Abstract
Despite the high inhibition of viral replication achieved by current anti-HIV drugs, many patients fail treatment, often with emergence of drug-resistant virus. Clinical observations show that the relationship between adherence and likelihood of resistance differs dramatically across drug class. We developed a mathematical model that explains these observations and makes novel predictions. Our model incorporates drug properties, fitness differences between susceptible and resistant strains, mutation, and adherence. We show that antiviral activity falls quickly for drugs with sharp dose-response curves and short half-lives, such as boosted protease inhibitors, limiting the time when resistance can be selected. We find that poor adherence to such drugs causes failure via growth of susceptible virus, explaining puzzling clinical observations. Furthermore, our model predicts that certain single-pill combination therapies can prevent resistance regardless of patient adherence. Our approach represents a first step for simulating clinical trials and may help select novel drug regimens for investigation.
Introduction
The prognosis of HIV infection has dramatically improved since the introduction of highly active antiretroviral therapy (HAART), which, when successful, can bring viral loads below the detection limit, improve immune function, and prevent progression to AIDS1. Although a complete understanding of how virologic, pharmacologic, and host factors interact to determine therapeutic outcome is still lacking, it is clear that a major obstacle to successful treatment is suboptimal drug adherence. Non-adherence can lead to virologic failure and the emergence of drug resistance2–5.
Because of their high antiviral activity, protease inhibitors (PIs) play a crucial role in HIV-1 treatment, and are used in three of the five recommended initial regimens and many salvage regimens6. Clinical trials have shown that for many drug combinations involving PIs, treatment failure occurs without resistance mutations in the protease gene7–10, though mutations conferring resistance to other drugs in the regimen are often found. It is generally believed that combination therapy works because it is unlikely that multiple mutations conferring resistance to all drugs in the combination will appear in the same viral genome. Thus, failure without PI resistance is puzzling because it appears to contradict this fundamental explanation for the success of HAART. It is commonly believed that PIs have a higher “barrier to resistance” than other drugs, meaning that clinically significant PI resistance requires the accumulation of multiple mutations in the protease gene11. PI resistance also typically occurs at a narrower range of adherence levels than resistance to other drug classes3;12. While these concepts are suggestive, no theory has been developed to explain why patients fail PI-based regimens without PI resistance.
A resistance mutation may exist prior to treatment in the latent or active viral populations, or may arise during treatment13. Drug resistance develops clinically if the mutant strain is selected over the wild-type strain. Selection depends on the fitness costs and benefits of the mutation, as well as on drug levels, which vary with the dosing interval, the drug half-life, and the patient’s adherence. Here we use a modeling approach to integrate these factors, enabling us to determine when a resistance mutation will be selected and to predict the outcome of therapy with different drugs. Our results explain the unique adherence-resistance relationship for PIs and show why patients fail PI-based therapy without PI resistance.
Results
Defining the mutant selection window
Antiretroviral drugs reduce viral fitness in a dose-dependent manner (Fig. 1a). Viral fitness can be summarized as a single parameter, the basic reproductive ratio R0, which encompasses all phases of the viral life-cycle14 (Supplementary Methods). The Hill dose-response curve describes the relationship between drug concentration and R0:
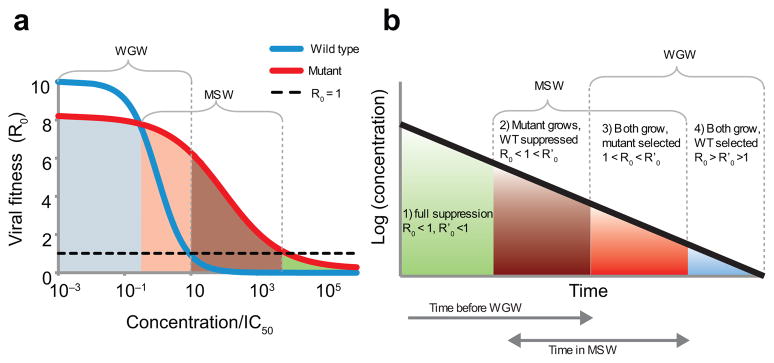
Figure 1.
Drug concentrations determine the relative fitness of the wild-type virus and a resistant mutant. (a) The fitness of the wild-type virus (R0, blue line) decreases with increasing drug concentration, following Equation (1). A drug-resistant strain (R′0, red line) is less fit than the wild type at low concentrations, but more fit at higher concentrations due to an increased IC50 or a reduced slope. The mutant selection window (MSW) is the range of concentrations where a resistant mutant, if present, will grow faster than the wild type and still has R′0> 1. The wild-type growth window (WGW) is the range of low concentrations where the wild type has R0> 1, leading to treatment failure without the need for resistance. For drug concentrations in the overlapping range of these windows, virologic failure (VF) can occur even without resistance, but will be hastened by the appearance of a faster-growing mutant. (b) As drug concentrations decay after the last dose is taken, the viral fitness passes through the four different selection ranges. Depending on the drug, dose level, and mutation, not all of these ranges may exist. The time spent in each selection window is also determined by the drug half-life.
Here D is drug concentration, IC50 is the concentration at which 50% inhibition occurs, and m is a parameter determining steepness of the curve15;16. The numerator R00 is baseline fitness in the absence of treatment.
A drug-resistant mutant is any viral variant that is less inhibited than the wild type for some drug concentration, described by the altered dose-response curve,
Mutations have a fitness cost, meaning that the drug-free fitness of the mutant virus is reduced by a fraction s (0 < s < 1). In the presence of the drug, the mutation confers a benefit, multiplying the IC50 by a factor ρ (the fold-change in IC50, ρ> 1). Many mutations also reduce the slope (m) of the dose-response curve by a fraction σ < 017.
Virologic failure (VF) occurs when treatment fails to prevent the growth of virus to high levels. A viral strain grows when R0> 1. The strain with highest R0 out-competes others14. The range of drug concentrations where a resistant mutant can cause VF is called the Mutant Selection Window (MSW)18;19. Above the MSW even replication of the mutant is suppressed (R′0(D) < 1), although toxicity may prevent these drug concentrations from being achieved clinically. We define the Wild-type Growth Window (WGW), where drug concentrations are so low that wild-type virus is not adequately suppressed and failure can occur even without resistance (R0(D) > 1).
