Kitada et al. look at the mechanism of variegated gene expression at telomeres. They characterize the histone marks associated with transcriptionally active and inactive telomeres and identify that H3K79me disrupts transcriptional silencing at active telomeres. Interestingly, maintenance of H3K79me depends on a transcription-mediated positive feedback loop. The study finds that epigenetic variegation is not due to the amount of Sir protein binding to nucleosomes, but rather the enrichment/depletion of H3K79me.
Keywords: epigenetics, position effect variegation, silencing, telomeres, histones, Sir complex
Abstract
Yeast contains heterochromatin at telomeres and the silent mating-type loci (HML/HMR). Genes positioned within the telomeric heterochromatin of Saccharomyces cerevisiae switch stochastically between epigenetically bistable ON and OFF expression states. Important aspects of the mechanism of variegated gene expression, including the chromatin structure of the natural ON state and the mechanism by which it is maintained, are unknown. To address this issue, we developed approaches to select cells in the ON and OFF states. We found by chromatin immunoprecipitation (ChIP) that natural ON telomeres are associated with Rap1 binding and, surprisingly, also contain known characteristics of OFF telomeres, including significant amounts of Sir3 and H4K16 deacetylated nucleosomes. Moreover, we found that H3K79 methylation (H3K79me), H3K4me, and H3K36me, which are depleted from OFF telomeres, are enriched at ON telomeres. We demonstrate in vitro that H3K79me, but not H3K4me or H3K36me, disrupts transcriptional silencing. Importantly, H3K79me does not significantly reduce Sir complex binding in vivo or in vitro. Finally, we show that maintenance of H3K79me at ON telomeres is dependent on transcription. Therefore, although Sir proteins are required for silencing, we propose that epigenetic variegation of telomeric gene expression is due to the bistable enrichment/depletion of H3K79me and not the fluctuation in the amount of Sir protein binding to nucleosomes.
Epigenetics is traditionally defined as “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence” (Riggs et al. 1996). Position effect variegation (PEV), discovered in the fruit fly Drosophila melanogaster, is a classic example of an epigenetic phenomenon (Girton and Johansen 2008). PEV is characterized by the reversible and stochastic switching of a gene positioned within heterochromatin between ON and OFF states. Telomere position effect (TPE), at the heterochromatin of telomeres in budding yeast, is a form of PEV (Supplemental Fig. S1; Gottschling et al. 1990; Mondoux and Zakian 2006). TPE involves the variegated expression of genes positioned near telomeres at the boundary of heterochromatin and euchromatin. Although TPE in yeast was discovered more than two decades ago (Gottschling et al. 1990), how the variegated gene expression pattern arises at telomeres is still poorly understood (Ptashne 2002; Mondoux and Zakian 2006; Madhani 2007).
The formation of telomeric and silent mating-type locus heterochromatin has been well characterized, and current data are consistent with a model in which yeast heterochromatin proteins assemble and spread along histones in a stepwise manner (Hecht et al. 1996; Rusche et al. 2003; Mondoux and Zakian 2006). In this process, Rap1 bound at the telomeric TG1–3 repeats (Buchman et al. 1988; Klein et al. 1992) recruits Sir4 through direct protein–protein interaction (Moretti et al. 1994; Hoppe et al. 2002; Luo et al. 2002). Sir4 in turn recruits Sir2 (Moazed et al. 1997; Strahl-Bolsinger et al. 1997), an NAD-dependent histone deacetylase (HDAC) with specificity for histone H4K16 acetylation (H4K16ac) (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000). Deacetylation of H4K16ac generates a high-affinity binding site for the Sir3 protein (Johnson et al. 1990; Liou et al. 2005), which in turn recruits more Sir4 and Sir2 (Hecht et al. 1996; Hoppe et al. 2002; Luo et al. 2002). Cycles of H4K16 deacetylation and Sir3 recruitment enable spreading of the Sir complex along telomeric heterochromatin. The spreading of the Sir complex is eventually blocked by H4K16ac in adjacent euchromatin by the histone acetyltransferase Sas2 (Kimura et al. 2002; Suka et al. 2002). Sas2-mediated acetylation of H4K16 is also thought to enhance the incorporation of the histone H2A variant Htz1/H2AZ (Shia et al. 2006), which may act as an additional barrier to Sir complex spreading (Meneghini et al. 2003).
Similarly, H3K4 methylation (H3K4me), H3K36me, and H3K79me have also been proposed to contribute to the boundary between heterochromatin and euchromatin, but the exact role that each modification plays in this process is less well defined (Verzijlbergen et al. 2009). It has been suggested, using histone point mutant and methyltransferase deletion strains, that the presence of H3K4me or H3K36me prevents ectopic binding of Sir proteins in euchromatin (Santos-Rosa et al. 2004; Tompa and Madhani 2007). More critically, the overexpression of the H3K79 methyltransferase Dot1 has been shown to disrupt gene silencing in vivo, and it has been proposed that H3K79me may block Sir complex binding to antagonize subtelomeric silencing in vivo (Singer et al. 1998; van Leeuwen et al. 2002; Ng et al. 2003; Katan-Khaykovich and Struhl 2005; Altaf et al. 2007; Fingerman et al. 2007; Onishi et al. 2007). Genetic, biochemical, and structural studies have shown that unmethylated H3K79 is a contact site for Sir3 and that methylation of H3K79 can disrupt that interaction between the H3K79 region and Sir3 in vitro (Ng et al. 2002; Altaf et al. 2007; Fingerman et al. 2007; Johnson et al. 2009; Martino et al. 2009; Armache et al. 2011; Ehrentraut et al. 2011). Moreover, removal of H3K79me has been shown to facilitate de novo establishment of silencing at the silent mating-type locus HML (Osborne et al. 2009). Although it has been reported that H3K79 methylation by Dot1 does not play a role in natural silencing at HML or at most subtelomeres (Takahashi et al. 2011), the study asked whether the genome-wide depletion of H3K79me would derepress heterochromatin silencing instead of directly addressing the function of H3K79me at heterochromatin per se.
The precise mechanism by which heterochromatin prevents the transcription of a gene is not known. However, it has been proposed that the Sir complex can prevent gene activation by either blocking the assembly of the preinitiation complex (PIC; general transcription factors and RNA polymerase II [RNAPII]) or regulating the transition between transcription initiation and RNAPII elongation (Sekinger and Gross 2001; Chen and Widom 2005; Gao and Gross 2008). Additionally, it has been shown that the abnormal lengthening of telomeres can increase the strength of gene silencing (Kyrion et al. 1993; Li and Lustig 1996; Mishra and Shore 1999; Park and Lustig 2000).
