Summary
The importance of the involvement of non-protein coding RNAs in biological processes has become evident in recent years along with the identification of the transcriptional regulatory mechanisms that allow them to exert their roles. MicroRNAs (miRNAs) are a novel class of small non-coding RNA that regulates messenger RNA abundance. The capacity of each miRNA to target several transcripts suggests an ability to build a complex regulatory network for fine tuning gene expression; a mechanism by which they are thought to regulate cell fate, proliferation and identity. The brain expresses more distinct miRNAs than any other tissue in vertebrates and it presents an impressive variety of cell types, including many different classes of neurons. Here we review more than 10 years of miRNA research, and discuss the most important findings that have established miRNAs as key regulators of neuronal development.
Keywords: Development, Differentiation, Gene expression, MicroRNA, Neurons
Introduction
A vast majority of the transcripts expressed from the mammalian genome do not code for any protein product; these RNA molecules are referred to as non-coding RNAs. Several classes of non-coding RNA have been identified; one of the most interesting classes comprises small (18–22 nucleotides) RNA molecules known as microRNAs (miRNAs). First identified in Caenorhabditis elegans, miRNAs are also expressed in vertebrates and other organisms, and their sequences are highly conserved.1 The actual number of miRNAs in the most studied organisms has still not been determined, with public databases for miRNAs constantly being updated.2 The latest release of miRbase reported the presence of 1527 miRNAs in the human genome, 741 in the mouse genome and 408 in the rat genome. It is thought that more than 1% of eukaryotic genes encode miRNAs.3
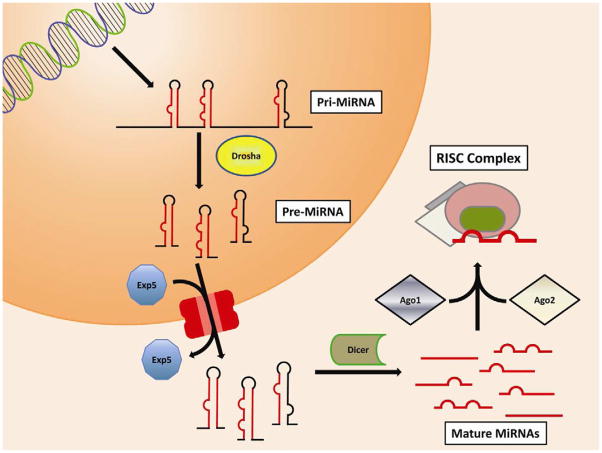
Even if most miRNAs are expressed as individual transcripts, loci are often organized as clusters of miRNAs expressed as one unique, long transcript, then processed into individual molecules.4 Transcription is usually mediated by RNA polymerase II, and transcripts are normally processed with the addition of a cap at the 5′ end and polyadenylation at the 3′ end. These primary transcripts (pri-miRNA) include a hairpin structure formed by the actual miRNA. The RNAse enzyme Drosha processes the transcripts, releasing the hairpin that constitutes the pre-miRNA that is then transported out of the nucleus by exportin-5. In the cytoplasm, pre-miRNAs are processed by the main actor in miRNA biogenesis: Dicer. Dicer is an RNA nuclease that cleaves double-stranded RNA. It recognizes the secondary structure of pre-miRNAs, and cleaves the loop at the extremity of the hairpin, releasing an RNA duplex containing the mature miRNA. One or both of the strands are then included in a protein complex called the RNA-induced silencing complex (RISC), whose main components are Argonaut 1 (AGO1) and 2 (AGO2) (Fig. 1).1
Figure 1.
The main MicroRNA biosynthetic pathway. MicroRNAs are often transcribed in multi-unit clusters known as pri-miRNAs. These long transcripts include several pre-miRNAs that become separated by the RNAse enzyme Drosha. Individual pre-miRNAs are actively transported to the cytoplasm by Exportin5. In the cytoplasm a second RNAse enzyme, Dicer, digests the loop between the two strands of the miRNA hairpin, releasing the miRNA duplex. Argonaut 2 recognizes one or both of the strands of the duplex and with the participation of Argonaut 1 mediates the inclusion in the RNA-induced silencing complex (RISC). [Fig. 1. Double column width, colour.]
