Abstract
Influenza causes more than 250,000 deaths annually in the industrialized world and bacterial infections frequently cause secondary illnesses during influenza outbreaks, including pneumonia, bronchitis, sinusitis, and otitis media. Here we demonstrate that cross-reactive immunity to mismatched influenza strains can reduce susceptibility to secondary bacterial infections, even though this fails to prevent influenza infection. Specifically, infecting mice with H3N2 influenza before challenging with mismatched H1N1 influenza reduces susceptibility to either gram-positive Streptococcus pneumoniae or gram-negative Klebsiella pneumoniae. Vaccinating mice with the highly conserved nucleoprotein of influenza also reduces H1N1-induced susceptibility to lethal bacterial infections. Both T cells and antibodies contribute to defense against influenza-induced bacterial diseases; influenza cross-reactive T cells reduce viral titers, whereas antibodies to nucleoprotein suppress induction of inflammation in the lung. These findings suggest that non-neutralizing influenza vaccines that fail to prevent influenza infection may nevertheless protect the public from secondary bacterial diseases when neutralizing vaccines are not available.
INTRODUCTION
Secondary bacterial infections often follow influenza infection and can lead to a variety of illnesses including pneumonia, bronchitis, sinusitis and otitis media (1, 2). Secondary bacterial pneumonia is a particularly serious consequence of influenza infection. It was the primary cause of death during the 1918 influenza pandemic (3) and was associated with significantly higher morbidity and mortality during the 2009 pandemic (4).
Vaccines are the mainstay of public health efforts to prevent influenza epidemics. These vaccines aim to prevent infection by eliciting neutralizing antibodies that bind the hemagglutinin and neuraminidase proteins on the surface of influenza virions. Unfortunately, mutations and reassortments in the surface proteins of influenza viruses allow new strains to emerge and evade neutralizing antibodies (5, 6). Consequently, each year, new vaccines are produced to “match” the most dangerous contemporary strains.
In animal models, mismatched influenza vaccines can prime non-neutralizing immunity that speeds viral clearance and reduces mortality, despite failing to prevent infection (7, 8). Mismatched vaccines may not prevent human pandemics, but they might lessen their severity when matched vaccines are not available. Indeed, several studies suggest humans may benefit from non-neutralizing immunity to influenza (9–12). A recent study demonstrated that the presence of influenza-specific memory in humans correlates with non-neutralizing immunity that significantly reduces the severity of illness (13). These researchers postulated that CD4 T cells confer protection by improving antibody responses to conserved internal viral proteins (13). Unfortunately, many factors confound the interpretation of human studies of influenza and public health campaigns to date have largely neglected the potential for non-neutralizing immunity to combat influenza outbreaks or the associated increase in secondary bacterial infections.
Data from mouse models suggest influenza infection increases susceptibility to secondary bacterial infections by suppressing neutrophil function, decreasing mucociliary flow, desensitizing innate immunity, and creating favorable environments for bacterial adherence and colonization (1). Cytokines, including interleukins and interferons, also affect susceptibility (14–16), suggesting that ongoing immune responses to influenza may facilitate bacterial colonization of the lung. Here, we investigate whether non-neutralizing, mismatched immunity to influenza impacts susceptibility to secondary bacterial infections.
MATERIALS AND METHODS
Mice
Wild type and B cell-deficient μMT C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Antibody-deficient AID.uS C57BL/6 mice were described previously (17, 18). All mice were bred in the specific pathogen free Trudeau Institute Animal Breeding Facility after embryo rederivation. Experimental mice were matched for age and sex, and cared for according to Trudeau Institute guidelines. Recumbent mice, and mice that lost more than 30% weight, were considered moribund and euthanized.
Viruses
In uenza virus A/HKx31 (H3N2), in uenza virus A/PR/8/34 (H1N1), cold-adapted influenza virus c.a.A/Alaska/72/CR9 (caH3N2) and the Enders strain of Sendai virus were grown, stored, and titered as previously described (19–21). Influenza infections and vaccinations were administered intranasally to anesthetized mice using 3000 EID-50 for H3N2, 400 EID-50 for H1N1, 350 TCID-50 for caH3N2, and 250 EID-50 for Sendai virus. The viral burden and level of inflammatory cytokines and chemokines in whole lung tissue was determined by real-time PCR measuring acid polymerase copy number (22).
Bacteria
Serotype 4 Streptococcus pneumoniae (ATCC strain 6304) grown on blood agar plates was used to inoculate Tryptic soy broth cultures, which were grown at 37°C without shaking in sealed tubes. After dilution to an OD600nm of 0.15, they were re-grown to an OD600nm of 0.45, washed with saline, and approximately 250 CFU were applied in a volume of 50 μl saline to the nares of lightly anesthetized mice. The number of bacteria in the inoculating dose was confirmed by plating. The intranasal median lethal dose of strain 6304 is approximately 1.5×104 CFU when grown as described above and administered to naïve mice. Additional studies also employed serotype 3 S. pneumoniae strain URF918 (23) and Klebsiella pneumoniae strain IA565 (24). Innocula of strain URF918 were prepared as described for ATCC strain 6304. Working stocks of strain IA565 were prepared by growing in Tryptic/Soy Broth to log phase (OD600=0.900), adding 20% glycerol and freezing aliquots at −70°C. To prepare inocula, 100 μl of working stock was streaked on Tryptic/Soy agar plates, grown overnight at 37°C, and scraped to seed Tryptic/Soy Broth cultures at OD600=0.050. Cultures were grown in 37°C incubator/shaker at 180rpm for approximately 2hr to log phase (OD600=0.900), spun down, and resuspended in PBS to a concentration of 108 CFU per 50 μl infection dose.
Treatments
T cell depletions were performed as described previously (19). The depletion protocols removed more than 90% of the targeted cells from spleen and bronchoalveolar lavage fluid, as determined by flow cytometric analyses of antibody-treated animals that were euthanized at day 5 after H1N1 infection (not shown). Recombinant A/PR/8/34 influenza NP was generated as a C terminal histidine-tagged protein in E. coli and isolated using the ProBond system (Invitrogen), as described (17). Immunizations contained 30 μg NP and employed 20 μg E. coli serotype 0111:B4 lipopolysaccharide (LPS; Enzo Life Sciences) plus alum as adjuvant (17). H3N2 immune serum was collected 21 days after infection with H3N2 and 350 ul was transferred to naïve mice by intraperitoneal injection on the day prior to H1N1 challenge. Passive immunization with mouse IgG2a NP-specific mAb H16-L10-4R5/HB-65 (25) was achieved by administering 350 ug intraperitoneal injections on the day of and the day prior to H1N1 challenge. Control mice received serum from naïve mice or isotype matched mAb C1.18.4. All mAb were Protein G purified and supplied by BioXcell, who reported <2 endotoxin units per mg.
Statistics
Survival curves were analyzed by Log rank tests. CFU and viral titer data that fell below the limit of detection were assigned a value below that limit and, thus, were analyzed by non-parametric Mann Whitney or Kruskal Wallis tests. Bacteremia was scored positive or negative and analyzed by Chi-square tests.