In contrast to the formation of the OFF state of telomeric heterochromatin, the chromatin structure of the natural ON state has not been well characterized. Potentially, the natural ON state could result from the absence of Rap1 binding to telomeric repeats or loss of interaction between the Sir complex and nucleosomes due to H4K16ac or H3K79me (Ng et al. 2003; Moazed 2011). However, this is not necessarily the case, as it has been shown that a telomeric gene can be derepressed in the presence of Sir complex binding in an H4K16R Sir2-345 catalytic mutant strain (Yang et al. 2008), an H3K56 mutant strain (Xu et al. 2007), an H3Δ4-30 tail deletion mutant strain (Sperling and Grunstein 2009), and a strain with a Gal4-Sir1 fusion protein artificially recruited to a synthetic HMR silent mating-type locus prior to the establishment of silencing (Kirchmaier and Rine 2006).
Therefore, to decipher the basis of epigenetic variegation, we sought to identify the molecular factors that determine the natural ON state of budding yeast TPE. To accomplish this, we first developed a method for isolating populations of cells with telomeres in the ON and OFF states. This approach is conceptually different from most previous studies in which mixed populations of cells with ON and OFF telomeres were compared with heterochromatin mutant strains with telomeres that are artificially ON (Rusche et al. 2003). We then assessed the structural differences in chromatin at the ON and OFF telomeres in vivo. Additionally, by in vitro reconstitution of heterochromatin, we asked whether any of the differences observed in vivo were sufficient to disrupt gene silencing using yeast nuclear extracts. Surprisingly, we found that Rap1 binding, Sir complex binding to nucleosomes, and H4K16 deacetylation were largely similar between the ON and OFF states in vivo. Instead, we demonstrate that H3K79me enables the disruption of gene silencing and inheritance of the natural ON state of the telomere by a transcription-mediated positive feedback loop despite the spreading of the Sir complex along nucleosomes. We conclude that H3K79me and not the difference in the amount of Sir complex binding to nucleosomes per se is the epigenetic basis for variegation at telomeres.
Results
Isolation of ON and OFF cells by medium selection
To determine the differences between the ON and OFF chromatin states, it was necessary to separate ON and OFF cells in bulk. To accomplish this, we employed a yeast strain harboring a URA3 reporter gene at a telomere at the left arm of chromosome VII (TEL07L). We isolated ON and OFF cells, respectively, by culturing the strain in medium lacking uracil (SD−ura) or medium containing the drug 5-FOA (SD+FOA), which is toxic to cells with Ura3 activity (Fig. 1A; Boeke et al. 1987). For comparison, YFR057W, a native gene located near a different telomere (native TEL06R), was monitored as a control.
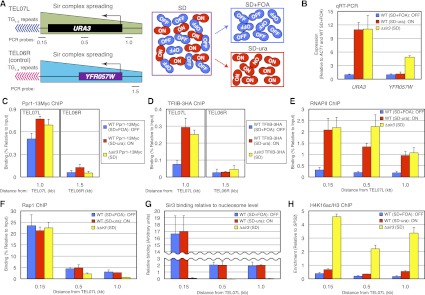
Figure 1.
Rap1 binding to DNA and Sir protein binding to nucleosomes are not different between ON and OFF telomeres. (A) Schematic of the medium selection approach to isolate ON and OFF telomeres. Probes were ∼0.15, 0.5, and 1.0 kb away from the telomeric repeats of URA3-TEL07L and ∼1.5 kb from native TEL06R. (B) qRT–PCR of URA3 at TEL07L and YFR057W at native TEL06R in wild-type (WT) SIR3 cells grown in SD+FOA (blue bars) and SD−ura (red bars) and Δsir3 cells grown is SD (yellow bars). Data are presented as mean ± standard deviation (SD). (C,D) ChIP of Ppr1-13Myc (C) and TFIIB-3HA (D) depicted as in B except Ppr1 and TFIIB were tagged with 13Myc and 3HA, respectively. (E–H) ChIP of RNAPII (E), Rap1 (F), Sir3 binding relative to nucleosome level (G), and H4K16ac/H3 (H) at URA3-TEL07L depicted as in B.
A recent study had shown that the URA3-FOA assay may identify false positive hits when used in screens for detecting silencing mutants, making it necessary to confirm the expression of URA3 using quantitative RT–PCR (qRT–PCR) (Rossmann et al. 2011). As shown in Figure 1B, the mRNA level of URA3 was low in cells cultured in SD+FOA and high in SD−ura when measured by qRT–PCR. In fact, the URA3 expression level of cells grown in SD−ura was comparable with that of a Δsir3 control strain in which heterochromatin is completely disrupted (Fig. 1B; Strahl-Bolsinger et al. 1997). Therefore, by the direct measurement of URA3 mRNA using qRT–PCR, we found that our medium-based selection approach is capable of separating ON and OFF cells in bulk.
TPE is regulated at the RNAPII PIC assembly step
Previous studies had reported, in a contradictory manner, that heterochromatin prevents transcription by blocking either PIC assembly (Chen and Widom 2005) or the transition between initiation of transcription and RNAPII elongation (Sekinger and Gross 2001; Gao and Gross 2008). Therefore, we wished to clarify which step of the transcription process differed in our wild-type ON and OFF cells separated by medium selection. To accomplish this, we measured the binding of the URA3 activator Ppr1 (Myc-tagged), general transcription factor TFIIB (HA-tagged), and RNAPII at URA3-TEL07L by chromatin immunoprecipitation (ChIP) in ON cells, OFF cells, and Δsir3 cells as a control. RNAPII and TFIIB are known to characterize PICs during gene activation (Hahn 2004; Kostrewa et al. 2009). As shown in Figure 1C, Ppr1 was enriched at the promoter of URA3 at a similar level in the ON and OFF states. In contrast, binding of TFIIB and RNAPII was observed at the ON but not OFF telomere (Fig. 1D,E). Thus, based on these results from our medium-selected ON and OFF cells, heterochromatin is permissive to activator binding but not PIC assembly. We conclude that the epigenetic variegation states of TPE are modulated at the PIC assembly step.
Histone methylation but not binding of heterochromatin proteins differentiates the ON and OFF telomeres
Differences in any of the steps of the heterochromatin assembly process could potentially explain how bistable ON and OFF chromatin states could exist at telomeres in wild-type yeast strains. To determine whether TPE can be explained by differences in the binding of key heterochromatin proteins, we measured the enrichment level of Rap1 and Sir3 at URA3-TEL07L in the medium-selected ON and OFF cells by ChIP. As shown in Figure 1F, binding of Rap1 to the ON and OFF telomeres was nearly identical. Similarly, and in contrast to previous models (Ng et al. 2003; Moazed 2011), we also observed that the level of Sir3 binding to nucleosomes along the subtelomeric region in the ON and OFF cells was essentially the same (Fig. 1G). Importantly, our measurements took into account the fact that the number of nucleosomes was expectedly reduced at ON telomeres compared with those that were OFF (Supplemental Fig. S2; Pokholok et al. 2005). Nevertheless, our data support the idea that epigenetic variegation at telomeres cannot simply be explained by Rap1 binding or the extent of Sir3 binding to nucleosomes.
