Abstract
Reprogramming of a somatic nucleus to an induced pluripotent state can be achieved in vitro through ectopic expression of Oct4 (Pou5f1), Sox2, Klf4 and c-Myc. While the ability of these factors to regulate transcription in a pluripotent context has been studied extensively, their ability to interact with and remodel a somatic genome remains underexplored. Several recent studies have begun to provide mechanistic insights that will eventually lead to a more rational design and improved understanding of nuclear reprogramming.
Introduction
With the report that ectopic expression of MyoD could induce a fibroblast to acquire myotube-like characteristics, many began to explore the effects of ectopic expression of lineage or cell type specific transcription factors (TFs) and their ability to induce cell state transitions [1–4]. TF mediated reprogramming experiments were complimented by studies using cell fusion and somatic cell nuclear transfer (SCNT) which demonstrated the ability to revert somatic nuclei to pluri- and totipotency, respectively [5–9]. However, the reprogramming field grew exponentially after 2006, when a seminal report by Takahashi and Yamanaka identified a combination of TFs whose ectopic expression was capable of reprogramming a somatic nucleus to a pluripotent state, now referred to as induced pluripotency or induced pluripotent stem cells (iPSCs)[10**]. Ectopic expression of Oct4 (Pou5f1), Sox2, Klf4 and c-Myc (OSKM) was shown to reprogram embryonic and adult fibroblast nuclei to a state resembling mouse embryonic stem cells (mESCs). Minor technical improvements to the original approach reported shortly thereafter enabled the establishment of iPSCs that were both molecularly and developmentally comparable to mESCs [11–14]. Successfully reprogrammed iPSCs exhibit the potential to contribute to each germ layer upon in vitro or in vivo differentiation [15–17], although at different efficiencies [18]. Despite notable variation that has been observed among iPSC cell lines [19*], their remarkable potential explains the continued excitement and therapeutic promise [20]. A recent perspective from Yamanaka suggested that the number of lines investigated in each study contributed notably to the respective conclusion whether or not differences exist between the tested ESC and iPSC lines [21].
In this review, we will discuss selected advances in the reprogramming field with a particular focus on the interaction of TFs and the (epi)genome. This includes a closer inspection of characteristics that may influence the ability of various TFs to induce cell state conversions in vitro.
Transcription Factors and the Genome: An Epigenetic Barrier?
Epigenetic regulation of transcription through mechanisms such as posttranslational histone modifications and DNA methylation is essential for maintaining cellular identity both in vivo [22, 23] and in vitro [24, 25]. During differentiation and reprogramming, remodeling of the epigenetic landscape is dictated by TFs [26, 27] and also non-coding RNAs [28, 29]. Any newly acquired state will exhibit distinct DNA methylation and histone modification patterns as compared to the starting population [26, 30]. As a result, it is not too surprising that reprogramming can be assisted by the manipulation of epigenetic modifiers. For example, it was found that chemical depletion of DNA methyltransferases [26], inhibition of histone deacetylases [31], or knockdown of several chromatin modifying enzymes [32] improved the overall reprogramming efficiency.
Closer inspection of all TFs used in the various studies provides relevant insights for dissecting their ability to facilitate reprogramming (Box 1). In contrast to the more limited capacity of OSKM, some TFs used during direct cell state conversions exhibit a relatively unique ability to access and remodel target chromatin in a monomeric, ATP-independent fashion [33–35]. These factors have been termed “pioneering transcription factors,” a group defined by their ability to access and remodel heterochromatic regions towards open chromatin [36, 37]. It is noteworthy that many lineage-specific TF reprogramming combinations include pioneer TFs [38–42] (Table 1). The basic helix-loop-helix domain (bHLH) containing factor MyoD, for example, can bind to DNA in a heterochromatic chromatin state and initiate chromatin remodeling [43, 44]. More recently, members of the Fox family have been used to induce the transition from a fibroblast to a hepatocyte-like cell [40, 41] and ectopic expression of various Gata family TFs has been used to induce a cardiomyocte-like state from fibroblasts [39]. As noted above, OSKM have not been shown to exhibit direct chromatin remodeling capabilities analogous to MyoD and the Fox/Gata families, but appear to cooperate with available chromatin regulators [45**].