After incorporation in the RISC complex, the miRNAs act in a peculiar way that differentiates them from other small non-coding RNAs, such as siRNAs (short interfering RNAs) or piRNAs (Piwi-interacting RNAs); the recognition of targets is mediated by a non-perfect matching of the sequence.5 Targeting is mainly determined by a core region of six to eight nucleotides, while the remaining portion modulates affinity to targets according to different properties.5 This complicated mechanism allows individual miRNAs to recognize multiple targets,5 just as individual transcripts can be targeted by several miRNAs.6 Sites recognized by miRNAs are referred to as MicroRNA responsive elements (MRE), which were initially thought to be in the 3′UTR region of mRNAs.1 More recently, it has been demonstrated that MREs can be located in the ORF and also the 5′UTR regions.7 Target recognition might result in inhibition of protein synthesis, but more often it mediates degradation of the transcript.8 The combinatorial possibilities among different miRNAs and their targets are vast, and generate complex regulatory networks involved in all biological processes that are only beginning to be appreciated.
MicroRNAs are essential for neuronal development
MiRNAs have been identified in all tissues in mammals, with their roles only recently being uncovered in various biological processes. In particular, miRNAs have been associated with processes of growth and development, with several of them having shown altered expression in situations of aberrant growth and in the de-differentiation that occurs during cancer.9 Neuronal development is no exception to this.
The importance of miRNAs in specific processes can be uncovered by disruption of the machinery responsible for their synthesis. Early studies in the nervous system evaluated the consequences of the depletion of the Dicer gene (which results in the absence of mature miRNAs) on neurogenesis. In Zebrafish, the complete absence of Dicer leads to critical defects in the general morphology of both the central nervous system (CNS) and peripheral nervous system (PNS), and impairment of neuronal differentiation.10
More information on the role of Dicer in development of specific areas of the CNS was gathered by more targeted deletions. Pioneering work by De Pietri Tonelli et al. selectively knocked out Dicer from the murine neocortex during development.11 The postnatal cortex decreased in size due to increased neuronal apoptosis and a deficit in cortical layering associated with an impairment in neuronal differentiation, even though no influence was observed in early differentiation, proliferation or cell cycle progression. Interestingly, when Dicer was depleted using a Wnt1-Cre strategy,12 malformations in the midbrain, cerebellum and PNS were observed, in addition to failure of differentiation in specific cell types such as dopaminergic neurons.
Selective depletion of Dicer from specific cell populations during differentiation has also been informative. Cuellar et al.13 analyzed the effects of Dicer deletion in dopaminergic cells. They found brain shrinkage and decreased cell size, but no neuronal degeneration. Interestingly, the authors observed an increased life span in these cells, in contrast to observations of Dicer knockout in other neuronal cell types, such as Purkinje cells14 or excitatory neurons of the cortex and hippocampus.15 Dicer depletion has been reported as negatively influencing survival of mouse cortical neural stem cells in very early stages of development, in addition to impairment of their differentiation process.16
Other enzymes are involved in the synthesis and function of miRNA. Two genes, FMR1 and FXR1, that interact with both Dicer and components of the RISC complex have been targeted. Pathological mutations in the FMR1 gene result in a form of mental retardation; mouse and Drosophila melanogaster knockout models of either of these proteins show aberrant development of the nervous system at the synaptic level.17 A third protein, Dgcr8, forms a complex with the Drosha enzyme and is fundamental for Drosha’s activity in pri-miRNA processing. When Dgcr08 is knocked out18 there are morphological abnormalities in the CNS and cognitive deficits, such as impaired spatial memory-dependent learning. At the cellular level, they observed alteration of dendritic development with a deficiency in the final complexity of the arborization.
Dicer, FMR1, FXR1 and Dgcr8 are not only involved in the biosynthesis of miRNAs, but are key players in the maturation of other classes of small RNAs such as siRNAs. Depleting the Dicer gene impairs not only miRNA biosynthesis, but other pathways that may be crucial for neuronal development. It is important to take this into consideration when interpreting the results of these experiments.
MicroRNAs exert specific roles in neuronal development
Disrupting the miRNA synthesis machinery does not reveal insights into the role of individual miRNAs in neuronal development. Evaluating the general role of miRNAs in the process of neuronal development has been a fundamental step in both miRNA research and developmental neuroscience. However, the need to understand the function of specific miRNAs has become increasingly evident. Therefore researchers have focused on evaluating the functional role of individual miRNAs and identifying regulators of nervous system development and various neuronal types.
Some studies approached this problem by rescuing phenotypes through overexpression of individual miRNAs. For example, injection of miR-430 into Zebrafish was enough to recover some of the brain defects caused by Dicer depletion.10 In other cases, phenotypes deriving from Dicer depletion were compared to phenotypes after knockdown of single miRNAs.