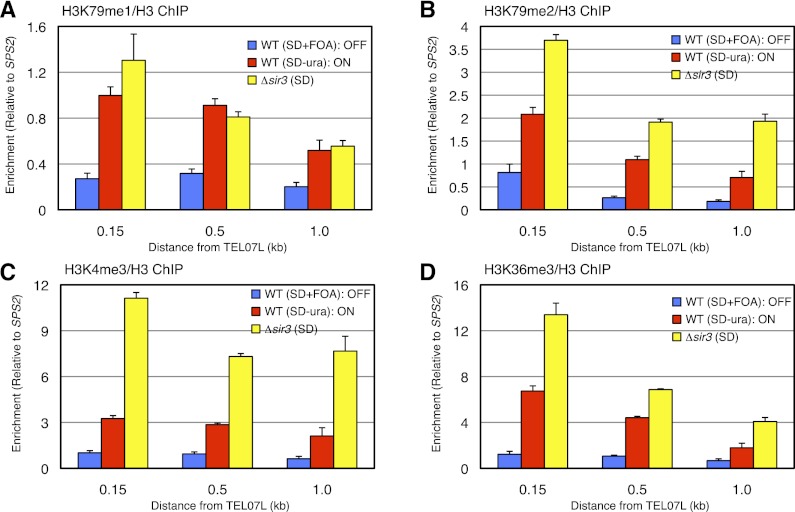
Since binding of Rap1 and Sir3 was similar between the ON and OFF telomeres, we next asked instead whether chromatin modifications antagonistic to silencing could be differentially enriched at these telomeres. To accomplish this, we performed ChIP at URA3-TEL07L in ON and OFF cells using antibodies specific to various chromatin modifications, including H4K16ac, H3K4me, H3K36me, and H3K79me. As expected from the efficient binding of Sir3, we found that H4K16, a key histone residue that regulates Sir3 spreading, was strongly hypoacetylated at both ON and OFF telomeres compared with Δsir3 (Fig. 1H; Supplemental Fig. S2). However, in contrast, we found that histone methylation was differentially enriched between the ON and OFF telomeres. Specifically, H3K79 monomethylation (H3K79me1), H3K79 dimethylation (H3K79me2), H3K4 trimethylation (H3K4me3), and H3K36me3 were enriched at the ON telomere (Fig. 2; Supplemental Fig. S2). We note that the enrichment levels of Htz1/H2AZ, H3K56ac, and H3K79me3, which are also capable of affecting gene silencing (Meneghini et al. 2003; Xu et al. 2007; Frederiks et al. 2008), were not obviously different between ON and OFF telomeres (Supplemental Fig. S2). The ChIP results for all of the above at the native TEL06R control locus are shown in Supplemental Figure S3. Therefore, our results argue that histone H3 methylation is enriched at ON telomeres and has the potential to disrupt gene silencing without affecting the amount of Sir3 binding to nucleosomes.
Figure 2.
Histone methylation is enriched at ON telomeres. (A–D) ChIP of H3K79me1/H3 (A), H3K79me2/H3 (B), H3K4me3/H3 (C), and H3K36me3/H3 (D) at URA3-TEL07L depicted as in Figure 1B.
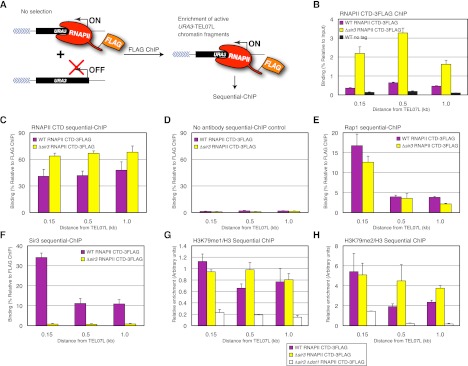
Sir proteins and RNAPII co-occupy chromatin in the ON state
As shown above, binding of the heterochromatin proteins Rap1 and Sir3 was similar between the ON and OFF telomeres. However, a ChIP assay measures the average level of protein binding or enrichment of a modification in a population of cells. Therefore, it was unclear whether the chromatin fragments with RNAPII binding that are responsible for gene activity were the same as those bound by heterochromatin proteins. To address this problem, we used sequential ChIP to determine whether RNAPII-bound telomere chromatin fragments were co-occupied by Rap1 or Sir3. The ON telomere fragments were first isolated by immunoprecipitation of Flag-tagged RNAPII using a Flag antibody, after which binding of Rap1 or Sir3 was measured by sequential ChIP (Fig. 3A). As shown in Figure 3B, RNAPII binding was low in wild-type SIR3 but high in Δsir3 control cells, as expected. Control sequential ChIP reactions with an RNAPII antibody or no antibody confirmed that RNAPII-bound chromatin fragments were enriched during the initial RNAPII-Flag ChIP (Fig. 3C,D). Importantly, sequential ChIP of Rap1 and Sir3 showed that these two proteins were indeed bound to the ON telomere (Fig. 3E,F). Furthermore, consistent with our ChIP experiments above using medium selection, we found that H3K79me1 and H3K79me2 are also enriched at ON telomeres (Fig. 3G,H; Supplemental Fig. S4). We conclude that RNAPII binding in the ON state is compatible with Rap1 or Sir3 binding.
Figure 3.
RNAPII-3Flag sequential ChIP assay confirms the co-occupancy of RNAPII and Rap1, Sir3, or H3K79me. (A) Schematic of the sequential ChIP approach to isolate ON telomeres. Rpb1, the subunit of the RNAPII complex containing the regulatory C-terminal domain (CTD), was C-terminally tagged with three tandem repeats of the Flag sequence and cultured in nonselective medium (YPD). ChIP was performed using an anti-Flag antibody to isolate chromatin fragments with RNAPII binding, including telomere fragments in the ON state. Probes were as in Figure 1A. (B) ChIP of RNAPII CTD-3Flag at URA3-TEL07L in wild-type (WT) SIR3 RNAPII CTD-3Flag (purple bars) and Δsir3 RNAPII CTD-3Flag (yellow bars) cells grown in nonselective medium (YPD). A wild-type SIR3 strain without a 3Flag tag (black bars) was used as a negative control. Data are presented as mean ± SD. (C–F) Sequential ChIP of RNAPII (C), Rap1 (E), and Sir3 (F) at URA3-TEL07L depicted as in B. (D) A mock sequential ChIP without an antibody was performed as a negative control. (G,H) Sequential ChIP of H3K79me1/H3 (G) and H3K79me2/H3 (H) at URA3-TEL07L, depicted as in C–F with the addition of Δsir3 Δdot1 RNAPII CTD-3Flag (white bars), which was used as a control strain that lacks H3K79me.
