFP-MALAT1_3′ transcript. Using extensive mutagenesis, we found that 51 of the 110 nt at the 3′ end of MALAT1 (Comp.14) (Fig. 2D) can be removed with little or no effect on cGFP-MALAT1_3′ RNA stability (Fig. 2G; Supplemental Fig. 5). Consistent with the evolutionary conservation patterns of MALAT1 (Fig. 1B) and MEN β (Fig. 1C), the well-conserved A- and U-rich motifs as well as the bottom half of the conserved stem–loop are required for cGFP-MALAT1_3′ stability (Figs. 2D, 3A). In contrast, more divergent regions, such as the sequences between U-rich motif 2 and the A-rich tract, either are dispensable or have only a minor supporting role in stabilizing the 3′ end of MALAT1 (Supplemental Fig. 5).
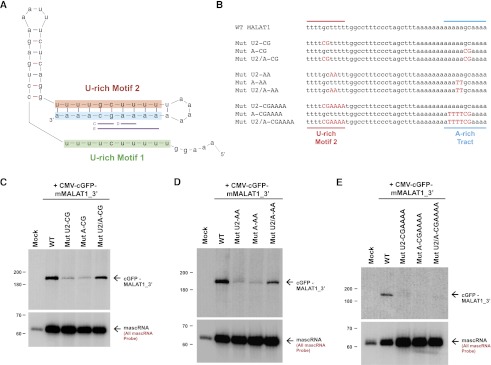
Figure 3.
Base-pairing between U-rich motif 2 and the A-rich tract is necessary but not sufficient for MALAT1 stability. (A) Predicted secondary structure of the 3′ end of the mature Comp.14 transcript. Denoted in purple are base pairs between U-rich motif 2 and the A-rich tract that were mutated in C–E. (B) Mutations (denoted in red) were introduced into the CMV-cGFP-mMALAT1_3′ expression plasmid. The full 174-nt mMALAT1_3′ region was present in these plasmids, although only the region between U-rich motif 2 and the A-rich tract is shown. (C–E) The wild-type (WT) or mutant plasmids were transfected into HeLa cells, and Northern blots were performed. RNase H treatment was performed prior to the Northern blots detecting cGFP-MALAT1_3′ RNA.
Secondary structure prediction of the minimal functional MALAT1 3′ end using Mfold indicated that the A-rich tract should base-pair with U-rich motif 2 (Fig. 3A). As these potential base pairs are perfectly conserved through evolution (Fig. 1B,C), we generated cGFP-MALAT1_3′ expression plasmids in which specific base pairs were disrupted (Fig. 3B; Supplemental Fig. 6). As shown in Figure 3, C and D, the cGFP-MALAT1_3′ RNA failed to accumulate when two mismatches were introduced in either U-rich motif 2 or the A-rich tract. When base-pairing was re-established by introduction of mutations in both motifs, a significant rescue in the level of cGFP-MALAT1_3′ RNA was detected (Fig. 3C,D). This indicates that base-pairing between U-rich motif 2 and the A-rich tract is critical for stabilizing the 3′ end of MALAT1. Interestingly, cGFP-MALAT1_3′ ending in a short homopolymeric poly(A) tail due to the GC in the A-rich tract being mutated to AA was also degraded in vivo (data not shown), indicating that a short poly(A) tail cannot functionally replace base-pairing at the 3′ end of MALAT1. As expected, when 6 base pairs (bp) were disrupted, the mutated cGFP-MALAT1_3′ transcript failed to accumulate in vivo (Fig. 3E). Unexpectedly, however, introduction of compensatory mutations that re-establish these 6 bp failed to rescue cGFP-MALAT1_3′ transcript levels (Fig. 3E), indicating that base-pairing between U-rich motif 2 and the A-rich tract is necessary but not sufficient for MALAT1 stability.
As U-rich motif 1 is also required for MALAT1 3′ end stability (Fig. 2E), is highly conserved (Fig. 1B,C), and is predicted to be in close proximity to U-rich motif 2 and the A-rich tract (Fig. 3A), we suspected that U-rich motif 1 may also interact with the duplex in, for example, a triple helix (Fig. 4A). Pioneering work by Felsenfeld, Davies, and Rich in 1957 (Felsenfeld et al. 1957) first described U-A•U triple helix structures where a poly(U) third strand forms Hoogsteen hydrogen bonds to the major groove of a Watson-Crick base-paired helix of poly(A)/poly(U) (Fig. 4B). Naturally occurring U-A•U RNA triple helix structures have recently been identified in telomerase RNA (Qiao and Cech 2008) and at the 3′ end of a noncoding RNA produced by Kaposi's sarcoma-associated herpesvirus and related γ-herpesviruses (Mitton-Fry et al. 2010; Tycowski et al. 2012). In the latter case, this structure was essential for stabilization of the RNA. C-G•C triple helices are structurally similar to U-A•U, although protonation of the cytosine in the third strand is required to fully stabilize the structure, making C-G•C triplexes favorable under acidic conditions (Fig. 4B). Importantly, at the 3′ ends of MALAT1 and MEN β, the U- and A-rich motifs are properly oriented to allow an intramolecular triple-helical structure to form by Hoogsteen hydrogen-bonding of U-rich motif 1 to the major groove of the Watson-Crick base-paired helix that is formed by U-rich motif 2 and the A-rich tract (Fig. 4A).
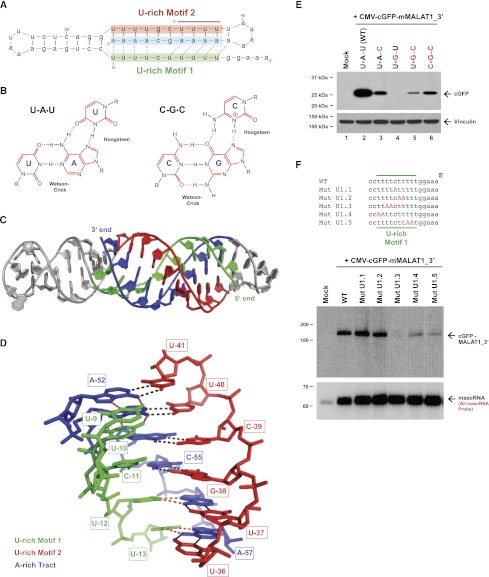
Figure 4.
