This work, related to that of Hahn et al. (this issue), addresses an important step of spliceosome assembly—that of unwinding the U4/U6 snRNA duplex during spliceosome catalytic activation. Lührmann and colleagues elucidate the underlying mechanism that regulates Brr2, the RNA helicase responsible for this unwinding. The Prp8 RNase H domain is found to bind U4/U6 snRNA, thereby competing with Brr2 and preventing its loading onto the duplex. Prp8 thus prevents premature unwinding of U4/U6 snRNA during splicing.
Keywords: pre-mRNA splicing, RNA helicase, RNA–protein complex, RNA–protein cross-linking, spliceosome catalytic activation
Abstract
The spliceosomal RNA helicase Brr2 catalyzes unwinding of the U4/U6 snRNA duplex, an essential step for spliceosome catalytic activation. Brr2 is regulated in part by the spliceosomal Prp8 protein by an unknown mechanism. We demonstrate that the RNase H (RH) domain of yeast Prp8 binds U4/U6 small nuclear RNA (snRNA) with the single-stranded regions of U4 and U6 preceding U4/U6 stem I, contributing to its binding. Via cross-linking coupled with mass spectrometry, we identify RH domain residues that contact the U4/U6 snRNA. We further demonstrate that the same single-stranded region of U4 preceding U4/U6 stem I is recognized by Brr2, indicating that it translocates along U4 and first unwinds stem I of the U4/U6 duplex. Finally, we show that the RH domain of Prp8 interferes with U4/U6 unwinding by blocking Brr2's interaction with the U4 snRNA. Our data reveal a novel mechanism whereby Prp8 negatively regulates Brr2 and potentially prevents premature U4/U6 unwinding during splicing. They also support the idea that the RH domain acts as a platform for the exchange of U6 snRNA for U1 at the 5′ splice site. Our results provide insights into the mechanism whereby Brr2 unwinds U4/U6 and show how this activity is potentially regulated prior to spliceosome activation.
Pre-mRNA splicing is catalyzed by a multisubunit RNA–protein enzyme, the spliceosome, which carries out two successive trans-esterification reactions that lead to removal of an intron and the ligation of its flanking exons. Spliceosomes are formed via the stepwise recruitment of small nuclear ribonucleoprotein particles (snRNPs) and numerous non-snRNP proteins to the pre-mRNA substrate (for review, see Wahl et al. 2009). Initially, U1 and U2 snRNPs bind the 5′ splice site (5′ SS) and the branch site (BS) of the pre-mRNA's intron, respectively. This is followed by the recruitment of the U4/U6.U5 tri-snRNP to the spliceosome, yielding the B complex, which does not yet have an active center. The U4 and U6 snRNAs are extensively base-paired in the tri-snRNP, thereby keeping U6 small nuclear RNA (snRNA) catalytically inert (Staley and Guthrie 1998). For catalytic activation of the spliceosome, U4 snRNA must be displaced from U6 snRNA, which allows the formation of new U2/U6 base-pairing interactions and a catalytically important U6 internal stem–loop (ISL) (Wahl et al. 2009). Concomitant with or prior to this, the base-pairing interaction between the U1 snRNA and the 5′ SS must be disrupted to allow the highly conserved ACAGAGA sequence of U6 snRNA to base-pair with the 5′ end of the intron. This newly formed U2–U6–pre-mRNA RNA interaction network is thought to comprise the heart of the catalytic center of the spliceosome (Madhani and Guthrie 1992; Villa et al. 2002).
Brr2 catalyzes the dissociation of the U4/U6 duplex and thus plays a key part in initiating catalytic activation (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998). Brr2 is one of at least eight conserved ATP-dependent DExD/H-box RNA/RNP remodeling enzymes that drive the multiple rearrangements of the spliceosome during its working cycle (Staley and Guthrie 1998). It is a member of the Ski2-like subfamily of helicase superfamily 2 (SF2) (Bleichert and Baserga 2007) with two tandem helicase cassettes. The structure of each cassette resembles that of the DNA helicase Hel308 with an additional immunoglobulin-like domain (Pena et al. 2009; Zhang et al. 2009; Santos et al. 2012), suggesting that Brr2 shares a similar helicase mechanism (i.e., translocation in a 3′-to-5′ direction along the RNA strand to which it binds). However, the mechanism whereby it unwinds U4/U6—in particular where it loads onto the U4/U6 di-snRNA, and thus which region of the Y-shaped U4/U6 duplex is first unwound—is currently unclear.
Most of the spliceosomal RNA helicases associate with the spliceosome only during the steps that require their activity. Brr2 is an exception in that it is an integral component of the U5 snRNP that interacts stably with other U5 snRNP proteins, including Prp8 and Snu114 (Achsel et al. 1998; van Nues and Beggs 2001; Liu et al. 2006). After its recruitment to the spliceosome during U4/U6.U5 tri-snRNP addition, Brr2 remains stably bound until the spliceosome ultimately dissociates (Small et al. 2006; Bessonov et al. 2008; Fabrizio et al. 2009). In addition to unwinding U4/U6, Brr2 is also required for dissociation of the spliceosome after splicing catalysis, where it appears to facilitate dissociation of U2 and U6 (Small et al. 2006). Thus, Brr2 needs to be tightly regulated at multiple steps—including, for example, suppressing its helicase activity in the tri-snRNP, where it coresides with its U4/U6 substrate, and also after the integration of the tri-snRNP into the spliceosome—to avoid premature disruption of U4/U6 base-pairing.
Two of Brr2's tight interacting partners, Prp8 and the EF-G-like GTPase Snu114, have been shown to modulate Brr2 activity. In its GDP-bound state, Snu114 represses Brr2 activity, whereas it promotes Brr2 activity when it is bound to GTP (Small et al. 2006). Based on genetic interaction studies, Prp8, one of the largest and most highly conserved spliceosomal proteins, appears to negatively regulate Brr2-mediated RNA unwinding events during splicing (Kuhn and Brow 2000; Kuhn et al. 2002). However, more recent studies have revealed that the C-terminal fragment (CTF) of Prp8 stimulates Brr2 helicase activity in vitro (Maeder et al. 2009; Pena et al. 2009) while suppressing Brr2's ATPase activity (Maeder et al. 2009). The Prp8 CTF interacts with both Brr2 (Maeder et al. 2009) and U4/U6 snRNA (Zhang et al. 2009); however, the mechanism whereby it regulates Brr2 activity is unclear. Ubiquitination of Prp8 has been proposed to play a role in regulating the activity of Brr2, but the mechanism whereby this is achieved is currently unknown (Bellare et al. 2008). A potential target is the C terminus of Prp8, which contains a Jab1/MPN-like domain (Pena et al. 2007; Zhang et al. 2007) that has lost its deubiquitination activity but retained its ability to bind ubiquitin (Bellare et al. 2006).
The Prp8 CTF also contains a domain with a three-dimensional (3D) fold resembling RNase H (RH), flanked on one side by a β-hairpin loop and on the other by an α-helical domain (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008). In the human B complex, a cross-link was identified between the 5′ SS and the RH domain of Prp8 (Reyes et al. 1996, 1999), leading to the proposal that it is involved in the handover of the 5′ SS from the U1 snRNA to the ACAGAGA box of the U6 snRNA. That the RH domain potentially provides a binding site for U4/U6 snRNAs is supported by genetic interaction studies. In particular, a triple nucleotide substitution in U4 that hyperstabilizes the U4/U6 interaction, leading to a cold-sensitive phenotype (U4-cs1) and a block in spliceosome activation, can be compensated by mutations in Prp8 (Kuhn et al. 1999; Kuhn and Brow 2000). Interestingly, some of these mutations map to the base of the β-hairpin loop of the RH domain, consistent with the idea that the RH domain might be involved in the regulation of U4/U6 duplex unwinding.
We demonstrate here that the RH domain of Saccharomyces cerevisiae Prp8 and the RNA helicase Brr2 both bind the single-stranded region of U4 preceding U4/U6 stem I (also denoted as the U4 central domain). Our data thus indicate that Brr2 translocates in a 3′-to-5′ direction along the U4 strand and unwinds U4/U6 stem I first. Finally, we demonstrate that the RH domain of Prp8 interferes with Brr2-mediated U4/U6 unwinding by preventing Brr2 loading onto the U4 snRNA. Our data thus elucidate a potential mechanism whereby Prp8 may negatively regulate Brr2 activity to prevent premature unwinding of the U4/U6 duplex during splicing.