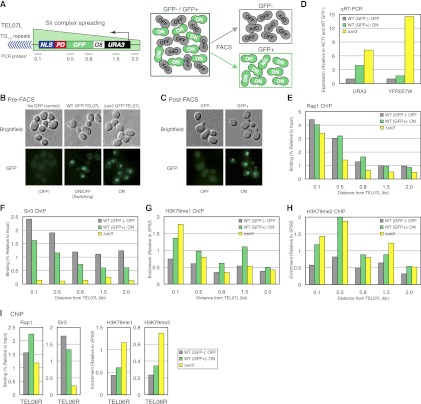
Fluorescence-activated cell sorting (FACS)-ChIP verification of the ON and OFF states
The sequential ChIP experiment described above showed that RNAPII and Rap1, Sir3, or H3K79me co-occupied the same chromatin fragments in the natural ON state of TPE. To further confirm this result and rule out the possibility that the medium-selection approach was causing an unexpected artifact, we wished to separate ON and OFF cells by FACS and compare the chromatin states of the ON and OFF telomeres using ChIP. To perform FACS-ChIP, we constructed a strain with a URA3-GFP fusion gene inserted at TEL07L (Fig. 4A). An octa-glycine (G8) linker was inserted between Ura3 and GFP so that GFP would not interfere with Ura3 function (Sabourin et al. 2007). To make the level of the GFP protein more accurately reflect the real-time expression state of the URA3-GFP gene, the half-life of Ura3-G8-GFP was reduced by attaching the Cln2 PEST domain (PD), a protein degradation sequence, to the C terminus of GFP (Xu et al. 2006). Last, to facilitate the visualization of Ura3-G8-GFP-PD, the fusion protein was concentrated in the nucleus using a nuclear localization signal (NLS). The variegated gene expression pattern of URA3-GFP in this strain was confirmed by fluorescence microscopy in pre-FACS cells (Fig. 4B). For FACS, exponentially growing cells were fixed using formaldehyde, and GFP-positive and GFP-negative cells were separated and confirmed by microscopy and qRT–PCR (Fig. 4C,D). Approximately 1 million sorted cells were used for ChIP analysis per protein or histone modification tested. As shown in Figure 4E, Rap1 bound well at URA3-GFP-TEL07L in both ON and OFF cells. Importantly, we observed a significant amount of Sir3 binding in the ON cells as well as the OFF cells (Fig. 4F). The slight drop in the absolute level of Sir3 binding at the ON telomere was likely due to the expected decrease in nucleosome density of a transcriptionally active locus, similar to the ChIP results observed in the medium-selected cells (Supplemental Fig. S2). Finally, H3K79me1 and H3K79me2 were enriched at the ON telomere compared with OFF (Fig. 4G,H). As controls, the binding of Rap1 and Sir3 and the enrichment of H3K79me1 and H3K79me2 at native TEL06R, which lacks integrated URA3, are shown in Figure 4I. We found very little change in any of these components at native TEL06R in the URA3 ON and OFF cells. Therefore, our FACS-ChIP data are consistent with the medium selection ChIP results above showing that Rap1, Sir3, H3K79me1, and H3K79me2 are enriched at the ON telomere of URA3-TEL07L.
Figure 4.
FACS-ChIP of URA3-GFP-TEL07L confirms that the ON and OFF states are differentiated by H3K79me. (A) Schematic of the FACS approach to isolate ON and OFF cells. URA3 regulated under its native promoter was fused to a G8 linker followed by yeast-enhanced GFP1, a CLN2 PD, and a NLS from SV40. Probes were ∼0.1, 0.5, 0.8, 1.5, and 2.0 kb away from the telomeric repeats of URA3-G8-GFP-PD-NLS-TEL07L. (B,C) Representative bright-field and fluorescence images of wild-type (WT) SIR3 and Δsir3 cells with URA3-G8-GFP-PD-NLS-TEL07L along with wild-type SIR3 cells with native TEL07L lacking GFP (negative control) before FACS (B), and GFP− and GFP+ wild-type SIR3 URA3-G8-GFP-PD-NLS-TEL07L cells after FACS (C). (D) qRT–PCR of URA3 at TEL07L and YFR057W at native TEL06R in wild-type SIR3 GFP− (gray bars) and GFP+ (green bars) cells and Δsir3 cells (yellow bars) grown in SD. Data are a representative result of three biological replicates. (E–H) ChIP of Rap1 (E), Sir3 (F), H3K79me1 (G), and H3K79me2 (H) at URA3-G8-GFP-PD-NLS-TEL07L, depicted as in D. (I) ChIP of Rap1, Sir3, H3K79me1, and H3K79me2 at native TEL06R using a probe ∼0.5 kb away from the telomeric repeats, depicted as in D.
H3K79me disrupts gene silencing without affecting Sir complex binding in vitro
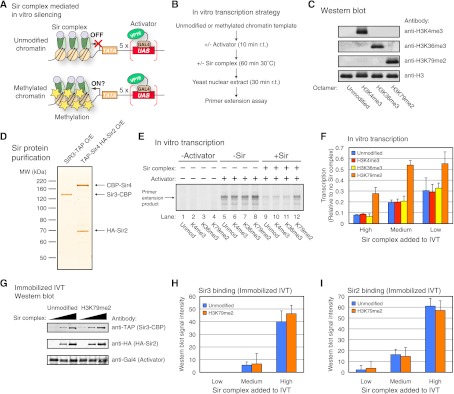
The methylation of histones has previously been implicated in disrupting gene silencing (van Leeuwen et al. 2002; Santos-Rosa et al. 2004; Altaf et al. 2007; Fingerman et al. 2007; Onishi et al. 2007; Tompa and Madhani 2007; Martino et al. 2009; Verzijlbergen et al. 2009). However, since histone methylation, particularly H3K4me and H3K36me, generally correlates with transcription in yeast (Millar and Grunstein 2006), it was possible that the enrichment of some of these methylation marks was merely a consequence of, rather than the cause for, the ON state of TPE. Therefore, we sought to distinguish the function of these modifications and test directly whether they would be sufficient to disrupt Sir complex-mediated silencing using a yeast in vitro transcription (IVT) system (Fig. 5A,B).
Figure 5.
H3K79me disrupts gene silencing without affecting Sir complex binding in vitro. (A) Schematic of the chromatin template and protein components involved in the in vitro silencing assay. (B) Outline of the in vitro silencing experiment. (C) Western blot of the MLA nucleosomes used for chromatin assembly. Anti-H3K4me3, anti-H3K36me3, anti-H3K79me2, and anti-H3 antibodies were used. (D) Silver-staining gel of the Sir proteins purified from yeast cells overexpressing Sir3-TAP or TAP-Sir4/HA-Sir2. (E) Representative phosphor screen image of a primer extension assay from the IVT experiment outlined in B. The signal represents the 32P end-labeled cDNA product generated by primer extension. (F) Quantification of phosphor screen images of the IVT experiments shown in E. Approximately 26 pmol of Sir3 and 8 pmol each of Sir2 and Sir4 were included in the reaction labeled “High.” Reactions labeled “Medium” and “Low” contained, respectively, one-half and one-fourth the amount of Sir proteins relative to “High.” Data are presented as mean ± SD. (G) Representative image of Sir protein binding from the immobilized IVT Western blot experiment. Anti-TAP, anti-HA, and anti-Gal4 antibodies were used. (H,I) Quantification of the immobilized IVT Western blot experiment shown in G. The binding levels of Sir3 (H) and Sir2 (I) are presented as mean ± SD.