The MSW explains therapy outcome patterns
To predict a drug’s vulnerability to resistant and susceptible strains, we computed the time during a treatment interruption that a patient spends in the MSW and WGW. During treatment interruption, both R0 and R′0 increase. Up to four selection ranges can be identified (Fig. 1b). Using pharmacokinetic and pharmacodynamic data16;17, we determined the time spent in these ranges for 66 drug-mutation pairs (Fig. 2a) based on their specific dose-response curves (Figs. 2b–e). For each pair, we show how soon after the most recent dose the mutant or wild-type virus starts to grow. This quantity is shorter than the expected time until VF, which requires the plasma HIV RNA to reach detectable levels and may also depend on the time until mutant virus appears. Throughout the paper we examine only single-point mutations that are fully characterized by their effect on the dose-response curve (2). In the Discussion, we address the extension to multiple mutations.
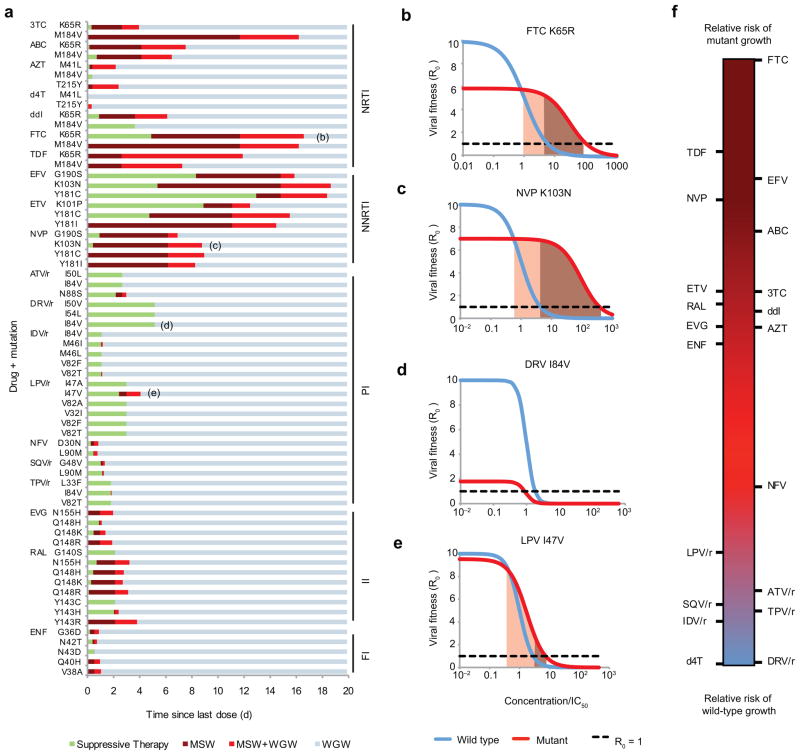
Figure 2.
Selection windows can be calculated for particular drug-mutation pairs. (a) The distance to the right along each horizontal bar is the time since the last dose, and the color corresponds to the selection window during that time interval (described in Fig. 1b) (b)–(e) Examples of dose-response curves for drug-mutation combinations indicated in (a). Shading indicates the MSW. If the cost of a mutation is too high or its benefit (ρ or σ) too low, it is possible that the MSW does not exist. (f) Rank of each drug for relative risk of wild-type versus mutant virus growth, independent of the overall risk of therapy failure. For each drug, we show a “synthetic,” worst-case, single-nucleotide mutation (Supplementary Methods, Supplementary Fig. 1). NRTI & NNRTI, (non)-nucleoside/nucleotide reverse transcriptase inhibitors; PI, protease inhibitors; FI, fusion inhibitors; II, integrase inhibitors; 3TC, lamivudine; ABC, abacavir; AZT, zidovudine; d4T, stavudine; ddI, didanosine, FTC, emtricitabine; TDF, tenofovir disoproxil fu-marate; EFV, efavirenz; ETV, etravirine; NVP, nevirapine; ATV, atazanavir; DRV, darunavir; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; SQV, saquinavir; TPV, tipranavir; EVG, elvitegravir; RAL, raltegravir; ENF, enfuvirtide. PIs are often “boosted” (co-formulated) with ritonovir (/r), which interferes with break-down in the liver and increases half-life.
Successful treatment must both minimize the time spent in the MSW and delay entry into the WGW. These two goals are in tension, as shortening the time spent in the MSW (e.g., by decreasing drug half-life) can also hasten entry into the WGW (Fig. 1b). Results from our model (Fig. 2a) suggest that NNRTIs are protected against failure via wild-type virus due to their long half-lives, but are vulnerable to mutation due to the time spent in the MSW. PIs are at the opposite end of the spectrum, with little time spent in the MSW but rapid entry into the WGW. This behavior is caused by high slope parameters (extreme sensitivity to changes in concentration) and short half-lives. These results explain the unique bargain of PI therapy: greater protection against the evolution of resistance, but vulnerability to wild type-based VF after short treatment interruptions. This trade-off is depicted schematically by plotting the drugs along a single axis, which measures the relative risk of mutant growth versus wild-type growth, independent of the overall risk of VF (Fig. 2f, Supplementary Methods).
Simulation of clinical outcomes
While the MSW and WGW concepts describe instantaneous growth of mutant and wild-type virus for a given drug concentration, VF depends on sustained growth and, therefore, drug concentrations over time. To explain clinical observations across drug classes and adherence levels, we developed a stochastic model of viral evolution (Fig. 3, Methods). Our model builds on the large body of previous work modeling HIV therapy14;20–23 by integrating new data on class-specific drug properties16 and realistic costs and benefits of mutations17. We also modified past approaches by allowing drug concentrations, and hence R0, to fluctuate instead of taking time-averages.
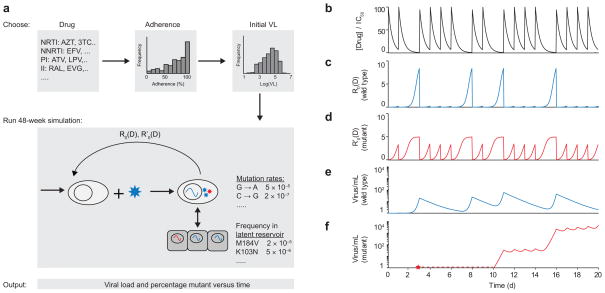
Figure 3.