Results
The RH, but not Jab1/MPN, domain of the Prp8 CTF binds U4/U6 snRNA
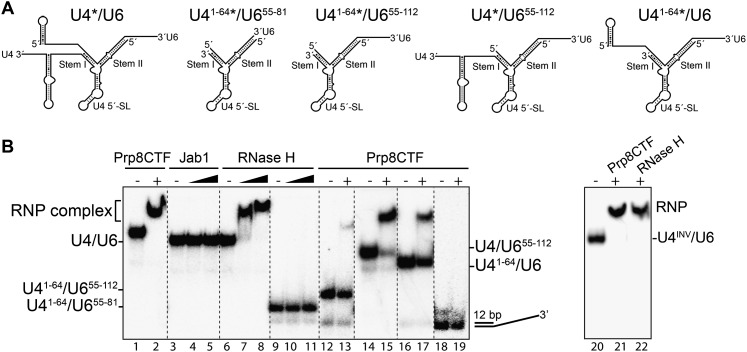
The yeast Prp8 CTF, comprised of an RH and Jab1/MPN domain (Supplemental Fig. S1A), was shown to bind yeast U4/U6 snRNA (Zhang et al. 2009). To elucidate which of these CTF subdomains possesses RNA-binding activity, we performed electrophoretic mobility shift assays (EMSAs) with recombinant CTF (Prp81836–2397), RH (Prp81836–2092), or Jab1/MPN (Prp82112–2413) domains of Prp8 (Supplemental Fig. S1A). At a concentration of 2 μM, both the Prp8 CTF and its RH domain bound and shifted all of the U4/U6 duplex to a slower-migrating complex (Fig. 1B, lanes 1,2,6–8). In contrast, the Jab1/MPN domain did not bind U4/U6 snRNA, even at an 8000-fold molar excess over RNA (Fig. 1B, lanes 3–5). A finer titration of the RH domain revealed an affinity for U4/U6 snRNA (apparent Kd = 1.4 ± 0.6 μM) (Supplemental Fig. S1B) similar to that previously reported for the CTF (Kd of 2.2 μM) (Zhang et al. 2009).
Figure 1.
Interaction of the S. cerevisiae Prp8 CTF and its Jab1/MPN and RH-like subdomains with U4/U6 snRNA. (A) Structure of the S. cerevisiae U4/U6 wild-type snRNA and truncation mutants used in EMSAs. (B) 32P-labeled wild-type U4/U6 snRNA (lanes 1–8); the truncated versions U41–64/U655–81 (lanes 9–11), U41–64/U655–112 (lanes 12,13), U4/U655–112 (lanes 14,15), U41-64/U6 (lanes 16,17), or U4INV/U6 (lanes 21,22); or a 12-bp linear RNA duplex with a 31-nt 3′ overhang (lanes 18,19) (0.5 nM each) were incubated alone (lanes 1,3,6,9,12,14,16,18,20) or in the presence of 2 uM (or, additionally, 4 uM in lanes 5,8,11) Prp8 CTF (lanes 2,13,15,17,19,21), RH domain (lanes 7,8,10,11,22), or Jab1/MPN domain (lanes 4,5). Escherichia coli tRNA (50 μg/mL) was included as competitor in all reactions. Complex formation was then analyzed by EMSA on a 6% native polyacrylamide gel, and RNA/RNP complexes were visualized by autoradiography.
The Prp8 RH domain requires ssRNA adjacent to U4/U6 stem I for efficient binding
Next we determined the minimum RNA requirement for efficient binding of the RH domain to U4/U6 snRNA using a series of truncation mutants of U4 and/or U6 snRNA. As the Jab1/MPN domain had little or no affinity for full-length U4/U6 snRNA, we used either a recombinant Prp8 CTF or the RH domain alone for these studies. Deleting the SLs and the single-stranded regions of either U6 or U4 snRNA preceding stem I (U4/U655–112 or U41–64/U6, respectively) reduced but did not abolish binding, with the single-stranded region of U4 snRNA playing a more important role (Fig. 1B, lanes 14–17). Deletion of both U4 and U6 overhangs adjacent to stem I (U41–64/U655–112) or additionally the single-stranded 3′ end of U6 snRNA downstream from stem II (U41–64/U655–81) completely abolished complex formation (Fig. 1B, lanes 9–13). The former suggests that the single-stranded 3′ end of U6 snRNA downstream from stem II does not contribute to the interaction of the RH domain with U4/U6. No binding was observed with a 12-base-pair (bp) dsRNA containing a 31-nucleotide (nt) 3′ overhang, suggesting that the sequence and/or structure of the U4/U6 duplex is critical for binding of the Prp8 RH domain (Fig. 1, lanes 18,19). However, replacement of U4 nucleotides 65–90 with the inverted sequence (U4INV/U6) had no effect on the interaction of the Prp8 CTF or RH domain (Fig. 1B, lanes 20–22), indicating that the latter interacts with the single-stranded region of U4 adjacent to stem I in a sequence-independent manner.
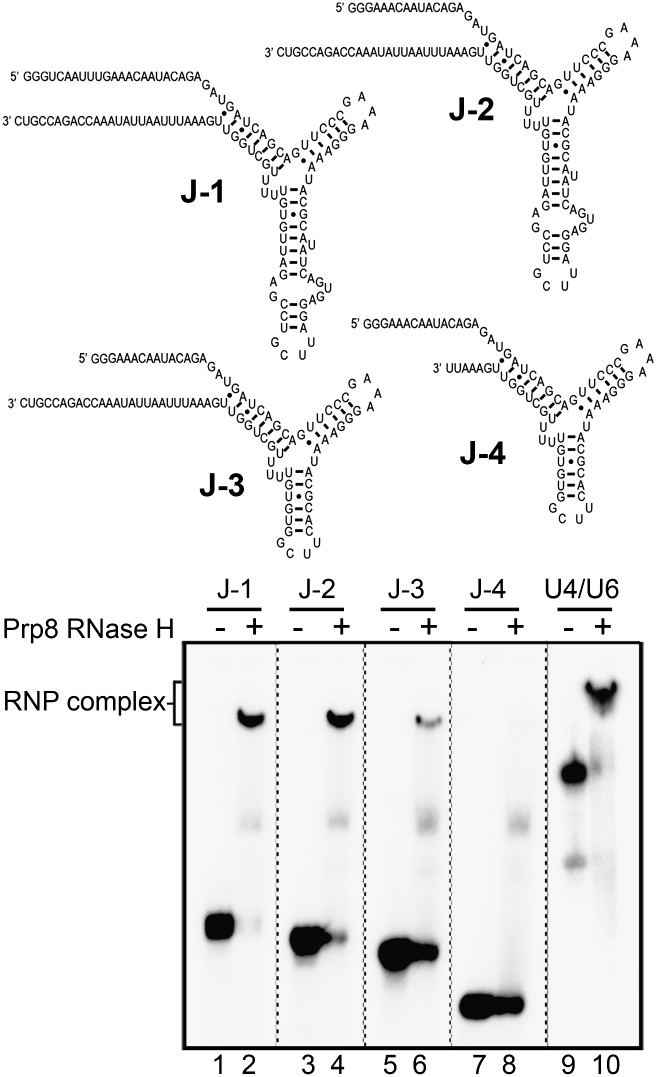
To assess the potential contribution of regions of the U4/U6 Y-shaped interaction domain, we assayed the binding of the RH domain to additional truncation mutants. Deletion of the 5′ SL of U6 and the 3′-terminal SL of U4 had little effect on complex formation, even if U4/U6 stem II was shortened by 11 bp and closed with a GAAA tetraloop (Fig. 2, cf. U4/U6 with the J-1 construct). Additional trimming of the 5′ single-stranded region of U6 (J-2) had only a minimal effect on binding (Fig. 2, lanes 3,4). Notably, a significant decrease in binding activity was observed when the U4 5′ SL was truncated and only the upper part of the U4 stem remained (construct J-3). This suggests that the lower region of the U4 5′ SL also contributes to the interaction of the Prp8 RH domain with U4/U6 snRNA. Additional trimming of the central domain of U4 snRNA (J-4) completely abolished complex formation, confirming the important contribution of this single-stranded region to RH domain binding.
Figure 2.
U4/U6 sequence and secondary structure requirements for Prp8 RH domain binding. (A) Sequence and predicted structure of the truncated U4/U6 constructs assayed for RH domain binding. (B) EMSAs were performed with the Prp8 RH domain (3 μM, +) and the indicated RNAs (1 nM) as in Figure 1.