Box 1. TFs utilized to induce cellular transitions.
Many combinations of TFs have been utilized to induce cellular transitions (Table 1). In contrast to the factors that reprogram somatic cells to iPSCs, the combinations of factors used for these experiments generally include a factor with known chromatin remodeling capabilities, such as Fox and Gata TFs (Table 1). Additionally, these experiments commonly use factors with the ability to bind DNA as monomers, which are not reported features of OSKM in the specific context of reprogramming to iPSCs (Table 2). TF binding abilities are presumably context dependent, therefore the information included in Table 2, which was often derived in vitro or using limited cell types, should be considered with these caveats in mind.
Table 1.
| Starting cell type | Induced state | TFs used |
|---|---|---|
| MEFs | neural precursor | Brn2, Sox2, FoxG1 [78] |
| MEFs | motor neuron | Ascl1, Brn2, Myt1l, Lhx3, Hb9, Isl1, Ngn2 [42] |
| MEFs | cardiomyocyte | Gata4, Mef2c, Tbx5 [39] |
| MEFs | hepatocyte-like | Hnf4α plus Foxa1, Foxa2 or Foxa3 [41] |
| MEFs | hepatocyte-like | Gata4, Hnf1α, Foxa3 [40] |
| B cells | macrophage | CEBPα [3] |
| pancreatic exocrine cells | β -cells | Ngn3, Pdx1 and Mafa [79] |
| MEFs | macrophage-like | PU.1, CEBPα, CEBPβ [38] |
| somatic cells | iPSC | Oct4, Sox2, Klf4, c-Myc [10] |
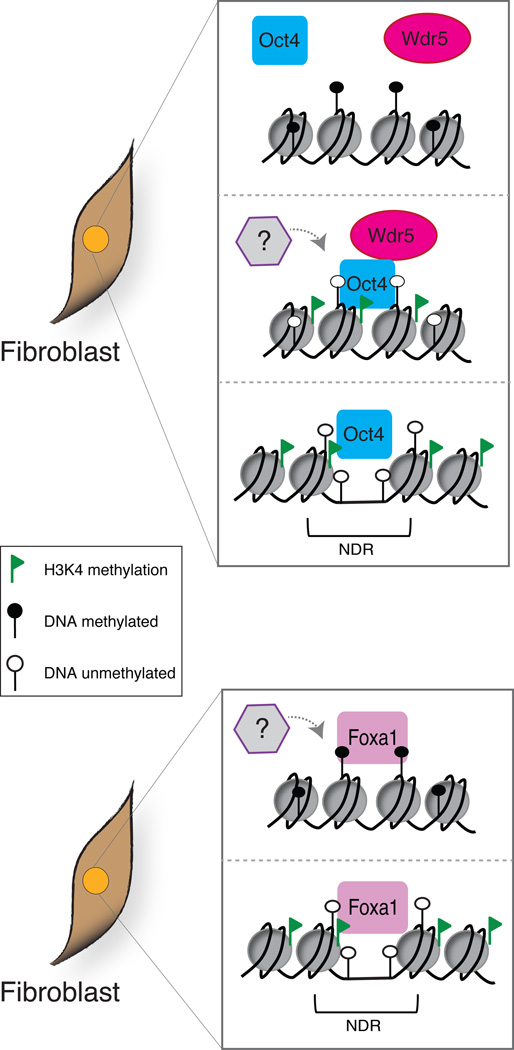
Myc, a non-essential reprogramming factor, is categorized as a transcriptional pause release factor and is unlikely to cause chromatin decondensation given that it associates with the Pol II pre-initiation complex, which cannot assemble without preexisting H3K4 methylation [46–49]. Furthermore, a recent report suggested that Oct4 could promote the establishment of nucleosome-depleted regions (NDRs), another requirement for transcriptional activation, only in the absence of DNA methylation [50*]. This highlights Oct4’s limited ability to bind in a preexisting heterochromatic environment and explains why only selected Oct4 target genes show immediate changes in H3K4 methylation [51**]. In contrast to the restricted binding of c-Myc and Oct4, chromatin immunoprecipitation (ChIP) results detected binding of the pioneer factor FoxA1 at a distal regulatory element that contains DNA methylation [52] (Figure 1). During subsequent differentiation, DNA methylation was eliminated proximal to FoxA1 binding and this region exhibited de novo gain of H3K4me1, a modification associated with active and poised distal regulatory elements. Further supporting the Fox family’s abilities to remodel heterochromatic regions, this factor was also used in combination with other TFs to induce the conversion of fibroblasts to hepatocyte-like cells [41]. More recent technical advances combining ChIP with bisulfite sequencing (ChIP-BS-Seq) will enable a more detailed characterization of the direct relationships between TFs and DNA methylation in this context [53, 54].