In this system, we used a DNA template containing Gal4 DNA-binding sites and a TATA box (Fig. 5; Tantin et al. 1996). This template was previously shown to be highly responsive to activator GAL4-VP16 derivatives in a yeast nuclear extract (Ohashi et al. 1994). We assembled the template into chromatin using either unmodified histone octamers or octamers containing H3K4me3, H3K36me3, or H3K79me2. Methylated histones were generated using the methyl-lysine analog (MLA) technique (Simon et al. 2007) and validated by Western blot (Fig. 5C) and mass spectrometry (data not shown). GAL4-VP16 was first prebound to the chromatinized templates, and purified Sir proteins (Sir2/Sir3/Sir4), sufficient for silencing in vitro (Johnson et al. 2009), were added to the reactions. Sir proteins (Sir3-TAP and TAP-Sir4/HA-Sir2) were purified using a yeast overexpression system described previously by Moazed and colleagues (Johnson et al. 2009) (Fig. 5D). Yeast nuclear extract was added to the reaction following the binding of Sir proteins to the chromatinized template along with nucleoside triphosphates (NTPs), and transcription was measured by primer extension. An outline of this in vitro silencing experiment is depicted in Figure 5B.
As shown in Figure 5E, transcription was strongly dependent on Activator (lanes 1–8) and was reduced by the addition of Sir proteins to the reaction (lanes 5–12). However, strikingly, when the chromatin template was dimethylated at H3K79, silencing was strongly reduced compared with the template with no modifications (Fig. 5E, lanes 9–12). This effect was specific to H3K79me2, as neither H3K4me3 nor H3K36me3 was able to disrupt silencing (Fig. 5E, lanes 9–12). Transcription increased by approximately twofold to threefold on the H3K79me2 chromatin template compared with the unmodified template in the presence of Sir proteins (Fig. 5F). Therefore, since H3K79me is found at subtelomeric chromatin selectively in the ON state and its presence on chromatin is sufficient to disrupt Sir protein-mediated silencing in vitro, we conclude that H3K79me plays a causal role in determining the natural epigenetic ON state.
We next sought to assess the amount of Sir protein binding to the unmodified and H3K79me2 chromatin templates during IVT. To accomplish this, we performed an IVT reaction in a manner similar to that used above but with biotinylated unmodified and H3K79me2 chromatin templates immobilized to streptavidin-coated magnetic beads (Lin and Carey 2012). The amount of Sir protein binding to the immobilized templates was determined by Western blot following IVT and washing (Fig. 5G). Critically, as quantified in Figure 5, H and I, binding of Sir3 and Sir2 did not differ between the unmodified and H3K79me2 templates. Similarly, we did not observe a significant difference in the binding of the Sir complex to the unmodified and H3K79me2 chromatin templates when the Sir complex–chromatin interaction was measured in the absence of Activator or yeast nuclear extract (Supplemental Fig. S5). We conclude that H3K79me2 can disrupt gene silencing without noticeably affecting the amount of binding of the Sir complex to nucleosomes in vitro.
The discrepancy between our results and those of a previous study in which H3K79me had been shown to block Sir complex binding to a chromatin template in vitro (Martino et al. 2009) may be due to differences in the experimental techniques used. While the previous study had used electrophoretic mobility shift assays (EMSAs) to determine the Sir complex–chromatin interaction (Martino et al. 2009), here we used an immobilized chromatin template assay to directly measure Sir protein binding by Western blot and showed that the amount of Sir complex bound to chromatin was largely not affected by H3K79me. In either case, methylation of H3K79 may disrupt the interaction between Sir3 and the region surrounding H3K79 (Altaf et al. 2007; Fingerman et al. 2007). We propose that this disruption alters the overall conformation of the Sir2/Sir3/Sir4–nucleosome complex and that this alteration in turn enables the epigenetic ON state.
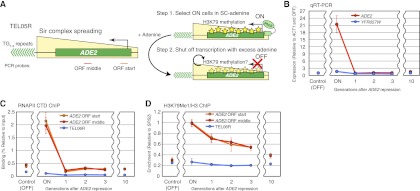
Maintenance of H3K79me is dependent on transcription in the epigenetic ON state
As demonstrated above, the key difference between ON and OFF telomeres is the enrichment of H3K79me, which is capable of disrupting gene silencing. We next addressed how H3K79me is maintained epigenetically through multiple cell generations at the ON telomere. Since the H3K79 methyltransferase Dot1 is recruited to chromatin through transcription (Shahbazian et al. 2005; Millar and Grunstein 2006), we hypothesized that the maintenance of H3K79me in the ON state may be dependent on transcription. To test this possibility, we asked whether H3K79me at the ON telomere would be lost upon inhibition of transcription. We monitored the chromatin state of ADE2-TEL05R, whose ON state could be selected by growing cells in medium lacking adenine (SC−ade). As a control, we examined YFR057W at native TEL06R. Similar to the ON state of URA3-TEL07L, the ADE2-TEL05R ON state was accompanied by an increase in H3K79me (Supplemental Fig. S6). After selecting for ADE2-TEL05R ON by growing cells in SC−ade, we specifically repressed ADE2 through negative feedback by adding excess adenine to the medium. This treatment causes the dissociation of the activator Pho2 from the promoter of ADE2 (Fig. 6A; Pinson et al. 2009). The same method cannot be used for repression of URA3-TEL07L, since adding excess uracil to the medium would be toxic to the cells (Gadsden et al. 1993). As shown in Figure 6, B and C, by qRT–PCR and RNAPII ChIP, ADE2 expression decreased rapidly to near-background level following adenine addition. In a corresponding manner, we found that H3K79me1 is gradually lost every cell cycle and eventually drops to near-background level (Fig. 6D; Supplemental Fig. S6). These results imply that a self-reinforcing feedback loop in which H3K79me both results from and is causal for transcription maintains the epigenetic ON state of TPE.
Figure 6.
Maintenance of H3K79me at the ON telomere depends on transcription. (A) Schematic of the ADE2 feedback repression experiment to monitor the level of histone methylation after inhibition of transcription. The ADE2 ORF middle and ORF start probes are ∼1.0 and 2.0 kb away, respectively, from the telomeric repeats of ADE2-TEL05R. (B) qRT–PCR of ADE2 at TEL05R (red lines) and YFR057W at native TEL06R (blue lines) before and after the addition of adenine. A culture continuously grown in the presence of excess adenine (>30 generations) was used as an OFF control. Data are presented as mean ± SD. (C,D) ChIP of RNAPII (C) and H3K79me1/H3 (D) at ADE2-TEL05R and native TEL06R using the cultures described in B. The ADE2 ORF middle (red lines) and ORF start (orange lines) probes were as described in A. The native TEL06R (blue lines) probe is ∼1.5 kb away from the telomeric repeats. Data are presented as mean ± SD.