The RH domain contacts the single-stranded regions of U4 and U6 snRNA adjacent to U4/U6 stem I
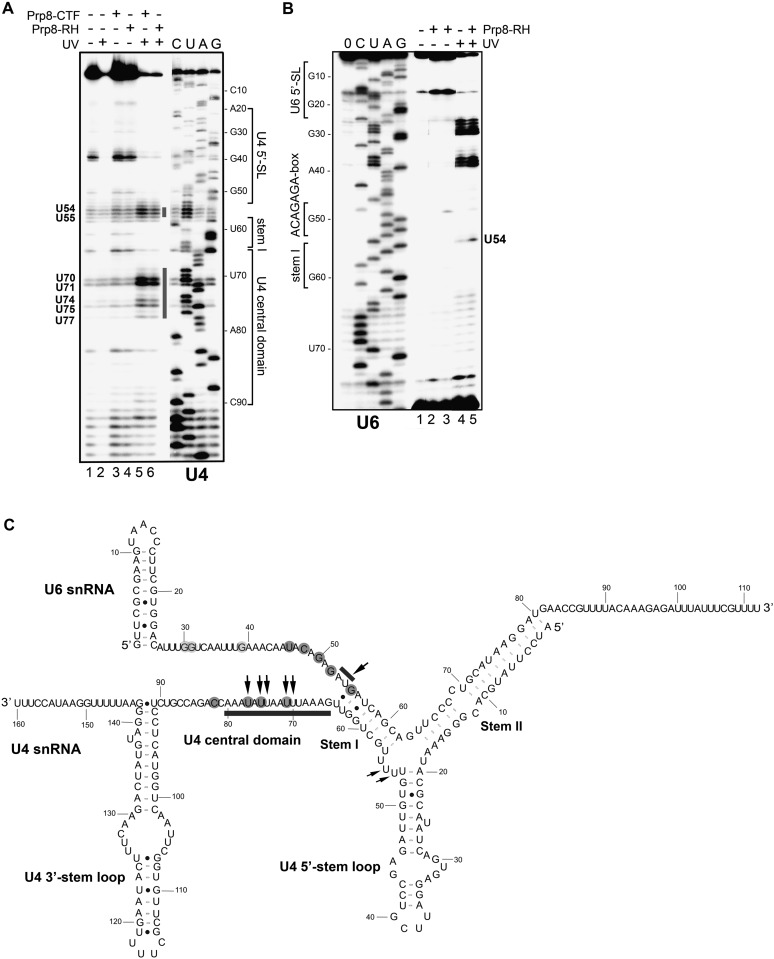
To learn more about the interaction sites of the Prp8 RH domain on the U4/U6 snRNA, we performed UV cross-linking with RNP complexes formed in vitro from wild-type U4/U6 snRNAs and the recombinant yeast Prp8 RH domain. After protease digestion, primer extension analysis was performed to identify those nucleotides cross-linked to the RH domain. As cross-linked nucleotides interfere with primer extension, strong reverse transcriptase stops induced by UV irradiation that are present in the RNP lanes but absent or less intense in the “RNA-only” lanes indicate the position of RNA–protein cross-links, with an actual interaction site situated 1 nt upstream of the stop site. Prp8 CTF and RH domain cross-link sites on the U4 snRNA were observed at nucleotides U70–U71, U74–U75, and U77 (Fig. 3A, cf. lanes 2 and 5,6); i.e., in the U4 central domain (Fig. 3C). Moreover, additional weak reverse transcriptase stops above background were observed at nucleotides U54/U55, which are in the U-rich internal U4 snRNA loop at the three-way junction. An RH domain cross-link on the U6 snRNA was observed at U54, just upstream of the last base pair of U4/U6 stem I (Fig. 3B,C).
Figure 3.
Identification of U4/U6 snRNA nucleotides cross-linked to the Prp8 RH domain. U4/U6 snRNA alone or incubated with protein (6–12 μM, +) (as indicated above each lane) was subjected to UV irradiation (or untreated) as indicated above (−/+ UV). RNA cross-linking sites were assayed by primer extension using 32P-labeled oligonucleotides complementary to U4 nucleotides 106–133 (A) or U6 nucleotides 77–102 (B) and were visualized by autoradiography. A sequencing ladder (C, U, A, and G on the left or right) was obtained by primer extension of in vitro transcribed U4 (A) or U6 (B) RNA in the presence of dideoxynucleotides. (Lane 0) Primer extension reaction in the absence of dideoxynucleotides. Those nucleotides that are cross-linked to the RH domain are labeled on the left or right. (C) Summary of U4/U6 cross-linked nucleotides and those protected from RNase T1/A digestion in the presence of the Prp8 RH domain. Nucleotides protected from RNase T1/A digestion are shaded gray, and light gray indicates less protection. Arrows indicate cross-linked nucleotides identified by primer extension, and bars indicate the likely positions of cross-linked nucleotides identified by MS.
We also carried out RNase T1 and/or RNase A enzymatic probing of U4/U6 snRNAs in the absence or presence of the RH domain, followed by primer extension of the cleaved products (Supplemental Fig. S2). In the absence of protein, major RNase T1 and/or RNase A cleavage sites were detected by reverse transcription stops in those regions of U4 and U6 preceding U4/U6 stem I but not within stem I, confirming that they are indeed single-stranded. Likewise, DMS (dimethylsulfate) and CMCT [1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate] chemical modification studies performed with the naked U4/U6 duplex also indicated that these regions of U4 and U6 are single-stranded (Supplemental Fig. S2). U6 nucleotides G50, G52, and G55, and to a lesser extent G30, G31, and G39, were partially protected from RNase T1 cleavage in the presence of the Prp8 RH domain, as evidenced by a reduction in the intensity of reverse transcription stops at these positions. In addition, a reduction in RNase A cleavage was observed at U46 and C48 (Supplemental Fig. S2A). U4 snRNA nucleotides protected from RNase A in the presence of the RH domain included U71, U75, U77, and C82 (Fig. 3C; Supplemental Fig. S2B). Taken together, these data reveal that the RH domain contacts single-stranded regions of both the U4 and U6 snRNA directly preceding stem I (Fig. 3B,C), consistent with our binding experiments showing that these regions of U4 and U6 are required for efficient interaction of the Prp8 RH domain with U4/U6 snRNA.
Identification of RH domain residues that contact the U4/U6 snRNA
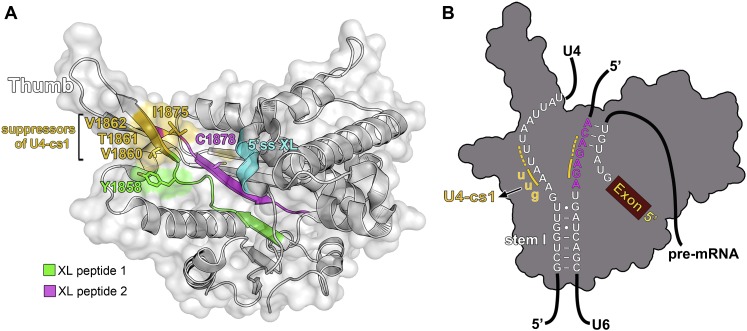
To determine which amino acids of the RH domain cross-link to the U4/U6 snRNA, we performed UV-induced protein–RNA cross-linking as described above and, after protease and nuclease digestion followed by titanium dioxide enrichment, we identified the cross-linked, RNA–peptide conjugates by mass spectrometry (MS). To aid the interpretation of which regions of U4 or U6 are cross-linked, we used the truncated U4/U6 snRNA construct J-1, which is still efficiently bound by the RH domain (Fig. 2). We identified and sequenced two cross-linked peptides that encompass residues L1850–R1859 (peptide 1) and A1874–K1885 (peptide 2) of the RH domain (Table 1). Inspection of the MS/MS fragment spectra of the peptide–RNA oligonucleotide conjugates indicates that Tyr1858 of peptide 1 and Cys1878 of peptide 2 (Fig. 4A) are the cross-linked amino acids (Supplemental Fig. S3). We monitored the effect of mutation of Y1858, which is located at the base of the thumb of the RH domain, or its neighboring amino acid, R1859, on yeast viability. For this purpose, we introduced plasmids encoding wild-type Prp8 or Prp8 containing Y1858A or R1859A mutations into a yeast strain harboring the wild-type PRP8 gene on a counter-selectable plasmid (Pena et al. 2008). The R1859A mutation had no significant effect on yeast growth, whereas mutation of Tyr1858 to alanine led to a temperature-sensitive phenotype with poor growth at 37°C (Supplemental Fig. S4). In addition, an accumulation of pre-U3 RNA was detected in the Y1858A strain (compared with the wild-type Prp8 strain) at an elevated temperature, as evidenced by primer extension analyses of total yeast RNA (Supplemental Fig. S4), demonstrating that the growth defect observed upon mutation of Y1858 arises from a defect in pre-mRNA splicing.
