Abstract
A copper-containing amine oxidase from the latex of Euphorbia characias was purified to homogeneity and the copper-free enzyme obtained by a ligand-exchange procedure. The interactions of highly purified apo- and holoenzyme with several substrates, carbonyl reagents, and copper ligands were investigated by optical spectroscopy under both aerobic and anaerobic conditions. The extinction coefficients at 278 and 490 nm were determined as 3.78 × 105 m−1 cm−1 and 6000 m−1 cm−1, respectively. Active-site titration of highly purified enzyme with substrates and carbonyl reagents showed the presence of one cofactor at each enzyme subunit. In anaerobiosis the native enzyme oxidized one equivalent substrate and released one equivalent aldehyde per enzyme subunit. The apoenzyme gave exactly the same 1:1:1 stoichiometry in anaerobiosis and in aerobiosis. These findings demonstrate unequivocally that copper-free amine oxidase can oxidize substrates with a single half-catalytic cycle. The DNA-derived protein sequence shows a characteristic hexapeptide present in most 6-hydroxydopa quinone-containing amine oxidases. This hexapeptide contains the tyrosinyl residue that can be modified into the cofactor 6-hydroxydopa quinone.
Copper-amine oxidases (amine oxygen oxidoreductase deaminating, copper containing; EC 1.4.3.6) are widespread enzymes oxidizing primary amines with the formation of the corresponding aldehyde, ammonia, and hydrogen peroxide:
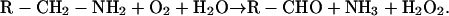 |
These enzymes are ubiquitous in nature, occurring in microorganisms (fungi and bacteria; Cooper et al., 1992), plants (Medda et al., 1995a), and mammals (McIntire and Hartmann, 1993). The crystal structures of copper-amine oxidases from Escherichia coli (Parson et al., 1995) and pea seedlings (Kumar et al., 1996) were recently determined. Amine oxidases are homodimers of 70- to 95-kD subunits. Each subunit contains a tightly bound Cu(II) center that is essential for the enzyme redox activity (Dooley et al., 1991; Medda et al., 1995b), and TPQ is formed by a posttranslational modification from a tyrosinyl residue in a copper-dependent, autocatalytic reaction (Janes et al., 1990; Brown et al., 1991; Cai and Klinman, 1994; Matsuzaki et al., 1994; Choi et al., 1995; Nakamura et al., 1996; Mure and Tanizawa, 1997).
The catalytic mechanism of amine oxidases has been reported previously (Medda et al., 1995b; Steinebach et al., 1996). The amine substrate binds to the organic cofactor of the resting oxidized enzyme Cu(II)-TPQ to form a Schiff base, Cu(II)-quinone ketimine; both of these forms are assumed to give the broad 498-nm absorption band. Transformation of the Cu(II)-quinone ketimine into a Cu(II)-quinolaldimine is accompanied by bleaching of the 498-nm absorption band (Bellelli et al., 1985, 1991). Oxidation of the bound substrate releases the aldehyde product and yields the Cu(II)-aminoresorcinol enzyme form, which contains the ammonia derived from substrate; this species is still colorless. The Cu(II)-aminoresorcinol species is to a variable extent in equilibrium with the yellow, electron-paramagnetic resonance-detectable Cu(I)-semiquinolamine radical characterized by absorption bands at 464, 434, and 360 nm (Finazzi Agró et al., 1984; Dooley et al., 1987, 1991; Pedersen et al., 1992). It is reasonable to assume that both reduced enzyme species may react with oxygen to release hydrogen peroxide and ammonia and to regenerate the Cu(II)-quinone species, but at least in the plant enzymes, the radical seems to be much more reactive than the Cu(II)-aminoresorcinol (Medda et al., 1998). Although discrepancies concerning the active TPQ:protein stoichiometry are present in the literature (Klinman and Mu, 1994; Agostinelli et al., 1997), most recent studies support an active TPQ:monomer ratio of 1:1 (Janes and Klinman, 1991; Padiglia et al., 1992; McGuirl et al., 1994).
An amine oxidase from the latex of Euphorbia characias, a perennial Mediterranean shrub, has been purified, but only a preliminary characterization of this enzyme was reported (Rinaldi et al., 1982). We present a detailed investigation of the Cu(II):TPQ:protein stoichiometry of highly purified E. characias latex amine oxidase upon interaction with substrates and carbonyl reagents containing a primary amino group. The results obtained provide clear evidence that this enzyme contains two active organic cofactors and two Cu(II) per dimer. An examination of the turnover of copper-free enzyme showed that copper-free E. characias latex amine oxidase can oxidize substrates under single-turnover conditions and releases 1 mol of aldehyde product per enzyme subunit. The isolation of DNA and sequence analysis of a DNA fragment showed a conserved region of 183 bp, with a hexapeptide containing the tyrosinyl residue that can give rise to the TPQ cofactor.
MATERIALS AND METHODS
Reagents
Putrescine, benzylamine hydrochloride, DABA hydrochloride, kynuramine dihydrobromide, BHY dihydrochloride, 4-hydroxyquinoline, SCA hydrochloride, and diethyldithiocarbamate were purchased from Sigma and used without further purification. TNM was from Aldrich, PHY hydrochloride was from Across (Geel, Belgium) and 4-(dimethylamino)benzaldehyde was from Fluka. All other chemicals were obtained as pure commercial products and used without further purification.
Purification of ELAO
The purification procedure (Table I) essentially follows the method described elsewhere (Rinaldi et al., 1982) with minor modifications as follows.
Table I.
Purification of ELAO
| Step | Total Protein | Total Activity | Specific Activity | Yield | Purification |
|---|---|---|---|---|---|
| mg | units | units/mg | % | -fold | |
| 1. Acetone powder | 7800 | 2470 | 0.32 | 100 | 1 |
| 2. Crude extract | 3150 | 2100 | 0.65 | 85 | 2 |
| 3. Ammonium sulfate fractionation | 800 | 2000 | 2.5 | 81 | 8 |
| 4. DEAE-cellulose column chromatography | 200 | 1800 | 9 | 73 | 28 |
| 5. Hydroxyapatite column chromatography | 80 | 1600 | 20 | 65 | 63 |
| 6. ω-Aminohexy-Sepharose 4B column chromatography | 40 | 1300 | 34 | 53 | 106 |
Starting material was 300 mL of latex.