Figure 1. Chromatin Remodeling induced by ectopic TF expression.
Schematic of two types of TFs (Oct and Fox) and their ability to bind DNA methylated regions when overexpressed in fibroblasts. Oct4 cannot bind until DNA methylation is removed. Once it binds it can recruit factors like Wdr5 to induce gain of H3K4 methylation. This is insufficient to cause transcriptional activation and likely requires additional factors/complexes (indicated by the question mark) to create nucleosome depleted regions (NDRs) and expression. In contrast a pioneering TF, such as FoxA1 has the ability to bind multiple chromatin states including DNA methylated regions directly (as a monomer) and will subsequently induce remodeling.
Many pioneering factors such as FoxA1 and Gata4, perform their functions as monomers [33, 55–57] (Table 2), unlike Oct4, which requires dimerization with Sox2 in the pluripotent context [58, 59]. Further insight into the role of dimerization and cofactors during reprogramming was provided by a recent study that showed Sox17 could be transformed into a reprogramming factor by changing specific residues that promoted interaction with Oct4 [60**]. While the DNA binding profile of the new dimer was not directly interrogated, the ability of multiple Sox, but not Oct, family members to serve as iPSC reprogramming factors may suggest distinct roles for these families. Oct4 appears critical for DNA binding specificity while Sox2 or other Sox factors likely recruit histone acetyltransferases, such as p300 to these targets [61, 62]. More generally, these findings suggest that one could construct TFs with novel chromatin remodeling functions and DNA binding preferences. Systematic dissection of the functions associated with specific domains that have been retained by evolutionarily conserved TFs will provide the necessary insights. Interestingly, Hiryai et al. recently affixed the transactivation domain (TAD) of MyoD to Oct4, and examined this engineered factor’s ability to promote the reprogramming of MEFs [63**]. Indeed they found that substituting the wildtype form of Oct4 with the TAD fusion construct accelerated the appearance of Oct4 positive colonies when ectopically expressed. This suggests that wildtype Oct4, in combination with the other reprogramming factors (SKM), has a limited ability to access and remodel heterochromatic regions, which is consistent with published observations [50*, 51**, 64]. However, it highlights that this constraint can be readily overcome by engineering improved reprogramming factors as shown for Oct4-TAD.
Table 2.
| TF | Conserved domains/families | Binding preference |
|---|---|---|
| Asl1 | HLH | homodimer/heterodimer [80] |
| Bm2 | homeodomain, POU | Homodimer [81] |
| c-Myc | HLH, MYC-N, MYC-LZ | Heterodimer [82] |
| CEBPa | bZIP | Homodimer [76] |
| CEBPb | bZIP | Heterodimer/Homodimer [83] |
| FoxA1 | FH, HNF_C | Monomer [33] |
| FoxA2 | FH, HNF_C | Monomer [33] |
| FoxA3 | FH, HNF_C | Monomer [84] |
| Gata4 | ZnF_Gata, GATA_N | Monomers [33] |
| Hb9/mnx1 | homeodomain | unknown |
| Hnf4a | NR_DBD | Homodimer [85] |
| Isil | homeodomain, LIM1, LIM2 | unknown |
| Klf4 | zf-H2C2_2, zf-C2H2 | unknown |
| Lhx3 | homeodomain, LIM1, LIM2 | Monomer [86] |
| Mafa | bZIP, Maf_N | monomer, homodimer [87] |
| Mef2c | MADS | homodimer, heterodimer [88] |
| MyoD | HLH, Myf5 | monomer, heterodimer [89] |
| Myt1l | MYT1, C2H2, TMF, SMC | unknown |
| Ngn2 | HLH | heterodimer [90] |
| Ngn3 | HLH | Heterodimer [91] |
| Pdx1 | homedomain | monomer, heterodimer, heterotrimer [92] |
| POU5f1 | homeodomain | homodimer, heterodimer [59] |
| PU.1 | ETS | unknown |
| Sox2 | HMG-box, SOXP | monomer, heterodimer [59] |
| Tbx5 | T-box | Monomer, heterodimer [93] |
Abbreviations: HLH (helix-loop-helix), FH (forkhead), HNF_C (Hepatocyte Nuclear Factor C terminal domain), GATA_N (Gata-type transcriptional activator, N terminus), NR_DBD (nuclear receptor, DNA binding domain), zf-H2C2_2 (Zinc-finger double domain), bZIP (basic region, leucine zipper), MYT1 (myelin TF 1), ZF-C2H2 (zinc-finger double domain), TMF (TATA element modulatory factor 1), NDR (Nucleosome Depleted Region)