RESULTS
Non-neutralizing immunity to influenza protects from secondary bacterial infection
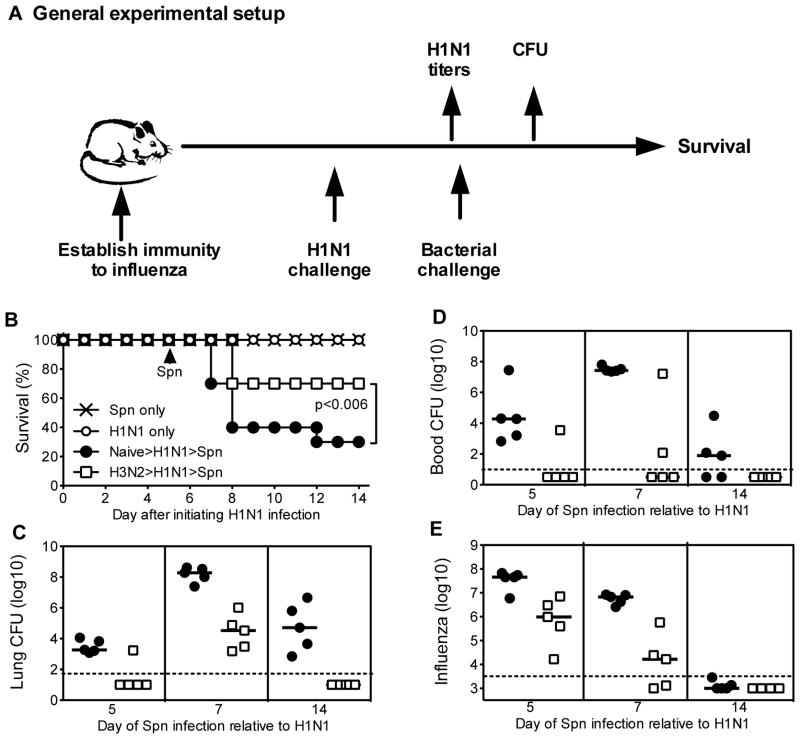
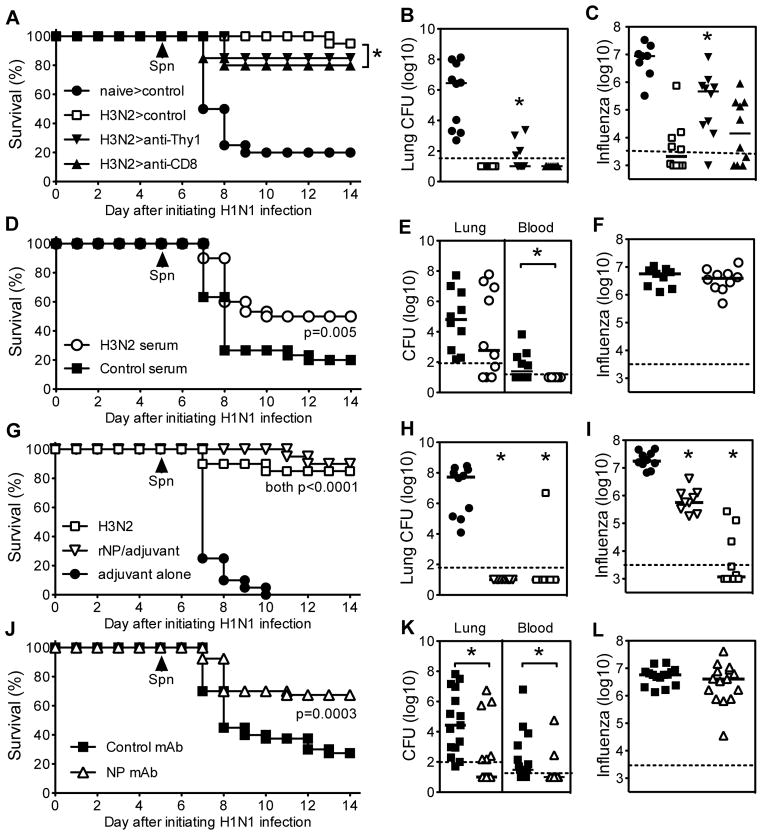
Fig 1A depicts our general experimental approach to assessing the impact of prior immunity to influenza on susceptibility to secondary bacterial infection. Naive mice readily survived low dose intranasal challenge with 250 CFU of S. pneumoniae (Fig. 1B). Naive mice also survived low dose intranasal challenge with H1N1 influenza (Fig. 1B). However, consistent with prior reports (26), we observed that mice succumbed to bacterial infection when challenged with low dose S. pneumoniae following a sublethal influenza challenge (Fig. 1B). To investigate the impact of non-neutralizing, mismatched immunity to influenza, we infected mice with low dose H3N2 influenza, challenged 5–6 months later with low dose H1N1 influenza, and then measured susceptibility to S. pneumoniae. We observed that prior exposure to H3N2 influenza improved survival (Fig. 1B), reduced pneumococcal colonization of lung tissue (Fig. 1C), and largely prevented bacteremia (Fig. 1D). Notably, susceptibility to bacterial infection did not simply correlate with viral titers at the time of challenge. For example, mice challenged with S. pneumoniae on days 5 and 14 after H1N1 infection exhibited similar bacterial burden (Fig. 1C), despite more than a 10,000-fold difference in viral titers at those time points (Fig. 1E).
Figure 1. Long-term cross-reactive immunity to influenza protects from secondary bacterial infection.
(A) General experimental approach followed for all studies; see each figure legend for specifics of treatments and timing. For B–E, C57BL/6 mice were infected with H3N2 influenza or left uninfected. After 5–6 months, the mice were challenged with H1N1 influenza, followed 5, 7, or 14 days later by infection with S. pneumoniae (Spn). (B) Survival of mice challenged with Spn on day 5 after H1N1 infection (n=10 mice/group). Mice infected previously with H3N2 showed significantly greater survival than control mice (p=0.006 by Log rank test). (C) Bacterial burden in the lung 24 hours after Spn infection. Mice exposed previously to H3N2 harbored significantly fewer bacteria than control mice when both groups were infected with Spn at days 5, 7, and 14 after H1N1 infection (all p<0.04 by Mann Whitney test). Although susceptibility peaked at day 7, we focused survival studies on day 5 because H1N1-infected naïve mice showed significantly greater weight loss than H3N2 immune mice on days 7 and 14, but not on day 5 (not shown). (D) Bacterial burden in the blood 24 hours after Spn infection. (E) Influenza burden at the time of Spn infection. Mice exposed previously to H3N2 harbored significantly less virus than control mice at days 5 and 7 after H1N1 infection (both p<0.02 by Mann Whitney test). The day 5 data and day 7/14 data were collected in two separate H1N1/Spn challenge studies using a single cohort of H3N2 exposed animals; dotted lines depicts limit of detection.