Discussion
The mechanism of heterochromatin spreading and gene silencing at the telomeres of Saccharomyces cerevisiae has been characterized extensively (Rusche et al. 2003; Mondoux and Zakian 2006). However, how variegated gene expression occurs at telomeres has been unclear. To address this problem, we separated the natural ON and OFF cells from a population of yeast undergoing TPE and directly compared the chromatin structure of the natural ON state with that of the OFF state. This is unlike previous studies in which mixed ON and OFF telomeres were compared with the disrupted telomeres of sir mutant strains, which made it impossible to characterize the natural ON state (Rusche et al. 2003). Our findings indicate that (1) the natural ON telomere is characterized by Sir complex binding to nucleosomes; (2) histone H4K16 is deacetylated at the ON telomere, which is consistent with the spreading of Sir3 through telomeric heterochromatin by its interaction with deacetylated H4K16; (3) H3K79 is methylated in the natural ON state and can disrupt silencing without affecting Sir complex binding in vitro; and (4) maintenance of H3K79me is dependent on a transcription-mediated positive feedback loop. Our results suggest that, since the ON telomere is characterized by Sir3 binding and H4K16 hypoacetylation, two factors that are normally associated with silencing, other factors must determine the ON state. We show that H3K79me is such a factor. This is in contrast to previous studies, which proposed that the variation in Sir complex binding to nucleosomes regulates TPE (Ng et al. 2003; Moazed 2011).
The deacetylation of H4K16, a major requirement of heterochromatin formation, in the ON state is of special interest. It argues that H4K16ac is not the determinant of epigenetic variegation. Thus, our study differentiates the function of two key histone modification marks at heterochromatin, where H4K16 deacetylation determines the distance of heterochromatin protein spreading from the telomere by virtue of its interaction with Sir3 (Johnson et al. 1990, 2009; Kimura et al. 2002; Suka et al. 2002; Liou et al. 2005; Onishi et al. 2007), and H3K79me regulates the actual ON/OFF expression state of a subtelomeric gene.
It had been proposed previously that H3K79me may disrupt the binding of the Sir complex to nucleosomes based on pull-down assays that measured the binding of the Sir3 protein to a peptide containing the H3K79 region (Altaf et al. 2007; Fingerman et al. 2007). Subsequently, it had been shown that binding of the whole Sir complex to a trinucleosomal chromatin template is also affected by H3K79me using a gel shift assay (Martino et al. 2009). However, we showed in vivo by ChIP and in vitro using an immobilized template assay in the presence of yeast nuclear extract that the overall binding level of the Sir proteins to the nucleosome was not significantly disrupted by H3K79me. Therefore, we favor instead a model in which the methylation-dependent loss of the Sir3–H3K79 interaction leads to a conformational change in the structure of the Sir protein–nucleosome complex, which results in disrupted gene silencing.
H3K79me and its methyltransferase, Dot1, are conserved in many organisms, including fruit flies, mice, and humans (Nguyen and Zhang 2011). It has been shown that mutations in the fruit fly DOT1 homolog grappa disrupts Polycomb group-mediated silencing as well as telomeric silencing in flies (Shanower et al. 2005). Similarly, knockout of the mouse DOT1 homolog Dot1L leads to the loss of heterochromatin-associated marks such as H3K9me from centromeric and telomeric heterochromatin in mouse embryonic stem (ES) cells (Jones et al. 2008). Thus, H3K79me and Dot1 are relevant to gene silencing and heterochromatin formation in organisms other than the budding yeast.
In contrast, there are no homologs of Dot1 or detectable levels of H3K79me in the fission yeast Schizosaccharomyces pombe (Sinha et al. 2010). Thus, while gene expression at the heterochromatin of S. pombe is also known to be regulated epigenetically, the mechanism inevitably cannot involve H3K79me. Allshire and colleagues (Ekwall et al. 1997) have shown that transient treatment of S. pombe cells with an HDAC inhibitor leads to a heritable hyperacetylated chromatin state accompanied by the loss of gene silencing at centromeric heterochromatin. Likewise, Grewal and colleagues (Nakayama et al. 2000) have shown that expression of a gene at the partially compromised centromeric heterochromatin of S. pombe is associated with hyperacetylation and lack of heterochromatin protein Swi6/HP1 binding. The epigenetic inheritance of gene expression in these studies could be explained by a positive feedback loop involving histone acetylation and lack of heterochromatin-binding proteins. This is in stark contrast to our findings at the telomeric heterochromatin of S. cerevisiae, which show that neither H4K16ac nor binding of heterochromatin proteins is a key regulator of gene variegation.
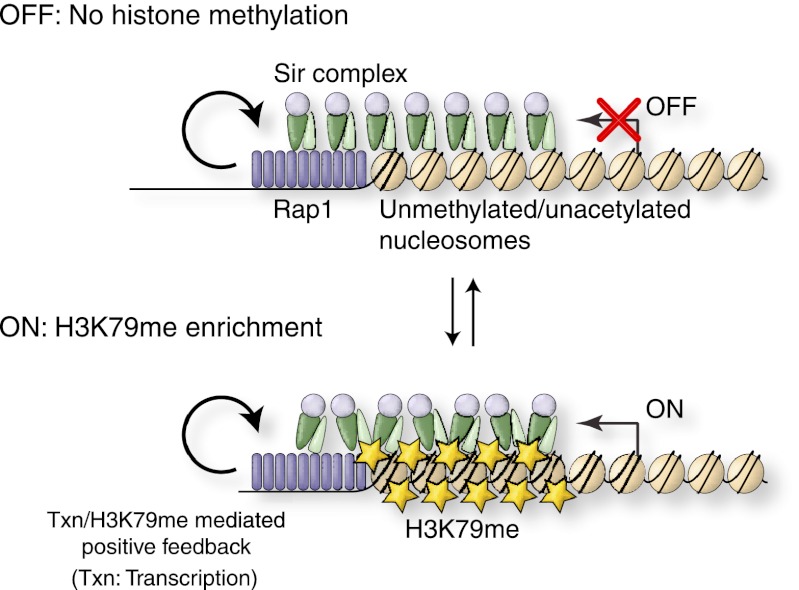
Instead, a positive feedback loop mediated by transcription and H3K79me is at the heart of our model regarding the mechanism of epigenetic variegation at S. cerevisiae telomeres, as described below (Fig. 7). In this model, the ON state is characterized by H3K79me. The maintenance of H3K79me is dependent on transcription, which had previously been shown to recruit the histone H3 Lys79 methyltransferase Dot1 (Shahbazian et al. 2005). H3K79me in turn disrupts the local interaction between Sir3 and the H3 core region surrounding Lys79 (Altaf et al. 2007; Fingerman et al. 2007). However, in contrast to previous models, the Sir complex as a whole can still spread along the subtelomere through its interaction with deacetylated H4K16. In this structure, the methylation of H3K79 enables PIC assembly and transcription, possibly by inducing a conformational change in the Sir protein–nucleosome complex, thus promoting a positive feedback loop. The possible absence of an H3K79 histone demethylase (Liang et al. 2007) may further enhance the stability of this continuous ON state. In contrast, the absence of transcription in the OFF state precludes Dot1 recruitment and ensures H3K79 hypomethylation. It had previously been shown that Sir3 binding to nucleosomes can prevent Dot1 from methylating chromatin (Altaf et al. 2007; Fingerman et al. 2007). Therefore, the lack of Dot1 recruitment and the prevention of Dot1 access to the H3K79 residue help establish a stable OFF state.