Although it appears that some of the direct conversion experiments do not require cell division for reprogramming, it is worth emphasizing that forkhead domain-containing factors remain bound to mitotic chromosomes [65], while Pou family members do not [66, 67]. The nuclear exclusion of Oct4 and Sox2 during mitosis, in combination with distinct Oct4-Sox2 stoichiometric requirements [68], which must be reestablished after each cell division, may therefore contribute to the low reprogramming efficiency.
Transcription Factor Induced Chromatin Dynamics and Remodeling
By isolating populations based on the number cell divisions post-OSKM induction, our lab previously reported that initial gene expression dynamics were predominantly observed at loci containing a preexisting euchromatic state as defined by H3K4 methylation and/or H3K27 methylation [51**]. However, a de novo or enhanced gain of H3K4me2 was observed at promoters of many developmental and pluripotency-associated genes prior to detectable transcription originating from these loci, suggesting that reprogramming cells transition through an early state of orchestrated epigenetic priming. The observed dynamics are in agreement with an independent study that found Oct4 to interact with Wdr5, a component of the MLL complex conferring H3K4 methylation [45**].
It appears that only parts of the somatic genome are amenable to immediate epigenetic remodeling as a result of ectopic OSKM expression. This is further supported by the observed correlation between the epigenetic state of the starting cell type and the reactivation of genes that are expressed in pluripotent cells [26]. Taberlay and colleagues found hat putative enhancer regions enriched for the repressive mark H3K27me3 that also contain a NDR nearby, permit Oct4 binding [69**]. Ectopic expression of OCT4 in human fibroblasts confirmed it’s binding to the NDR within the H3K27me3-enriched enhancer of MYOD. This resulted in subsequent gain of H3K4me3 at the promoter while maintaining enrichment of H3K27me3 and transcriptional silencing [69**]. Examining the nucleosome deposition and DNA methylation dynamics during in vitro differentiation in more detail shows that genomic regions silenced via histone modifications remain generally more amenable to TF binding and subsequent activation, while regions that have already gained DNA methylation are typically not reactivated within the same time frame [50*]. Interestingly, multiple-epithelial related genes such as Cdh1 and Ocln that are activated during the early to intermediate stages of reprogramming [70, 71] contain CpG island (CGI)-promoters, and remain free of DNA methylation in MEFs and are instead silenced via H3K27 methylation. This permissive chromatin state explains why certain loci show more dynamic chromatin changes upon induction of OSKM than others [50*, 51**, 69**].
Facilitating the Elimination of Heterochromatin
In order to successfully convert one cell type into another, the original gene expression program needs to be silenced and the new transcriptional program must be established in such a way that it can be stably propagated. To systematically screen for novel factors that may facilitate reprogramming, Singhal et al. conducted a genome-wide shRNA screen and found that ectopic expression of Brg1 and Baf155 had a notable effect [72**]. The results suggest that nucleosome remodeling is a key factor in the cell state transition and that core components of the BAF complex can facilitate the establishment of euchromatin. Ectopic expression of these factors enhanced binding of Oct4 at multiple promoters [72**], which is consistent with the prior observations that its binding is generally limited by heterochromatic regions and DNA methylation [69**] (Figure 1). Recent unrelated DNA methylation studies in mESCs further support the antagonistic relationship between TF binding and DNA methylation on a global scale [73**, 74**]. The notion that TF binding protects regions from DNA methylation has been established by classic studies [75, 76] and is further supported by more recent evidence including FoxD3 binding at the Alb1 enhancer in mESCs which is responsible for maintaining an unmethylated, primed state at this locus [77]. It seems that the creation of NDRs during reprogramming could facilitate loss of DNA methylation at discrete sites by allowing TFs to bind the DNA, consequently protecting regions from further inheritance of DNA methylation during ensuing cell divisions and thereby facilitate eventual activation.