Steps 1 through 4
The eluate from a DEAE-cellulose column (see Table I) was made 70% saturated with solid ammonium sulfate with constant stirring at 4°C for 30 min and centrifuged at 9000 rpm for 30 min. The precipitate was dissolved in 30 mL of 1 mm KPi buffer, pH 7.0, and dialyzed against 10 L of the same buffer at 4°C for 12 h. The insoluble material was removed by centrifugation.
Step 5
The supernatant from fraction 5 was loaded onto a column (2 × 8 cm) of hydroxyapatite equilibrated with 1 mm KPi buffer, pH 7.0. The column was washed with the same buffer until the A280 became ≤0.01. The ELAO was then eluted with 50 mm KPi buffer, pH 7.0. The fractions showing enzymatic activity were pooled.
Step 6
The material resulting from step 6 was loaded onto a column (2 × 10 cm) of ω-aminohexyl-Sepharose 4B equilibrated with 50 mm KPi buffer, pH 7.0. The column was washed with the same buffer until the A280 became ≤0.01. The ELAO was then eluted with 100 mm KPi buffer, pH 7.0. The fractions showing enzymatic activity were pooled and concentrated by filtration under a vacuum.
Copper-Free Enzyme
Copper-free ELAO was prepared as follows: ELAO solution (10 mg/mL) was dialyzed against double-distilled water containing 1 mm sodium cyanide at 4°C for 6 h. The dialyzed solution was brought to 10 mm in sodium diethyldithiocarbamate and allowed to stand at 4°C for 6 h, then centrifuged at 105,000g for 2 h. The supernatant was dialyzed against double-distilled water and then centrifuged at 15,000 rpm for 30 min. The pink supernatant was stored at −20°C when not used immediately.
Determination of Metals
Copper and manganese contents were determined by atomic absorption using an IL 951 atomic absorption spectrometer Instrumentation Laboratory, Wilmington, DE). The spectral line chosen was 324.7 nm for copper and 285.2 nm for manganese.
Protein Concentration
Protein concentrations were measured by the Lowry method, as modified by Hartree (1972), with BSA as a standard.
Analytical PAGE and Determination of Relative Molecular Mass
Electrophoresis under nondenaturing conditions was performed as described previously (Gabriel, 1971). Continuous SDS-PAGE was carried out according to the method of Weber and Osborne (1969). Preparation of protein samples for SDS-PAGE was by two methods. First, the protein samples were incubated for 48 h at 4°C with 0.2% SDS and 100 mm 2-mercaptoethanol in 100 mm Tris-HCl, pH 8.0. Second, the protein samples were heated at 100°C for 5 min in 10 mm NaPi buffer, pH 7.0, containing 1% SDS and 100 mm 2-mercaptoethanol. No difference was observed with the two methods. For molecular mass determination the standards used were myosin (205 kD), β-galactosidase (116 kD), phosphorylase B (97.4 kD), BSA (66 kD), ovalbumin (43 kD), and carbonic anhydrase (31 kD) as high standards; and phosphorylase B (97.4 kD), BSA (66 kD), ovalbumin (43 kD), carbonic anhydrase (31 kD), and trypsin inhibitor (20.1 kD) as low standards.
Estimation of Mr under nondenaturing conditions was performed by gel filtration using a Sephadex G-200 (fine grade) column (2.5 × 100 cm) equilibrated and eluted at 4°C with 100 mm KPi buffer, pH 7.0, containing 300 mm KCl. The distribution coefficient was obtained as described previously (Gelotte, 1960) using blue dextran to measure the void volume and Tyr to measure the total volume. The standards used were β-amylase (200 kD), alcohol dehydrogenase (150 kD), BSA (66 kD), carbonic anhydrase (31 kD), and Cyt c (12.4 kD).
Spectrophotometric Methods
Absorption spectra were recorded at 25°C with a Cary 2300 spectrophotometer (Varian, Victoria, Australia). Anaerobic experiments were made after several cycles of vacuum followed by flushing with O2-free argon at 25°C in a Thunberg-type spectrophotometer cuvette (Vetroscientifica, Rome, Italy) in which anaerobic additions of various reagents could be made through a rubber cap with a syringe. Fluorescence spectra were obtained using a Perkin-Elmer LS-3 spectrofluorimeter.
Determination of TPQ Stoichiometry
Native ELAO was titrated anaerobically in 100 mm KPi buffer, pH 7.0, at 25°C with putrescine, benzylamine, DABA, and kynuramine as the substrates. The aromatic aldehyde production from benzylamine was measured by the A250 increase using an ε250 of 12,800 m−1 cm−1 (Neumann et al., 1975). The aldehyde production from DABA was measured by the A350 increase using an ε350 of 31,200 m−1 cm−1 (Ehrlich's reagent; Medda et al., 1995b). The reaction with kynuramine was followed photometrically at 326 and 314 nm (formation of 4-hydroxyquinoline) or at 356 and 255 nm (disappearance of kynuramine; Padiglia et al., 1997). The amount of reaction product formed was calculated as the mean of at least three different measurements. ELAO was also titrated with carbonyl-containing compounds in the presence and absence of oxygen. Titrations were performed by stepwise addition of 5 μL of freshly prepared reagent, and the optical density changes were measured at the appropriate wavelengths (see ResultsDiscussion) on a Cary 2300 spectrophotometer at 25°C. At each addition the optical density of the reaction mixture was checked at the appropriate time (see ResultsDiscussion) and aliquots of the reaction mixture were tested for activity. The activity of the enzyme was measured polarographically with an oxygraph (Gilson Medical Electronics, Middleton, WI) at 37°C. The incubation mixture (1 mL) contained 2 μg of purified ELAO in 100 mm KPi buffer, pH 7.0. The reaction was started by the addition of 50 μL of putrescine solution (17 mm) after at least 10 min of preincubation.