A triple helix forms at the 3′ end of MALAT1. (A) Base triples (denoted by dashed lines) form at the 3′ end of the mature Comp.14 transcript. This structure is similar to that shown in Figure 3A except that the orientation of the conserved stem–loop has been rotated by 90°. The U-A•U base triples that were mutated in E are denoted in purple. (B) U-A•U and C-G•C base triples form via Hoogsteen hydrogen bonds to the major grove of a Watson-Crick base-paired helix. (C) Rosetta model of the MALAT1 Comp.14 3′ end in cartoon representation. Bases 1–5 are not included to achieve modeling convergence. As in A, U-rich motif 1 is in green, U-rich motif 2 is in red, and the A-rich tract is in blue. Remaining bases are in gray. (D) Close-up view of the triple helix surrounding the nonbonded base C-11 (numbering as in A). Bases are shown in stick representation with Watson-Crick hydrogen bonds in black and Hoogsteen hydrogen bonds in red. (E) Four of the U-A•U base triples were progressively converted to C-G•C base triples in the CMV-cGFP-mMALAT1_3′ expression plasmid. In the name of each construct, the asterisk represents the Hoogsteen hydrogen bonds. The wild-type (WT) or mutant plasmids were then transfected into HeLa cells, and Western blots were performed to detect cGFP protein expression. Vinculin was used as a loading control. (F) Mutations (denoted in red) were introduced into the CMV-cGFP-mMALAT1_3′ expression plasmid. The full 174-nt mMALAT1_3′ region was present in these plasmids, although only the region around U-rich motif 1 is shown. Note that the 5′ end of each transcript is on the right side to allow a direct comparison with the structure in A. The wild-type (WT) or mutant plasmids were then transfected into HeLa cells, and Northern blots were performed.
To assess the ability of the 3′ end of MALAT1 to form a triple helix, we used fragment assembly of RNA with full atom refinement, known as FARFAR (Das et al. 2010). This Rosetta-based algorithm predicts low-energy tertiary RNA structures de novo to near-atomic resolution (Das and Baker 2007). As shown in Figure 4C, the 59-nt Comp.14 mMALAT1_3′ region is predicted to be able to fold into a barbell-like structure with loops at each end of a continuous Watson-Crick base-paired helix, part of which further forms a triple-helical structure with U-rich motif 1 binding in the major groove. Nine U-A•U base triples are able to form by base-pairing between the Hoogsteen face of the A nucleotides in the A-rich tract and the Watson-Crick face of the U nucleotides of U-rich motif 1 (Fig. 4C,D; Supplemental Fig. 7). We note that our modeling does not support the formation of a C-G•C triple. Although FARFAR does not allow modeling of a protonated cytidine residue at the Hoogsteen base (Fig. 4D), other steric constraints may preclude formation of this C-G•C triple. The predicted structure lacks chain breaks and has reasonable stereochemistry, indicating that there are no structural constraints blocking the formation of the triple helix. Nicely, nucleotides that are not critical for MALAT1 3′ end stability and thus are deleted from the Comp.14 transcript (Fig. 2D) are all predicted to be in loop regions at the ends of the barbell-like structure, physically separated from the core triple helix (Fig. 4A,C). Furthermore, the structural model indicates that the 3′-terminal nucleotide of MALAT1 is part of the core triple helix and thus is well protected from addition of either nontemplated nucleotides or exonucleases. It is likely that significant free energy would be necessary to unwind this triple helix. Consistent with this model, 3′ RACE revealed that 3′–5′ exonucleases often pause within this structure (Fig. 2F).
Although the structural model predicts that the triple helix structure can form, it does not prove that the triple helix does form in vivo. Nevertheless, we have several independent lines of evidence that support the existence and functional significance of the triple helix in vivo. First, all of the base triples are nearly perfectly conserved through evolution at the 3′ ends of both MALAT1 (Fig. 1B) and MEN β (Fig. 1C). Second, the mutational analysis in Figure 3 revealed that base-pairing between U-rich motif 2 and the A-rich tract is necessary but not sufficient for stabilizing the 3′ end of MALAT1. Of particular interest is the Mut U2/A-CGAAAA transcript (Fig. 3B,E), in which nucleotides that form six of the base pairs between U-rich motif 2 and the A-rich tract were swapped across the helix. These nucleotide swaps should not alter the structural integrity of the double helix but should eliminate the potential to form base triples, providing indirect support for this structure in the stabilization of MALAT1. Third, to directly test for the presence of the triple helix in vivo, we investigated the effect of converting four of the U-A•U base triples at the 3′ end of MALAT1 (denoted in purple in Fig. 4A) to C-G•C base triples (Fig. 4E). Mutating the four consecutive A nucleotides to G (Fig. 4E, lane 4) caused the cGFP-MALAT1_3′ transcript to be unstable and not translated in vivo (see below for further information about translation). Compared with a transcript only able to form a double helix with C-G base pairs (Fig. 4E, lane 5), significantly greater protein expression was observed when C-G•C base triples were able to form (Fig. 4E, lane 6). This is strong evidence that a functional triple helix forms in vivo.
To then investigate whether the entire triple helix structure is necessary for stabilizing the 3′ end of MALAT1 in vivo, we generated cGFP-MALAT1_3′ expression plasmids in which select base triples were disrupted by mutating U-rich motif 1 (Fig. 4F). Interestingly, we found that mutations in the middle of U-rich motif 1 (Mut U1.1 and Mut U1.2) had no effect on cGFP-MALAT1_3′ transcript levels (Fig. 4F). This result is consistent with data from Figure 3, C and D, where base-pairing between U-rich motif 2 and the A-rich tract in this middle region was necessary for RNA stability, but the identities of the nucleotides on either side of the double helix (and thus the ability to form a base triple or not) were not critical. In contrast, base triples at both ends of the triple helix are critical for cGFP-MALAT1_3′ to be stable (Mut U1.3 to Mut U1.5) (Fig. 4F). These results support a model in which U-A•U base triples at each end of the MALAT1 triple helix ensure the structural stability of the overall structure and prevent transcript degradation by 3′–5′ exonucleases.