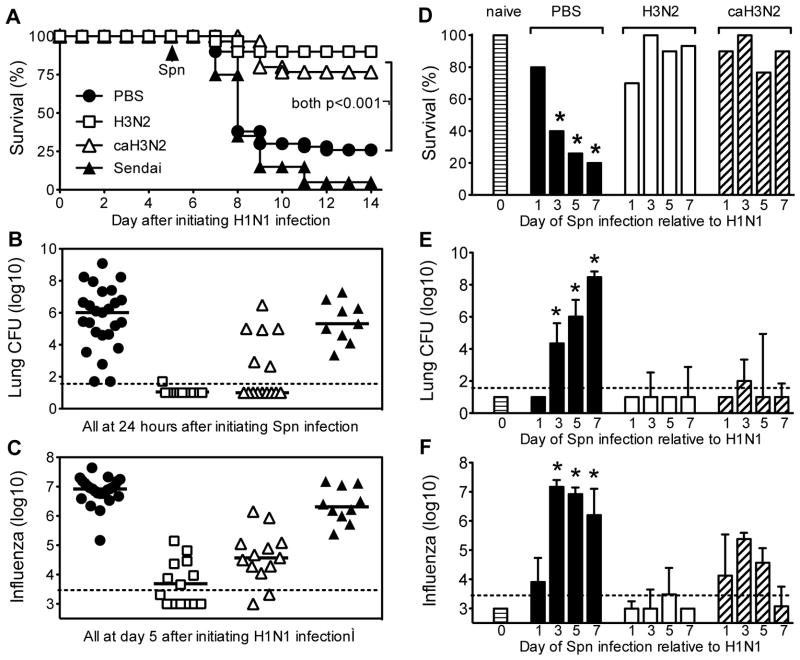
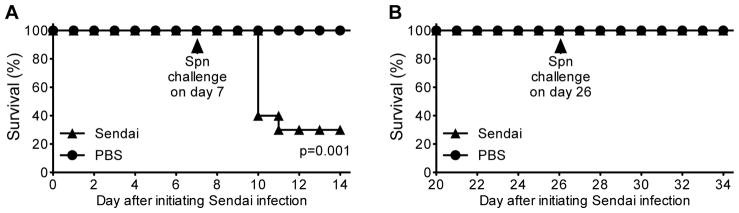
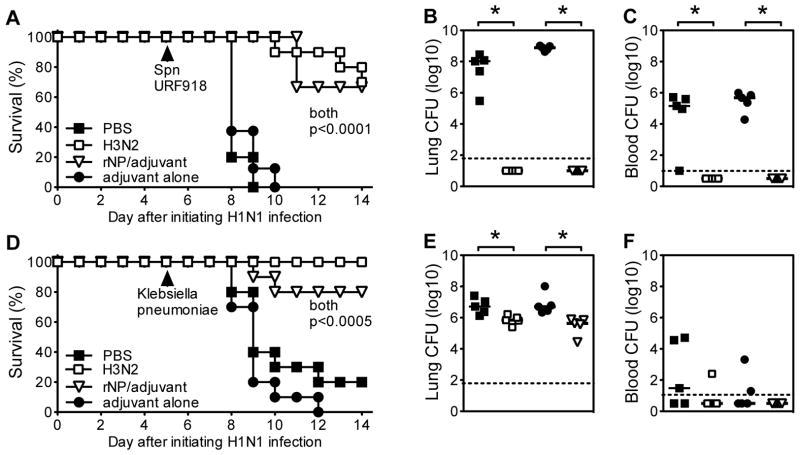
To investigate the specificity of the H3N2-induced protection from secondary bacterial infection, we evaluated protection conferred by Sendai virus, a parainfluenza virus that causes an acute pulmonary infection similar to influenza, but does not prime cross-reactive immunity to influenza (20). In parallel, we examined protection conferred by a cold-adapted H3N2 (caH3N2) vaccine strain (19, 21, 27, 28). We found that exposure to either the H3N2 influenza virus or the live attenuated caH3N2 vaccine protected against H1N1-induced susceptibility to pneumococcal infection as early as 3 weeks after exposure (Fig. 2A). The protection was associated with reduced bacterial burden in the lungs (Fig. 2B) and reduced H1N1 titers (Fig. 2C). In contrast, prior exposure to Sendai virus had no significant impact on H1N1-induced susceptibility to pneumococcal infection (Fig. 2A). Infection with Sendai virus, like H1N1 infection, induced susceptibility to S. pneumoniae when challenged on day 7 (29), but this susceptibility waned by 26 days after infection (Fig. 3A and B), indicating that residual impacts of primary Sendai infection did not account for the pneumococcal susceptibility observed when Sendai-exposed mice were infected with H1N1 influenza. Thus, specific cross-reactive immunity to influenza, not just conditioning of the lung by any viral infection, reduces susceptibility to secondary bacterial infection. Importantly, the cross-reactive immunity to influenza reduced susceptibility to diverse types of bacterial infections: the bacterial challenge studies described above used a serotype 4 strain of gram-positive S. pneumoniae but similar results were observed when mice were challenged with serotype 3 S. pneumoniae (Fig. 4A, B, C) or Klebsiella pneumoniae (Fig. 4D, E, F), a gram-negative bacterium.
Figure 2. Short-term cross-reactive immunity to influenza specifically protects from secondary bacterial infection.
C57BL/6 mice were infected intranasally with H3N2 influenza, Sendai virus or attenuated cold-adapted H3N2 influenza (caH3N2); controls were mock infected with saline (PBS) or left untreated (naïve). After 21 days, mice were challenged intranasally with H1N1 influenza, followed by Spn 5 days later (A–C), or 1, 3, 5, or 7 days later (D–F). (A) Survival (n=20 mice/group for Sendai, 30 for H3N2, 30 for caH3N2, and 50 for PBS; data is pooled from three independent experiments). Mice infected with H3N2 or caH3N2, but not mice infected with Sendai virus, showed significantly greater survival than PBS-treated mice (p<0.001 by Log rank test). (B) Bacterial burden in the lung 24 hours after Spn infection and (C) influenza burden at the time of Spn infection. Mice infected with H3N2 or caH3N2, but not mice infected with Sendai virus, showed significantly reduced bacterial and influenza burden as compared with PBS-treated mice (p<0.001 by Kruskal Wallis test). In B/C, each symbol depicts data for an individual mouse; bar depicts group median; dotted line depicts limit of detection. (D) Survival at day 14 after Spn infection (n=10 or more mice/group). * indicates p<0.05 compared with naïve using Fisher’s exact test. (E) Bacterial burden in lung 24 hours after Spn infection and (F) influenza burden at the time of Spn infection. In E/F, bars depict median and interquartile range (n=5 or more mice/group); dotted line depicts limit of detection. * indicates p<0.01 by Kruskal Wallis test when comparing data from each day with the naïve mice challenged with Spn.
Figure 3. Sendai virus infection induces susceptibility to secondary bacterial infection but the susceptibility wanes by day 26 after infection.
In Figure 2A–C, we demonstrated that exposure to Sendai virus, unlike exposure to H3N2 influenza, does not reduce the capacity of a subsequent (21 days later) H1N1 infection to induce susceptibility to S. pneumoniae (Spn) on day 5 after the H1N1 infection. To demonstrate that Sendai virus itself was not causing the observed susceptibility to Spn at 26 days after prior Sendai virus infection, C57BL/6 mice were infected with Sendai virus or treated with PBS vehicle and then challenged with Spn after 7 (A) or 26 days (B). Consistent with prior report (29), Sendai virus infection increased susceptibility to Spn significantly when mice were challenged on day 7 (p=0.001 by Log rank test; n=10 mice/group). However, this susceptibility was no longer evident when they were challenged on day 26.