Schematic of algorithm for simulating viral dynamics in a patient undergoing treatment. (a) A single simulated patient takes a particular drug (or drug combination) with a designated adherence level, starting with a chosen initial viral load. Over a 48-week clinical trial, drug levels fluctuate and viral load levels are simulated according to a viral dynamics model. (b) Drug levels fluctuate according to patient’s dosing pattern and pharmacokinetics (dose size, half-life, bioavailability); gaps show missed doses (figure shows single drug). (c) Wild-type viral fitness (R0) fluctuates in response to drug concentration depending on the dose-response curve. (d) Fitness of drug-resistant strain (R′0) depends on an altered dose-response curve; at high drug concentrations, mutant fitness exceeds that of the wild type. (e) Wild-type viral load depends on viral dynamics equations, which account for active replication, exit from the latent reservoir, and competition between strains. (f) A mutant virus may appear (red star) but be below the threshold for detection (dotted red line) before eventually leading to virologic failure.
We first simulated 48-week trials of single agents in a cohort of patients. The results are presented in two ways: as outcome versus patient adherence, at the trial endpoint (Fig. 4a), and as outcome versus time, for a distribution of patient adherence levels (Fig. 4b–c).
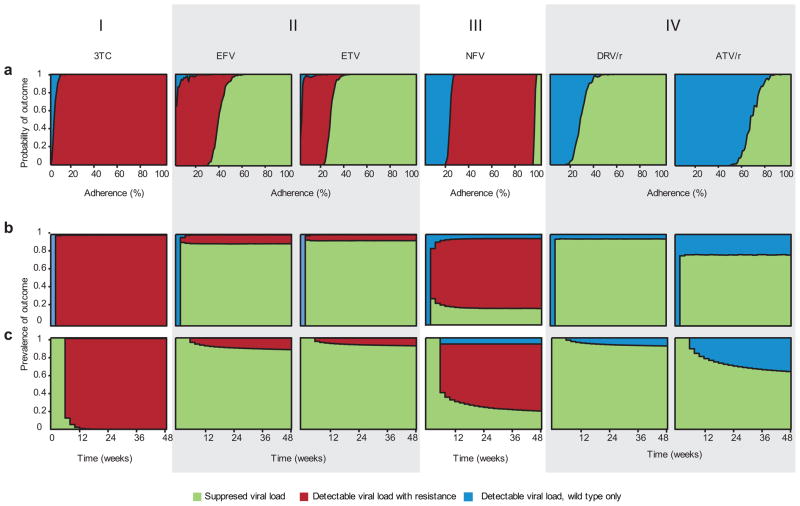
Figure 4.
Outcomes for simulated patients in a clinical trial. The height of the area shaded indicates probability of the corresponding outcome at a given adherence level (a) or time-point (b and c). (a) Adherence (x-axis) is defined as the fraction of scheduled doses taken. These are maintenance trials (see Methods). (b–c) Time is on the x-axis; measurements are taken every 2 weeks for simulated patients with a distribution of adherence levels (Supplementary Methods, Supplementary Fig. 2b). (b) Suppression trials (see Methods). (c) Maintenance trials. (I) 3TC therapy (pattern includes AZT, ABC, d4T, ENF, EVG, FTC, NVP, RAL, TDF). (II) EFV and ETV therapy. (III) NFV therapy (pattern includes ddI). (IV) DRV/r and ATV/r therapy (pattern includes ATV, TPV/r; variation on this pattern described in text includes LPV/r, SQV, SQV/r IDV, IDV/r).
Consistent with previous meta-analysis of combination therapy clinical trials24, our model predicts that the level of adherence necessary for mutant VF differs by drug class (Fig. 5). Specifically, for the NNRTIs efavirenz (EFV) and etravirine (ETV), the risk of mutant VF is greatest at low adherence levels; for unboosted PIs, the risk peaks at a higher adherence level and remains substantial up to 100% adherence; and for boosted PIs, resistance occurs infrequently and at intermediate adherence levels. Researchers have previously argued that drug half-life and fitness costs of mutations are important factors explaining these general trends3;12. By incorporating these factors as parameters, our model formalizes this argument.
Figure 5.
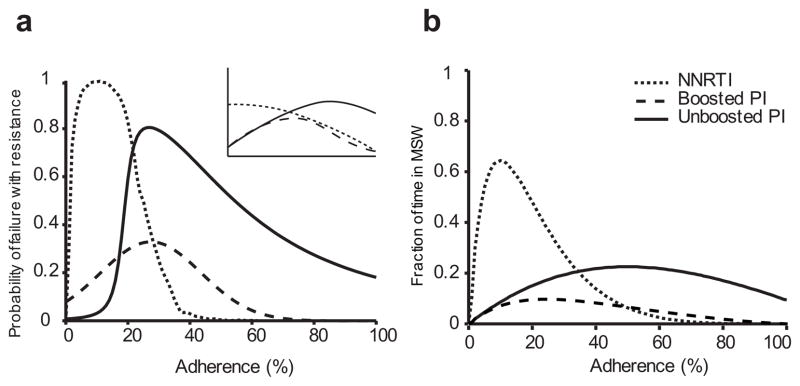
Our calculated adherence-resistance relations are in agreement with those observed in clinical trials. (a) Adherence versus simulated probability of resistance in a 48-week suppression trial for a PI, a boosted PI and an NNRTI. The inset shows a qualitative summary of results from a meta-analysis of clinical trials24, which agrees with our simulations. (b) Adherence versus fraction of time spent in the MSW for the same drugs. Adherence-resistance trends demonstrate that “time in MSW” is a good proxy for the risk of mutant-based virologic failure (VF). For both plots, curves were generated by averaging over all boosted PIs, all unboosted PIs, and the NNRTIs EFV and ETV. PI curves in (a) were fitted to skewed-T distributions to smooth step-like behavior. NVP, which was excluded from this figure, displays a different pattern from the other two NNRTIs; specifically, mutant VF can occur even for perfect adherence (Supplementary Figs. 3, 4).
Examining simulations of each drug individually (Supplementary Figs. 3–9), we find four qualitative patterns of outcome, which correspond closely – but not exactly – to drug class (Fig. 4).
For most NRTIs, the IIs, the fusion inhibitor (FI), and the NNTRI NVP, even perfect adherence leads to mutant VF in all simulated patients. As adherence declines, some wild-type VF occurs. VF and resistance occur soon after the trials are started. These results are consistent with the notion that monotherapy often leads to rapid evolution of resistance.