Carbanion Detection
Reaction of TNM with enzyme alone or with enzyme and putrescine as the substrates was monitored by the time-dependent increase in A350 (Medda et al., 1993). The reaction was initiated by adding the substrates to a reaction mixture (1 mL) containing TNM and known quantities of ELAO. The rates of nitroform production reported are initial rates and were determined by the slope of the tangent to the absorbance versus time curve 6 to 8 s after addition of the substrate.
Preparation of Genomic DNA from Leaf Tissue
Frozen Euphorbia characias leaf tissue was completely powdered using a pestle-and-mortar grinding method. DNA was isolated using a plant DNA-isolation kit (Boehringer Mannheim), according to the manufacturer's instructions.
PCR amplification of a partial ELAO sequence was based on corresponding segments in the cDNA sequences of LSAO and PSAO. The peptide sequence VRTIVT near the VGNyDNV sequence, which contains the tyrosinyl residue (y) modified into TPQ, and a FYIYYLD peptide sequence localized after the IGIYHDH sequence were chosen to design sense and antisense primers for PCR using E. characias DNA as the template. In both LSAO and PSAO these sequences delimit an identical fragment of 222 bp. The sense primer was GTA/AGA/ACC/ATA/GTG/ACG, whereas the antisense primer was ATC/AAG/ATA/GTA/AAT/ATA/GAA. The PCR amplification was performed in a solution composed of 1.5 mm MgCl2, 50 mm KCl, and 200 μm deoxyribonucleotide triphosphate mixture; 1 μm sense primer; 1 μm antisense primer; 1 μg of E. characias template DNA; and 1 to 3 units of Ampli Taq Gold DNA polymerase (Perkin-Elmer) in 100 mm Tris-HCl buffer, pH 8.3. The thermal cycles of amplification were carried out in a Perkin-Elmer DNA thermal cycler using this program: step 1, 94°C for 9 min; step 2, 94°C for 30 s, 42°C to 45°C (variable annealing temperatures) for 30 s, 72°C for 1 min; and step 3, 72°C for 3 min. Step 2 was repeated 40 times. Reactions were performed in 50- to 100-μL reaction volumes using 0.2-mL reaction tubes. Samples were subsequently analyzed on 1% agarose gels or 6% polyacrylamide gels and stained with ethidium bromide using φX174 HaeIII digest as a molecular mass marker (Sigma). In a large number of reactions a clear band was observed that corresponded approximately to a 200-bp product; this product was purified from 1% agarose gel using the QIAquick gel-extraction kit protocol (Qiagen, Chatsworth, CA).
Cycle Sequencing of Purified Fragment
DNA-sequence analysis of fragments was performed with the Digoxigenin Taq DNA Sequencing Kit (Boehringer Mannheim). Forward and reverse nonradioactive digoxigenin-sequencing primers labeled with digoxigenin at the 5′ end were synthesized with the same sequences as the PCR primers. Cycling conditions were the same as those used in PCR amplification. After the sequencing reaction DNA fragments were separated on the sequencing gel. As they reached the bottom of the gel, the fragments were transferred directly to a nylon membrane (Sartorius, Edgewood, NY). The transfer was completed after approximately 8 to 10 h, and the cross-linking of the transferred DNA was done using a standard UV-light transilluminator for about 3 min. Colorimetric detection of the blotted sequencing ladders was performed using a Digoxigenin Nucleic Acid Detection Kit (Boehringer Mannheim) with additional required reagents. The color precipitate started to form within a few minutes and the reaction was usually complete after 16 h. The reaction was stopped by washing the membrane for 5 min with 10 mm Tris-HCl buffer, pH 8.0, containing 1 mm EDTA.
RESULTS AND DISCUSSION
Molecular Properties
The enzyme preparations used in our studies were assayed for copper content by atomic absorption spectrometry. The purified enzyme contained 0.090% (w/w) of copper. On this basis, a minimum Mr of 70,000 can be calculated. Because the crystal structure of PSAO (Kumar et al., 1996) revealed a second site with a metal tentatively identified as manganese, we looked for this metal with atomic absorption spectrometry, but none was detected.
Relative Molecular Mass Determination
The purified ELAO showed a single symmetrical peak of molecular mass at 145 ± 2 kD in gel-filtration chromatography (results not shown), and a single band in SDS-PAGE (Fig. 1). The molecular mass of the ELAO monomer as determined by SDS-PAGE was found to be 72 ± 1 kD (Fig. 1). These results are in good agreement with the preliminary data reported (Rinaldi et al., 1982).
Figure 1.
SDS-PAGE pattern of ELAO. Lane a, ELAO; lane b (from top to bottom) ELAO with low standard proteins: phosphorylase B (97.4 kD), BSA (66 kD), ovalbumin (43 kD), carbonic anhydrase (31 kD), and trypsin inhibitor (20.1 kD).
Reaction of ELAO with TNM
ELAO catalyzes the reaction of putrescine with TNM, giving rise to the nitroform anion. No production of nitroform was seen during 2 to 3 min of preincubation of LSAO with 250 μm TNM at 25°C in 10 mm KPi buffer, pH 7.0. Moreover, the rate of nitroform production was negligible in an enzyme-free reaction mixture containing TNM, putrescine, ammonia, hydrogen peroxide, and aldehyde product, indicating that the nitroform generation occurs only in the reaction of TNM with an ELAO-substrate complex. Therefore, no correction of the rates was required. The initial rates for nitroform production were linearly related to the concentration of active sites of ELAO present in each assay (not shown). The amount of nitroform that accumulated in 10 min was 14 to 20 times the content of enzyme active sites, consistent with the requirement of enzyme turnover to release nitroform. The increase at 350 nm was immediate when putrescine was added. A double-reciprocal plot of the data A350 min−1/TNM concentration at saturating concentration of putrescine was linear. The Km value obtained was 1.25 mm, whereas the maximal rate of nitroform production was 11.4 nmol min−1.
Spectroscopic Properties and Titration of ELAO
In addition to the protein absorbance maximum at 278 nm, the enzyme showed a maximum at 490 nm in the visible region.