Most studies have approached the issue by identifying miRNAs that are highly expressed in the nervous system, and then disrupting them in the hope of illuminating their roles in development. A prime example of this is the study of miRNA-124. Mir-124 is the most highly expressed miRNA in the murine brain, found in differentiating and mature neurons, accounting for 25–48% of all brain miRNAs.19 Human, mouse and rat present three homologs of the mir-124 gene in the genome: mir-124-1, mir-124-2 and mir-124-3. Mir-124-1 is completely conserved from C. elegans to the human.
MiRNA 124-1 expression in developing neurons has been tested in many different systems and organisms. This miRNA exhibits increased expression during differentiation, under the control of the transcription factor REST.20 Microarrays of HeLa cells revealed that overexpression of mir-124 induced a gene expression profile similar to that of neurons, with several non-neuronal transcripts downregulated.21 In P19 cells mir-124 overexpression induced differentiation into a neuronal phenotype with axonal outgrowth.22
A role for miRNA-124 in neurogenesis in the adult murine nervous system has recently been demonstrated in vivo. MiR124 is found in progenitor cells in the subventricular zone (SVZ) and influences maturation of neuronal precursors towards specific neuronal phenotypes.23 Overexpression of mir-124 in the developing cerebral cortex induces an increased neuronogenesis in vivo.24 In the sea-squirt, Ciona intestinalis, mir-124 was shown to function in the development of PNS neurons by acting on unique targets, such as the transcription factor Notch, in addition to some that are conserved across species, including SCP1.25
Another miRNA found to be highly expressed in the nervous system and highly conserved among species, exhibiting 100% similarity between D. Melanogaster and vertebrates, is miRNA-9.19 Flies deficient for miR-9a have ectopic and supernumerary sensory neurons in these organisms, suggesting that mir-9a inhibits neuronal fate by downregulating Sens gene expression.26
MiR-9 overexpression in the developing murine brain alters migration and proliferation of neural precursors, inducing premature differentiation.27 A similar gain-of-function approach revealed that miR-9 negatively controls the generation of Cajal–Retzius neurons in the medial pallidum.28 It is known that there are three different isoforms of mir-9 (1, 2 and 3) in different brain regions during development. Overexpression of mir-9-2 in the cerebral cortex induced premature neural differentiation. At the same time, the number of Reelin- and p73-positive cells, markers that identify Cajal–Retzius cells, is reduced. The researchers suggested that transcriptional regulation mediated by mir-9-2 and 9-3 involves downregulation of several transcription factors, such as FoxG1, Elavl2, Gsh2, Nr2e1 and Pax6, and the utilization of various mechanisms.
Another important role of miRNA-9 is seen in its involvement in neuronal subtype development in spinal cord motor neurons.29 Mir-9 is expressed transiently in the chick spinal cord; in particular, it is expressed in the motor neurons that form the lateral motor columns (LCM). Overexpression of miRNA-9 decreased the number of LCM. It also altered axonal projections with reduced targeting to the wing muscles and increasing fibers innervating axial muscles. This was mediated by mir-9 altering expression of the transcription factor Foxp1. In human embryonic stem cells, mir-9 is strongly expressed when the cells commit to a specific neuronal phenotype, with its absence leading to the suppression of neural progenitor cell proliferation, even as migration was enhanced.30 These data demonstrate that mir-9 is involved in the development and organization of the nervous system but also promotes differentiation and targeting of specific subtypes of neuronal cells.