The triple helix structure also functions as a translational enhancer element
As the cGFP-MALAT1_3′ reporter mRNA is stable and efficiently exported to the cytoplasm (Fig. 2B), we investigated whether the cGFP ORF is translated. Surprisingly, similar levels of protein expression were observed from the cGFP transcripts ending in the mMALAT1_3′ region as compared with those ending in a poly(A) tail (Fig. 5A). This suggests that the 3′ end of MALAT1 may also function to promote translation. The 3′ end of MEN β similarly supported significant cGFP protein expression (Supplemental Fig. 3C). These results are particularly surprising considering that endogenous MALAT1 and MEN β are nuclear-retained transcripts and thus are not thought to interact with the translation machinery.
Figure 5.
The MALAT1 triple helix functions as a translational enhancer element. (A) Plasmids expressing cGFP transcripts ending in the designated 3′ end sequences were transfected into HeLa cells. The mMALAT1_3′ region and the polyadenylation signals were inserted in either the sense or antisense direction as denoted. Western blots were performed to detect cGFP protein expression. Vinculin was used as a loading control. (B) Schematic of the two-color fluorescent reporter expression system. (C) The two-color expression plasmids were transiently transfected into HeLa cells, and flow cytometry was used to measure mCherry and eYFP protein expression in single cells. Shown are box plots of the ratios of mCherry to eYFP protein expression measured in individual transfected cells ([horizontal line] median; [box] 25th–75th percentile; [error bars] 1.5× interquartile range) from a representative experiment (n = 3). (D) qPCR was used to measure the ratio of mCherry mRNA to eYFP mRNA in populations of cells transfected with the two-color expression plasmids. The data were normalized to the polyadenylated construct and are shown as mean and standard deviation values of three independent experiments. (E) Mutations or deletions (denoted in red) were introduced into the mMALAT1_3′ region of the CMV-cGFP-mMALAT1_3′ expression plasmid. (F) The wild-type (WT) or mutant plasmids were then transfected into HeLa cells, and Northern blots were performed. RNase H treatment was performed prior to the Northern blot that detects cGFP-MALAT1_3′ RNA. (G) Western blotting was used to detect cGFP expression in the transfected HeLa cells. (H) Transfected HeLa cells were fractionated to isolate nuclear and cytoplasmic total RNA, which was then subjected to Northern blot analysis. (I) Nucleotides that function in promoting translation (denoted in purple) flank the triple-helical region at the 3′ end of MALAT1.
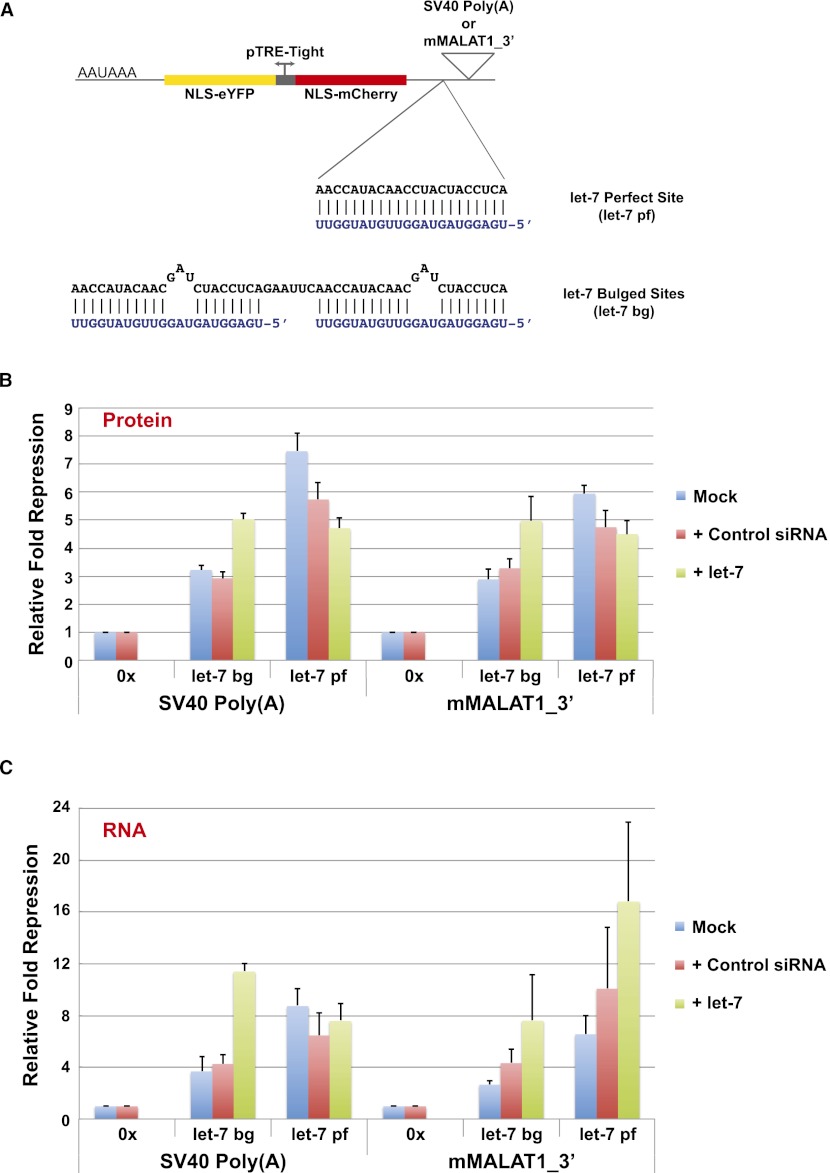
To better quantitate the translational output obtained from a transcript ending in the MALAT1 3′ end versus that obtained from a transcript ending in a poly(A) tail, we took advantage of a two-color fluorescent reporter system recently developed by our laboratory that allows measurements of gene expression in single mammalian cells (Mukherji et al. 2011). This construct consists of a bidirectional Tet-inducible promoter that drives expression of the fluorescent proteins mCherry and enhanced yellow fluorescent protein (eYFP) tagged with nuclear localization sequences (Fig. 5B). In the 3′ UTR of mCherry, we inserted either the SV40 polyadenylation signal or the mMALAT1_3′ region. In contrast, the 3′ UTR of eYFP always ended with the SV40 polyadenylation signal, allowing eYFP expression to serve as an internal normalization control, as it is a sensitive reporter of transcriptional and translational activity from the bidirectional promoter. Using flow cytometry to monitor protein expression in single cells, we first compared the levels