Reaction of ELAO with Hydrazines
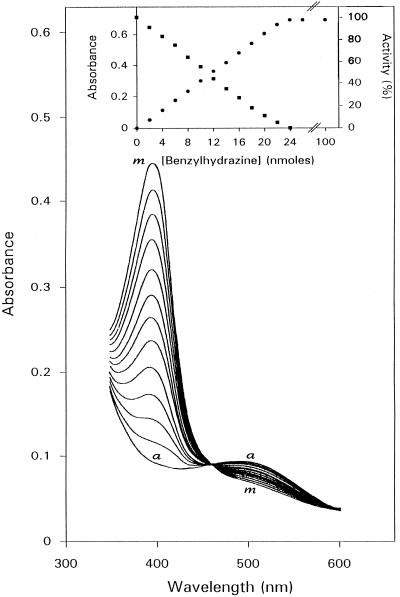
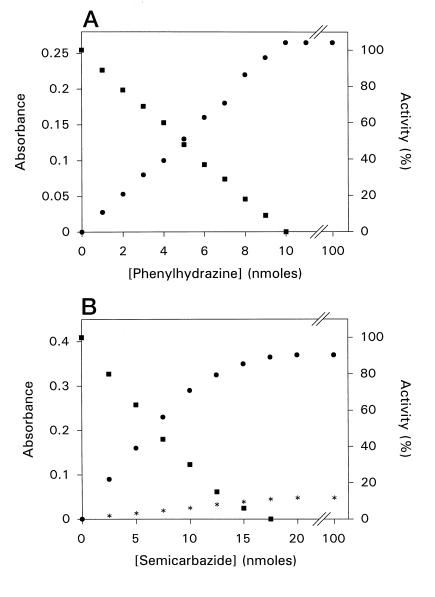
Chemical modification of ELAO was made by addition of PHY or BHY to enzyme solutions (1 mL) in 100 mm KPi buffer, pH 7.0, both in air and anaerobically. The addition of PHY or BHY to ELAO was followed by the formation of strong absorption bands at 430 nm (ε430 = 55,000 m−1 cm−1) and 380 nm (ε380 = 60,000 m−1 cm−1), respectively. Both derivatives were nonfluorescent. The reaction of ELAO with either PHY or BHY was irreversible. Formation of the BHY-modified enzyme was concomitant with the disappearance of the absorption band at 490 nm, and a clean isosbestic point at 460 nm was observed upon titration of ELAO with BHY (Fig. 2). Analogous changes could be obtained upon titration with PHY, with an isosbestic point observed at 514 nm (data not shown). A titration end point was obtained at a ratio BHY:ELAO dimer (Fig. 2, inset) or PHY:ELAO dimer (Fig. 3A) of 2. The spectral changes of ELAO with PHY and BHY were also studied as a function of time. The reaction was very fast for both PHY and BHY, and the end point for maximum spectral change at high amounts of inhibitor was 10 min.
Figure 2.
Half-titration of ELAO with BHY. ELAO (24-nmol active sites) in 1 mL of 100 mm KPi buffer, pH 7.0, was treated with aliquots (5 μL = 1 nmol) of BHY. Spectra were recorded before (a) and after (m) (second spectra) each addition of BHY. Inset, Correlation between spectrum modification and inhibition of enzymic activity of ELAO by BHY. Absorbance readings were taken at 380 nm (•) and 5-μL portions of the reaction mixture were taken simultaneously for activity assay (▪).
Figure 3.
A, Correlation between spectrum modification and inhibition of enzymatic activity of ELAO by PHY. For each aliquot (5 μL of 1-nmol PHY to 10-nmol ELAO active sites in 1 mL of 100 mm KPi buffer, pH 7.0) absorbance readings were taken at 430 nm (•) and 5-μL portions of the reaction mixture were taken from the cuvette for activity assay (▪). B, Correlation between spectrum modification and inhibition of enzymatic activity of ELAO by SCA. For each aliquot (5 μL of 2.5-nmol SCA to 17-nmol ELAO active sites in 1 mL of 100 mm KPi buffer, pH 7.0) absorbance readings were taken at 480 nm (*) and 345 nm (•), and 5 μL of the reaction mixture was removed from the cuvette for activity assay (▪).
Reaction of ELAO with SCA
The addition of SCA to the incubation mixture was followed by the formation of two absorption bands at 480 nm (ε480 = 5,000 m−1 cm−1) and 345 nm (ε345 = 23,000 m−1 cm−1). The formation of these new absorption bands was always concomitant with the disappearance of the absorption band at 490 nm. The reaction was very fast and the total time required for maximum spectral change with increasing amount of SCA was 10 min. Because of the irreversibility of the reaction, it was possible to titrate ELAO with SCA (Fig. 3B). An end point at a ratio SCA:ELAO dimer of 2 was always obtained.
Reaction of ELAO with Putrescine
Putrescine is the best substrate for ELAO. In 100 mm KPi buffer, pH 7.0, the turnover number was 2300 min−1. When putrescine was added to ELAO in anaerobiosis, the broad absorption band at 490 nm disappeared instantaneously, indicating the rapid conversion of the TPQ cofactor to a bleached species, presumably the quinolaldimine. Then the solution turned yellow as a result of the formation of new absorption bands centered at 430 and 460 nm, indicative of a free radical intermediate species generated upon reaction in the absence of oxygen (Finazzi Agró et al., 1984; Dooley et al., 1987; Medda et al., 1995b). Precisely 20 nmol of substrate was required to completely reduce 10 nmol of ELAO (results not shown).
Reaction of ELAO with Benzylamine
Benzylamine is the best substrate for benzylamine oxidase from pig or beef plasma (Janes and Klinman, 1991; Morpurgo et al., 1991). Also, ELAO was able to oxidize benzylamine, with a Km value of 0.4 mm, which is similar to that found for putrescine (Rinaldi et al., 1982). The turnover number at saturating benzylamine concentrations was found to be 10.5 min−1. The anaerobic titration of 10 nmol of ELAO with benzylamine required 20 nmol of substrate (not shown), and the formation of 1.98 ± 0.2 mol of benzaldehyde per mol of enzyme dimer was directly observable by the increase in A250.