Most of the work on miRNAs involved in neuronal development has focused on these two miRNAs, miRNA-124 and miRNA-9, which are highly expressed in the nervous system. Current evidence suggests that they are critical actors in the differentiation of pluripotent cells towards a neuronal phenotype. Yoo et al.31,32 have shown they both target the BAF53a gene. BAF53a and BAF45a are substituted by two homologs, BAF53b and BAF45, in the neural-progenitor-specific BAF complex (npBAF), which is involved in ATP-dependent chromatin remodeling. This switch is critical in allowing for stem cells to exit their multipotency and differentiate towards a neuronal phenotype, for regulating proliferation of progenitor cells, and for the correct formation of the morphology of the nervous system. Expression of BAF53a represses the expression levels of BAF53b. In order to differentiate into neuronal types, the level of BAF53a in progenitor cells has to decrease. Downregulation of BAF53a is mediated by the two miRNAs, thereby regulating the neuronal fate of embryonic progenitor cells, and thus triggering dendritic outgrowth in neuronal cells. In the pluripotent state the REST transcription inhibitor keeps the levels of the two miRNAs low, allowing BAF53a transcripts to be present in the cells, thus inhibiting transcription of the neuronal-specific BAF53b.32 Overexpression of miRNA-9 and -124 in human fibroblasts resulted in a transformation into a neuronal phenotype. Addition of neurogenic transcription factors, such as NeuroD2, ASCL1 or MYT1L, enhanced the transformation, but overexpression of the transcription factors alone did not induce a neuronal phenotype.31
Although mir-9 and -124 may be sufficient for neuronal fate determination, other miRNAs have been identified as either sharing this role in the early development of the nervous system, or in the differentiation of individual cell types. A perfect example is the determination of the left (ASEL) and right (ASER) asymmetric ASE chemosensory neurons in C. elegans.33,34 These neurons are identified by different expression of chemoreceptors: Gcy-7 in ASEL and Gcy-5 in ASER. Specific expression is regulated by a network orchestrated by miRNA Lsy-6 in the ASEL, which targets the Cog1 transcription factor,33 and by miR-273 in the ASER hypothetically through the regulation of Die-1.34 MiR-128 has been shown to be involved in neuronal differentiation in amphibians, chickens and mammals. The mature form of the miRNA is strongly induced during CNS development in vivo. It promotes neural differentiation in vitro of murine neural stem cells by targeting genes involved in the ‘non-sense mediated decay’ pathway, including UPF1 and MLN51.35
It is clear that neuronal differentiation and nervous system formation are complex processes that require the regulation of proliferation, cell death, differentiation and maturation. The miRNAs mentioned above exert major and minor roles in the network of regulating events, together with others that have been identified, including mir-134,36 mir-29b,37 mir-137,38 miR-200b39 and mir-34a.40 We still lack a complete understanding of the mechanisms and network of regulation that lie behind them. Considering that it has been calculated that almost 1% of the genes code for miRNAs,3 it is reasonable to hypothesize that other miRNAs play roles in the developing nervous system. It is thus important to identify them and characterize the mechanisms through which they regulate development.
MicroRNAs and neuronal morphology
MiRNAs are not involved only in early stages of development or neuronal differentiation. In order to be functional neurons need to extend their axons and dendrites and reach their targets. Regulation of mRNA abundance and translation is fundamental for controlling neurite genesis, outgrowth and synapse formation. In many cases this regulation happens at a local level at distal domains of the neurons.41 Considering the nature of miRNA activity it’s easy to imagine a role for them in rapid regulation of local translation by targeting transcripts that are present at dendritic termini. Data from a differential expression profile of small RNAs between somas and dendrites reinforce this hypothesis42 revealing that most of the miRNAs in neurons are located in the dendritic compartment. Moreover they showed gradients of expression between the soma and the dendrite.
The two miRNAs that have been studied most extensively for their roles in neurite extension and synapse formation are mir-134 and mir-132.
Mir-134 is a brain-specific miRNA enriched in dendrites, and more specifically in synapses. Its expression level increases during development in the hippocampus, and peaks 13 days after birth, both in vivo and in vitro, and it is induced by neuronal activity.43 At the central level, neuronal stimulation through BDNF or through depolarization induces the expression of the mir-379–410 cluster that includes mir-134.44 This induction is mediated by the Mef2 transcription factor. Mir-134 targets Pumilio 2, an RNA-binding protein that suppresses protein translation and has been implicated in dendrite morphogenesis.45 In dendritic spines local translation of Limk1 is important to determine the size of the synaptic spines. MiRNA-134 targets transcripts encoding Limk1 to repress spine growth during development. However, BDNF stimulation is able to overcome this inhibition repristinating Limk1 local translation.43 These mechanisms occur in different compartments and act in opposite directions: this could permit an individually stimulated spine to grow, keeping constant the general excitability of the cell.44
Expression of miRNA-132 in neuronal cells in culture happens under the control of the cAMP responsive element binding protein (CREB). This transcription factor is a key regulator of neuronal plasticity, maturation and development and its activation in neurons has been shown to be rapidly induced by stimuli associated with neuronal potentiation.46 Activity-induced CREB induction of mir-132 promotes neurite outgrowth by targeting the GTPase activating protein p250.47 The mechanism demonstrated in vitro also operates in vivo, where it acts to mediate the integration of the newborn neurons into the synaptic circuitry by downregulating many targets associated with the inflammatory response.48
New miRNAs involved in synaptic development, spine formation and potentiation are constantly emerging. A good example are the two miRNAs 29a and 29b that reduce spine formation, inducing dendritic growth by remodeling the actin cytoskeleton targeting the Arpc3 protein.49 Given the nascent state of this field, many more are certain to be discovered.