Figure 4. Cross-reactive immunity to influenza protects from secondary infection with both gram-positive and gram-negative bacteria.
(A–C) C57BL/6 mice were infected with H3N2 influenza or immunized intraperitoneally with recombinant NP (rNP) using LPS/alum adjuvant; controls were mock immunized with PBS or adjuvant alone, respectively. After 21 days, mice were challenged intranasally with H1N1 influenza, followed 5 days later with serotype 3 S. pneumoniae (Spn) strain URF918 (23). (A) Survival (n=10 mice/group). (B) Bacterial burden in the lung 48 hours after Spn infection. (C) Bacterial burden in the blood 48 hours after Spn infection. Infection with H3N2 or immunization with rNP significantly increased survival (both p<0.0001 by Log rank tests) and decreased bacterial burden in lung and blood (all p=0.008 by Mann Whitney tests). (D–F) C57BL/6 mice were infected with H3N2 influenza or immunized intraperitoneally with recombinant NP (rNP) using LPS/alum adjuvant; controls were mock immunized with PBS or adjuvant alone, respectively. After 21 days, mice were challenged intranasally with H1N1 influenza, followed 5 days later with Klebsiella pneumoniae clinical isolate strain IA565 (24). (D) Survival (n=10 mice/group). (E) Bacterial burden in the lung 48 hours after Spn infection. (F) Bacterial burden in the blood 48 hours after Spn infection. Infection with H3N2 or immunization with rNP significantly increased survival (both p<0.0005 by Log rank tests), and significantly decreased bacterial burden in lung (p=0.02 and 0.008, respectively, by Mann Whitney tests).
It has been shown that non-neutralizing immunity to influenza can accelerate viral clearance (7, 8, 30). Thus, preexisting immunity to influenza may have shifted the period of H1N1-induced susceptibility to pneumococcal infection, such that mice became susceptible prior to day 5 after H1N1 challenge. To investigate this possibility, we examined the kinetics of susceptibility in greater detail. We found that mice were susceptible to H1N1-induced pneumococcal infection when bacteria were administered on days 3, 5 and 7, but not day 1, after H1N1 infection, and that prior infection or vaccination with H3N2 suppressed pneumococcal susceptibility at these same times (Fig. 2D). Again, susceptibility correlated with increased bacterial burden (Fig. 2E) and higher viral titers (Fig. 2F). Thus, prior exposure to H3N2 influenza did not accelerate the time of susceptibility. Rather, preexisting mismatched immunity to influenza reduced overall susceptibility to pneumococcal infection.
Cross-reactive T cells and antibody contribute to protection
Cross-reactive CD8 T cells can facilitate non-neutralizing protection against mismatched influenza strains (7, 8), and influenza cross-reactive memory T cells produce interferon-gamma, one of the cytokines that contributes to H1N1-induced susceptibility to pneumococcal infection (15). Thus, cellular immunity to influenza might be predicted to exacerbate susceptibility to pneumococcal disease. However, depletion of all T cells (anti-Thy1 treatment) or depletion of only CD8 T cells from H3N2-immune mice immediately prior to H1N1 challenge modestly diminished the protection conferred by prior exposure to H3N2 (Fig. 5A) and slightly elevated both the bacterial burden (Fig. 5B) and viral titer (Fig. 5C). Thus, the presence of cross-reactive memory T cells did not exacerbate pneumococcal infection and, rather, contributed to cross-reactive defense against bacterial disease, at least in part, by reducing H1N1 titers.
Figure 5. Cross-reactive T cells and antibody both contribute to protection from secondary bacterial infection.
(A–C) C57BL/6 mice were infected with H3N2 influenza or left untreated (naïve). On day 21, mice were challenged intranasally with H1N1 influenza, followed 5 days later with Spn. On days 20 and 22, mice were treated with Thy1 mAb to deplete all T cells or CD8 mAb to deplete CD8 T cells; controls received a rat IgG2b control mAb. (A) Survival (n=20 mice/group; data pooled from two independent studies). (B) Bacterial burden in the lung 24 hours after Spn infection and (C) influenza burden at the time of Spn infection. In comparison with H3N2 infected mice treated with control mAb, mice treated with anti-CD8 exhibited significantly decreased survival (p=0.04 by Log rank test), and mice treated with anti-Thy1 showed significantly increased bacterial and viral burden (both p<0.05 by Kruskal Wallis test comparing all H3N2 infected mice). (D–F) C57BL/6 mice received passive immunizations with H3N2-immune serum or control serum. The next day, they were challenged intranasally with H1N1 influenza. After 5 days, all mice were challenged with Spn. (D) Survival (n=30 mice/group; data pooled from three independent studies). (E) Bacterial burden in the lung and blood 24 hours after Spn infection, and (F) influenza burden at the time of Spn infection. Passive immunization with H3N2-immune serum significantly increased survival (p=0.005 by Log rank test) and decreased the incidence of bacteremia (p=0.003 by Chi-square test; n=10 mice/group), but did not significantly impact pulmonary bacterial or viral burden. (G–I) C57BL/6 mice were infected with H3N2 influenza or immunized intraperitoneally with recombinant NP (rNP) using LPS/alum adjuvant; controls were mock immunized with adjuvant alone. After 21 days, mice were challenged intranasally with H1N1 influenza, followed 5 days later with Spn. (G) Survival (n=20 mice/group; data pooled from two independent studies). (H) Bacterial burden in the lung 24 hours after Spn infection. (I) Influenza burden at the time of Spn infection. Infection with H3N2 or immunization with rNP significantly increased survival (both p<0.0001 by Log rank tests), decreased bacterial burden (both p<0.001 by Kruskal Wallis test), and decreased viral burden (p<0.001 and p<0.05, respectively by Kruskal Wallis test). (J–L) C57BL/6 mice were passively immunized with NP-specific mAb or isotype-matched control mAb (Mouse IgG2a) and then challenged intranasally with H1N1 influenza. After 5 days, all mice were challenged with Spn. (J) Survival (n=40 mice/group; data pooled from four independent studies). (K) Bacterial burden in the lung and blood 24 hours after Spn infection. (L) Influenza burden at the time of Spn infection. Passive immunization with NP mAb significantly increased survival (p=0.0003 by Log rank test), decreased the pulmonary bacterial burden (p=0.004 by Mann Whitney; n=15 mice/group) and incidence of bacteremia (p=0.003 by Chi-square test; n=15 mice/group), but did not significantly impact viral burden.
Antibodies to conserved viral proteins also contribute to cross-reactive immunity to influenza (7, 17, 18, 31, 32). To test the role of antibody in defense against H1N1-induced pneumococcal susceptibility, we administered H3N2-immune serum or control serum to naïve mice prior to infection with H1N1 influenza. The mismatched H3N2-immune serum significantly decreased susceptibility to secondary pneumococcal infection (Fig. 5D). Despite conferring significant protection from lethality, passive immunization with mismatched serum did not significantly reduce bacterial burden (Fig. 5E) or viral titers (Fig. 5F) in the lung. However, H3N2-immune serum did significantly reduce levels of bacteremia and the number of mice with detectable bacteria in blood cultures (Fig. 5E).