Oxidation of DABA
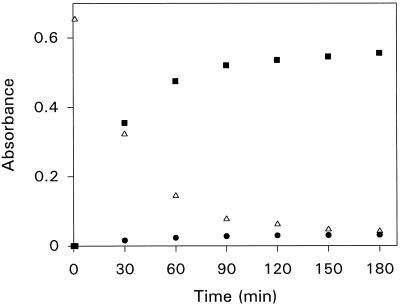
DABA was a poor substrate for ELAO. The turnover number with DABA was 0.028 min−1, a small value compared with the corresponding number of 2300 min−1 found for putrescine. However, when DABA was added to ELAO in anaerobiosis, the broad absorption band at 490 nm disappeared instantaneously. Together with the formation of the quinolaldimine, a new band centered at 400 nm appeared. This band was assigned to the protonated tautomeric form of the quinolaldimine, as reported for DABA oxidation by LSAO (Medda et al., 1995b). Under anaerobic conditions this species decayed slowly, in parallel with formation of the yellow radical intermediate (Fig. 4). These spectral changes occurred simultaneously with the liberation of 1.98 ± 0.2 mol of the corresponding aldehyde directly observable by the increase in A350. In the presence of air, the radical species reacted rapidly with oxygen to restore the oxidized enzyme and to liberate ammonia and hydrogen peroxide (data not shown).
Figure 4.
Time course of the reaction of 18-nmol ELAO active sites with 2 mm DABA in 1 mL of 100 mm KPi buffer, pH 7.0, in anaerobic conditions, measured at 460 nm (•), 400 nm (▵), and 350 nm (▪).
Oxidation of Kynuramine
Kynuramine is an endogenous amine that is formed from tryptamine through indole 2,3-dioxygenase and formamidase, or from decarboxylation of kynurenine (Johnson and Clarke, 1981). Kynuramine is an excellent substrate used in assays for the measurement of both types A and B of monoamine oxidase activity. In fact, this compound is converted by monoamine oxidase to an aldehyde that spontaneously rearranges to 4-hydroxyquinoline, which fluoresces strongly in alkaline solutions (Morinan and Garratt, 1985). Kynuramine is also a substrate for pig plasma copper amine oxidase and its fluorescence was used to probe the catalytic site of this enzyme (Massey and Churchich, 1977). However, kynuramine behaves as a competitive inhibitor of the SCA-sensitive copper amine oxidases (Elliott et al., 1988). ELAO was able to oxidize kynuramine to 4-hydroxyquinoline. At pH 7.0, in 100 mm KPi buffer, the turnover number for kynuramine was 0.24 min−1. At the same pH value the substrate inhibition was apparent at lower concentrations than when the substrate was putrescine (2 mm for kynuramine and 17 mm for putrescine; data not shown).
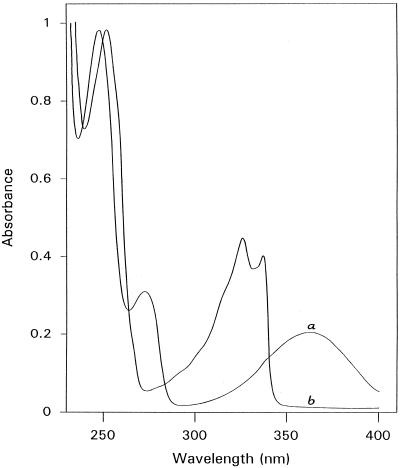
Kynuramine showed three distinct absorption maxima at 356 nm (ε356 = 3,900 m−1 cm−1), 255 nm (ε255 = 6,000 m−1 cm−1), and 224 nm (ε224 = 19,900 m−1 cm−1) in the UV spectrum, whereas 4-hydroxyquinoline showed maxima at 326 nm (ε326 = 7,980 m−1 cm−1), 314 nm (ε314 = 8,960 m−1 cm−1), and 228 nm (ε228 = 19,900 m−1 cm−1) (Fig. 5). When kynuramine was added to ELAO in aerobic conditions, the absorption bands at 356 and 255 nm disappeared in parallel with formation of bands centered at 326 and 314 nm (not shown). Further changes in the UV region were difficult to check because of the very high absorption of the protein. The reaction was also followed fluorimetrically using an excitation wavelength of 314 nm and an emission wavelength of 350 nm to monitor the formation of the 4-hydroxyquinoline. In alkaline solution (pH = 10.0) the emission wavelength shifted to 380 nm with a 15-fold increase in sensitivity. Addition of kynuramine under anaerobic conditions resulted in the rapid disappearance of the absorption at 490 nm, and the solution turned yellow as a result of the formation of the free radical intermediate species (not shown). These spectral changes occurred simultaneously with the liberation of the aldehyde; in fact, formation of the product was directly observable by the increases in A326 and A314. After subtraction of the appropriate blanks the formation of 1.98 ± 0.2 mol of aldehyde per mol of enzyme dimer was determined.
Figure 5.
UV-visible spectra of 50 μm kynuramine (a) and 50 μm 4-hydroxyquinoline (b) in 100 mm KPi buffer, pH 7.0.
The results obtained by irreversible inhibitors and substrates indicated unequivocally the presence of two quinones per protein dimer and showed that both quinones were active toward the substrates. Using these data, the extinction coefficient was determined to be 6000 m−1 cm−1 at 490 nm and 3.76 × 105 m−1 cm−1 at 278 nm for the purified enzyme (two copper ions and Mr 145,000). The ratio A278:A490 of the purified ELAO was 62; this ratio is traditionally used to characterize amine oxidases (McGuirl et al., 1994).
The Radical Species
The spectrum of the radical species is independent of the substrate used (putrescine, benzylamine, DABA, or kynuramine), in analogy with other plant amine oxidases (Medda et al., 1996).
The Effect of Cyanide
Cyanide is known to trap the semiquinone form by stabilizing Cu(I), and the yield of the radical increases in Arthrobacter P1 and porcine kidney amine oxidases by addition of cyanide (McGuirl et al., 1997). When cyanide (2 mm) was added to anaerobically reduced ELAO using benzylamine as the substrate, the spectrum of the radical form showed a 70% increase in intensity.