Table 1.
Identification of cross-linked U4/U6 RNA nucleotides and RH domain peptides by MS
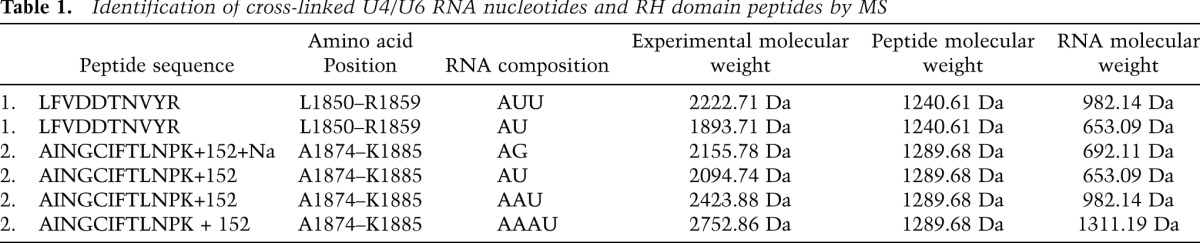
Figure 4.
Location of amino acids of the Prp8 RH domain that cross-link to U4 and/or U6 snRNA. (A) Ribbon diagram of the S. cerevisiae Prp8 RH domain (Protein Data Bank ID 3E9O) structure in which the residues of cross-linked peptide 1 (amino acids L1850–R1859) and peptide 2 (amino acids A1874–K1885) identified by MS are colored green and magenta, respectively, and the putative cross-linked amino acids Y1858 and C1878 are shown as sticks. The 310 helix (η5) corresponding to the region of human Prp8 that can be cross-linked to the 5′ SS is highlighted in cyan. The surface-exposed amino acids at the base of the β finger that, when mutated, suppress the cold-sensitive U4-cs1 mutation, are colored gold. V1860 and I1875 that reside adjacent to peptide 1 or within peptide 2, respectively, are shown as sticks (B) Model of the potential position of the U4/U6 stem I and the U6/5′ SS duplex shown on an outline of the RH domain. The invariant U6 ACAGAGA box is colored magenta. Mutations in the conserved AAA sequence of U4 that result in the U4-cs1 phenotype are indicated by gold lowercase letters, and the nucleotides that form additional base pairs between nucleotides of U4 and U6 are underlined in gold.
Peptide 1 is cross-linked to an oligonucleotide that contains one A and one or two U residues (Table 1; Supplemental Fig. S3A). Considering the major binding site of the RH domain described above and the position of cross-linked U4 and U6 nucleotides identified by primer extension, it is likely that this peptide is cross-linked to U4 snRNA in the region between nucleotides A69 and A76 (which contains multiple AU, UA, AUU, or UUAs) or potentially to A53–U54 on the U6 snRNA (Fig. 3C). Peptide 2 is cross-linked to an RNA oligonucleotide containing three As and one U, which is found in the J-1 U4/U6 construct at U4 nucleotides 77–80 or 66–69 and at U6 nucleotides 44–47 (Table 1; Supplemental Fig. S3B), and also to a GA dinucleotide (Supplemental Fig. S3C). However, since all of the cross-linked nucleotides found attached to peptide 2 (a combination of three As and one U as well as a GA dinucleotide) are found solely between nucleotide positions 65 and 69 on the U4 snRNA and as UV cross-links were detected by primer extension only at position U54 of U6 snRNA (which is not directly flanked by AAAU or corresponding nucleotide combinations), it is most likely that peptides 1 and 2 are both cross-linked within positions 65–80 of U4 snRNA (Fig. 3C). However, it is important to note that cysteine cross-links to RNA nucleotides do not necessarily cause reverse transcriptase stops, probably due to the labile nature of these cross-links during denaturing RNA sequencing analyses (Kuhn-Holsken et al. 2010; H. Urlaub, pers. comm.). Thus, we cannot entirely rule out that peptide 2 is cross-linked to the U6 snRNA.
The RH domain consists of an N-terminal subdomain (amino acids 1836–1992) with an RH fold and protruding β-hairpin (β1a and β1b) and a C-terminal α-helical domain (amino acids 1993–2092) (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008). In the 3D structure of the Prp8 RH domain, peptide 1 consists of amino acids found in the β1 and β1a strands and their connecting linker (Fig. 4; Supplemental S1A). Peptide 2 lies immediately next to this region and contains sequences of β2 and a small portion of β1b. The apparent cross-linked amino acids Y1858 (peptide 1) and C1878 (peptide 2) are both found at the base of the β-hairpin loop. In our model, which depicts the 3D structure as a “left-handed mitten” (Pena et al. 2008), this corresponds to the base of the thumb (Fig. 4A,B). In this model, the thumb and fingers frame a channel across the palm (comprised of the RH fold), and this channel has a width compatible with the accommodation of an RNA duplex. Thus, it is conceivable that in the U4/U6–RH RNP complex, U4/U6 stem I and the three-way junction are accommodated in the channel, while the single-stranded region of U4 snRNA is in contact with the thumb region (Fig. 4C). This model is supported by the surface accessibility of cross-linked amino acids in peptides 1 and 2 (Fig. 4A) and with the presence of patches of positive surface potential that carpet regions around the base of the thumb (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008). We note that certain Prp8-cat alleles that suppress the U4-cs1 phenotype (Kuhn and Brow 2000) also map close to cross-linked sites at the base of the thumb (Fig. 4A). For example, one allele has a V1860D mutation that is located directly adjacent to peptide 1, while another suppressor mutation, I1875T, is located in the β1b strand and is the second residue of the cross-linked peptide 2 (Fig. 4A). Both of these mutated sites are also surface-exposed below the thumb. Interestingly, EMSAs revealed a significantly lower binding affinity of the Prp8 CTF containing a V1860D mutation for U4/U6 as compared with the wild-type Prp8 CTF (Supplemental Fig. S5), consistent with this region of the RH domain playing a major role in U4/U6 snRNA binding.
Brr2 unwinds U4/U6 snRNAs by loading onto the single-stranded region of U4 adjacent to stem I
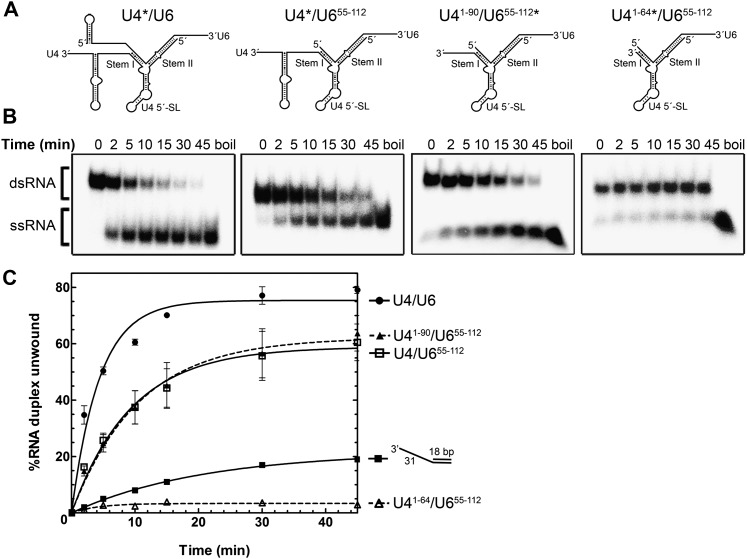
The interaction site of the RH domain (i.e., with the fork region preceding stem I of U4/U6 snRNA) could potentially overlap with that of Brr2. That is, as a member of the SF2 helicase family, Brr2 should unwind RNA duplexes in a 3′–5′ direction and use a 3′ single-stranded RNA as a loading strand (Bleichert and Baserga 2007). During spliceosome activation, Brr2 could thus either interact with the single-stranded region of U4 snRNA (i.e., the U4 central domain) adjacent to U4/U6 stem I and begin with unwinding of stem I or, alternatively, bind the single-stranded 3′ end of U6 and begin with unwinding of stem II. To distinguish between these possibilities, we used various U4/U6 snRNA truncation mutants and tested their ability to bind and/or be unwound by Brr2. In these studies, Brr2 was used at a 100-fold excess over RNA under multiple enzymatic turnover conditions. A comparison of the time course of Brr2-ATP-mediated unwinding of wild-type U4/U6 with that of U4/U655–112, in which the 5′ single-stranded overhang of U6 snRNA was deleted, revealed a twofold reduction in the rate of unwinding (Fig. 5). This is consistent with the idea that Brr2 requires a forked RNA structure for efficient unwinding, analogous to the DNA helicase Hel308, which more efficiently unwinds DNA forks as opposed to linear duplex DNA (Guy and Bolt 2005). An identical rate and extent of unwinding were observed when the 3′ SL of U4 snRNA was additionally removed (U41–90/U655–112) (Fig. 5). However, deletion of the remaining 3′ single-stranded stretch of U4 snRNA (i.e., yielding a blunt stem I [U41–64/U655–112]) abolished Brr2-catalyzed unwinding of the U4/U6 snRNAs despite the fact that this construct still contained the 3′ overhang of the U6 snRNA (Fig. 5C). This demonstrates that in vitro, Brr2 requires the central domain of U4 snRNA preceding stem I to initiate unwinding of U4/U6 snRNAs. Brr2 also unwound a generic RNA duplex (i.e., an 18-bp dsRNA containing a 31-nt 3′ overhang); however, the rate and extent of unwinding compared with U4/U6 was significantly reduced.