Influenza cross-reactive non-neutralizing immunity typically recognizes conserved, internal proteins of influenza, and immunity to nucleoprotein (NP), a highly conserved internal protein, confers significant protection from lethal influenza challenge (7, 8, 17, 18, 32–38). We observed that vaccinating mice with purified recombinant NP conferred robust protection from H1N1-induced susceptibility to secondary pneumococcal infection (Fig. 5G). NP vaccination markedly reduced pneumococcal burden in the lung (Fig. 5H), despite only modestly reducing viral titers (Fig. 5I). NP vaccination also significantly reduced H1N1-induced susceptibility to Klebsiella pneumoniae and serotype 3 S. pneumoniae (Fig. 4).
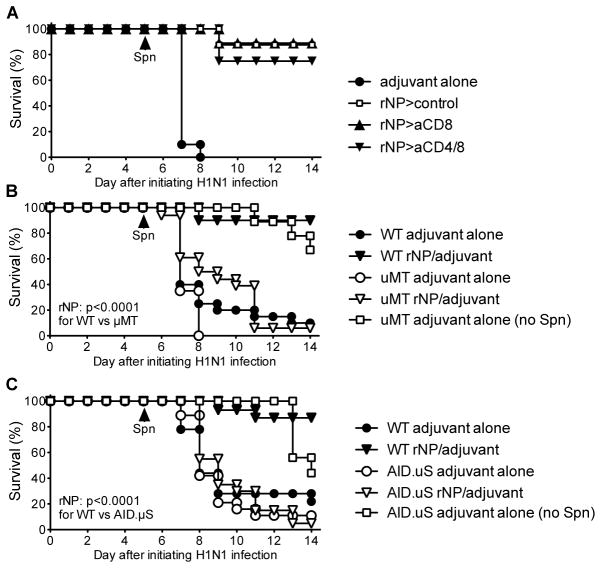
Depletion of CD8 T cells alone, or both CD4 and CD8 T cells, at the time of H1N1 challenge did not significantly impair the protection conferred by NP vaccination (Fig. 6A), suggesting the involvement of alternative effector mechanisms. Other studies have demonstrated that antibodies to NP can confer significant protection from lethal influenza challenge (17, 18, 32). Consistent with a critical role for antibodies, NP vaccination poorly protected B cell-deficient μMT mice and antibody-deficient AID/μS mice from death following influenza and S. pneumoniae infections (Fig. 6B and C). To definitively assess the protective capacity of NP-specific antibodies, we passively transferred NP-specific mAb into naïve mice and then challenged with influenza followed by S. pneumoniae. Remarkably, administration of NP-specific mAb conferred robust protection from H1N1-induced secondary pneumococcal disease (Fig. 5J). As with passive immunization with H3N2-immune serum, NP-specific mAb did not significantly reduce viral titers in the lung (Fig. 5L), but did reduce the number of bacteremic mice and levels of bacteremia (Fig. 5K). Notably, passive immunization with NP-specific mAb did not did not suppress bacterial infection directly since it did not affect the median lethal dose of S. pneumoniae in naïve mice (data not shown) and only reduced pneumococcal lethality in mice infected previously with influenza.
Figure 6. The immunity to influenza NP that protects from secondary bacterial infection is compromised in mice lacking B cells or circulating antibody, but not T cells.
(A) Wild type mice were immunized intraperitoneally with rNP using LPS/alum adjuvant; controls were mock immunized with adjuvant alone. After 21 days, mice were challenged intranasally with H1N1 influenza, followed 5 days later with Spn. On days 20 and 22, mice were treated with CD8 mAb or a combination of CD4 and CD8 mAb; controls received a rat IgG2b control mAb. Survival was not compromised significantly in mice treated with CD8 or CD4 and CD8 mAb (n=8–10 mice/group). (B/C) Wild type (WT) and B cell-deficient μMT mice (B) or circulating antibody-deficient AID.μS mice (C) were immunized intraperitoneally with rNP using LPS/alum adjuvant; controls were mock immunized with adjuvant alone. After 21 days, mice were challenged intranasally with H1N1 influenza, followed 5 days later with Spn. Among mice immunized with rNP, WT mice showed significantly increased survival when compared to either μMT or AID.μS mice (both p<0.0001 by Log rank tests; n=10 mice/group).
Immunity to NP reduces lung inflammation
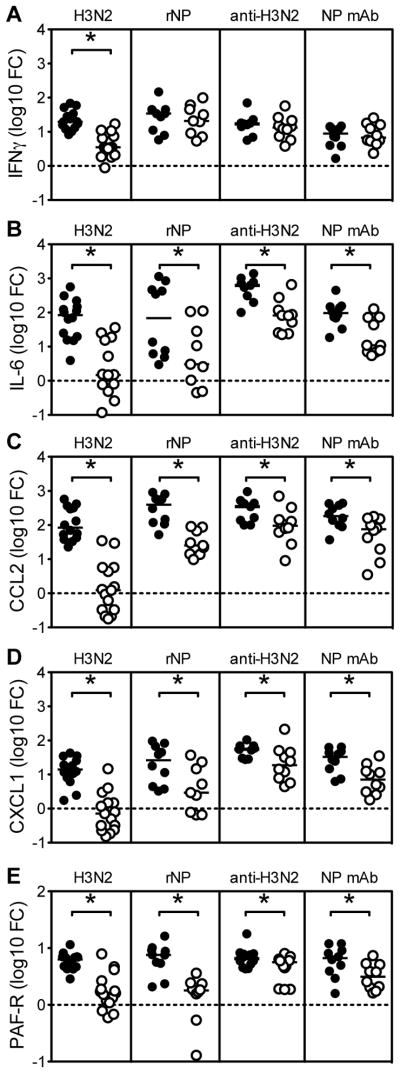
NP-specific antibodies can help to reduce H1N1 titers in lethal influenza challenge models (17, 18, 31, 32), but did not appear to reduce H1N1 titers in our model of sublethal influenza challenge. Antibodies are known to play diverse roles during host defense, including the suppression of inflammation (39). Indeed, in our studies, passive immunization with NP-specific mAb markedly suppressed levels of inflammatory cytokines and chemokines in lung tissue of H1N1-infected mice, including IL-6, CCL2/MCP-1, and CXCL1/KC (Fig. 7B, C and D). Notably, levels of interferon-gamma were not affected (Fig. 7A). Treatment with H3N2 serum likewise reduced levels of pulmonary inflammation, as did active vaccination with H3N2 virus or NP (Fig. 7B, C and D). In addition, the non-neutralizing immunity induced by infection, immunization or antibody transfer also significantly reduced the expression of platelet activating factor receptor (Fig 7E). Elevated expression of platelet activating factor receptor has been associated with enhanced inflammation and lung pathology during influenza infection (40, 41). These findings suggest non-neutralizing immunity, and cross-reactive NP-specific antibodies in particular, may reduce susceptibility to bacterial infection by reducing inflammation and pathological damage to lung epithelium, events that facilitate colonization of the lung by bacteria (1).
Figure 7. Immunity to NP suppresses induction of inflammatory cytokines and chemokines.