Figure 7.
Model to explain the variegated gene expression pattern of TPE. See the text for details.
How is it then possible for a gene in one expression state to escape these feedback loops and convert to the other state? One possibility may be that changes in the length of telomeres (elongation/shortening) lead to the interconversion of epigenetic expression states. Previous studies from Lustig and colleagues (Kyrion et al. 1993; Park and Lustig 2000) have shown that elongated telomeres are associated with stronger subtelomeric gene silencing. Since the length of telomeres naturally fluctuates within a cell (Shore and Bianchi 2009), some telomeres may become abnormally shortened, and this may lead to a compromised heterochromatin structure susceptible to transcription. In contrast, abnormal lengthening may cause a structural change at an ON telomere that can overcome the anti-silencing effect of H3K79me and dampen gene expression until methylation is passively lost. In any case, whether the natural variation in telomere length is sufficient to induce epigenetic switching is still unknown. Changes in H3K79me are shown here to regulate the maintenance of the variegated ON/OFF expression states at telomeric heterochromatin. However, the rare transient upstream events that initiate switching between the ON and OFF states remain to be observed and determined.
Materials and methods
Yeast strains, plasmids, and oligonucleotide probes
Yeast strains, plasmids, and oligonucleotide probes used in this study are listed in the Supplemental Material. Plasmid and PCR product-based genetic manipulations were performed using standard yeast transformation techniques (Gietz and Woods 2002). Full details are provided in the Supplemental Material.
Protein purification
Xenopus laevis histones and histone mutants (H3, H3C110AK4C, H3C110AK36C, H3C110AK79C, H4, H2A, and H2B) were purified as described previously (Luger et al. 1997). GAL4-VP16 was purified as described previously (Tantin et al. 1996). Sir proteins were purified as described previously (Tanny et al. 2004; Johnson et al. 2009) with some modifications to the protocol. Full details are provided in the Supplemental Material.
MLA histone preparation
H3K4me3, H3K36me3, and H3K79me2 MLA histones were generated from H3C110AK4C, H3C110AK36C, and H3C110AK79C histone mutants, respectively, as described previously (Simon et al. 2007).
qRT–PCR
RNA was extracted using the hot acid phenol extraction method (Bookout et al. 2006). The extracted RNA samples were treated with DNase I (Qiagen), purified, and reverse-transcribed using random primers and M-MLV reverse transcriptase (Invitrogen). qPCR was performed and analyzed using the ΔΔCt method (Bookout et al. 2006). Full details are provided in the Supplemental Material.
Western blot
Western blot assays were performed using the ODYSSEY infrared imaging system (LI-COR) following the manufacturer's protocol. Full details are provided in the Supplemental Material.
ChIP
Standard ChIP assays were performed as described previously (Hecht et al. 1996; Suka et al. 2001) with minor modifications to the protocol. Full details are provided in the Supplemental Material.
Sequential ChIP
Sequential ChIP was performed as described elsewhere (Kao et al. 2004) with minor modifications to the protocol. Briefly, chromatin lysate was immunoprecipitated overnight with anti-Flag M2 agarose beads (Sigma-Aldrich). The agarose beads were washed, and the chromatin fragments were eluted off the beads with 3x Flag peptide (Sigma-Aldrich). Part of the eluate was saved and used as the input control DNA for the second (sequential) ChIP. Sequential ChIP assays were performed using the same protocol as standard ChIP. Full details are provided in the Supplemental Material.
FACS-ChIP
FACS was performed using BD FACSAria II (BD Biosciences) according to the manufacturer's manual. Full details are provided in the Supplemental Material.
IVT/silencing
IVT was performed as described previously (Lin and Carey 2012) with minor modifications to the protocol. The DNA template containing five Gal4 DNA-binding sites and an adenovirus E4 promoter (G5E4T) (Tantin et al. 1996) was assembled into chromatin by salt dilution as described previously (Steger et al. 1997). Following prebinding of GAL4-VP16 to the template, Sir proteins were added to the IVT reaction. Yeast nuclear extract, prepared as described previously (Rani et al. 2004), was added to the reaction, and primer extension was performed to measure the amount of transcription. Full details are provided in the Supplemental Material.
Immobilized chromatin template
The immobilized chromatin template assays were performed essentially as described previously (Lin and Carey 2012) with some modifications to the protocol. Buffer conditions and DNA/protein components were as described above for the IVT/silencing experiments except that GAL4-VP4, a variant of GAL4-VP16 containing four tandem repeats of the activation domain, was used (Ohashi et al. 1994). We confirmed that the results of the IVT/silencing experiments described above were reproducible when GAL4-VP4 was substituted for GAL4-VP16 in the reaction (data not shown). Briefly, biotinylated chromatin templates were immobilized on M280 streptavidin beads, and IVT reactions were incubated by rotation. The beads were washed twice with reaction buffer and eluted with Laemmli buffer. Western blot was performed as described above and quantified using ImageQuant TL software.
Acknowledgments
We thank the present and past members of the laboratories of M.G., M.C. (especially Lynn Lehmann for GAL4-VP16, Justin Lin for H3K4me3 octamer, and Jason Gehrke), and Siavash Kurdistani (especially Maria Vogelauer) for providing materials, valuable suggestions, and constructive criticisms during the course of this work. We also thank Daniel Gottschling, James Broach, Danesh Moazed, and Ivan Sadowski for plasmids, strains, and antibodies; Aaron Johnson for sharing his TAP-Sir4/HA-Sir2 purification protocol; Rachelle Crosbie for sharing her fluorescence microscope; and Jamie Marshall for microscopy advice. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility, which is supported by National Institutes of Health awards CA-16042 and AI-28697 and by the JCCC, the UCLA AIDS Institute, the David Geffen School of Medicine at UCLA, and the UCLA Chancellor's Office. T.K. was supported by a fellowship from the Nakajima Foundation. B.G.K. was supported by Ruth L. Kirschstein National Research Service Award GM007185. This work was supported by National Institutes of Health grants GM23674, GM42421 (to M.G.), GM074701, and GM085002 (to M.C).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.201095.112.