Concluding Remarks
More than three decades of TF mediated reprogramming studies, including hundreds of recent papers focused on OSKM reprogramming, have begun to provide mechanistic insights that make the outcome of ectopic TF expression more predictable. Here, we have summarized selected reports regarding the role of some TFs and the extent to which they can function within a nuclear reprogramming context as illustrated in Figure 1. The distinct abilities of various TFs to induce epigenetic changes including chromatin remodeling have highlighted the need for methodical examination of domain-specific functions, their global- and context-specific abilities to access binding sites, the influence of context-specific co-factors and the presence of epigenetic remodeling complexes. The growing number of detailed epigenetic maps for a broad range of cell types will enable further integration and eventually lead to more targeted manipulations. In time, these analyses will provide the insight necessary to promote rationally designed reprogramming experiments, develop more powerful reprogramming factors and create more advanced methods for the production of desired cell types in vitro.
Acknowledgements
We would like to thank Michael Ziller, Camille Sindhu, Zachary Smith, David Kelley and Cole Trapnell for critical reading of the manuscript. AM is supported by the Pew Charitable Trusts and NIH grants (U01ES017155 and P01GM099117).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 2.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86(14):5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–765. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 4.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 5.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182(4627):64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 6.Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16(21):6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 8.Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415(6875):1035–1038. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- 9.Pereira CF, Terranova R, Ryan NK, Santos J, Morris KJ, Cui W, Merkenschlager M, Fisher AG. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet. 2008;4(9):e1000170. doi: 10.1371/journal.pgen.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. This paper is the seminal study that reported the generation of induced pluripotent stem (iPSCs) using Oct4, Sox2, Klf4 and c-Myc. The factors were identified through an elaborate screen using initially two-dozen ectopically expressed TFs.
- 11.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1(1):55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25(10):1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 13.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322(5903):949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 14.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2(1):10–12. doi: 10.1016/j.stem.2007.12.001. 15. [DOI] [PubMed] [Google Scholar]
- 15.Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5(2):135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, Wang L, Zeng F, Zhou Q. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461(7260):86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 17.Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo- Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480(7378):547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(3):279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439–452. doi: 10.1016/j.cell.2010.12.032. This paper describes epigenetic and transcriptional variation between human pluripotent cell lines (hESC and iPSC). One key conclusion is that ESCs and iPSCs exihibit as much varition between them as among them and therefore no general difference exists between the two types of pluripotent cells. The study then goes on and presents a scorecard metric for assessing ESC/iPSC differentiation potential.
- 20.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481(7381):295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10(6):678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 23.O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21(13):4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149(2):467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins-Taylor K, Schroeder DI, Lasalle JM, Lalande M, Xu RH. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics. 2012;7(1) doi: 10.4161/epi.7.1.18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, Salbert G, Eeckhoute J. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21(4):555–565. doi: 10.1101/gr.111534.110. This report evaluates the ability of specific TFs to remodel chromatin. They find that the gain of FoxA1 binding at enhancers is associated with local DNA demethylation followed by gain of H3K4me1.
- 28.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNARoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. In this paper, the authors performed shRNA-directed knockdown experiments in mESCs to investigate the role that lincRNAs play in the maintenance of pluripotency. Many lincRNAs affect the gene expression program of mESCs when depleted and several directly interact with chromatin modifying complexes.