of mCherry and eYFP protein obtained when both transcripts terminated in a canonical poly(A) tail. By calculating the ratio of mCherry to eYFP protein detected in each analyzed cell, we found that the expression of the fluorescent proteins is, as expected, highly correlated (ratio of 0.91 ± 0.05) (Fig. 5C; Supplemental Fig. 8A). This correlation was mirrored on the transcript level when measured across the population of cells by quantitative PCR (qPCR) (Fig. 5D). We then compared the levels of mCherry and eYFP proteins and mRNAs obtained when the mMALAT1_3′ region was inserted downstream from mCherry. Consistent with the results with the cGFP reporter in Figure 5A, the mMALAT1_3′ region supported strong translation (mCherry/eYFP protein ratio of 1.00 ± 0.05) (Fig. 5C,D; Supplemental Fig. 8B). Northern blots confirmed that the mCherry transcript ended in the mMALAT1_3′ region as generated by RNase P, thus eliminating the possibility that a cryptic polyadenylation signal was responsible for the efficient translation observed (Supplemental Fig. 8E).
To determine the sequence elements in the mMALAT1_3′ region required for efficient translation, we mutated the cGFP-MALAT1_3′ Comp.14 transcript (Fig. 2D), which contains the minimal elements required for RNA stability (Figs. 2G, 5F) and efficient translation (Fig. 5G), to test whether a transcript that is stable but poorly translated could be identified. By mutating every nucleotide at the 3′ end of MALAT1 not present in the core triple-helical region (while maintaining base-pairing in the conserved stem–loop) (Comp.15) (Fig. 5E), a cGFP transcript that is stable (Fig. 5F) but poorly translated (Fig. 5G) was identified. The Comp.15 transcript was exported to the cytoplasm as efficiently as Comp.14, indicating that this decrease in translational efficiency is not due to increased nuclear retention of the transcript (Fig. 5H). Confirming these results, when the mMALAT1_3′ Comp.15 region was placed downstream from mCherry in the two-color fluorescent reporter system (Fig. 5B), an approximately fivefold decrease in translational efficiency was observed when compared with the wild-type mMALAT1_3′ region, while the level of mRNA decreased only approximately twofold (Fig. 5C,D; Supplemental Fig. 8C,D).
Additional mutagenesis was then performed to determine which of the 27 mutations present in the Comp.15 region were required for this decrease in translational efficiency (Fig. 5E). Interestingly, this analysis revealed that certain subsets of the 27 mutations (Comp.27) (Fig. 5E) caused the transcript to no longer be stable (Fig. 5F). Nevertheless, we were able to identify other subsets of mutations (Comp.25 and Comp.26) (Fig. 5E) that generated a stable cGFP transcript (Fig. 5F) that was poorly translated (Fig. 5G). This indicates that the nucleotides immediately flanking each side of the core triple-helical region have critical roles in promoting translation (Fig. 5I).
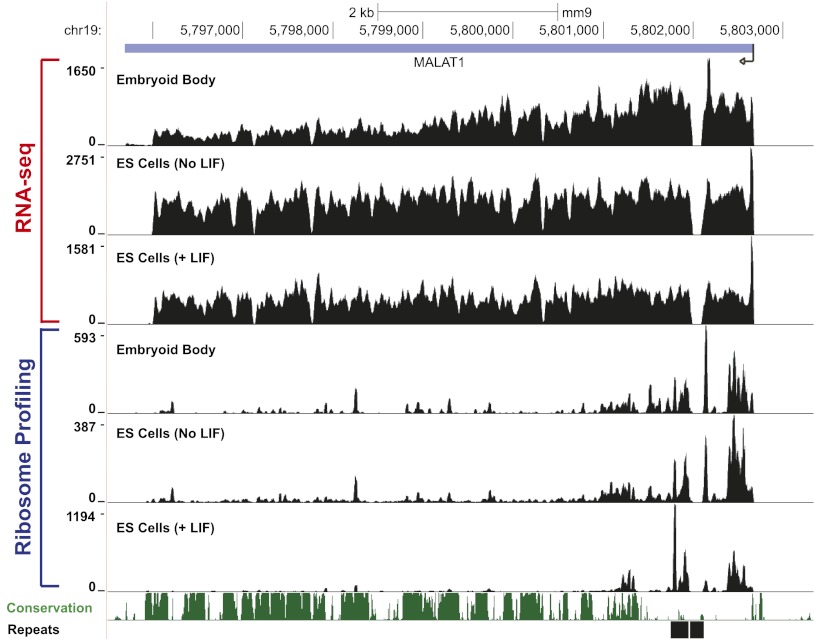
As these results indicate that a strong translational enhancer element is present at the 3′ end of MALAT1 and MEN β, we investigated whether there was any evidence of translation of these endogenous noncoding RNAs. Although the MEN β transcript is lowly expressed in mouse embryonic stem (ES) cells (data not shown), MALAT1 is highly expressed, and ribosome profiling (Ingolia et al. 2011) suggests that reproducible and nonrandom regions near the 5′ end of MALAT1 are protected by ribosomes (Fig. 6). We were unable to identify obvious well-conserved ORFs in these regions, although it may be that species-specific short peptides are produced from the 5′ end of MALAT1, as there are potential start codons in mice near several of the regions where ribosomes are concentrated.