Conclusions and future perspectives
The studies surveyed here make it clear that miRNAs exert an important influence in the molecular networks that regulate the formation of the nervous system. MiRNAs modulate gene expression at each step in the development of neuronal and glial cells, from early differentiation to synaptogenesis and myelination. Whereas a small number of miRNAs are critical regulators of proliferation and cell fate determination, many miRNAs appear to have subtler functions. This might be due to targeting of different gene sets, and/or to different effects on similar groups of targets. Although miRNAs may be predicted to target hundreds of transcripts, typically they exert strong control only over a few primary targets, whereas the degree of control of other targets is less strict.50
At present, our understanding of the identities of miRNA targets in the developing nervous system is still in its infancy. Most work has focused on the identification of MREs in the 3′UTR of mRNAs, but it is known that targeting can also happen in the coding region or in the 5′UTR.7 Moreover, most studies have focused on single isoforms of genes, whereas it is now evident that many (or even most) genes have multiple isoforms, likely with distinct functions. Alternative start sites or splicing can produce different transcripts that could be targeted by distinct miRNAs. New sequencing technologies allow identification of the complete pool of isoforms expressed in a cell type or tissue. This opens up a whole new territory for identifying regulatory interactions between miRNAs and individual transcripts.
Finally, other non-coding RNAs can interact with miRNAs, either as targets to be regulated or as regulators of miRNA activity. New theories suggest that different kinds of transcripts – short and long, coding and non-coding – act in a complex synergistic way to ensure a delicate balance of gene expression.51 How this ‘non-coding revolution’ will change our approach to studying the process of neuronal development and developmental disorders is still not clear, but it will challenge genomic and development biology research for many years to come.
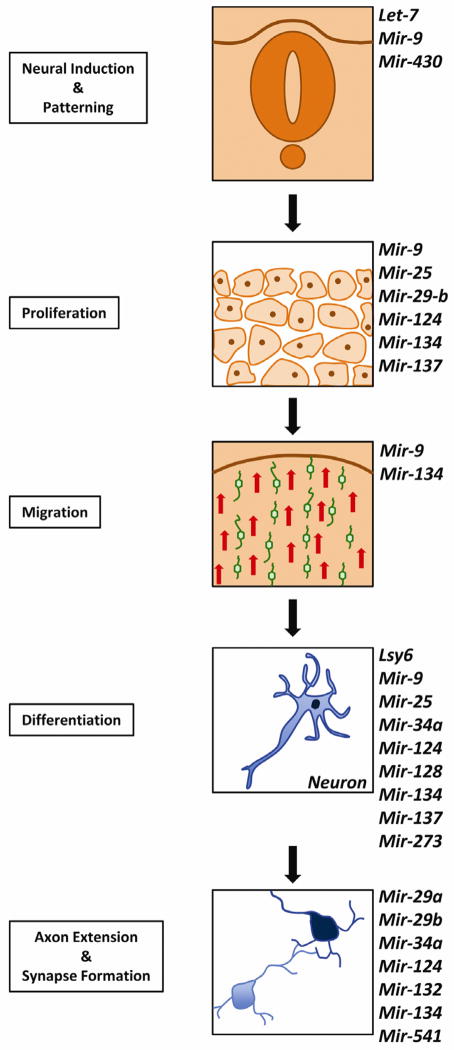
Figure 2.
S in development of the nervous system and differentiation of different neuronal cells. Next to each panel representing the different steps are listed individual microRNAs that have been demonstrated to play a role in the process. [Fig. 2. Single column width, colour.]
Practice points.
Numerous studies in animal models demonstrate that microRNAs play essential roles in controlling neuronal proliferation, migration and precursor fates.
Several individual microRNAs exert major or minor roles in specific processes in brain development.
Specific examples in humans show that disruption of microRNA function is associated with brain dysfunction and psychiatric disease.
Research directions.
Gain-of-function and loss-of-function experiments on individual microRNAs will help identify factors in the organization of neuronal development and define their roles.