The Effect of Azide
Azide is known to bind to Cu(II), generating a Cu(II)-N3− complex that absorbs at 400 nm. In 100 mm KPi buffer, pH 7.0, and at 20°C, the Keq of the Cu(II)-azide complex for oxidized ELAO was determined to be 34 ± 5 m−1. When ELAO was first reduced by 2 mm benzylamine to form a mixture of Cu(I)-semiquinolamine and Cu(II)-aminoresorcinol, and then azide was added to the sample, the semiquinolamine radical spectrum decreased, probably because of conversion to the aminoresorcinol form (results not shown). However, in ELAO, as in other amine oxidases (McGuirl et al., 1997), the reduction of TPQ cofactor by the substrate had no effect on the magnitude of Keq for azide binding to Cu2+.
Temperature Effect
The Cu(I)-semiquinolamine radical form of ELAO was monitored as a function of temperature. As reported for the pea enzyme (Turowski et al., 1993), the concentration of the yellow species increased with the temperature in the range 10°C to 30°C and was constant from 30°C to 40°C, the highest temperature investigated (results not shown).
Reaction of Copper-Free ELAO
Copper could be removed from native ELAO by treatment with cyanide and diethyldithiocarbamate (see Methods). The residual copper, measured by atomic absorption spectroscopy, was 0.2 ± 0.2% of the original content. Copper-free ELAO showed a broad absorption peak at 480 nm (Fig. 6, inset), shifted toward shorter wavelengths with respect to the native enzyme, but with similar intensity (ε480 = 6000 m−1 cm−1). The addition of DABA to the copper-free enzyme caused the formation of the peak at 400 nm, which later decayed in parallel with the release of aldehyde. Within the experimental error the absorption changes observed at 350 and 400 nm were identical to those seen for the holoenzyme in anaerobiosis. However, the complete absence of bands at 434 and 464 nm showed that no formation of the free radical intermediate occurred (results not shown).
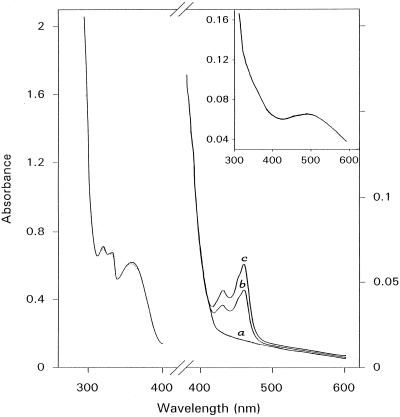
Figure 6.
Absorption spectra of copper-free ELAO upon reaction with kynuramine. The sample contained 11 μm copper-free enzyme under anaerobic conditions in 100 mm KPi buffer, pH 7.0. Spectrum a, Copper-free ELAO after addition of 160 μm kynuramine; spectrum b, spectrum a 5 min after the addition of CuCl2 (200 μmol); spectrum c, spectrum a after 10 min. The dotted line represents the dilution effect after addition of CuCl2. Inset, Spectrum of 11 μm ELAO copper-free enzyme.
Reaction of the apoenzyme with kynuramine in aerobiosis as well as in anaerobiosis resulted in permanent bleaching of the 480-nm band, with the concomitant release of the aldehyde (Fig. 6). From the disappearance of the 356-nm band as a result of oxidation of kynuramine, and the parallel appearance of two bands at 326 and 314 nm attributable to the formation of the 4-hydroxyquinoline, the aldehyde product may be quantified to 1.95 ± 0.2 mol per mol of enzyme dimer, as for the native enzyme. Notice that rearrangement of the aldehyde to hydroxyquinoline demonstrates that the product has effectively been released from the enzyme. Copper was essential for the reoxidation step. In fact, the copper-free enzyme did not recover its color after reacting with the substrate, and did not form the yellow intermediate in anaerobiosis, but only the aminoresorcinol species. However, the addition of 200 μm CuCl2 to the incubation mixture in anaerobiosis caused the formation of the radical species absorbing at 460 and 430 nm, whereas the absorbance of kynuramine at 356 nm and the absorbance of the 4-hydroxyquinoline at 326 and 314 nm showed only a small decrease attributable to dilution (Fig. 6), indicating an equilibrium between aminoresorcinol and radical species, without further release of the aldehyde. Addition of oxygen eventually restored the 490-nm band, demonstrating the reoxidation of the enzyme.
PCR Amplification and Sequencing Reactions
Alignment of amino acid sequences of all known amine oxidases shows that the homology is in the range of 20% to 99% and that the important residues determining the enzyme activity are conserved. The homology is high for amine oxidases obtained from related organisms, as for LSAO and PSAO (>92%), but becomes very low when comparing plant and mammal enzymes (25%; Frébort and Adachi, 1995). Comparison of the amino acid sequence around the TPQ cofactor in several amine oxidases obtained from either cDNA or direct sequencing of the protein shows a much higher homology for the C than for the N terminus. TPQ bound in the typical sequence NyD/E is located somewhere in the middle. The motif HXH (X = Q, D, or T), found in the amine oxidases located at the positions around 40 to 50 residues from the TPQ position toward the C terminus, is conserved in all amine oxidases (with a single exception, the amine oxidase from Aspergillus niger). Moreover, two conserved histidyl residues were found in all enzymes 30 residues from TPQ, one toward the N terminus and one near the C terminus. It was shown that three histidyl residues, two of them in the HXH motif and one at the C terminus, are ligands of copper. The DNA sequence of the ELAO active site and the amino acid sequence shown in Figure 7 are very similar to the corresponding sequences of PSAO and LSAO, in which the position of TPQ has been demonstrated to be the Tyr located between an Asn and an Asp. Therefore, we propose that in ELAO this tyrosinyl residue can be modified to TPQ in the mature protein.
Figure 7.
Sequence of the ELAO (E) DNA compared with cDNA of LSAO (L) and PSAO (P). Circles indicate the Tyr that is modified to TPQ and two conserved His residues. Differences are underlined.
CONCLUSIONS
ELAO has been prepared to homogeneity by a new method of purification. Highly purified ELAO has a specific activity of 34 μmol substrate min−1 mg−1. The extinction coefficients determined by several substrates and inhibitors yield values of ε490 = 6000 m−1 cm−1 and ε278 = 3.76 × 105 m−1 cm−1. Active-site titration by substrates and inhibitors shows that ELAO contains two active TPQs per protein dimer. Copper-free ELAO, obtained by a simple method, can oxidize a substrate under single-turnover conditions and release 1 mol of aldehyde per mol of enzyme subunit. The amino acid sequence shows a region that could represent a consensus sequence for TPQ, which is formed by a posttranslational modification of a Tyr residue, as already proposed for other amine oxidases. The determination of the complete cDNA-derived amino acid sequence of ELAO is now in progress.