Figure 5.
Time course of unwinding of wild-type and truncated U4/U6 by Brr2 under multiple turnover conditions. (A) Structure of the wild-type U4/U6 snRNA and the truncation mutants thereof used in unwinding assays. (B) After preincubation of S. cerevisiae Brr2 (end concentration 100 nM) with the indicated RNAs (0.5 nM) (asterisks indicate the radiolabeled strand) for 5 min at 20°C, the reaction was started by adding 1 mM ATP/MgCl2 and then incubated for the indicated times at 20°C. U4/U6 unwinding was analyzed by native PAGE and visualized by autoradiography. (C) Data were fit to a single exponential equation: % duplex unwound = A[1 − exp(− ku t)], where A is the amplitude of the reaction, ku is the apparent first-order rate constant for unwinding, and t is time. The resulting kinetic parameters were as follows: A(U4/U6) = 76.0 ± 3.7; ku(U4/U6) = 0.24 ± 0.05 min−1; A(U4/U655–112) = 70.8 ± 2.5; ku(U4/U655–112) = 0.10 ± 0.01 min−1; A(U41-90/ U655–112) = 71.2 ± 2.0; and ku(U41-90/U655–112) = 0.10 ± 0.01 min−1.
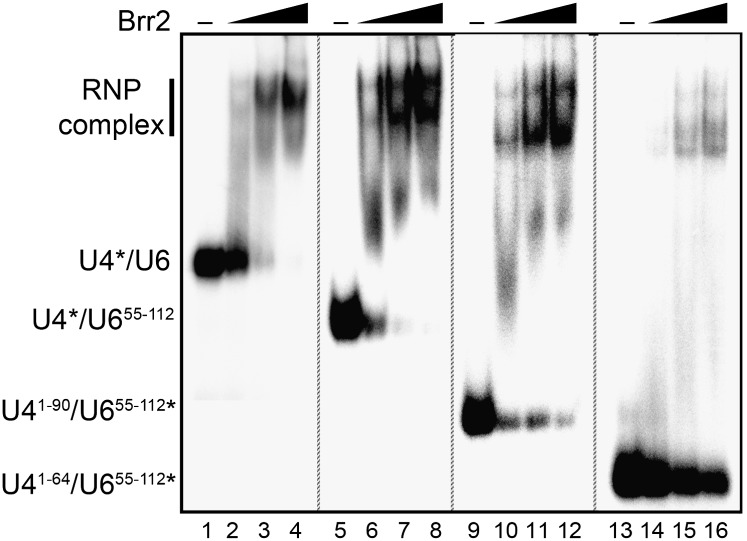
We next investigated by EMSA whether truncation of the U4/U6 snRNAs also alters Brr2 binding. We previously showed that Brr2 binds U4/U6 with high affinity and that, unlike other DExH/D-box proteins that require NTPs for RNA binding, Brr2 interaction with U4/U6 is not dependent on the presence of NTPs (Supplemental Fig. S6; Pena et al. 2009); in contrast, U4/U6 unwinding by yeast Brr2 was observed only in the presence of ATP and not with any other NTPs (Supplemental Fig. S6). As shown in Figure 6 (lanes 1–8), in the absence of ATP, Brr2 forms a complex with wild-type U4/U6 and U4/U655–112 to a similar extent (i.e., in both cases, with an apparent Kd < 50 nM), indicating that the 5′ region of U6 upstream of stem I is not important for Brr2 interaction. We note that the interaction of Brr2 with U4/U6 and the truncation mutants thereof leads to a double band, the basis of which is not clear but may reflect multiple conformations of the RNP complexes formed. Partial truncation of the 3′ end of U4 (U41–90/U655–112) had a minimal effect on binding (Fig. 6, lanes 9–12), whereas deletion of the entire single-stranded region of U4 directly before stem I (U41–64/U655–112) severely reduced Brr2–U4/U6 complex formation (Fig. 6, lanes 13–16). Thus, the latter single-stranded region of U4 is essential for efficient Brr2 loading onto U4/U6, and its absence leads to reduced U4/U6 duplex unwinding (see above).
Figure 6.
Interaction of Brr2 with wild-type and truncated versions of U4/U6 snRNA. 32P-labeled U4/U6 snRNA (final concentration 1 nM) was incubated alone (lanes 1,5,9,13) or in the presence of 20 nM (lanes 2,6,10,14), 50 nM (lanes 3,7,11,15), or 80 nM (lanes 4,8,12,16) Brr2 in the absence of ATP as described in the Materials and Methods. Brr2–RNA complex formation was then analyzed by EMSA on a 5% native polyacrylamide gel, and RNA/RNP complexes were visualized by autoradiography.
The Prp8 RH domain interferes with Brr2-mediated unwinding of U4/U6 snRNA in vitro
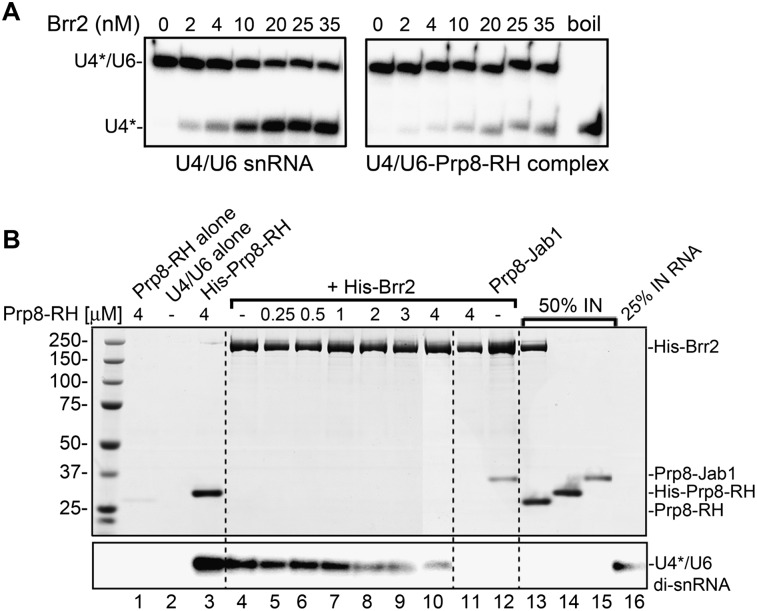
Our findings suggest that Brr2 and the RH domain of Prp8 have overlapping binding sites on the U4/U6 snRNA, which raises the possibility that the RH domain may inhibit U4/U6 unwinding by Brr2 by competing for its binding site on U4/U6. To test this, we preincubated U4/U6 snRNA with or without the Prp8 RH domain and then assayed for U4/U6 unwinding in the presence of Brr2. Compared with Brr2 alone, U4/U6 was clearly less efficiently unwound in the presence of the RH domain (Fig. 7A, cf. left and right panels). As previous studies demonstrated that the RH domain does not bind Brr2 (Weber et al. 2011), inhibition of unwinding most likely does not arise via direct interaction of the RH domain with Brr2, but rather by the interaction of the RH domain with the U4/U6 snRNA, which could either block Brr2 binding or prevent its translocation on the U4/U6 snRNA. Indeed, inhibition of Brr2-mediated unwinding was first observed at RH domain concentrations near or above its Kd for U4/U6 binding (i.e., at 0.5 μM) (Supplemental Fig. S7).