Carefully designed next-generation sequencing experiments will enable identification of target genes and the network of events leading to correct development of the nervous system.
In-vivo experiments trying to re-establish the correct functionality of these networks, when disrupted, will help determine whether microRNAs can be therapeutic targets in different conditions of pathologically altered nervous system development.
Acknowledgments
Funding sources
This work was supported by National Institutes of Health grants HD057632 and NS059866, US Army (W81XWH-05-1-0061), the Walter G. Ross foundation and the Buoniconti Foundation. The sponsors had no involvement in the preparation of this manuscript.
Footnotes
Conflict of interest statement
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP, Lee R, Feinbaum R. MicroRNAs: genomics, biogenesis, mechanism, and function genomics: the miRNA genes. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.John B, Enright AJ, Aravin A, et al. Human microRNA targets. PLoS Biology. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha M, Ghose J, Bhattarcharyya NP. Micro RNA -214,-150,-146a and-125b target Huntingtin gene. RNA Biology. 2011;8(6) doi: 10.4161/rna.8.6.16035. [DOI] [PubMed] [Google Scholar]
- 7.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 8.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–87. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 10.Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–8. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 11.De Pietri Tonelli D, Pulvers JN, Haffner C, et al. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–21. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Molec Cell Biol. 2010;2:152–63. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- 13.Cuellar TL, Davis TH, Nelson PT, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105:5614–9. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer A, O’Carroll D, Tan CL, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–8. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis TH, Cuellar TL, Koch SM, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–30. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawase-Koga Y, Low R, Otaegi G, et al. RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J Cell Sci. 2010;123(Pt 4):586–94. doi: 10.1242/jcs.059659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–46. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark KL, Xu B, Bagchi A, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nature Genet. 2008;40:751–60. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 20.Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 22.Yu J-Y, Chung K-H, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–33. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng L-C, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nature Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiorano NA, Mallamaci A. Promotion of embryonic cortico-cerebral neuronogenesis by miR-124. Neural Dev. 2009;4:40. doi: 10.1186/1749-8104-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JS, Pedro MS, Zeller RW. miR-124 function during Ciona intestinalis neuronal development includes extensive interaction with the Notch signaling pathway. Development. 2011;138:4943–4953. doi: 10.1242/dev.068049. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Wang F, Lee J-A, Gao F-B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nature Struct Molec Biol. 2009;16:365–71. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–22. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31:809–18. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaloy C, Liu L, Lee J-A, et al. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–35. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460(7255):642–6. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo AS, Sun AX, Li L, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston RJ, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci USA. 2005;102:12449–54. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang S, Johnston RJ, Frøkjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430(7001):785–9. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 35.Bruno IG, Karam R, Huang L, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Molec Cell. 2011;42:500–10. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cerebral Cortex. 2011 Aug;:1857–69. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- 37.Kole AJ, Swahari V, Hammond SM, Deshmukh M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011;25:125–30. doi: 10.1101/gad.1975411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smrt RD, Szulwach KE, Pfeiffer RL, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–70. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hertzano R, Elkon R, Kurima K, et al. Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis. PLoS Genetics. 2011;7:e1002309. doi: 10.1371/journal.pgen.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aranha MM, Santos DM, Solá S, Steer CJ, Rodrigues CMP. miR-34a regulates mouse neural stem cell differentiation. PLoS One. 2011;6:e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giuditta A, Kaplan BB, van Minnen J, Alvarez J, Koenig E. Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci. 2002;25:400–4. doi: 10.1016/s0166-2236(02)02188-4. [DOI] [PubMed] [Google Scholar]
- 42.Kye M-J, Liu T, Levy SF, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT–PCR. RNA. 2007;13:1224–34. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schratt GM, Tuebing F, Nigh E, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 44.Fiore R, Khudayberdiev S, Christensen M, et al. Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye B, Petritsch C, Clark IE, et al. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–21. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 46.Sakamoto K, Karelina K, Obrietan K. CREB: a multifaceted regulator of neuronal plasticity and protection. J Neurochem. 2011;116:1–9. doi: 10.1111/j.1471-4159.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wayman GA, Davare M, Ando H, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105:9093–8. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luikart BW, Bensen AL, Washburn EK, et al. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PloS One. 2011;6:e19077. doi: 10.1371/journal.pone.0019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lippi G, Steinert JR, Marczylo EL, et al. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J Cell Biol. 2011;194:889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–3. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 51.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]