Abbreviations:
- BHY
benzylhydrazine
- DABA
p-(dimethylamino)benzylamine
- εx
extinction coefficient at x nm
- ELAO
Euphorbia characias latex amine oxidase
- LSAO
Lens esculenta seedling amine oxidase
- PHY
phenylhydrazine
- PSAO
Pisum sativum seedling amine oxidase
- putrescine
1,4-diaminobutane dihydrochloride
- SCA
semicarbazide
- TNM
tetranitromethane
- TPQ
6-hydroxydopa quinone (2,4,5-trihydroxyphenylalanine)
Footnotes
This study was partially supported by Ministero Università Ricerca Scientifica e Technologica funds (60%) and by the European Commission (European Social Funds).
LITERATURE CITED
- Agostinelli E, De Matteis G, Sinibaldi A, Mondovì B, Morpurgo L. Reactions of the oxidized organic cofactor in copper-depleted bovine serum amine oxidase. Biochem J. 1997;324:497–501. doi: 10.1042/bj3240497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellelli A, Brunori M, Finazzi Agró A, Floris G, Giartosio A, Rinaldi A. Transient kinetics of copper-containing lentil (Lens culinaris) seedling amine oxidase. Biochem J. 1985;232:923–926. doi: 10.1042/bj2320923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellelli A, Finazzi Agró A, Floris G, Brunori M. On the mechanism and rate of substrate oxidation by amine oxidase from lentil seedlings. J Biol Chem. 1991;266:20654–20657. [PubMed] [Google Scholar]
- Brown DE, McGuirl MA, Dooley DM, Janes SM, Mu D, Klinman JP. The organic functional group in copper-containing amine oxidases. J Biol Chem. 1991;266:4049–4051. [PubMed] [Google Scholar]
- Cai D, Klinman JP. Evidence for a self-catalytic mechanism of 2,4,5-trihydroxyphenylalanine quinone biogenesis in yeast copper amine oxidase. J Biol Chem. 1994;269:32039–32042. [PubMed] [Google Scholar]
- Choi YH, Matsuzaki R, Fukui T, Shimizu E, Yorifuji T, Sato H, Ozaki Y, Tanizawa K. Copper topa quinone containing histamine oxidase from Arthrobacter globiformis: molecular cloning and sequencing, overproduction of precursor enzyme, and generation of topa quinone cofactor. J Biol Chem. 1995;270:4712–4720. doi: 10.1074/jbc.270.9.4712. [DOI] [PubMed] [Google Scholar]
- Cooper RA, Knowles PF, Brown DE, McGuirl MA, Dooley DM. Evidence for copper and 3,4,6-trihydroxyphenylalanine quinone cofactors in an amine oxidase from the Gram-negative bacterium Escherichia coli K-12. Biochem J. 1982;288:337–340. doi: 10.1042/bj2880337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley DM, McGuirl MA, Brown DE, Turowski PN, McIntire WS, Knowles PF. A Cu(I)-semiquinone state in substrate-reduced amine oxidases. Nature. 1991;349:262–264. doi: 10.1038/349262a0. [DOI] [PubMed] [Google Scholar]
- Dooley DM, McGuirl MA, Peisach J, McCracken J. The generation of an organic free radical in substrate-reduced pig kidney diamine oxidase-cyanide. FEBS Lett. 1987;214:274–278. doi: 10.1016/0014-5793(87)80069-8. [DOI] [PubMed] [Google Scholar]
- Elliott J, Callingham BA, Sharman DF . Semicarbazide-sensitive amine oxidase (SSAO) of the rat aorta: interactions with some naturally occurring amines and their structural analogues. Biochem Pharmacol. 1988;38:1507–1515. doi: 10.1016/0006-2952(89)90191-3. [DOI] [PubMed] [Google Scholar]
- Finazzi Agró A, Rinaldi A, Floris G, Rotilio G. A free radical intermediate in the reduction of plant Cu-amine oxidases. FEBS Lett. 1984;176:378–380. [Google Scholar]
- Frébort I, Adachi O. Copper/quinone-containing amine oxidases, an exciting class of ubiquitous enzymes. J Ferment Bioeng. 1995;80:625–632. [Google Scholar]
- Gabriel O. Analytical disc gel electrophoresis. Methods Enzymol. 1971;22:565–578. [Google Scholar]
- Gelotte B. Studies on gel filtration sorption properties of the bed material Sephadex. J Chromatogr. 1960;3:330–342. [Google Scholar]
- Hartree F. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal Biochem. 1972;48:422–427. doi: 10.1016/0003-2697(72)90094-2. [DOI] [PubMed] [Google Scholar]
- Janes SM, Klinman JP. An investigation of bovine serum amine oxidase active site stoichiometry: evidence for an aminotransferase mechanism involving two carbonyl cofactors per enzyme dimer. Biochemistry. 1991;30:4599–4605. doi: 10.1021/bi00232a034. [DOI] [PubMed] [Google Scholar]
- Janes SM, Mu D, Wemmer D, Smith AJ, Kaur S, Maltby D, Burlingame AL, Klinman JP. A new redox cofactor in eukaryotic enzymes: 6-hydroxydopa at the active site of bovine serum amine oxidase. Science. 1990;248:981–987. doi: 10.1126/science.2111581. [DOI] [PubMed] [Google Scholar]
- Johnson TD, Clarke DE . An α-adrenoceptor inhibitory action of kynuramine. Eur J Pharmacol. 1981;72:351–356. doi: 10.1016/0014-2999(81)90574-4. [DOI] [PubMed] [Google Scholar]
- Klinman JP, Mu D . Quinoenzymes in biology. Annu Rev Biochem. 1994;63:299–344. doi: 10.1146/annurev.bi.63.070194.001503. [DOI] [PubMed] [Google Scholar]
- Kumar V, Dooley DM, Freeman HC, Guss JM, Harvey I, McGuirl MA, Wilce MCJ, Zubak V. Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 Å resolution. Structure. 1996;4:943–955. doi: 10.1016/s0969-2126(96)00101-3. [DOI] [PubMed] [Google Scholar]