- 30.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28(10):1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 31.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483(7391):598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 34.Cuesta I, Zaret KS, Santisteban P. The forkhead factor FoxE1 binds to the thyroperoxidase promoter during thyroid cell differentiation and modifies compacted chromatin structure. Mol Cell Biol. 2007;27(20):7302–7314. doi: 10.1128/MCB.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatta M, Cirillo LA. Chromatin opening and stable perturbation of core histone:DNA contacts by FoxO1. J Biol Chem. 2007;282(49):35583–35593. doi: 10.1074/jbc.M704735200. [DOI] [PubMed] [Google Scholar]
- 36.Smale ST. Pioneer factors in embryonic stem cells and differentiation. Curr Opin Genet Dev. 2010;20(5):519–526. doi: 10.1016/j.gde.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105(16):6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13(3):215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 40.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 41.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(7356):390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 42.Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2012;9(3):205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollenberg SM, Cheng PF, Weintraub H. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc Natl Acad Sci U S A. 1993;90(17):8028–8032. doi: 10.1073/pnas.90.17.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11(4):436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 45. Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–197. doi: 10.1016/j.cell.2011.03.003. This study reports a direct interaction of Wdr5 with Oct4 and shows recruitment to its target genes thereby providing a mechanistic explanation for the observed gain of K4 methylation in the early stages of reprogramming (see Ref 51). Ecoptic expression of Wdr5 can improve reprogramming of efficiency of MEFs to iPSCs.
- 46.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8(7):764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 47.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28(4):665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 49.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci U S A. 2011;108(35):14497–14502. doi: 10.1073/pnas.1111309108. This paper utilizes NOMe-seq to study nucleocome positioning and concludes that de novo DNA methylation occurs after nucleosome deposition. They also report that OCT4 can only maintain or create NDRs in the absence of methylated DNA.
- 51. Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8(1):96–105. doi: 10.1016/j.stem.2010.12.001. Using isolated populations based on the number of cell divisions post OSKM induction, this report shows that early gene expression changes (up and down) occur mostly in euchromatic regions. However, many additional loci including key developmental and pluripoetncy related genes show changes in their K4 methylation state prior to any detectable expression.
- 52.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132(6):958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A, Stunnenberg HG. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012 doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Statham AL, Robinson MD, Song JZ, Coolen MW, Stirzaker C, Clark SJ. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIPseq) directly informs methylation status of histone-modified DNA. Genome Res. 2012 doi: 10.1101/gr.132076.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13(7):4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13(7):3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15(5):2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on proteinprotein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17(11):6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17(16):2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jauch R, Aksoy I, Hutchins AP, Ng CK, Tian XF, Chen J, Palasingam P, Robson P, Stanton LW, Kolatkar PR. Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells. 2011;29(6):940–951. doi: 10.1002/stem.639. This paper alters parts of Sox17 which allow it to now dimerize with Oct4 during reprogramming of MEFs. It establishes that TF specificity and function can be tailored by combining atttributes of different factors.
- 61.Nowling TK, Johnson LR, Wiebe MS, Rizzino A. Identification of the transactivation domain of the transcription factor Sox-2 and an associated co-activator. J Biol Chem. 2000;275(6):3810–3818. doi: 10.1074/jbc.275.6.3810. [DOI] [PubMed] [Google Scholar]
- 62.Tsuda M, Takahashi S, Takahashi Y, Asahara H. Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem. 2003;278(29):27224–27229. doi: 10.1074/jbc.M303471200. [DOI] [PubMed] [Google Scholar]
- 63. Hirai H, Tani T, Katoku-Kikyo N, Kellner S, Karian P, Firpo M, Kikyo N. Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells. 2011;29(9):1349–1361. doi: 10.1002/stem.684. The authors attach the transactivation domain (TAD) of MyoD directly to Oct4, which improves the efficiency of reprogramming and demonstrates further that designing TFs for specific functions is possible and effective.
- 64.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136(2):364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan J, Xu L, Crawford G, Wang Z, Burgess SM. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol. 2006;26(1):155–168. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; 64 Roberts SB, Segil N, Heintz N. Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science. 1991;253(5023):1022–1026. doi: 10.1126/science.1887216. [DOI] [PubMed] [Google Scholar]
- 66.Roberts SB, Segil N, Heintz N. Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science. 1991;253(5023):1022–1026. doi: 10.1126/science.1887216. [DOI] [PubMed] [Google Scholar]
- 67.Segil N, Roberts SB, Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254(5039):1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- 68.Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, Creyghton MP, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9(6):588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 69. Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147(6):1283–1294. doi: 10.1016/j.cell.2011.10.040. This report characterizes enhancer remodeling as a result of ectopic expression of MYOD1, and found that enrichment of H3K27me3 at NDR-containing enhancers demarcates enhancers competent for TF binding and subsequent transcriptional activation.