For most PIs and the NNRTIs EFV and ETV, however, perfect adherence results in treatment success in simulations. Control of viral replication has been observed in a substantial fraction of patients in PI monotherapy trials25, but similar trials with EFV and ETV have not been carried out. In simulations, declining adherence affects performance of these two drug classes differently.
For the NNRTIs EFV and ETV, there is a large range of low-to-intermediate adherence for which mutant VF is likely. Below this range, wild-type VF becomes increasingly likely, while above this range patients succeed. The size of this range is explained by low fitness costs of drug-resistant mutations and long half-lives of NNRTIs, which allow the patient to remain within the MSW for a substantial duration (suggested in 26).
The PI nelfinavir (NFV) and the NRTI didanosine (ddI) show a large range of intermediate adherence leading to mutant VF. Near-perfect adherence is required for treatment success. Under most clinical settings (adherence < 95%), our model predicts that these drugs perform similarly to monotherapy with other NRTIs, typically leading to mutant VF.
For many PIs, a decline from perfect adherence leads abruptly from success to wild-type VF, with little or no intermediate range for mutant VF. This result explains the outcomes of clinical studies, which have shown that VF in many boosted PI-based regimens (including monotherapy) does not require the evolution of resistance7–9. Variations on this pattern exist for some PIs: lopinavir (LPV/r), saquinavir (SQV, SQV/r), and indinavir (IDV, IDV/r) do admit mutant VF at low and moderate adherence levels, mainly for suppression trials. Still, like all the PIs simulated except NFV, as adherence declines from the successful range, the first failing outcome observed is wild-type VF (Supplementary Figs. 3, 4).
We also examined the sensitivity of our results to changes in the baseline viral fitness, R00 (Supplementary Figs. 10, 11). Since intracellular half-lives of several NRTIs are not definitively established, we tested a range of half-lives for 3TC, AZT, d4T, ddI, and TDF (Supplementary Fig. 12).
Explaining outcomes of combination therapy
Equipped with a model of drug interaction, we can extend the simulations to combination therapy (Supplementary Methods, Supplementary Fig. 13). For proof-of-concept, we use a two-drug combination of the boosted PI darunavir (DRV/r) with the II raltegravir (RAL). The combined effect of these two drugs is given by a Bliss-independent27 interaction pattern28, which describes drugs acting on different targets, therefore reducing viral replication multiplicatively. In a recent DRV/r-RAL clinical trial8 patients experiencing VF had their plasma viral population genotyped. While 17% of patients tested positive for RAL-resistance mutations in the integrase gene, no patients tested positive for DRV resistance in protease8. Our simulation is consistent with this study: treatment failure occurs without DRV resistance (Fig. 6a).
Figure 6.
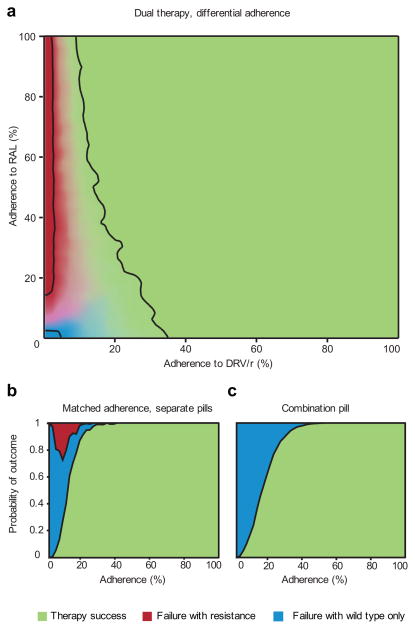
Outcomes of DRV/r + RAL dual suppression therapy simulations, considering resistant mutants for both drugs. (a) Each drug is taken independently and adherence may differ between them. The brightness of each color at a particular point indicates the probability of the corresponding outcome, with the black contours showing where each outcome occurs 95% of the time. Success depends largely on adherence to DRV/r (almost certain if > 50%), while the type of failure is determined by adherence to RAL (resistance almost certain if > 30%). All failure via resistance is due to RAL mutant-based VF. DRV mutant-based virologic failure (VF) never occurs in the simulations. (b)–(c) Drugs are taken with equal average adherence. The height of the area shaded indicates probability of the corresponding outcome at that adherence level. (b) Drugs are taken as separate pills. Average adherence is the same but pills are taken independently. (c) Drugs are packaged as a combination pill and are always taken together. Mutant VF occurs only when the two drugs are given in separate pills; combination pills eliminate mutant VF but increase the adherence required for near-certain success.
RAL-resistant mutants are selected only when the concentration of DRV/r is low and the concentration of RAL is moderate-to-high. This particular state can occur if the drugs are administered as separate pills. If, however, dual therapy were administered as a combination pill, then the two concentrations would rise and fall roughly together, reducing the chance that they reach the discordant levels that select for resistance. Simulation of dual therapy as a single combination pill verifies this hypothesis. However, this protection from resistance comes at a cost: higher adherence is required to prevent wild-type VF. For example, to ensure a 95% chance of success in the simulation, a patient taking separate pills must be 25% adherent to each pill (Fig. 6b), but 35% adherent to a combination pill (Fig. 6c). We expect this trend to apply to other drug combinations.
Discussion
Recent efforts to quantify pharmacodynamics16;17;29, combined with insights into patients’ drug-taking behavior30, have enabled us to develop the first explanatory model of virologic failure in agreement with clinical trials. All parameters in our model have direct physical interpretations, and their values were taken directly, or derived from, previous literature. The model was not fit or trained to match clinical data. Despite our model’s simplicity, it can explain the clinically observed drug-class-specific relationship between adherence and outcome24 (Fig. 5). Even without full viral dynamic simulations, a straightforward analysis of the mutant selection window can explain why certain drugs are more likely to select for resistance (Figs. 2f, 5b).
In addition, we address a long-standing puzzle of antiretroviral therapy. Even when failure of PI-based regimens is documented, mutations that confer resistance to the PI appear infrequently7–10. While it is possible that mutations may occur outside the protease gene31–34 and escape routine detection, our model provides a more straightforward explanation: due to the sharp slope of PI dose-response curves16, even relatively strong PI resistance mutations are selected only in a narrow range of drug concentrations. Moreover, as PI concentrations decay rapidly compared to other drugs, they traverse this narrow range quickly, leaving little time for a resistant strain to grow before wild type-based VF. We predict that patients who fail PI therapy with wild-type virus should be able to re-suppress virus if the same drug is taken with improved adherence. Kempf et al.12 observed this outcome in patients who failed LPV/r without detectable resistance. Even with PIs that are more susceptible to resistance, only wild-type virus is detectable when adherence dips below the level guaranteeing success, providing an “anti-resistance buffer” that may warn clinicians of resistance risk. NFV is the sole exception to this pattern, owing to it having the lowest slope and second highest IC50 of the PIs, and consistent with its documented vulnerability to resistance12.