Levels of mRNA encoding interferon-gamma (A), IL-6 (B), CCL2 (C), and CXCL1 (D) in lung tissue from C57BL/6 mice infected with H1N1 influenza for 5 days. Graphs show the fold change (FC) from uninfected controls. As indicated, mice were previously exposed to H3N2 influenza, vaccinated with rNP, passively immunized with H3N2 serum, or passively immunized with NP mAb (open circles). Control mice (closed circles) were previously exposed to PBS (control for H3N2), vaccinated with adjuvant alone (control for rNP), passively immunized with non-immune serum (control for H3N2 serum), or passively immunized with irrelevant mouse IgG2a mAb (control for NP mAb). Active and passive immunity to NP significantly suppressed levels of mRNA encoding IL-6, CCL2, and CXCL1, but not interferon-gamma (all p<0.05 by student’s t test versus respective controls).
DISCUSSION
Secondary bacterial infections are a common complication of influenza infection and cause significant morbidity and mortality (1, 2). Because of this important clinical problem, we have employed a murine model to examine the influence of influenza infection on susceptibility to secondary bacterial infections and how this can be prevented. Using this model, we have shown that the susceptibility to secondary infection applies to several strains of bacteria and results in significant lung colonization, bacterial dissemination and death. Consistent with prior studies (1, 14–16), we also found that the window of susceptibility to secondary infection begins at day 3 of influenza infection and extends until at least day 14, when the vast majority of virus has been cleared. These results suggest that susceptibility to secondary infection is not just due to influenza infection but may also be associated with the immune response to the virus, including inflammation in the lungs. In fact, it has been postulated that the extensive mortality observed in the 1918 influenza pandemic was the result of extensive immunopathology that increased susceptibility to secondary bacterial pneumonia (42).
This model has also allowed us to explore how this susceptibility to secondary infections can be overcome. The most common way to manipulate immunity to influenza is via vaccination. Neutralizing immunity to influenza, which is the goal of the yearly influenza vaccine, can prevent infection and illness. While this is the most desirable goal of vaccination, it is not always possible due to rapid antigenic changes in the virus or the appearance of new viral strains. Prior studies have demonstrated that non-neutralizing immunity can reduce influenza illness and mortality in mice. Our observations demonstrate that non-neutralizing immunity to influenza also confers remarkable protection from secondary bacterial infections. Non-neutralizing immunity can be conferred by mismatched live attenuated vaccines (such as Flumist) or by prior influenza infection (19, 43). It can also be conferred by vaccination with conserved internal proteins from the influenza virus, such as NP (17, 18, 32). This non-neutralizing immunity protects from secondary bacterial infection by reducing lung colonization, bacterial dissemination and death.
Our results demonstrate that antibody is a main effector mechanism of the non-neutralizing immunity that protects from bacterial infection. Vaccination of mice that lack B cells or secreted antibody could not confer protection from secondary bacterial infection. In contrast, passive transfer of serum collected from mice that had been previously infected with a mismatched, heterosubtypic influenza strain, or a monoclonal antibody to influenza NP, could protect from secondary bacterial infection. The presence of non-neutralizing antibodies was associated with significant reduction in inflammatory molecules in the lungs of influenza-infected mice. As discussed in a recent review (45), the ability of antibodies to modulate inflammation during an immune response has been appreciated for a long while. Under certain conditions, antibodies can dampen the inflammatory response and they have been used clinically as an anti-inflammatory agent. While the mechanism of this action most likely involves Fc receptors, precisely how this activity works has yet to be elucidated. Importantly, the use of prophylactic antibody treatment to reduce an inflammatory response is not unprecedented in the clinic (46, 47).
The efficient generation of high affinity NP-specific antibodies presumably requires CD4 T helper cells specific for influenza NP (48). A recent study in humans demonstrated an important role for CD4 T cells specific for internal influenza proteins in protection from severe influenza-induced illness (13). In individuals with preexisting CD4 T cells with specificities for NP or matrix protein, there was less illness following influenza challenge. The CD4 T cells identified following viral challenge could respond to a number of different influenza strains, indicating that they would be useful during a mismatched, heterosubtypic infection. While it could not be formally demonstrated in these human studies, the authors speculated that these influenza-specific CD4 T cells were exerting this protective influence by acting as helper cells for a humoral response directed at internal influenza proteins. Moreover, in concordance with our findings, the authors of the human study proposed that the reduction in illness resulted from a reduction in immunopathology, which should result in a reduced susceptibility to secondary bacterial infections (13).
While antibody could suffice to provide significant protection in our mouse model, more robust protection was observed after H3N2 infection (Fig 3A) or rNP vaccination (Fig 3G), suggesting that additional components of the immune system, presumably T cells, also contributed to an optimal protective response. Indeed, we observed that depleting T cells modestly impacted H1N1 titers (Fig 5C), bacterial burden (Fig 5B), and survival (Fig 5A and 6A). Another study using a similar mouse model of secondary bacterial infection recently suggested dominant protective roles for CD4 T cells (44). Specifically, that study demonstrated that seasonal FluMist vaccine could protect against mismatched H1N1 influenza infection and secondary bacterial infection, with CD4 T cells participating in the control of viral titers. Notably, the authors of that study concluded that antibody did not contribute to protection. They came to that conclusion after observing that immune serum from FluMist immunized mice could not neutralize the infectivity of the mismatched H1N1 virus. We obtained analogous results after passive transfer of H3N2 immune serum (Fig 5F) or NP-specific mAb (Fig 5L). Nevertheless, we found that the H3N2 immune serum and NP-specific mAb significantly improved survival and reduced burden after secondary bacterial infection, despite their failure to impact viral titers. Thus, our study reveals a previously unappreciated mechanism of protection in this model, namely that non-neutralizing cross-reactive antibody to influenza can provide significant protection from secondary bacterial infection.
Our decisive findings in a well-controlled animal model substantially strengthen the conclusions of prior studies reporting that mismatched immunity to influenza confers clinical efficacy (11, 12, 49–51). Together, these clinical and animal studies provide compelling evidence that certain mismatched vaccines may benefit public health when matched vaccines are not available. Moreover, our studies suggest that boosting NP immunity may suffice to provide clinical benefit. Boosting NP immunity may require use of live attenuated influenza vaccines or NP-containing subunit vaccines since classical inactivated influenza vaccines only contain low quantities of NP and weakly boost NP antibody responses (18). Passive immunotherapy using NP-specific antibody also may be useful, particularly for those who respond inadequately to active immunization regimen, such as the immunocompromised and elderly. Finally, by demonstrating that preexisting immunity to influenza NP impacts susceptibility to secondary pneumococcal infection, this report opens new lines of investigation for those studying the pathology, epidemiology, treatment and prevention of pneumonia, bronchitis, sinusitis, otitis media, and other bacterial diseases commonly associated with influenza infections (1, 3, 6, 52–54).
Acknowledgments
The authors thank Drs. Markus Mohrs, Marcia Blackman, and Elizabeth Leadbetter for reading our manuscript and the employees of the Trudeau Institute Animal Breeding and Maintenance Facilities for care of the mice used in these studies. We also thank Drs. Richard Dutton and Brian Murphy for providing influenza strain c.a.A/Alaska/72/CR9, and Drs. Kazuyoshi Kawakami and Thomas Moore for S. pneumoniae strain URF918 and Klebsiella pneumoniae strain IA565, respectively.