Figure 6.
Ribosome footprints are observed near the 5′ end of MALAT1 in mouse ES cells. The RNA sequencing (RNA-seq) and ribosome footprint profiles of MALAT1 in mouse embryoid bodies and mouse ES cells (grown in the presence or absence of leukemia inhibitory factor [LIF]) as determined by Ingolia et al. 2011 are shown. The MALAT1 transcription start site is denoted by an arrow on the right side of the figure.
A transcript ending in the MALAT1 triple helix is efficiently repressed by microRNAs
As most long transcripts lacking a poly(A) tail are rapidly degraded in cells, it has generally been difficult to define regulatory roles for the poly(A) tail or poly(A)-binding protein (PABP) in vivo. Now, the expression system built around the 3′ end of MALAT1 represents a unique and valuable tool to address these issues, as it generates in vivo stable transcripts that lack a poly(A) tail. It is unlikely that transcripts with mMALAT1_3′ sequences at their 3′ ends interact with PABP, since this protein requires at least 12 consecutive A residues for binding (Sachs et al. 1987). To demonstrate the utility of our system for investigating how nonpolyadenylated transcripts are regulated in vivo, we asked whether microRNAs repress a transcript ending in the MALAT1 triple helix as efficiently as they do a polyadenylated transcript. MicroRNAs function as part of RISC (RNA-induced silencing complex) and bind to partially complementary sites in target mRNAs, causing translational repression and/or transcript degradation (for review, see Bartel 2009). As the core RISC protein component GW182 can directly interact with PABP as well as deadenylases (Braun et al. 2011), a model has emerged in which an interaction between RISC and PABP is required for maximum repression by microRNAs (Fabian et al. 2009; Huntzinger et al. 2010; Moretti et al. 2012). However, the functional importance of these interactions has been debated (Fukaya and Tomari 2011; Mishima et al. 2012).
To investigate the role of the poly(A) tail in microRNA-mediated repression in vivo, we took advantage of the two-color fluorescent reporter system and inserted microRNA-binding sites into the 3′ UTR of mCherry upstream of either the SV40 polyadenylation signal or the mMALAT1_3′ region (Fig. 7A). In particular, we inserted either two bulged let-7-binding sites (denoted as let-7 bg) or a sequence that is perfectly complementary to let-7 (denoted as let-7 pf), thus converting the interaction between target and microRNA into a catalytic, RNAi-type repression. Using HeLa cells that naturally express let-7 microRNA, we found that when mCherry ended in a canonical poly(A) tail, the addition of two bulged let-7-binding sites caused 3.2 ± 0.2-fold repression as measured by protein expression (Fig. 7B; Supplemental Fig. 9A). Surprisingly, 2.9 ± 0.4-fold repression was observed when two bulged let-7 sites were added to mCherry ending in the MALAT1 triple helix (Fig. 7B; Supplemental Fig. 9B), a level of repression that is not statistically different from that obtained when a poly(A) tail was present. Regardless of whether mCherry ended in a poly(A) tail or in the MALAT1 triple helix, these effects were mirrored on the transcript level (Fig. 7C), indicating that the effects on protein production are likely at least partially due to decreased RNA levels. Upon transfecting synthetic let-7 to increase the level of the microRNA in HeLa cells, the levels of microRNA-mediated repression observed increased consistently regardless of the 3′-terminal sequence present (Fig. 7B,C). These results suggest that a poly(A) tail is not necessary for maximum repression by microRNAs in vivo and thus that microRNAs may also efficiently target nonpolyadenylated transcripts in cells. Although the mechanism by which nonpolyadenylated transcripts are degraded in response to microRNAs is unclear, these results suggest that deadenylation may not always be required for efficient microRNA-mediated silencing.
Figure 7.
A transcript ending in the MALAT1 triple helix is efficiently repressed by microRNAs in vivo. (A) Inserted into the 3′ UTR of mCherry was either a sequence perfectly complementary to let-7 or two bulged let-7-binding sites. The let-7 microRNA sequence is shown in blue. (B) HeLa cells were transfected with two-color fluorescent reporter plasmids ending in either the SV40 polyadenylation signal or the mMALAT1_3′ region with or without (denoted 0x) microRNA-binding sites. In addition, 40 nM control siRNA or exogenous let-7 microRNA was cotransfected as indicated. Flow cytometry was then used to measure mCherry and eYFP protein levels. Relative fold repression was calculated as the ratio of the mean mCherry to the mean eYFP signal of the targeted construct normalized to the equivalent ratio for the nontargeted (0x) reporter. Data are shown as mean and standard deviation values of three independent experiments. (C) qPCR was used to measure mCherry and eYFP transcript levels across the population of cells, and the relative fold repression of mCherry RNA expression was calculated analogously to above. Data are shown as mean and standard deviation values of three independent experiments.
Discussion
Despite lacking a poly(A) tail, the long noncoding RNA MALAT1 is a stable transcript that is expressed at a level comparable with or higher than many protein-coding genes in vivo (Wilusz et al. 2008; Zhang et al. 2012). In the present study, we demonstrated that the 3′ end of MALAT1 is protected from degradation by an evolutionarily conserved triple helix. We further identified a highly similar triple-helical structure that stabilizes the 3′ end of the MEN β long noncoding RNA. Surprisingly, these triple-helical regions also function as strong translational enhancer elements, allowing a nonpolyadenylated mRNA to be translated as efficiently as an mRNA with a canonical poly(A) tail. Transcripts ending in a triple helix are efficiently repressed by microRNAs, arguing that a poly(A) tail is not required for efficient microRNA-mediated silencing in vivo. Our data provide new insights into how MALAT1, MEN β, and likely other transcripts that lack poly(A) tails are stabilized and regulated in vivo.