The tradeoff between protection from resistant and susceptible strains occurs not only between drug classes, but also between different formulations of the same drugs. We predict that a novel combination pill containing DRV/r and RAL would not lead to resistance, even though the current separate-pill formulation does. This result suggests that some combination pills may be “resistance-proof,” but their known benefit of increasing patient adherence must be weighed against the fact that they require higher adherence to prevent wild type-based VF. This tradeoff results from the possibility that a patient who is prescribed multiple pills may at times take only some of them35, providing partial protection from the virus but allowing entry into a “zone of monotherapy”26 that can select for resistance.
We can extend our model to a broader range of combination therapies once interactions between drugs28 are characterized; these interactions affect the evolution of resistance36. Our monotherapy results are a first step for examining how pharmacokinetics and pharmacodynamics determine treatment outcomes. These results can inform innovations in lower-cost maintenance therapy among highly adherent patients, for whom monotherapy shows promise, but also poses resistance risks25. Specifically, based on our simulations, we propose that EFV and ETV monotherapy may be promising avenues for further study, despite disheartening performance of monotherapy with the first approved NNRTI, NVP37, and ambiguous performance of ETV-based HAART for patients with resistance to the NRTI backbone38.
Maintenance and suppression therapy generally showed similar outcomes; however, for several drugs, failure with resistance was more likely during the suppression phase. Such differences are often attributed to the presence of pre-existing mutants when viral load is high39–42. However, in our model frequent reactivation from the latent reservoir provides a sufficient source of mutants during both phases (Supplementary Tables 5, 6), and ongoing replication is an additional common cause of resistance (Supplementary Figs. 8, 9). The key difference between the two phases is in how VF is defined. Since patients remained in suppression simulations until the predefined endpoint, wild-type growth sometimes preceded (and contributed to) growth of the mutant. More frequent measurement of viral load in maintenance simulations improved the chance that VF was diagnosed before resistance reached detectable levels, consistent with clinical meta-analysis43. Also consistent with clinical observations44, continuation of maintenance trials after rebound allowed the possibility of re-suppression, but sometimes led to emergence of resistance (Supplementary Fig. 7).
It is difficult to quantitatively compare our simulations to clinical trials, since adherence is rarely precisely known. We suspect that our results are biased towards success for several reasons. First, we considered only single-point mutations, but strains with multiple mutations may lead to failure at higher adherence levels. Second, we considered neither correlations between consecutive missed doses nor variations in the time of day when a dose is taken, both factors which lead to longer treatment interruptions and increase the chance of VF20;45–48. Third, as is common in models of viral dynamics, we assumed that the virus population is homogeneous and well-mixed. Actual infections may include subpopulations that grow faster (higher R0, e.g., due to cell-to-cell transmission49) or that reside in tissues that drugs do not fully penetrate50–52. For example, the concentration of EFV in the cerebrospinal fluid is only 0.5% of plasma concentrations53. Since our predictions rely on plasma drug concentrations, they may be optimistic in the case of EFV. In the absence of strong evidence for these effects, suboptimal adherence is the most likely cause of treatment failure.
Patients experiencing VF may not respond to a similar regimen in the future45;54;55, but the precise reasons for this are not clear. The simplest explanation is that growth of a resistant strain during prior treatment makes it more likely this strain will exist in the future56. This explanation assumes that, in the absence of prior growth, most resistant mutants are relatively rare. If the diversity (“effective population size”) of the latent reservoir is not severely depleted over time, then our calculations contradict this assumption for single mutations: even in the absence of prior treatment, a majority of mutations exit the reservoir every few weeks. Resistance is then available to be selected regardless of prior growth. The occurrence of multiple mutations within the same viral genome is unlikely, however, without prior growth. To explain generally how prior VF undermines future treatment, we need to model the long-term accumulation of multi-step mutations in the viral population57;58. To build such models, it will be important to understand interactions between mutations (including compensatory mutations) and account for recombination59.
Throughout this paper, we have emphasized the variable nature of anti-HIV drug resistance. Common practice classifies a genotype as “resistant” if it is associated with VF in a meta-analysis of clinical outcomes; otherwise it is “sensitive.” This categorization is misleading: a mutation’s ability to promote viral growth depends on all drugs in a regimen, adherence, and the other mutations present. As standards of care evolve and study populations change, a mutation may gain or lose “resistant” status due to shifts in these confounding variables. Our model provides a rigorous alternative for evaluating resistance, by using mechanistic parameters to predict clinical outcomes. Our framework can help researchers prioritize drugs for clinical trials and select regimens for personalized HIV treatment.
Methods
Pharmacokinetics, pharmacodynamics, and the mutant selection window
Viral fitness followed Equation (1) with parameters R00, IC50, and m. Fitness of resistant mutants followed Equation (2) with parameters s, ρ, and σ. (Supplementary Tables 1, 3–4). Relative wild-type and mutant viral fitness values R0(D)/R00 and R′0(D)/R00 were measured using in vitro assays and were fit to Hill curves to determine the parameters IC50, m, σ, ρ, and s; these values were reported previously16;17. We estimated absolute in vivo viral fitness in the absence of drugs (R00) using measurements from previous studies (Supplementary Methods). We modeled drug concentration as instantaneously increasing after a dose to the steady state peak concentration (Cmax), and then decaying exponentially (with half-life T1/2) to the trough concentration (Cmin) before the subsequent dose. When doses were missed (representing suboptimal adherence) the concentration continued to decay, and a subsequent dose increased the concentration by ΔC = Cmax − Cmin.
We determined the bounds of the Mutant Selection Window (MSW) by solving for D in R0(D) = R′0(D) and R′0(D) = 1. We determined the upper bound of the Wild-type Growth Window (WGW) by solving R0(D) = 1. We computed the time after a single dose when a particular concentration D was reached by solving for t in D = Cmax × 2t/T1/2.