Figure 7.
U4/U6 snRNA unwinding by Brr2 is inhibited by the Prp8 RH domain. (A) U4/U6 duplex (1 nM) was preincubated alone (left panel) or with 3 μM Prp8 RH domain (right panel). Brr2 was added to 0–35 nM (as indicated above each lane), and the reaction was started by the addition of 1 mM ATP/MgCl2. U4/U6 unwinding was analyzed as described in the legend for Figure 5. (B) The RH domain inhibits the association of Brr2 with the U4/U6 snRNA. 32P-labeled U4/U6 snRNA (2 nM, U4-labeled) was preincubated alone (lanes 2,4), in the presence of His-tagged RH domain (as a control for interaction with U4/U6) (lane 3), or in the presence of 0.25–4 μM (as indicated above each lane) RH domain lacking a His tag (lanes 5–10). (Lanes 4–10) His-Brr2 (0.8 μM) was then added. Alternatively, Brr2 was incubated in the absence of U4/U6 with the RH domain (lane 11) or the Prp8 Jab1/MPN domain (lane 12), or the RH domain was incubated alone (lane 1). (Lanes 1–12) A pull-down was subsequently performed with nickel-agarose beads, and precipitated/coprecipitated proteins were analyzed by SDS-PAGE and visualized by staining with Coomassie (shown in the rtop panel). (Bottom panel) Coprecipitated U4/U6 was visualized by autoradiography. Lanes 13–16 show the following inputs (IN): 50% of His-Brr2 and the highest concentration of the RH domain (lane 13), 50% of His-tagged RH domain (lane 14), 50% of the Prp8 Jab 1 domain (lane 15), or 25% input of U4/U6 snRNA (lane 16).
The Prp8 RH domain blocks Brr2 interaction with U4/U6 snRNA in vitro
To provide direct evidence that the RH domain blocks Brr2 loading onto the U4/U6 RNA, we first incubated U4/U6 snRNA with increasing amounts of the RH domain (0.25–4 μM), and then added 0.8 μM His-tagged Brr2. Brr2 association with the U4/U6 snRNA was subsequently monitored by His pull-down assays (Fig. 7B). Efficient pull-down of the U4/U6 snRNA was observed with Brr2 alone (Fig. 7B, lane 4). However, at an RH domain concentration approximately twofold above its Kd for U4/U6 binding, there was a clear decrease in the amount of U4/U6 coprecipitated with Brr2 (Fig. 7B, lane 8). The RH domain did not coprecipitate with Brr2 alone even at a fivefold molar excess (Fig. 7B,top panel, lane 11), consistent with previous results demonstrating that these two proteins do not interact with each other (Weber et al. 2011). In contrast, the Jab1/MPN domain of the Prp8 CTF, which was previously shown by gel filtration experiments to interact with Brr2 (Weber et al. 2011), was precipitated together with Brr2 (Fig. 7B, top panel, lane 12). Furthermore, the RH domain was not coprecipitated together with Brr2 in the presence of U4/U6 snRNA (Fig. 7B, bottom panel, lanes 4–10), consistent with the idea that they have mutually exclusive RNA-binding sites. These data indicate that the Prp8 RH domain inhibits U4/U6 unwinding by Brr2 in vitro by blocking Brr2 loading onto the U4 snRNA.
Discussion
We showed that the RH domain of Prp8 and the RNA helicase Brr2 interact in vitro with the single-strand region of the U4 snRNA preceding U4/U6 stem I. Our data thus indicate that Brr2 translocates along the U4 snRNA, initially unwinding stem I of the U4/U6 di-snRNA. We further demonstrate that the Prp8 RH domain negatively regulates Brr2 unwinding of the U4/U6 snRNA in vitro by blocking Brr2's single-stranded loading site on the U4/U6 duplex. Interaction of Prp8 with the U4/U6 snRNA thus potentially plays a key role in preventing premature U4/U6 dissociation within the U4/U6.U5 tri-snRNP and also in the precatalytic spliceosomal B complex.
The isolated Prp8 RH domain interacts with the U4/U6 snRNA
We demonstrate that the RH domain of Prp8 interacts with the U4/U6 snRNAs in vitro with micromolar affinity and that the single-stranded regions of the U4 and U6 snRNAs adjacent to U4/U6 stem I are the major RH domain-binding sites (Figs. 1–3; Table 1). Previously, only weak RNA-binding activity (Kd of ∼20 μM) for an RNA construct mimicking a U2/U6–5′ SS four-helix junction was reported for the isolated RH domain (Ritchie et al. 2008). Additional structural elements of the U4/U6 di-snRNA also appear to contribute to complex formation, including the lower region of the U4 5′ SL (the binding site for Snu13) and, conceivably, also the three-way junction of the U4/U6 Y-shaped interaction region.
Previous cross-linking studies demonstrated that Prp8 contacts multiple RNAs within the U4/U6.U5 tri-snRNP and the spliceosome, including the nucleotides of the pre-mRNA involved in catalysis (i.e., the 5′ SS, 3′ SS, and BS), as well as the U5 and U6 snRNAs (for review, see Grainger and Beggs 2005). In reconstituted yeast tri-snRNPs, Prp8 was shown to cross-link to thiouridine-substituted U54 of the U6 snRNA, directly preceding U4/U6 stem I (Vidal et al. 1999). Consistent with this observation, we also detected a UV-induced U6–RH domain cross-link at this position (Fig. 3). To date, Prp8 cross-links to U4 snRNA have not been reported. Here we demonstrate that in a binary system, the RH domain of Prp8 contacts the single-stranded region of U4 adjacent to U4/U6 stem I. Structure probing of purified human and yeast tri-snRNP complexes indicated that this U4 single-stranded region is either inaccessible (in the case of humans) or somewhat less accessible (in the case of yeast), which is at least consistent with Prp8 interacting with this region of the U4 snRNA in the U4/U6.U5 tri-snRNP (Mougin et al. 2002). As mentioned above, the cold-sensitive U4 mutation (U4-cs1), which stabilizes the U4/U6 interaction that must be disrupted during activation of the spliceosome, is suppressed by several Prp8 mutations, including those mapping to its RH domain. These genetic interactions are consistent with physical interactions occurring between Prp8's RH domain and U4/U6 snRNA in vivo. Based on our data, it is tempting to surmise that the suppression of the U4-cs1 phenotype via mutations in the RH domain may be due to reduced affinity of the latter for the U4/U6 snRNA. This idea was supported by our finding that the RH domain of one of these mutants (V1860D) has a lower affinity for U4/U6 snRNA than the wild-type RH domain (Supplemental Fig. S5).
Identification of Prp8 residues contacting U4 and/or U6 snRNA
MS revealed cross-links between U4/U6 snRNA and two Prp8 peptides (amino acids 1850–1859 and 1874–1885) at the base of the β-hairpin loop of the RH domain, with Y1858 and C1878 the apparent cross-linked amino acids (Table 1; Fig. 4). Cross-linking studies with the complex formed between the RH domain and an RNA construct mimicking a U2/U6–5′ SS four-helix junction also implicated amino acids located in a similar region (i.e., spanning amino acids 1869–1914) to form RNA cross-links (Ritchie et al. 2008). Based on our cross-link information and the previously observed cross-link between the RH domain and the pre-mRNA's 5′ SS (Reyes et al. 1996, 1999), we modeled the U4/U6 stem I and the three-way junction in the channel formed by the β-hairpin loop and C-terminal α-helical domain, with the single-stranded region of U4 snRNA contacting the thumb region (Fig. 4B). As depicted in this new model, the U6/5′ SS duplex that is first formed after disruption of the U1/5′ SS could also be envisioned to lie near the amino acid that cross-links to the 5′ SS in the human B complex (Fig. 4; Reyes et al. 1996, 1999). The idea that the single-stranded regions of both U4 and U6 snRNA are close to the 5′ SS within the spliceosome, as suggested by our model, is supported by 4-thiouridine cross-linking studies performed with a yeast trans-splicing system; here, cross-links between the 5′ SS and U75 of U4 or C43 of U6 as well as the U6 ACAGAGA box were observed prior to the first catalytic step of splicing (Johnson and Abelson 2001).