Abbreviations used
- ca
cold adapted influenza
- NP
influenza nucleoprotein
- Spn
Streptococcus pneumoniae
Footnotes
This work was supported by funds from The Trudeau Institute and National Institutes of Health grants AI071295 (S.T.S.), AG02160 (L.H.), AI072689 (T.D.R), AI83610 (J.E.K), AI67967 (D.L.W), and AI76499 (D.L.W.) and Department of Defense contract W911QY-10-C-0184 (L.H.).
References
- 1.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clinical Microbiology Reviews. 2006;19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stohr K. Preventing and treating influenza. BMJ. 2003;326:1223–1224. doi: 10.1136/bmj.326.7401.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brundage JF, Shanks GD. Deaths from bacterial pneumonia during 1918–19 influenza pandemic. Emerging Infectious Diseases. 2008;14:1193–1199. doi: 10.3201/eid1408.071313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice TW, Rubinson L, Uyeki TM, Vaughn FL, John BB, Miller RR, 3rd, Higgs E, Randolph AG, Smoot BE, Thompson BT. Critical illness from 2009 pandemic influenza A virus and bacterial coinfection in the United States*. Crit Care Med. 2012;40:1487–1498. doi: 10.1097/CCM.0b013e3182416f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subbarao K, Murphy BR, Fauci AS. Development of effective vaccines against pandemic influenza. Immunity. 2006;24:5–9. doi: 10.1016/j.immuni.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Fiore AE, Bridges CB, Cox NJ. Seasonal influenza vaccines. Current Topics in Microbiology and Immunology. 2009;333:43–82. doi: 10.1007/978-3-540-92165-3_3. [DOI] [PubMed] [Google Scholar]
- 7.Grebe KM, Yewdell JW, Bennink JR. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes and Infection. 2008;10:1024–1029. doi: 10.1016/j.micinf.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein SL, Price GE. Cross-protective immunity to influenza A viruses. Expert Review Vaccines. 2010;9:1325–1341. doi: 10.1586/erv.10.123. [DOI] [PubMed] [Google Scholar]
- 9.Slepushkin AN. The effect of a previous attack of A1 influenza on susceptibility to A2 virus during the 1957 outbreak. Bulletin of the World Health Organization. 1959;20:297–301. [PMC free article] [PubMed] [Google Scholar]
- 10.Sonoguchi T, Naito H, Hara M, Takeuchi Y, Fukumi H. Cross-subtype protection in humans during sequential, overlapping, and/or concurrent epidemics caused by H3N2 and H1N1 influenza viruses. Journal of Infectious Diseases. 1985;151:81–88. doi: 10.1093/infdis/151.1.81. [DOI] [PubMed] [Google Scholar]
- 11.Nichol KL, Mendelman PM, Mallon KP, Jackson LA, Gorse GJ, Belshe RB, Glezen WP, Wittes J. Effectiveness of live, attenuated intranasal influenza virus vaccine in healthy, working adults: a randomized controlled trial. JAMA. 1999;282:137–144. doi: 10.1001/jama.282.2.137. [DOI] [PubMed] [Google Scholar]
- 12.Epstein SL. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. Journal of Infectious Diseases. 2006;193:49–53. doi: 10.1086/498980. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu XN. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 14.van der Sluijs KF, van Elden LJ, Nijhuis M, Schuurman R, Pater JM, Florquin S, Goldman M, Jansen HM, Lutter R, van der Poll T. IL-10 is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. Journal of Immunology. 2004;172:7603–7609. doi: 10.4049/jimmunol.172.12.7603. [DOI] [PubMed] [Google Scholar]
- 15.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nature Medicine. 2008;14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 16.Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, Deng JC. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. Journal of Clinical Investigation. 2009;119:1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carragher DM, Kaminski DA, Moquin A, Hartson L, Randall TD. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. Journal of Immunology. 2008;181:4168–4176. doi: 10.4049/jimmunol.181.6.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamere MW, Lam HT, Moquin A, Haynes L, Lund FE, Randall TD, Kaminski DA. Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. Journal of Immunology. 2011;186:4331–4339. doi: 10.4049/jimmunol.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell TJ, Strutt T, Reome J, Hollenbaugh JA, Roberts AD, Woodland DL, Swain SL, Dutton RW. Priming with cold-adapted influenza A does not prevent infection but elicits long-lived protection against supralethal challenge with heterosubtypic virus. Journal of Immunology. 2007;178:1030–1038. doi: 10.4049/jimmunol.178.2.1030. [DOI] [PubMed] [Google Scholar]
- 20.Kohlmeier JE, Miller SC, Smith J, Lu B, Gerard C, Cookenham T, Roberts AD, Woodland DL. The chemokine receptor CCR5 plays a key role in the early memory CD8+ T cell response to respiratory virus infections. Immunity. 2008;29:101–113. doi: 10.1016/j.immuni.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy BR, Chanock RM, Clements ML, Anthony WC, Sear AJ, Cisneros LA, Rennels MB, Miller EH, Black RE, Levine MM, Betts RF, Douglas RG, Jr, Maassab HF, Cox NJ, Kendal AP. Evaluation of A/Alaska/6/77 (H3N2) cold-adapted recombinant viruses derived from A/Ann Arbor/6/60 cold-adapted donor virus in adult seronegative volunteers. Infection and Immunity. 1981;32:693–697. doi: 10.1128/iai.32.2.693-697.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jelley-Gibbs DM, Dibble JP, Brown DM, Strutt TM, McKinstry KK, Swain SL. Persistent depots of influenza antigen fail to induce a cytotoxic CD8 T cell response. Journal of Immunology. 2007;178:7563–7570. doi: 10.4049/jimmunol.178.12.7563. [DOI] [PubMed] [Google Scholar]
- 23.Kawakami K, Yamamoto N, Kinjo Y, Miyagi K, Nakasone C, Uezu K, Kinjo T, Nakayama T, Taniguchi M, Saito A. Critical role of Valpha14+ natural killer T cells in the innate phase of host protection against Streptococcus pneumoniae infection. Eur J Immunol. 2003;33:3322–3330. doi: 10.1002/eji.200324254. [DOI] [PubMed] [Google Scholar]
- 24.Lau HY, Clegg S, Moore TA. Identification of Klebsiella pneumoniae genes uniquely expressed in a strain virulent using a murine model of bacterial pneumonia. Microb Pathog. 2007;42:148–155. doi: 10.1016/j.micpath.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yewdell JW, Frank E, Gerhard W. Expression of influenza A virus internal antigens on the surface of infected P815 cells. Journal of Immunology. 1981;126:1814–1819. [PubMed] [Google Scholar]
- 26.McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. Journal of Infectious Diseases. 2002;186:341–350. doi: 10.1086/341462. [DOI] [PubMed] [Google Scholar]
- 27.Armerding D, Rossiter H, Ghazzouli I, Liehl E. Evaluation of live and inactivated influenza A virus vaccines in a mouse model. J Infect Dis. 1982;145:320–330. doi: 10.1093/infdis/145.3.320. [DOI] [PubMed] [Google Scholar]