The MSW concept as applied here to antiretroviral therapy was adapted from the extensive literature on antibiotic resistance. Both in vitro and in vivo, drug concentrations that fluctuate within the MSW lead to the development of resistance, but those outside it do not (reviewed in19). While some studies of antibiotic-resistant E. coli have found no upper limit to the MSW60, no such results are known for antiretroviral resistance. The definition of the MSW most commonly used in antibiotic work is slightly different from the one we use, with the lower limit defined as R0(D) = 1 due to experimental constraints18. We have chosen to modify this definition, since selection for the mutant can occur even at lower drug concentrations where R0(D)> 1 61. The MSW and WGW can be described for each drug during combination therapy (Supplementary Methods).
Simulation of the viral dynamics model
Our model for HIV dynamics during antiretroviral drug treatment uses equations common in the literature14. These equations track the number of uninfected CD4+ cells, amount of free virus, and number of infected CD4+ cells. A constant number of uninfected cells are produced each day, and they die at a constant rate. Cells are infected at a rate proportional to the number of uninfected cells, the amount of virus, and the viral fitness. Virion production from infected cells is described by the burst rate, and virions are cleared at a constant rate. Infected cells have a higher death rate than uninfected cells. Additionally, we include a population of long-lived infected cells in the latent reservoir, which activate at a constant daily rate regardless of viral fitness. Because we are only interested in viral dynamics during treatment and at the initial stages of failure, we have ignored effects of the immune response. Viral fitness, and hence the rate of infection of new CD4+ cells, is determined by the baseline R0 and the drug concentration. All equations and parameters are given in the Supplementary Methods and Supplementary Table 7. In the Supplementary Methods, we also derive a simplified form of HIV dynamics that requires fewer parameters and only one state variable per viral strain; we used this simplified model to design our simulations. More detailed models that explicitly track multiple stages of the viral lifecycle may more accurately reflect some short-term dynamics, such as lags in viral growth during acute infection or lags in viral decay during the early days of treatment62;63. Summarizing viral fitness by a single parameter (R0) smooths out these dynamics.
There may be multiple strains of virus (wild-type and mutants) and consequently multiple types of infected cells. Even in the absence of drug, mutations will arise due to random errors in replication, though they will be selected against due to their fitness cost (s). Each mutation appears at a rate u that depends on the particular nucleotide changes required to effect the desired amino acid substitution (Supplementary Tables 2–4). The balance between these two processes results in all mutations being present in the population at an expected low level u/s, called mutation-selection equilibrium64;65. We assume that the plasma virus population reaches this equilibrium in each patient before treatment (i.e., that sufficient time has passed between initial infection and treatment initiation, and that no prior treatment has selected for resistance to the particular drug being studied) and that the population in the latent reservoir is representative of the plasma population (Supplementary Tables 5, 6). De novo mutations occur with a probability u during replication.
We used stochastic simulations to study the dynamics of the system described. Many mutations have been characterized for each drug, and to model a realistic worst-case scenario, we considered a single “synthetic” mutant defined as having the highest benefits (ρ, negative σ), lowest cost (s), highest mutation rate, and highest equilibrium frequency (due to mutation-selection balance) of all the single-nucleotide mutants known for that drug. Each monotherapy simulation therefore tracked only two strains, wild-type and mutant. For dual therapy, we considered three strains: wild-type, resistant to Drug 1, and resistant to Drug 2. Simulations modeled 48-week trials, using discrete timesteps of Δt = 30 minutes. All simulations were done in Matlab R2010b. The full details of the algorithm for simulating a single patient are given in the Supplementary Methods.
In maintenance trials patients begin with full viral suppression (2 c ml−1) and undergo monotherapy for 48 weeks or until virologic failure (VF), whichever occurs first. VF is defined as “confirmed rebound”: two consecutive weekly measurements (starting at week 5) with viral load above 200 c ml−1. In suppression trials, patients begin with a realistic distribution of treatment-naive viral loads (between 3000 and 106 c ml−1) (Supplementary Fig. 2a) and undergo monotherapy for a full 48 weeks. We tracked measurements every 2 weeks. VF is defined as a viral load above 50 c ml−1 at week 48. In both types of trials, VF is classified as “with resistance” if at least 20% of the viral population at the time of detection is mutant.
We simulated imperfect adherence by allowing each dose to be missed with a constant probability given by the expected adherence level parameter. In reporting outcomes versus time, we simulated patients with a distribution of adherence levels taken from a study using unannounced pill counts30. For simulations with two drugs, the value of adherence may be different for each drug, allowing for “differential adherence” – which has been observed in many studies35. Even when adherence to the two drugs has the same average value, the drugs can be simulated as two separate pills (allowing each pill to be taken or forgotten independently) or as a single combination pill (causing the two drug concentrations to rise and fall in lockstep).
Supplementary Material
Acknowledgments
We thank T. Antal, I. Božić, F. Fu, M. Sampah, and L. Shen for discussion during the conception of this work, and we thank J. Gallant, J.-B. Michel, P. Pennings, and three anonymous reviewers for their comments on the manuscript. We thank D. Bangsberg of Massachusetts General Hospital for supplying adherence data from the REACH study (supported by US National Institutes of Health grant R01 MH54907). Simulations were run on the Odyssey cluster supported by the FAS Sciences Division Research Computing Group of Harvard University. We are grateful for support from the US National Institutes of Health (R01 AI081600 (R.F.S., S.A.R.), R01 GM078986 (M.A.N., A.L.H.)), the Bill & Melinda Gates Foundation (M.A.N., A.L.H.), a Cancer Research Institute Fellowship (S.A.R.), a US National Science Foundation Graduate Research Fellowship (D.I.S.R.), the Howard Hughes Medical Institute (R.F.S., S.A.R.), a Canada National Science and Engineering Research Council Post-Graduate Scholarship (A.L.H.), the John Templeton Foundation (M.A.N.), and J. Epstein (M.A.N.).
Footnotes
Author Contributions
D.I.S.R., A.L.H. and S.A.R. designed the models and conducted the simulations. D.I.S.R., A.L.H., S.A.R., R.F.S. and M.A.N. conceived the study and wrote the manuscript.