The channel of the RH domain appears to interact only transiently with the U4/U6 duplex, presumably at a very early stage of splicing, such as within the tri-snRNP or until tri-snRNP addition to the spliceosome. Previously, the RH domain was proposed to provide an RNA-binding platform that is involved in the handover of the 5′ SS from U1 snRNA to U6 snRNA (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008). Our data provide additional support for this model. In particular, the interaction of the RH domain with U4 could help position the U6 snRNA for its interaction with the 5′ SS and thereby promote the exchange of U6 for U1 at the 5′ SS prior to activation. We note that 4-thiouridine cross-links between the pre-mRNA's 5′ SS and the Prp8 RH domain were not detected in yeast spliceosomes formed in vitro, although cross-links with other regions of Prp8 were found (Turner et al. 2006). However, both intermediates and products of the splicing reaction accumulated under the reaction conditions used in the aforementioned study, suggesting that mainly spliceosomes that had already undergone activation and splicing catalysis were likely present. The apparent absence of an RH domain–5′ SS cross-link at these later stages could indicate that this region of Prp8 is rearranged upon activation of the spliceosome.
Brr2 loads onto a region of the U4 snRNA preceding U4/U6 stem I
We demonstrate here that Brr2 interacts with high affinity (Kd of ∼30 nM) with the U4/U6 snRNA in vitro (Fig. 6), consistent with previous results from our laboratory (Pena et al. 2009), with the single-stranded central domain of U4 adjacent to stem I also playing an essential role in Brr2 binding. Consistent with the central domain serving as a binding site for both the RH domain and Brr2, this region of U4 was recently shown to play an essential role in yeast pre-mRNA splicing (Hayduk et al. 2012). While we note that the RH domain has a lower affinity for U4/U6 than Brr2, we expect the local concentration of each protein within the tri-snRNP or spliceosome to exceed their respective U4/U6 dissociation constants. Furthermore, our studies examined an isolated fragment of the Prp8 protein, and it is conceivable that in the context of full-length Prp8 and also in the presence of other proteins, its affinity for U4/U6 is enhanced. During tri-snRNP formation, the interaction of the RH domain with the U4 and U6 single-stranded regions preceding stem I must be favored to prevent Brr2 interaction. Thus, additional mechanisms may be at work to reduce Brr2 affinity for U4/U6 within the tri-snRNP and/or the B complex.
Our data indicate that first stem I of the U4/U6 Y-shaped interacting region is unwound by Brr2. The results of cross-linking and sequencing (CRAC) studies mapping Brr2–RNA contacts in yeast cells (Hahn et al. 2012) likewise indicate that Brr2 translocates on U4 and first unwinds stem I. This would allow U6 to theoretically form U2/U6 helix 1a (which involves nucleotides of U6 that initially base-pair with U4 in U4/U6 stem I) even before stem II of the U4/U6 duplex has been unwound. These results are consistent with previous studies showing that in the U12-dependent spliceosome in which a homologous RNA/RNA interaction network is formed, stem I of the U4atac/U6atac snRNAs (which interact in a manner analogous to U4/U6) is also unwound prior to stem II during spliceosome activation (Frilander and Steitz 2001). It is not clear why Brr2 does not load onto the single-stranded 3′ end of U6 in the absence of other proteins; a simple explanation would be that the high percentage of uridines or potential internal base-pairing at the 3′ end of U6 may be unfavorable for Brr2 interaction. Within the U4/U6.U5 tri-snRNP, this region of the U6 snRNA is bound by the Lsm 2–8 proteins (Achsel et al. 1999; Mayes et al. 1999), which could also prevent Brr2 interaction and unwinding of U4/U6 stem II prior to stem I.
A novel mechanism whereby Prp8 potentially regulates Brr2 unwinding activity
Brr2 and Prp8 are stable components of the U5 snRNP, and upon U5 interaction with U4/U6 snRNP to form the U4/U6.U5 tri-snRNP, Brr2 must be prevented from unwinding the U4/U6 di-snRNA prematurely. We demonstrate here that the isolated Prp8 RH domain inhibits Brr2-mediated unwinding of U4/U6 snRNA in vitro (Fig. 7A; Supplemental Fig. S7) by hindering Brr2 loading onto U4 (Fig. 7B). Although Brr2 interaction was not completely inhibited under the tested conditions, the residual interaction of Brr2 with U4/U6 may occur in part via Brr2 contacts with other regions of U4/U6 not bound by the RH domain. As Brr2 pull-down experiments failed to detect complexes containing Brr2–U4/U6 and the RH domain, our data suggest that Brr2 and the RH domain cannot stably interact simultaneously with the U4/U6 duplex, indicating that the RH domain does not prevent Brr2 translocation after it loads onto the central domain of U4 preceding U4/U6 stem I. This also suggests that the RH domain does not additionally decrease the presumed processivity of the Brr2-mediated U4/U6 unwinding reaction by stabilizing an unfavorable conformation of the RNA substrate (i.e., stabilizing duplex formation), as this would also involve simultaneous interactions of Brr2 and the RH domain with U4/U6. It should be noted, however, that the in vitro approach used here limits the possible outcomes and only describes the properties of Brr2 and a fragment of Prp8 in isolation; within the context of the spliceosome or tri-snRNP, other proteins or even other domains of Prp8 may also contribute to the regulation of Brr2.
Our results are in line with previous genetic studies in yeast that suggested that Prp8 acts as a negative regulator of Brr2 helicase activity in vivo (Kuhn and Brow 2000; Kuhn et al. 2002). However, previous in vitro studies indicated that the Prp8 CTF, which is comprised of both an RH and a Jab1/MPN domain, stimulates Brr2 unwinding of U4/U6 but inhibits Brr2's RNA-dependent ATPase activity (Maeder et al. 2009). The mechanism whereby the CTF modulates Brr2 activity is unclear. At a first glance, our results potentially would explain the reduction in RNA-dependent ATPase activity of Brr2 in the presence of the Prp8 CTF; namely, by interfering with Brr2–U4/U6 binding. However, the previously reported inhibitory and stimulatory effects of the CTF were observed under conditions where only little interaction with U4/U6 would be expected (namely, at 0.25 μM, which is well below the CTF's and RH domain's U4/U6 dissociation constants [∼1–2 μM]); thus, the previously observed effects of the CTF are likely mediated by direct interaction with Brr2 via the Jab1/MPN domain.
Prior to activation, the interaction between the RH domain and, minimally, the single-stranded region of U4 must be disrupted to allow efficient Brr2 loading and subsequent U4/U6 unwinding. The trigger for this proposed remodeling event is unknown, but potentially could involve post-translational modification of Prp8. Interestingly, the Jab1/MPN domain, which is located adjacent to the RH domain in the CTF of Prp8, can bind ubiquitin (Bellare et al. 2006). Significantly, ubiquitination of Prp8 appears to down-regulate Brr2 activity within tri-snRNPs (Bellare et al. 2008); when ubiquitination is prevented, U4/U6 unwinding is accelerated, leading to tri-snRNP dissociation. It is thus tempting to speculate that deubiquitination of Prp8 within the spliceosome might help to trigger the release of Prp8 from the U4/U6 snRNA, allowing Brr2 interaction and subsequent U4/U6 unwinding. It will thus be interesting to investigate in the future whether ubiquitination/deubiquitination affects the interaction of the Prp8 CTF with U4/U6. It is also conceivable that the action of the GTPase Snu114, which also has been shown to regulate Brr2 activity, might trigger release of the RH domain and thereby allow Brr2 interaction with the U4/U6 snRNA. Taken together, our data reveal new insights into the mechanism of Brr2-mediated U4/U6 unwinding. They also suggest multiple roles for the Prp8 RH domain during splicing, including acting as an RNA-binding platform that promotes RNA–RNA base-pairing rearrangements and also contributing to the regulation of Brr2 unwinding activity prior to the catalytic activation of the spliceosome.
Materials and methods
Protein expression and purification
The S. cerevisiae Prp8 CTF (residues 1836–2397), Prp8 CTFV1860D, RH domain (residues 1836–2092), or Jab1/MPN domain (residues 2012–2413) were expressed in Escherichia coli and further purified as described previously (Pena et al. 2009; Weber et al. 2011). Yeast Brr2 containing an N-terminal His6 tag was purified as described in the Supplemental Material.