- 28.Tannock GA, Paul JA. Homotypic and heterotypic immunity of influenza A viruses induced by recombinants of the cold-adapted master strain A/Ann Arbor/6/60-ca. Arch Virol. 1987;92:121–133. doi: 10.1007/BF01310067. [DOI] [PubMed] [Google Scholar]
- 29.Alymova IV, Portner A, Takimoto T, Boyd KL, Babu YS, McCullers JA. The novel parainfluenza virus hemagglutinin-neuraminidase inhibitor BCX 2798 prevents lethal synergism between a paramyxovirus and Streptococcus pneumoniae. Antimicrob Agents Chemother. 2005;49:398–405. doi: 10.1128/AAC.49.1.398-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulman JL, Kilbourne ED. Induction of Partial Specific Heterotypic Immunity in Mice by a Single Infection with Influenza a Virus. Journal of Bacteriology. 1965;89:170–174. doi: 10.1128/jb.89.1.170-174.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangel-Moreno J, Carragher DM, Misra RS, Kusser K, Hartson L, Moquin A, Lund FE, Randall TD. B cells promote resistance to heterosubtypic strains of influenza via multiple mechanisms. Journal of Immunology. 2008;180:454–463. doi: 10.4049/jimmunol.180.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamere MW, Moquin A, Lee FE, Misra RS, Blair PJ, Haynes L, Randall TD, Lund FE, Kaminski DA. Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance. J Virol. 2011;85:5027–5035. doi: 10.1128/JVI.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrew ME, Coupar BE, Ada GL, Boyle DB. Cell-mediated immune responses to influenza virus antigens expressed by vaccinia virus recombinants. Microbial Pathogenesis. 1986;1:443–452. doi: 10.1016/0882-4010(86)90006-9. [DOI] [PubMed] [Google Scholar]
- 34.Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, Gromkowski SH, Deck RR, DeWitt CM, Friedman A, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 35.Tamura S, Miyata K, Matsuo K, Asanuma H, Takahashi H, Nakajima K, Suzuki Y, Aizawa C, Kurata T. Acceleration of influenza virus clearance by Th1 cells in the nasal site of mice immunized intranasally with adjuvant-combined recombinant nucleoprotein. Journal of Immunology. 1996;156:3892–3900. [PubMed] [Google Scholar]
- 36.Epstein SL, Kong WP, Misplon JA, Lo CY, Tumpey TM, Xu L, Nabel GJ. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine. 2005;23:5404–5410. doi: 10.1016/j.vaccine.2005.04.047. [DOI] [PubMed] [Google Scholar]
- 37.Roy S, Kobinger GP, Lin J, Figueredo J, Calcedo R, Kobasa D, Wilson JM. Partial protection against H5N1 influenza in mice with a single dose of a chimpanzee adenovirus vector expressing nucleoprotein. Vaccine. 2007;25:6845–6851. doi: 10.1016/j.vaccine.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price GE, Soboleski MR, Lo CY, Misplon JA, Pappas C, Houser KV, Tumpey TM, Epstein SL. Vaccination focusing immunity on conserved antigens protects mice and ferrets against virulent H1N1 and H5N1 influenza A viruses. Vaccine. 2009;27:6512–6521. doi: 10.1016/j.vaccine.2009.08.053. [DOI] [PubMed] [Google Scholar]
- 39.Casadevall A, Pirofski LA. A new synthesis for antibody-mediated immunity. Nat Immunol. 2011;13:21–28. doi: 10.1038/ni.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia CC, Russo RC, Guabiraba R, Fagundes CT, Polidoro RB, Tavares LP, Salgado AP, Cassali GD, Sousa LP, Machado AV, Teixeira MM. Platelet-activating factor receptor plays a role in lung injury and death caused by Influenza A in mice. PLoS Pathog. 2010;6:e1001171. doi: 10.1371/journal.ppat.1001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinojosa E, Boyd AR, Orihuela CJ. Age-associated inflammation and toll-like receptor dysfunction prime the lungs for pneumococcal pneumonia. J Infect Dis. 2009;200:546–554. doi: 10.1086/600870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shanks GD, Brundage JF. Pathogenic responses among young adults during the 1918 influenza pandemic. Emerg Infect Dis. 2012;18:201–207. doi: 10.3201/eid1802.102042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanthier PA, Huston GE, Moquin A, Eaton SM, Szaba FM, Kummer LW, Tighe MP, Kohlmeier JE, Blair PJ, Broderick M, Smiley ST, Haynes L. Live attenuated influenza vaccine (LAIV) impacts innate and adaptive immune responses. Vaccine. 2011;29:7849–7856. doi: 10.1016/j.vaccine.2011.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun K, Ye J, Perez DR, Metzger DW. Seasonal FluMist vaccination induces cross-reactive T cell immunity against H1N1 (2009) influenza and secondary bacterial infections. J Immunol. 2011;186:987–993. doi: 10.4049/jimmunol.1002664. [DOI] [PubMed] [Google Scholar]
- 45.Casadevall A, Pirofski LA. A new synthesis for antibody-mediated immunity. Nat Immunol. 2012;13:21–28. doi: 10.1038/ni.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nimmerjahn F, Ravetch JV. The antiinflammatory activity of IgG: the intravenous IgG paradox. J Exp Med. 2007;204:11–15. doi: 10.1084/jem.20061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anthony RM, Kobayashi T, Wermeling F, Ravetch JV. Intravenous gammaglobulin suppresses inflammation through a novel T(H)2 pathway. Nature. 2011;475:110–113. doi: 10.1038/nature10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, Davies DH, Johnston RJ, Peters B, Rafii-El-Idrissi Benhnia M, Hoffmann J, Su HP, Singh K, Garboczi DN, Head S, Grey H, Felgner PL, Crotty S. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohmit SE, Victor JC, Rotthoff JR, Teich ER, Truscon RK, Baum LL, Rangarajan B, Newton DW, Boulton ML, Monto AS. Prevention of antigenically drifted influenza by inactivated and live attenuated vaccines. N Engl J Med. 2006;355:2513–2522. doi: 10.1056/NEJMoa061850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichol KL, Nordin JD, Nelson DB, Mullooly JP, Hak E. Effectiveness of influenza vaccine in the community-dwelling elderly. N Engl J Med. 2007;357:1373–1381. doi: 10.1056/NEJMoa070844. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Garcia L, Valdespino-Gomez JL, Lazcano-Ponce E, Jimenez-Corona A, Higuera-Iglesias A, Cruz-Hervert P, Cano-Arellano B, Garcia-Anaya A, Ferreira-Guerrero E, Baez-Saldana R, Ferreyra-Reyes L, Ponce-de-Leon-Rosales S, Alpuche-Aranda C, Rodriguez-Lopez MH, Perez-Padilla R, Hernandez-Avila M. Partial protection of seasonal trivalent inactivated vaccine against novel pandemic influenza A/H1N1 2009: case-control study in Mexico City. BMJ. 2009;339:b3928. doi: 10.1136/bmj.b3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. Journal of Infectious Diseases. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palacios G, Hornig M, Cisterna D, Savji N, Busetti AV, Kapoor V, Hui J, Tokarz R, Briese T, Baumeister E, Lipkin WI. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS ONE. 2009;4:e8540. doi: 10.1371/journal.pone.0008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.2009 Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1) - United States. MMWR Morbidity and Mortality Weekly Report. 2009 May-Aug;58:1071–1074. [PubMed] [Google Scholar]