References
- 1.Palella FJ, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 2.Vanhove GF, Schapiro JM, Winters MA, Merigan TC, Blaschke TF. Patient compliance and drug failure in protease inhibitor monotherapy. J Am Med Assoc. 1996;276(24):1955–1956. [PubMed] [Google Scholar]
- 3.Gardner EM, Burman WJ, Steiner JF, Anderson PL, Bangsberg DR. Antiretroviral medication adherence and the development of class-specific antiretroviral resistance. AIDS. 2009;23(9):1035–1046. doi: 10.1097/QAD.0b013e32832ba8ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maggiolo F, et al. Effect of adherence to HAART on virologic outcome and on the selection of resistance-conferring mutations in NNRTI- or PI-treated patients. HIV Clin Trials. 2007;8(5):282–292. doi: 10.1310/hct0805-282. [DOI] [PubMed] [Google Scholar]
- 5.Harrigan PR, et al. Predictors of HIV drug-resistance mutations in a large antiretroviral- naive cohort initiating triple antiretroviral therapy. J Infect Dis. 2005;191(3):339–347. doi: 10.1086/427192. [DOI] [PubMed] [Google Scholar]
- 6.US Department of Health and Human Services. Panel on Antiretroviral Guidelines for Adults & Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. 2011 http://aidsinfo.nih.gov/contentfiles/AdultandAdolescentGL.pdf.
- 7.Arribas JR, et al. The MONET trial: darunavir/ritonavir with or without nucleoside analogues, for patients with HIV RNA below 50 copies/ml. AIDS. 2010;24:223–230. doi: 10.1097/QAD.0b013e3283348944. [DOI] [PubMed] [Google Scholar]
- 8.Taiwo B, et al. Efficacy of a nucleoside-sparing regimen of darunavir/ritonavir plus raltegravir in treatment-naive HIV-1-infected patients (ACTG a5262) AIDS. 2011;25(17):2113–2122. doi: 10.1097/QAD.0b013e32834bbaa9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havlir DV, et al. Drug susceptibility in HIV infection after viral rebound in patients receiving indinavir-containing regimens. J Am Med Ass. 2000;283(2):229–234. doi: 10.1001/jama.283.2.229. [DOI] [PubMed] [Google Scholar]
- 10.Pulido F, Arribas J, Hill A, Moecklinghoff C. No evidence for evolution of genotypic resistance after three years of treatment with darunavir/ritonavir, with or without nucleoside analogues. AIDS Res Hum Retroviruses. 2012 doi: 10.1089/AID.2011.0256. [DOI] [PubMed] [Google Scholar]
- 11.Condra JH. Resistance to HIV protease inhibitors. Haemophilia. 1998;4:610–615. doi: 10.1046/j.1365-2516.1998.440610.x. [DOI] [PubMed] [Google Scholar]
- 12.Kempf DJ, et al. Incidence of resistance in a double-blind study comparing lopinavir/ritonavir plus stavudine and lamivudine to nelfinavir plus stavudine and lamivudine. J Infect Dis. 2004;189:51–60. doi: 10.1086/380509. [DOI] [PubMed] [Google Scholar]
- 13.Noe A, Plum J, Verhofstede C. The latent HIV-1 reservoir in patients undergoing HAART: an archive of pre-HAART drug resistance. J Antimicrob Chemother. 2005;55:410–412. doi: 10.1093/jac/dki038. [DOI] [PubMed] [Google Scholar]
- 14.Nowak MA, May RMC. Virus Dynamics: Mathematical principles of immunology and virology. Oxford University Press; USA: 2000. [Google Scholar]
- 15.Chou TC. Derivation and properties of Michaelis-Menten type and Hill type equations for reference ligands. J Theor Biol. 1976;59(2):253–276. doi: 10.1016/0022-5193(76)90169-7. [DOI] [PubMed] [Google Scholar]
- 16.Shen L, et al. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med. 2008;14(7):762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampah MES, Shen L, Jilek BL, Siliciano RF. Dose-response curve slope is a missing dimension in the analysis of HIV-1 drug resistance. Proc Natl Acad Sci USA. 2011;108(18):7613–7618. doi: 10.1073/pnas.1018360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drlica K. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother. 2003;52(1):11–17. doi: 10.1093/jac/dkg269. [DOI] [PubMed] [Google Scholar]
- 19.Drlica K, Zhao X. Mutant selection window hypothesis updated. Clin Infect Dis. 2007;44(5):681–688. doi: 10.1086/511642. [DOI] [PubMed] [Google Scholar]
- 20.Wahl LM, Nowak MA. Adherence and drug resistance: predictions for therapy outcome. Proc R Soc B. 2000;267(1445):835–843. doi: 10.1098/rspb.2000.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, et al. Modeling long-term HIV dynamics and antiretroviral response: effects of drug potency, pharmacokinetics, adherence, and drug resistance. J Acquir Immune Defic Syndr. 2005;39(3):272–283. doi: 10.1097/01.qai.0000165907.04710.da. [DOI] [PubMed] [Google Scholar]
- 22.Smith RJ. Adherence to antiretroviral HIV drugs: how many doses can you miss before resistance emerges? Proc R Soc B. 2006;273(1586):617–624. doi: 10.1098/rspb.2005.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rong L, Feng Z, Perelson AS. Emergence of HIV-1 drug resistance during antiretroviral treatment. Bull Math Biol. 2007;69(6):2027–2060. doi: 10.1007/s11538-007-9203-3. [DOI] [PubMed] [Google Scholar]
- 24.Bangsberg DR, Moss AR, Deeks SG. Paradoxes of adherence and drug resistance to HIV antiretroviral therapy. J Antimicrob Chemother. 2004;53(5):696–699. doi: 10.1093/jac/dkh162. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Valero I, Arribas JR. Protease inhibitor monotherapy. Curr Opin Infect Dis. 2011;24:7–11. doi: 10.1097/QCO.0b013e3283422cdf. [DOI] [PubMed] [Google Scholar]
- 26.Bangsberg DR, Kroetz DL, Deeks SG. Adherence-resistance relationships to combination HIV antiretroviral therapy. Curr HIV/AIDS Rep. 2007;4(2):65–72. doi: 10.1007/s11904-007-0010-0. [DOI] [PubMed] [Google Scholar]
- 27.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26(3):585–615. [Google Scholar]
- 28.Jilek BL, et al. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat Med. 2012;18(3):446–451. doi: 10.1038/nm.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen L, et al. A critical subset model provides a conceptual basis for the high antiviral activity of major HIV drugs. Sci Transl Med. 2011;3(91):91ra63. doi: 10.1126/scitranslmed.3002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bangsberg DR, et al. Adherence-resistance relationships for protease and non-nucleoside reverse transcriptase inhibitors explained by virological fitness. AIDS. 2006;20(2):223–231. doi: 10.1097/01.aids.0000199825.34241.49. [DOI] [PubMed] [Google Scholar]