A triple helix can functionally replace a poly(A) tail
The poly(A) tail at the 3′ ends of long RNA Pol II transcripts functions to ensure that the mature RNA is stable, exported to the cytoplasm, and efficiently translated (for review, see Zhao et al. 1999). It has long been known that a stem–loop structure functionally replaces the poly(A) tail at the 3′ ends of replication-dependent histone mRNAs (for review, see Marzluff et al. 2008). Our work has demonstrated that the triple-helical structures at the 3′ ends of the MALAT1 and MEN β long noncoding RNAs can likewise functionally replace a poly(A) tail. In addition to supporting transcript stability, these triple helices support efficient export (Fig. 2B) and translation (Fig. 5) of a reporter transcript. The endogenous noncoding RNAs are not exported, however, as nuclear retention signals elsewhere in the transcripts (Fig. 2C) somehow override any export signals present at the 3′ ends. Interestingly, the various functions ascribed to the triple-helical region can be separated from one another, as we were able to identify mutations that generate a stable and exported transcript that is not efficiently translated. Identifying the mechanism by which the triple-helical region promotes translation may reveal important new modes of translational control.
PAN (polyadenylated nuclear) RNA, an abundant long noncoding RNA generated by Kaposi's sarcoma-associated herpesvirus, has previously been shown to also have a triple helix at its 3′ end (Mitton-Fry et al. 2010). Unlike MALAT1 and MEN β, PAN RNA is subjected to canonical cleavage/polyadenylation and binds PABP (Borah et al. 2011). Nevertheless, five consecutive U-A•U base triples form between part of the PAN RNA poly(A) tail and a U-rich region ∼120 nt upstream of the poly(A) tail (Mitton-Fry et al. 2010). Formation of this triple helix inhibits RNA decay and has been proposed to be required for nuclear retention of PAN RNA. In contrast, we found that the MALAT1/MEN β triple helices are not critical for nuclear retention (Fig. 2B). Using the PAN RNA triple helix structure as a guide, recent computational work identified six additional transcripts that likely form triple helices, although two of them were simply PAN RNA homologs in related γ-herpesviruses (Tycowski et al. 2012). The MALAT1 and MEN β triple helices were not identified in this study, likely due to the subtle differences in these structures compared with the PAN RNA triple helix. Considering that base triples can be formed by nucleotides far away from one another in a transcript's primary sequence (or even be encoded on separate independent transcripts), additional functional RNA triple helices likely form in vivo and await discovery.
In addition to the histone stem–loop structure and the MALAT1/MEN β triple helices, other RNA structural motifs may be able to functionally replace a poly(A) tail. For example, it is known that tRNA-like structures stabilize the 3′ ends of several ssRNA viruses, such as turnip yellow mosaic virus and bacteriophage Qβ (for review, see Fechter et al. 2001). To begin to screen for other stabilizing RNA structures, we modified our CMV-cGFP-mMALAT1_3′ expression plasmid by replacing the region of MALAT1 upstream of the RNase P cleavage site with the sequences of various riboswitches, RNA elements that bind cellular metabolites and often fold into elaborate structures (Supplemental Fig. 10; for review, see Serganov and Patel 2012). As the mascRNA tRNA-like structure is present immediately downstream from the 3′ end of the riboswitch, RNase P cleavage generates a mature cGFP transcript ending in the riboswitch sequence in vivo. Interestingly, the Thermoanaerobacter tengcongensis glmS catalytic riboswitch, which senses glucosamine-6 phosphate (Klein and Ferre-D'Amare 2006), was able to stabilize the 3′ end of the cGFP message and support translation, although the effects were much weaker than that obtained with the MALAT1 triple helix (Supplemental Fig. 10). Nevertheless, these results suggest that there are indeed likely additional RNA sequences that are sufficient to stabilize the 3′ ends of nonpolyadenylated transcripts. We believe that this expression system provides an ideal method for in vivo screening for these sequences, as our approach efficiently generates transcripts that have a well-defined 3′ end and lack a poly(A) tail in vivo.
A growing role for uridylation in RNA degradation
Disrupting the integrity of the MALAT1 triple helix causes the transcript to be efficiently degraded. We surprisingly found numerous cGFP-MALAT1_3′ transcripts ending in post-transcriptionally added short U-rich tails when the transcript was undergoing degradation (Fig. 2F; Supplemental Fig. 4C), implicating uridylation in the decay process. Oligouridylation has been linked to the degradation of numerous classes of small RNAs, including tRNAs (Supplemental Fig. 1), microRNA precursors (Heo et al. 2009), mature microRNAs (Li et al. 2005), and transcription start site-associated RNAs (Choi et al. 2012). Although there is currently less evidence for U-tails on long transcripts in vivo, uridylation can promote mRNA decapping (Song and Kiledjian 2007; Rissland and Norbury 2009), and U-tails have been observed on the products of microRNA-directed cleavage (Shen and Goodman 2004). Interestingly, histone mRNAs are subjected to uridylation and degradation following the completion of DNA synthesis (Mullen and Marzluff 2008). Analogous to what we observed at the highly structured 3′ end of MALAT1 (Fig. 2F; Supplemental Fig. 4C), Mullen and Marzluff (2008) observed short U-tails on histone mRNAs that appeared to have been shortened previously by 3′–5′ exonucleases. We further observed a similar phenomenon at the 3′ end of a mutant mascRNA transcript targeted for degradation (Supplemental Fig. 1). These results suggest that oligouridylation may play a much more significant role in the degradation of regions of extensive RNA secondary structure than we currently appreciate. In particular, we suggest that when a 3′–5′ exonuclease stalls at a region of extensive secondary structure, an oligo(U) tail can be added to provide a single-stranded tail that is subsequently recognized by decay factors and used to restart the degradation process.