- 70.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMPdriven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7(1):64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 71.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 72. Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141(6):943–955. doi: 10.1016/j.cell.2010.04.037. Various components of the SWI/SNF machinery were ectopically expressed during reprogramming to show that enhanced nucleosome remodeling capabilities could improve reprogramming efficiency.
- 73. Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43(11):1091–1097. doi: 10.1038/ng.946. This paper systematically inserted various promoter sequences into a defined mESC locus, and describes the minimal features to maintain and/or establish correct DNA methylation patterns during differentiation. These methylation determining regions (MDRs) are combining several features such as CG density and TF binding sites in line with earlier studies from the Bird and Cedar labs (Refs 75 and 76).
- 74. Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. This report utilizes whole genome bisulfite sequencing and highlights CpG poor regions within distal regulatory elements that exhibit low DNA methylation levels (LMRs). Consistent with their other study, they find these LMRs to contain TF binding motifs and additionally report that ectopic expression of a TF can induce the creation of a LMR in specific sequence contexts.
- 75.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8(19):2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 76.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371(6496):435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 77.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissuespecific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23(24):2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A. 2012;109(7):2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455(7213):627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henke RM, Meredith DM, Borromeo MD, Savage TK, Johnson JE. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev Biol. 2009;328(2):529–540. doi: 10.1016/j.ydbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smit DJ, Smith AG, Parsons PG, Muscat GE, Sturm RA. Domains of Brn-2 that mediate homodimerization and interaction with general and melanocytic transcription factors. Eur J Biochem. 2000;267(21):6413–6422. doi: 10.1046/j.1432-1327.2000.01737.x. [DOI] [PubMed] [Google Scholar]
- 82.Walhout AJ, Gubbels JM, Bernards R, van der Vliet PC, Timmers HT. c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res. 1997;25(8):1493–1501. doi: 10.1093/nar/25.8.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Williams SC, Cantwell CA, Johnson PF. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991;5(9):1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 84.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 85.Bogan AA, Dallas-Yang Q, Ruse MD, Jr, Maeda Y, Jiang G, Nepomuceno L, Scanlan TS, Cohen FE, Sladek FM. Analysis of protein dimerization and ligand binding of orphan receptor HNF4alpha. J Mol Biol. 2000;302(4):831–851. doi: 10.1006/jmbi.2000.4099. [DOI] [PubMed] [Google Scholar]
- 86.Bridwell JA, Price JR, Parker GE, McCutchan Schiller A, Sloop KW, Rhodes SJ. Role of the LIM domains in DNA recognition by the Lhx3 neuroendocrine transcription factor. Gene. 2001;277(1–2):239–250. doi: 10.1016/s0378-1119(01)00704-1. [DOI] [PubMed] [Google Scholar]
- 87.Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003;23(17):6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Olson EN, Perry M, Schulz RA. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172(1):2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 89.Mitsui K, Shirakata M, Paterson BM. Phosphorylation inhibits the DNA-binding activity of MyoD homodimers but not MyoD-E12 heterodimers. J. Biol Chem. 1993;268(32):24415–24420. [PubMed] [Google Scholar]
- 90.Vosper JM, Fiore-Heriche CS, Horan I, Wilson K, Wise H, Philpott A. Regulation of neurogenin stability by ubiquitin-mediated proteolysis. Biochem J. 2007;407(2):277–284. doi: 10.1042/BJ20070064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Breslin MB, Wang HW, Pierce A, Aucoin R, Lan MS. Neurogenin 3 recruits CBP co-activator to facilitate histone H3/H4 acetylation in the target gene INSM1. FEBS Lett. 2007;581(5):949–954. doi: 10.1016/j.febslet.2007.01.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y, MacDonald RJ, Swift GH. DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas-specific basic helix-loop-helix complex. J Biol Chem. 2001;276(21):17985–17993. doi: 10.1074/jbc.M100678200. [DOI] [PubMed] [Google Scholar]
- 93.Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28(3):276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]