- 31.Nijhuis M, et al. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007;4(1):e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parry CM, et al. Gag determinants of fitness and drug susceptibility in protease Inhibitor- Resistant human immunodeficiency virus type 1. J Virol. 2009;83(18):9094–9101. doi: 10.1128/JVI.02356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dam E, et al. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 2009;5(3):e1000345. doi: 10.1371/journal.ppat.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RK, et al. Full length HIV-1 gag determines protease inhibitor susceptibility within in vitro assays. AIDS (London, England) 2010;24(11):1651–1655. doi: 10.1097/qad.0b013e3283398216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardner EM, et al. Differential adherence to combination antiretroviral therapy is associated with virological failure with resistance. AIDS. 2008;22(1):75–82. doi: 10.1097/QAD.0b013e3282f366ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel JB, Yeh PJ, Chait R, Moellering RC, Kishony R. Drug interactions modulate the potential for evolution of resistance. Proc Natl Acad Sci USA. 2008;105:14918–14923. doi: 10.1073/pnas.0800944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheeseman SH, et al. Phase I/II evaluation of nevirapine alone and in combination with zidovudine for infection with human immunodeficiency virus. J Acquir Immune Defic Syndr. 1995;8(2):141–151. [PubMed] [Google Scholar]
- 38.Ruxrungtham K, et al. Impact of reverse transcriptase resistance on the efficacy of TMC125 (etravirine) with two nucleoside reverse transcriptase inhibitors in protease inhibitor-nave, nonnucleoside reverse transcriptase inhibitor-experienced patients: study TMC125-C227. HIV Medicine. 2008;9(10):883–896. doi: 10.1111/j.1468-1293.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 39.Bonhoeffer S, Nowak MA. Pre-existence and emergence of drug resistance in HIV-1 infection. Proc R Soc B. 1997;264(1382):631–637. doi: 10.1098/rspb.1997.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paredes R, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201:662–671. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jourdain G, et al. Association between detection of HIV-1 DNA resistance mutations by a sensitive assay at initiation of antiretroviral therapy and virologic failure. Clin Infect Dis. 2010;50:1397–1404. doi: 10.1086/652148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simen BB, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 43.Gupta RK, et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under who guidelines: a systematic review and meta-analysis. Lancet Infect Dis. 2009;9:409–417. doi: 10.1016/S1473-3099(09)70136-7. [DOI] [PubMed] [Google Scholar]
- 44.Hoffmann CJ, et al. Viremia, resuppression, and time to resistance in human immunod-eficiency virus (HIV) subtype c during first-line antiretroviral therapy in south africa. Clin Infect Dis. 2009;49:1928–1935. doi: 10.1086/648444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parienti JJ, et al. Not all missed doses are the same: sustained NNRTI treatment inter-ruptions predict HIV rebound at low-to-moderate adherence levels. PLoS ONE. 2008;3(7):e2783. doi: 10.1371/journal.pone.0002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parienti JJ, et al. Average adherence to boosted protease inhibitor therapy, rather than the pattern of missed doses, as a predictor of HIV RNA replication. Clin Infect Dis. 2010;50(8):1192–1197. doi: 10.1086/651419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H, et al. Repeated measures analyses of dose timing of antiretroviral medication and its relationship to HIV virologic outcomes. Stat Med. 2007;26(5):991–1007. doi: 10.1002/sim.2592. [DOI] [PubMed] [Google Scholar]
- 48.Kastrissios H, et al. Characterizing patterns of drug-taking behavior with a multiple drug regimen in an AIDS clinical trial. AIDS. 1998;12(17):2295. doi: 10.1097/00002030-199817000-00011. [DOI] [PubMed] [Google Scholar]
- 49.Sigal A, et al. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature. 2011;477:95–98. doi: 10.1038/nature10347. [DOI] [PubMed] [Google Scholar]
- 50.Kepler TB, Perelson AS. Drug concentration heterogeneity facilitates the evolution of drug resistance. Proc Natl Acad Sci USA. 1998;95(20):11514–11519. doi: 10.1073/pnas.95.20.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J Virol. 2010;84:2395–2407. doi: 10.1128/JVI.01863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Marle G, et al. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology. 2007;4:87. doi: 10.1186/1742-4690-4-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Best B, et al. Efavirenz concentrations in CSF exceed IC50 for wild-type HIV. J Antimicrob Chemother. 2011;66(2):354. doi: 10.1093/jac/dkq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirsch M, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society–USA panel. J Am Med Ass. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 55.Lima VD, et al. Differential impact of adherence on long-term treatment response among naive HIV-infected individuals. AIDS. 2008;22:2371–2380. doi: 10.1097/QAD.0b013e328315cdd3. [DOI] [PubMed] [Google Scholar]
- 56.Boltz VF, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci USA. 2011;108(2):9202–9207. doi: 10.1073/pnas.1105688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nowak MA, et al. Antigenic diversity thresholds and the development of AIDS. Science. 1991;254:963–969. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- 58.Shankarappa R, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73(12):10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neher RA, Leitner T. Recombination rate and selection strength in HIV intra-patient evolution. PLoS Comput Biol. 2010;6(1):e1000660. doi: 10.1371/journal.pcbi.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao X, Drlica K. Restricting the selection of antibiotic-resistant mutant bacteria: measurement and potential use of the mutant selection window. J Infect Dis. 2002;185(4):561–565. doi: 10.1086/338571. [DOI] [PubMed] [Google Scholar]
- 61.Gullberg E, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7(7):e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sedaghat AR, Dinoso JB, Shen L, Wilke CO, Siliciano RF. Decay dynamics of HIV-1 depend on the inhibited stages of the viral life cycle. Proc Natl Acad Sci USA. 2008;105(12):4832. doi: 10.1073/pnas.0711372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ribeiro RM, et al. Estimation of the initial viral growth rate and basic reproductive number during acute HIV-1 infection. J Virol. 2010;84(12):6096–6102. doi: 10.1128/JVI.00127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ribeiro RM, Bonhoeffer S, Nowak MA. The frequency of resistant mutant virus before antiviral therapy. AIDS. 1998;12(5):461. doi: 10.1097/00002030-199805000-00006. [DOI] [PubMed] [Google Scholar]
- 65.Hartl DL, Clark AG. Principles of Population Genetics. Sinauer Associates; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.