Implications of the triple helix for the functions of MALAT1 and MEN β
Unlike many long noncoding RNAs that are rapidly degraded and thus expressed at near undetectable levels (Wyers et al. 2005; Preker et al. 2008), MALAT1 and MEN β are stable transcripts with half-lives of >12 h (Wilusz et al. 2008; Sunwoo et al. 2009). By preventing degradation from the 3′ ends of these noncoding RNAs, the triple helices play a critical role in not only ensuring RNA stability but also allowing these transcripts to perform important cellular functions. For example, the MEN β noncoding RNA is an essential structural component of paraspeckles in the nucleus (Sunwoo et al. 2009). When MEN β is depleted from cells, this subnuclear domain is no longer observed, and paraspeckle-associated proteins and RNAs instead are dispersed. The exact cellular function of MALAT1 is currently a matter of contention, as conflicting results have been published (Tripathi et al. 2010; Yang et al. 2011b; Eissmann et al. 2012; Zhang et al. 2012). Nevertheless, MALAT1 is commonly overexpressed in many cancers, suggesting a possible role in a malignant phenotype.
Our finding that the MALAT1 and MEN β triple helices function as strong translational enhancer elements adds an additional unexpected twist to how these nuclear-retained transcripts may function. Considering that the Xist long noncoding RNA evolved from a protein-coding gene (Duret et al. 2006), it may be that the same is true for MALAT1 and MEN β, and thus their associated translation control elements are simply relics of their evolutionary pasts. Alternatively, these noncoding RNAs may interact with ribosomes, possibly producing short peptides, as has been shown for other transcripts that were once considered noncoding (Galindo et al. 2007; Ingolia et al. 2011). Reproducible and nonrandom ribosome footprints were found on MALAT1 in mouse ES cells (Fig. 6), although no obvious well-conserved ORFs were identified. It is also possible that MALAT1 and MEN β may simply interact with components of the translation machinery, thereby serving as a “sponge” that prevents the binding of these factors to mRNAs.
Finally, the availability of expression vectors that produce stable cytoplasmic mRNAs without a poly(A) tail will allow the in vivo testing of mechanisms for translational control involving this nontemplate-encoded structure. For example, we found that microRNAs regulate the expression of target mRNAs with a poly(A) tail as efficiently as mRNAs ending with the MALAT1 triple helix.
In summary, we identified highly conserved triple-helical structures at the 3′ ends of the nonpolyadenylated MALAT1 and MEN β long noncoding RNAs, which function to prevent RNA decay. When placed downstream from an ORF, the triple helices additionally function to promote efficient translation. Our findings thus reveal novel paradigms for how transcripts that lack a canonical poly(A) tail can be stabilized, regulated, and translated. Considering the complexity of the human transcriptome and the presence of many other long transcripts that may lack a poly(A) tail, it is likely that triple helices and other RNA structural elements may have additional unappreciated roles in ensuring transcript stability and regulating gene expression.
Materials and methods
Expression plasmid construction
To generate the CMV-cGFP-mMALAT1_3′ expression constructs, we modified a previously described plasmid (Gutschner et al. 2011) in which the CMV promoter and the cGFP ORF were cloned into the multicloning site of the pCRII-TOPO vector (Life Technologies). Downstream from cGFP, we inserted the mMALAT1_3′ region (nucleotides 6581–6754 of GenBank accession no. EF177380) into the NotI cloning site in the sense direction (to generate CMV-cGFP-mMALAT1_3′ sense) or in the antisense direction (to generate CMV-cGFP-mMALAT1_3′ antisense). The NotI cloning site was similarly used to generate CMV-cGFP expression plasmids ending in the SV40 polyadenylation signal, the bGH polyadenylation signal, the mMEN β_3′ region, and all of the mutant mMALAT1_3′ regions. To generate the CMV-SpeckleF2-mMALAT1_3′ expression plasmid, nucleotides 1676–3598 of mouse MALAT1 was inserted into the EcoRV and BstEII cloning sites of the CMV-cGFP-mMALAT1_3′ sense plasmid. The sequences of the inserts for all plasmids are provided in Supplemental Table 1.
Transfections and RNA analysis
HeLa cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing high glucose (Life Technologies), supplemented with penicillin–streptomycin and 10% fetal bovine serum. CMV-cGFP expression plasmids were transfected using Lipofectamine 2000 (Life Technologies), and total RNA was isolated using Trizol (Life Technologies) as per the manufacturer's instructions. Northern blots were performed as previously described (Wilusz et al. 2008). For RNase H treatments, 9 μg of total RNA was first mixed with 20 pmol of antisense oligo and heated for 10 min to 65°C. After the antisense oligos were allowed to anneal by slow cooling, the RNA was treated with RNase H (New England Biolabs) for 30 min at 37°C and then subjected to Northern blot analysis. Nuclear and cytoplasmic fractionation was performed as described previously (Wilusz et al. 2008). All oligonucleotide probe sequences are provided in Supplemental Table 2. 3′ RACE PCR using microRNA Cloning Linker 3 (Integrated DNA Technologies) was performed as previously described (Wilusz et al. 2008).
Protein analysis
Western blots were performed using the Nu-PAGE Bis-Tris electrophoresis system (Life Technologies) as per the manufacturer's instructions. The cGFP antibody was obtained from GenScript, and the Vinculin antibody was obtained from Sigma-Aldrich.
Two-color fluorescent reporter system
The two-color fluorescent reporter vector was previously described (Mukherji et al. 2011) and contains the SV40 polyadenylation signal in the 3′ UTR of mCherry. To replace this polyadenylation signal with the mMALAT1_3′ region, the EcoRV and AatII cloning sites that flank the SV40 polyadenylation signal were used. Target sites for let-7 were inserted into the 3′ UTR of mCherry using HindIII and SalI cloning sites, and the sequences are provided in Supplemental Table 1. HeLa cells were seeded at 175,000 cells per well of a 12-well plate for 20 h prior to transfection of equivalent amounts (250 ng) of the reporter plasmid and rtTA using Lipofectamine 2000. At the time of transfection, the medium was changed to complete DMEM supplemented with 2 μg/mL doxycycline (Sigma). Where indicated, control siRNA (siGENOME nontargeting siRNA #2, Dharmacon) or an siRNA equivalent of murine let-7g (Dharmacon) were cotransfected at final concentrations of 40 nM. Flow cytometry or RNA isolation was performed 18–20 h post-transfection. Flow cytometry, qPCR, and raw data processing were performed as previously described (Mukherji et al. 2011) and are further described in the Supplemental Material.