- Massey JB, Churchich JE . Kynuramine, a fluorescent substrate and probe of plasma amine oxidase. J Biol Chem. 1977;252:8081–8084. [PubMed] [Google Scholar]
- Matsuzaki R, Fukui T, Sato H, Ozaki Y, Tanizawa K. Generation of the topa quinone cofactor in bacterial monoamine oxidase by cupric ion-dependent autooxidation of a specific tyrosyl residue. FEBS Lett. 1994;351:360–364. doi: 10.1016/0014-5793(94)00884-1. [DOI] [PubMed] [Google Scholar]
- McGuirl MA, Brown DE, Dooley DM. Cyanide as a copper-directed inhibitor of amine oxidases: implications for the mechanism of amine oxidation. J Biol Inorg Chem. 1997;2:336–342. [Google Scholar]
- McGuirl MA, McCahon CD, McKeown KA, Dooley DM. Purification and characterization of pea seedling amine oxidase for crystallization studies. Plant Physiol. 1994;106:1205–1211. doi: 10.1104/pp.106.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire W, Hartmann C (1993) Copper containing amine oxidases. In VL Davidson, ed, Principles and Applications of Quinoprotein. Marcel Dekker, New York, pp 97–171
- Medda R, Padiglia A, Bellelli A, Sarti P, Santanchè S, Finazzi Agró A, Floris G. Intermediates in the catalytic cycle of lentil seedling copper-containing amine oxidase. Biochem J. 1998;332:431–437. doi: 10.1042/bj3320431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medda R, Padiglia A, Floris G. Plant copper-amine oxidases. Phytochemistry. 1995a;39:1–9. [Google Scholar]
- Medda R, Padiglia A, Pedersen JZ, Floris G. Evidence for α-proton abstraction and carbanion formation involving a functional histidine residue in lentil seedling amine oxidase. Biochem Biophys Res Commun. 1993;196:1349–1355. doi: 10.1006/bbrc.1993.2401. [DOI] [PubMed] [Google Scholar]
- Medda R, Padiglia A, Pedersen JZ, Lorrai A, Floris G. Substrate specificity of lentil seedling amine oxidase. Biochem Mol Biol Int. 1996;40:629–637. doi: 10.1080/15216549600201223. [DOI] [PubMed] [Google Scholar]
- Medda R, Padiglia A, Pedersen JZ, Rotilio G, Finazzi Agró A, Floris G. The reaction mechanism of copper amine oxidase: detection of intermediates by the use of substrates and inhibitors. Biochemistry. 1995b;34:16375–16381. doi: 10.1021/bi00050a018. [DOI] [PubMed] [Google Scholar]
- Morinan A, Garratt HM . An improved fluorimetric assay for brain monoamine oxidase. J Pharmacol Methods. 1985;13:213–223. doi: 10.1016/0160-5402(85)90021-x. [DOI] [PubMed] [Google Scholar]
- Morpurgo L, Agostinelli E, Mondovì B, Avigliano L, Silvestri R, Stefanich G, Artico M. Bovine serum amine oxidase: half-site reactivity with phenylhydrazine, semicarbazide, and aromatic hydrazides. Biochemistry. 1991;31:2615–2621. doi: 10.1021/bi00124a023. [DOI] [PubMed] [Google Scholar]
- Mure M, Tanizawa K. Chemical and biochemical characteristics of topa quinone. Biosci Biotechnol Biochem. 1997;61:410–417. [Google Scholar]
- Nakamura N, Matsuzaki R, Choi Y-H, Tanizawa K, Sanders-Loehr J. Biosynthesis of topa quinone cofactor in bacterial amine oxidases. J Biol Chem. 1996;271:4718–4724. doi: 10.1074/jbc.271.9.4718. [DOI] [PubMed] [Google Scholar]
- Neumann R, Hevey R, Abeles RH. The action of plasma amine oxidase on β-haloamines. J Biol Chem. 1975;250:6362–6367. [PubMed] [Google Scholar]
- Padiglia A, Medda R, Floris G. Lentil seedling amine oxidase: interaction with carbonyl reagents. Biochem Int. 1992;28:1097–1107. [PubMed] [Google Scholar]
- Padiglia A, Medda R, Lorrai A, Congiu D, Pedersen JZ, Floris G. Oxidation of kynuramine by lentil seedling copper amine oxidase: demonstration of a single turnover mechanism in the apoenzyme. Phytochem Anal. 1998;117:1–9. [Google Scholar]
- Parson MR, Convery MA, Wilmot CM, Yadav KDS, Blakeley V, Corner AS, Phillips SEV, McPherson MJ, Knowles PF. Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 Å resolution. Structure. 1995;3:1171–1184. doi: 10.1016/s0969-2126(01)00253-2. [DOI] [PubMed] [Google Scholar]
- Pedersen JZ, El-Sherbini S, Finazzi Agró A, Rotilio G. A substrate-cofactor free radical intermediate in the reaction mechanism of copper amine oxidase. Biochemistry. 1992;31:2–6. doi: 10.1021/bi00116a002. [DOI] [PubMed] [Google Scholar]
- Rinaldi A, Floris G, Finazzi Agró A. Purification and properties of diamine oxidase from Euphorbia latex. Eur J Biochem. 1982;127:417–422. doi: 10.1111/j.1432-1033.1982.tb06888.x. [DOI] [PubMed] [Google Scholar]
- Steinebach V, de Vries S, Duine JA. Intermediates in the catalytic cycle of copper-quinoprotein amine oxidase from Escherichia coli. J Biol Chem. 1996;271:5580–5588. doi: 10.1074/jbc.271.10.5580. [DOI] [PubMed] [Google Scholar]
- Turowski PN, McGuirl MA, Dooley DM. Intramolecular electron transfer rate between active-site copper and TOPA quinone in pea seedling amine oxidase. J Biol Chem. 1993;268:17680–17682. [PubMed] [Google Scholar]
- Weber K, Osborne M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]