RNA preparation and EMSA
RNAs were transcribed in vitro or, in the case of the short 12- and 18-bp duplexes, purchased from IBA GmbH, and the duplexes were generated as described previously (Santos et al. 2012). EMSAs were performed by combining the CTF of yeast Prp8 (residues 1836–2397; Prp8 CTF), its RH domain (residues 1836–2092), or Brr2 at the indicated concentrations with 0.5–1 nM in vitro transcribed, 32P-labeled wild-type or truncated U4/U6 or combining a linear 12-bp RNA duplex with a 31-nt 3′ overhang in binding buffer (40 mM Tris-HCl at pH 7.5, 50 mM NaCl, 8% [w/v] glycerol, 2.5 mM MgCl2, 1.5 mM DTT, 100 ng/μL acetylated BSA), where indicated, in the presence of 50 μg/mL E. coli tRNA. Samples were incubated for 25–30 min at 20°C, and RNP complex formation was then analyzed by 6% (Prp8) or 5% (Brr2) native PAGE.
Analysis of protein–RNA interactions via pull-down
Pull-down experiments for analysis of protein–RNA interactions were performed with 2 nM 32P-labeled wild-type U4/U6 preincubated with the indicated concentrations of the RH domain (without a His tag) in buffer containing 40 mM Tris-HCl (pH 7.5), 50 mM NaCl, 8% (w/v) glycerol, 0.5 mM MgCl2, 1.5 mM DTT, and 600 ng/μL acetylated BSA for 15 min at 20°C. His6-tagged Brr2 was added to a final concentration of 0.8 μM, followed by additional incubation for 15 min at 20°C. Brr2–RNA complexes were captured by incubation with Ni-NTA agarose beads (Qiagen) for 40–50 min at 4°C; washed with buffer containing 40 mM Tris-HCl (pH 7.5), 50 mM NaCl, 8% (w/v) glycerol, and 15 mM imidazole; and then analyzed by SDS-PAGE. After pull-down, U4/U6 was visualized by autoradiography.
RNA–protein cross-linking and primer extension mapping of RNA cross-links
Wild-type U4/U6 (0.58 pmol) purified by native PAGE was incubated in the presence or absence of a saturating amount of the Prp8 CTF or Prp8 RH domain for 20 min at 20°C. The RNA/RNP complexes were UV-irradiated at 254 nm for 45–90 sec on ice. Irradiated and nonirradiated samples were ethanol-precipitated and digested with Proteinase K (0.5–1 mg/mL) for 20 min at 40°C. After PCI extraction and ethanol precipitation, the pellet was dried and dissolved in water. Primer extension analysis was performed using oligodeoxynucleotide primers complementary to nucleotides 77–102 of U6 and 106–133 of U4 as described (Rasche et al. 2012).
Identification of UV-induced cross-linking sites by MS
Two nanomoles of the Prp8 RH domain and 4 nmoles of U4/U6 J-1 RNA were incubated in 100 μL of buffer (40 mM Tris-HCl at pH 7.5, 100 mM NaCl, 2% glycerol [w/v], 2.5 mM MgCl2, 1.5 mM DTT) at 20°C for 30 min (to allow complex formation), and UV cross-linking was then performed at 254 nm for 10 min. After digestion with trypsin and RNases A and T1, the cross-linked sites were identified by Nano-LC-ESI-MS as previously described (Kramer et al. 2011; Schmitzová et al. 2012).
U4/U6 snRNA unwinding assays
Helicase assays were performed at 20°C as previously described (Santos et al. 2012). For U4/U6 unwinding assays in the presence of the Prp8 RH domain, RH–U4/U6 RNP complexes were first formed by preincubation for 20 min at 20°C, followed by an additional 5 min of incubation in the presence of Brr2.
Acknowledgments
We thank Klaus Hartmuth for helpful discussions concerning RNA structure probing, Reinhard Rauhut and Berthold Kastner for critically reading the manuscript, and Gabi Heyne for help in the analysis of in vivo splicing. This work was supported by the Deutsche Forschungsgemeinschaft (grants SFB 860 [to R.L. and H.U.] and SFB 740/2 [to M.C.W.]). S.M-J. has been a doctoral student of the PhD program “Molecular Biology” at the International Max Planck Research School and the Göttingen Graduate School for Neurosciences and Molecular Biosciences (GGNB) (DFG grant GSC 226/1) at the Georg August University Göttingen.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.200949.112.
References
- Achsel T, Ahrens K, Brahms H, Teigelkamp S, Lührmann R 1998. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol Cell Biol 18: 6756–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J 18: 5789–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ 2006. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA 12: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ 2008. A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol 15: 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Bleichert F, Baserga SJ 2007. The long unwinding road of RNA helicases. Mol Cell 27: 339–352 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608 [DOI] [PubMed] [Google Scholar]
- Frilander MJ, Steitz JA 2001. Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol Cell 7: 217–226 [DOI] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD 2005. Prp8 protein: At the heart of the spliceosome. RNA 11: 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CP, Bolt EL 2005. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res 33: 3678–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D, Kudla G, Tollervey D, Beggs JD 2012. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev (this issue). doi: 10.1101/gad.199307.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayduk AJ, Stark MR, Rader SD 2012. In vitro reconstitution of yeast splicing with U4 snRNA reveals multiple roles for the 3′ stem–loop. RNA 18: 1075–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TL, Abelson J 2001. Characterization of U4 and U6 interactions with the 5′ splice site using a S. cerevisiae in vitro trans-splicing system. Genes Dev 15: 1957–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer K, Hummel P, Hsiao H, Luo X, Wahl M, Urlaub H 2011. Mass-spectrometric analysis of proteins cross-linked to 4-thio-uracil- and 5-bromo-uracil-substituted RNA. Int J Mass Spectrom 304: 184–194 [Google Scholar]
- Kuhn AN, Brow DA 2000. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics 155: 1667–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AN, Li Z, Brow DA 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol Cell 3: 65–75 [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Reichl EM, Brow DA 2002. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc Natl Acad Sci 99: 9145–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn-Holsken E, Lenz C, Dickmanns A, Hsiao HH, Richter FM, Kastner B, Ficner R, Urlaub H 2010. Mapping the binding site of snurportin 1 on native U1 snRNP by cross-linking and mass spectrometry. Nucleic Acids Res 38: 5581–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Lührmann R 1998. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci 95: 4188–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Rauhut R, Vornlocher HP, Luhrmann R 2006. The network of protein–protein interactions within the human U4/U6.U5 tri-snRNP. RNA 12: 1418–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Maeder C, Kutach AK, Guthrie C 2009. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol 16: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes AE, Verdone L, Legrain P, Beggs JD 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J 18: 4321–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougin A, Gottschalk A, Fabrizio P, Lührmann R, Branlant C 2002. Direct probing of RNA structure and RNA-protein interactions in purified HeLa cell's and yeast spliceosomal U4/U6.U5 tri-snRNP particles. J Mol Biol 317: 631–649 [DOI] [PubMed] [Google Scholar]
- Pena V, Liu S, Bujnicki JM, Luhrmann R, Wahl MC 2007. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell 25: 615–624 [DOI] [PubMed] [Google Scholar]
- Pena V, Rozov A, Fabrizio P, Lührmann R, Wahl MC 2008. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 27: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena V, Jovin SM, Fabrizio P, Orlowski J, Bujnicki JM, Lührmann R, Wahl MC 2009. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell 35: 454–466 [DOI] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8: 847–855 [DOI] [PubMed] [Google Scholar]
- Rasche N, Dybkov O, Schmitzová J, Akyildiz B, Fabrizio P, Lührmann R 2012. Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre. EMBO J 31: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Kois P, Konforti BB, Konarska MM 1996. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA 2: 213–225 [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Gustafson EH, Luo HR, Moore MJ, Konarska MM 1999. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA 5: 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM 2008. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol 15: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Santos K, Jovin SM, Weber G, Pena V, Lührmann R, Wahl MC 2012. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci doi: 10.1073/pnas.1208098109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzová J, Rasche N, Dybkov O, Kramer K, Fabrizio P, Urlaub H, Lührmann R, Pena V 2012. Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein. EMBO J 31: 2222–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 23: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Turner IA, Norman CM, Churcher MJ, Newman AJ 2006. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA 12: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Beggs JD 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157: 1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal VP, Verdone L, Mayes AE, Beggs JD 1999. Characterization of U6 snRNA-protein interactions. RNA 5: 1470–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T, Pleiss JA, Guthrie C 2002. Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell 109: 149–152 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Weber G, Cristao VF, de L Aves F, Santos KF, Holton N, Rappsilber J, Beggs JD, Wahl MC 2011. Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev 25: 1601–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhang L, Xu T, Heroux A, Zhao R 2008. Crystal structure of the β-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci 105: 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Shen J, Guarnieri MT, Heroux A, Yang K, Zhao R 2007. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci 16: 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R 2009. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol 16: 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]