Structural model prediction
De novo RNA folding was carried out using Rosetta version 3.4 (http://www.rosettacommons.org) (Das and Baker 2007; Das et al. 2010). For a converging model of the 3′ end of the MALAT1 Comp.14 transcript, the first 5 nt (AAGGG) were removed. Suspected helical interactions were defined (all nine U-A•U base triples were defined), and 2000 models were calculated. The model converged between 3 and 4 Å (see Supplemental Fig. 7C). The full-length (59-nt) Comp.14 3′ end was subjected to the same procedure, although convergence could not be achieved due to high flexibility of the 5′ end.
Acknowledgments
We thank Sven Diederichs for providing expression plasmids, Rhiju Das for aid with the de novo RNA folding, Xuebing Wu for help with the ribosome profiling data, the Koch Institute Flow Cytometry Facility, and members of the Sharp and Joshua-Tor laboratories for discussions and advice. J.E.W. is a fellow of the Leukemia and Lymphoma Society. C.-D.K. is supported by a Jane Coffin Childs Memorial Fund for Medical Research Fellowship. This work was supported by NIH grants R01-GM34277 (to P.A.S.) and R01-CA133404 (to P.A.S.) as well as partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute (to P.A.S.). L.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.204438.112.
References
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S, Darricarrere N, Darnell A, Myoung J, Steitz JA 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 7: e1002300 doi: 10.1371/journal.ppat.1002300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Tang W, Baumann P 2008. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature 456: 910–914 [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Choi YS, Patena W, Leavitt AD, McManus MT 2012. Widespread RNA 3′-end oligouridylation in mammals. RNA 18: 394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Das R, Baker D 2007. Automated de novo prediction of native-like RNA tertiary structures. Proc Natl Acad Sci 104: 14664–14669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Karanicolas J, Baker D 2010. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods 7: 291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, et al. 2003. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci 100: 6051–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655 [DOI] [PubMed] [Google Scholar]
- Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, et al. 2012. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 9: 1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. 2012. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechter P, Rudinger-Thirion J, Florentz C, Giege R 2001. Novel features in the tRNA-like world of plant viral RNAs. Cell Mol Life Sci 58: 1547–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Davies DR, Rich A 1957. Formation of a three-stranded polynucleotide molecule. J Am Chem Soc 79: 2023–2024 [Google Scholar]
- Fukaya T, Tomari Y 2011. PABP is not essential for microRNA-mediated translational repression and deadenylation in vitro. EMBO J 30: 4998–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5: e106 doi: 10.1371/journal.pbio.0050106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Baas M, Diederichs S 2011. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res 21: 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J 29: 4146–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39 doi: 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. 2003. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041 [DOI] [PubMed] [Google Scholar]
- Kirsebom LA 2007. RNase P RNA mediated cleavage: Substrate recognition and catalysis. Biochimie 89: 1183–1194 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferre-D'Amare AR 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313: 1752–1756 [DOI] [PubMed] [Google Scholar]
- Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, Schuuring E, Schoenmakers EF, van Kessel AG 2003. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet 12: 1661–1669 [DOI] [PubMed] [Google Scholar]
- Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS 2011. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol 29: 1810–1816 [DOI] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS 2007. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26: 851–858 [DOI] [PubMed] [Google Scholar]
- Lutz CS, Moreira A 2011. Alternative mRNA polyadenylation in eukaryotes: An effective regulator of gene expression. Wiley Interdiscip Rev RNA 2: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Fukao A, Kishimoto T, Sakamoto H, Fujiwara T, Inoue K 2012. Translational inhibition by deadenylation-independent mechanisms is central to microRNA-mediated silencing in zebrafish. Proc Natl Acad Sci 109: 1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA 2010. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science 330: 1244–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, et al. 2012. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18: 738–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CL, Sharp PA 1985. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell 41: 845–855 [DOI] [PubMed] [Google Scholar]
- Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW 2012. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol 19: 603–608 [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A 2011. MicroRNAs can generate thresholds in target gene expression. Nat Genet 43: 854–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV 2012. Malat1 is not an essential component of nuclear speckles in mice. RNA 18: 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854 [DOI] [PubMed] [Google Scholar]
- Proudfoot N 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16: 272–278 [DOI] [PubMed] [Google Scholar]
- Qiao F, Cech TR 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15: 634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram V, Knezevich S, Bove KE, Perry A, Pfeifer JD 2007. DNA sequence of the translocation breakpoints in undifferentiated embryonal sarcoma arising in mesenchymal hamartoma of the liver harboring the t(11;19)(q11;q13.4) translocation. Genes Chromosomes Cancer 46: 508–513 [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Davis RW, Kornberg RD 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol 7: 3268–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ 2012. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys 41: 343–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM 2004. Uridine addition after microRNA-directed cleavage. Science 306: 997. [DOI] [PubMed] [Google Scholar]
- Song MG, Kiledjian M 2007. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13: 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA 2012. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep 2: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Spector DL 2010. An unexpected ending: Noncanonical 3′ end processing mechanisms. RNA 16: 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL 2008. 3′ End processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL 2009. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Whipple JM, Phizicky EM, Sharp PA 2011. tRNAs marked with CCACCA are targeted for degradation. Science 334: 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng Y, Zhou T, Zhang MQ, Wu CI, Wang SM 2008. Poly A- transcripts expressed in HeLa cells. PLoS ONE 3: e2803 doi: 10.1371/journal.pone.0002803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL 2011a. Genomewide characterization of non-polyadenylated RNAs. Genome Biol 12: R16 doi: 10.1186/gb-2011-12-2-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG 2011b. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]