References
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell 28: 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334: 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF 2006. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol 73: 15.8.1–15.8.28 [DOI] [PubMed] [Google Scholar]
- Buchman AR, Kimmerly WJ, Rine J, Kornberg RD 1988. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol 8: 210–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Widom J 2005. Mechanism of transcriptional silencing in yeast. Cell 120: 37–48 [DOI] [PubMed] [Google Scholar]
- Ehrentraut S, Hassler M, Oppikofer M, Kueng S, Weber JM, Mueller JW, Gasser SM, Ladurner AG, Ehrenhofer-Murray AE 2011. Structural basis for the role of the Sir3 AAA+ domain in silencing: Interaction with Sir4 and unmethylated histone H3K79. Genes Dev 25: 1835–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Fingerman IM, Li HC, Briggs SD 2007. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: Identification of a new trans-histone pathway. Genes Dev 21: 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiks F, Tzouros M, Oudgenoeg G, van Welsem T, Fornerod M, Krijgsveld J, van Leeuwen F 2008. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat Struct Mol Biol 15: 550–557 [DOI] [PubMed] [Google Scholar]
- Gadsden MH, McIntosh EM, Game JC, Wilson PJ, Haynes RH 1993. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J 12: 4425–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Gross DS 2008. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol Cell Biol 28: 3979–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Girton JR, Johansen KM 2008. Chromatin structure and the regulation of gene expression: The lessons of PEV in Drosophila. Adv Genet 61: 1–43 [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Hahn S 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol 11: 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22: 4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci 87: 6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D 2009. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell 35: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus G, Kadam S, Zhai H, Valdez R, et al. 2008. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4: e1000190 doi: 10.1371/journal.pgen.1000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev 18: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J 24: 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet 32: 370–377 [DOI] [PubMed] [Google Scholar]
- Kirchmaier AL, Rine J 2006. Cell cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol 26: 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF, Schweizer D, Gasser SM 1992. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Biol Chem 117: 935–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P 2009. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462: 323–330 [DOI] [PubMed] [Google Scholar]
- Kyrion G, Liu K, Liu C, Lustig AJ 1993. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev 7: 1146–1159 [DOI] [PubMed] [Google Scholar]
- Landry J, Slama JT, Sternglanz R 2000. Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun 278: 685–690 [DOI] [PubMed] [Google Scholar]
- Li B, Lustig AJ 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev 10: 1310–1326 [DOI] [PubMed] [Google Scholar]
- Liang G, Klose RJ, Gardner KE, Zhang Y 2007. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat Struct Mol Biol 14: 243–245 [DOI] [PubMed] [Google Scholar]
- Lin JJ, Carey M 2012. In vitro transcription and immobilized template analysis of preinitiation complexes. Curr Protoc Mol Biol 97: 12.14.1–12.14.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol 272: 301–311 [DOI] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M 2002. Rap1–Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD. 2007 From a to α: Yeast as a model for cellular differentiation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM 2009. Reconstitution of yeast silent chromatin: Multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Millar CB, Grunstein M 2006. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol 7: 657–666 [DOI] [PubMed] [Google Scholar]
- Mishra K, Shore D 1999. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol 9: 1123–1126 [DOI] [PubMed] [Google Scholar]
- Moazed D 2011. Mechanisms for the inheritance of chromatin States. Cell 146: 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: A SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci 94: 2186–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondoux MA, Zakian VA 2006. Telomere position effect: Silencing near the end. In Telomeres, Cold Spring Harbor Monograph Series 45 (ed. T de Lange et al.), pp. 261–316. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI 2000. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101: 307–317 [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 16: 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc Natl Acad Sci 100: 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y 2011. The diverse functions of Dot1 and H3K79 methylation. Genes Dev 25: 1345–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Brickman JM, Furman E, Middleton B, Carey M 1994. Modulating the potency of an activator in a yeast in vitro transcription system. Mol Cell Biol 14: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell 28: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Osborne EA, Dudoit S, Rine J 2009. The establishment of gene silencing at single-cell resolution. Nat Genet 41: 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Lustig AJ 2000. Telomere structure regulates the heritability of repressed subtelomeric chromatin in Saccharomyces cerevisiae. Genetics 154: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson B, Vaur S, Sagot I, Coulpier F, Lemoine S, Daignan-Fornier B 2009. Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev 23: 1399–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Ptashne M. 2002 Genes & signals. CSHL Press, Cold Spring Harbor, NY. [Google Scholar]
- Rani PG, Ranish JA, Hahn S 2004. RNA polymerase II (Pol II)–TFIIF and Pol II–mediator complexes: The major stable Pol II complexes and their activity in transcription initiation and reinitiation. Mol Cell Biol 24: 1709–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD, Martienssen RA, Russo VEA 1996. Introduction. In Epigenetic mechanisms of gene regulation, Cold Spring Harbor Monograph Series 32 (ed. VEA Russo et al.), pp. 1–4. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Rossmann MP, Luo W, Tsaponina O, Chabes A, Stillman B 2011. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol Cell 42: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Fisher TS, Zakian VA 2007. A flexible protein linker improves the function of epitope-tagged proteins in Saccharomyces cerevisiae. Yeast 24: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Bannister AJ, Dehe PM, Geli V, Kouzarides T 2004. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J Biol Chem 279: 47506–47512 [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Gross DS 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105: 403–414 [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell 19: 271–277 [DOI] [PubMed] [Google Scholar]
- Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P 2005. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia WJ, Li B, Workman JL 2006. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev 20: 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D, Bianchi A 2009. Telomere length regulation: Coupling DNA end processing to feedback regulation of telomerase. EMBO J 28: 2309–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM 2007. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha I, Buchanan L, Ronnerblad M, Bonilla C, Durand-Dubief M, Shevchenko A, Grunstein M, Stewart AF, Ekwall K 2010. Genome-wide mapping of histone modifications and mass spectrometry reveal H4 acetylation bias and H3K36 methylation at gene promoters in fission yeast. Epigenomics 2: 377–393 [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, et al. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci 97: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling AS, Grunstein M 2009. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc Natl Acad Sci 106: 13153–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Owen-Hughes T, John S, Workman JL 1997. Analysis of transcription factor-mediated remodeling of nucleosomal arrays in a purified system. Methods 12: 276–285 [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83–93 [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32: 378–383 [DOI] [PubMed] [Google Scholar]
- Takahashi YH, Schulze JM, Jackson J, Hentrich T, Seidel C, Jaspersen SL, Kobor MS, Shilatifard A 2011. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol Cell 42: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D 2004. Budding yeast silencing complexes and regulation of Sir2 activity by protein–protein interactions. Mol Cell Biol 24: 6931–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D, Chi T, Hori R, Pyo S, Carey M 1996. Biochemical mechanism of transcriptional activation by GAL4-VP16. Methods Enzymol 274: 133–149 [DOI] [PubMed] [Google Scholar]
- Tompa R, Madhani HD 2007. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175: 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Verzijlbergen KF, Faber AW, Stulemeijer IJ, van Leeuwen F 2009. Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae. BMC Mol Biol 10: 76 doi: 10.1186/1471-2199-10-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR 2006. Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell 23: 219–229 [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell 27: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Britton J, Kirchmaier AL 2008. Insights into the impact of histone acetylation and methylation on Sir protein recruitment, spreading, and silencing in Saccharomyces cerevisiae. J Mol Biol 381: 826–844 [DOI] [PubMed] [Google Scholar]