Abstract
Regenerative medicine, relying on human embryonic stem cell (hESC) technology, opens promising new avenues for therapy of many severe diseases. However, this approach is restricted by limited production of the desired cells due to the refractory properties of hESC growth in vitro. It is further hindered by insufficient control of cellular stress, growth rates, and heterogeneous cellular states under current culture conditions. In this study, we report a novel cell culture method based on a non-colony type monolayer (NCM) growth. Human ESCs under NCM remain pluripotent as determined by teratoma assays and sustain the potential to differentiate into three germ layers. This NCM culture has been shown to homogenize cellular states, precisely control growth rates, significantly increase cell production, and enhance hESC recovery from cryopreservation without compromising chromosomal integrity. This culture system is simple, robust, scalable, and suitable for high-throughput screening and drug discovery.
Introduction
The capacity of human embryonic stem cells (hESCs) for self-renewal, propagation, and maintenance of the pluripotent state in vitro offers the potential to utilize hESC technology for therapy of many severe human diseases as well as cell-based assays (Klimanskaya et al., 2005; Mallon et al., 2006; Rosler et al., 2004; Thomson et al., 1998; Xu et al., 2001). However, many negative factors contribute to currently inefficient culture systems for hESCs, which have limited the implementation of such a therapy. The ineffectiveness of hESC culture systems is due to (i) low plating efficiency when cells are seeded as single cells or small clumps (Androutsellis-Theotokis et al., 2006; Watanabe et al., 2007), (ii) very low recovery rates when cells are thawed after cryopreservation (Li et al., 2009), and (iii) acquired heterogeneous cellular states owing to various cellular stresses under excessive apoptotic or differentiation signals (Adewumi et al., 2007; Hartung et al., 2010). In addition, xenogeneic contaminants from any non-human feeder cells or foreign components of the culture system may also impede future clinical application (Mallon et al., 2006).
To solve these problems, we need to establish a robust and reliable system for hESC culture and assay. Standard colony-aggregated culture exhibits slow expansion and often gives rise to heterogeneous cells (Adewumi et al., 2007; Hartung et al., 2010) and frequent chromosomal abnormalities (Baker et al., 2007; Draper et al., 2004; Lefort et al., 2008; Maitra et al., 2005; Spits et al., 2008). Hence, a non-colony type culture is preferable. The use of JAK inhibitor I (JAKi) and the Rho-kinase inhibitor Y-27632 (ROCKi) has been shown to significantly improve single-cell plating efficiency in both neural stem cell and hESC cultures (Androutsellis-Theotokis et al., 2006; Chen et al., 2010; Li et al., 2009; Ohgushi et al., 2010; Pakzad et al., 2010; Watanabe et al., 2007).
Intuitively, hESCs with high single-cell plating efficiency using these small molecules could enable us to propagate the cells in a single-cell based non-colony type monolayer (NCM) culture, which would greatly improve the current culture conditions. Various defined substrates have been reported to support hESC culture as colonies under feeder- or xeno-free conditions (Klim et al., 2010; Mallon et al., 2006; Melkoumian et al., 2010; Rodin et al., 2010; Villa-Diaz et al., 2010). The use and characterization of a NCM method as an independent culture system for the maintenance of undifferentiated hESC lines under defined substrate conditions have not been reported. In this study, we report such a hESC culture system for facilitating pluripotent stem cell growth and assays.
Materials and methods
Human ES cell lines
The hESC lines used in this study include: hESBGN-01 (NIH Code: BG01), hESBGN-02 from BresaGen Inc. (Athens, GA); hES-1 and hES-4 (NIH codes: ES01 and ES04) from ES International (Singapore); I-3 (NIH code: TE03) from Technion-Israel Institute of Technology (Haifa, Israel); HSF-6 (NIH Code: UC06) from (University of California at San Francisco, San Francisco, CA); H1 (NIH Code: WA01), H7 (WA07), H9 (WA09), H13 (WA13), and H14 (WA14) from Wisconsin Alumni Research Foundation (WiCell Research Institute, Madison, WI); and SA001 (NIH code: SA01) from CellArtis AB (Göteborg, Sweden).
Induced pluripotent stem cell (iPSC) lines
We generated the iPSC line SCU-i10 by reprogramming bone marrow stromal cells using lentiviral transduction of the cells with the STEMCCA vector (Millipore, Billerica, MA), which contains the four transcription factors Oct-4, Klf4, SOX-2, and c-Myc (Kozhich et al., 2012). The BC1 iPSC line was provided by Dr. Guokai Chen (The National Heart, Lung, and Blood Institute, Bethesda, MD) (Chou et al., 2011).
Adaptation to single-cell based non-colony type monolayer (NCM) culture of hESCs
Human ES cells, initially grown as colonies on X-ray irradiated mouse embryonic fibroblasts (MEFs), were dissociated by collagenase IV for 15 to 30 min. The cell pellets were washed once in D-PBS and then incubated with 1X Accutase™ (Innovative Cell Technologies, Inc., San Diego, CA) for 15 min. The cell suspension was briefly resuspended in MEF-conditioned medium (MEF-CM) and centrifuged at 220×g for 5 min. Dissociated single cells were filtered through 40-μm BD Falcon™ Cell Strainer (BD Biosciences) to eliminate cell aggregates. Approximately 1.3 to 2×106 hESCs were seeded in one well (1.35 to 2.1×105 cells/cm2) of a 6-well plate coated with 7.5% hESC-qualified Matrigel (BD Biosciences) in MEF-CM supplemented with 100 ng/ml of FGF-2 and 10 μM ROCKi (or 1 μM JAKi) to facilitate the initial 24-hour single-cell plating. After 24 h, the medium was replaced with 50% regular hESC medium and 50% MEF-CM containing 4 ng/ml of FGF-2. Approximately, 85% single-cell plating efficiency was achieved. The cells were allowed to grow as a single-cell-formed monolayer for 3 days with medium change daily. At day 3 or day 4 (depending on the confluence of the culture), the cells are dissociated in Accutase™ for the second passage. Once the hESCs are adapted to the NCM condition, the number of cells plated and time for cell passage can be fixed to achieve reproducible cultures and consistent growth rates. Alternatively, hESCs can be grown under the above NCM method on 2.5% Matrigel in mTeSR ™1 medium with the aid of 10 μM ROCKi or 1 μM JAKi for the initial 24-hour plating.
Cytogenetic karyotyping and fluorescence in situ hybridization (FISH)
Cytogenetic analysis using G-banding was performed with 20 metaphases of hESCs to detect clonal chromosomal abnormalities by Cell Line Genetics (Madison, WI). Since the most frequent chromosomal changes in hESCs cultured on MEF feeder layers are trisomy 17 and trisomy 12 (Baker et al., 2007), we also monitored alterations of both chromosomes 17 and 12 by FISH in 200 to 300 interphase nuclei of hESCs. The detailed procedures for both G-banding and FISH were recently documented (Meisner and Johnson, 2008).
Immunofluorescence microscopy
Human ES cells were grown in 6-well plates, rinsed with D-PBS three times, and fixed in 4% paraformaldehye at room temperature for 20 min. The cells were blocked with 10% normal goat serum in the presence or absence of 0.1% Triton X-100 (in D-PBS) at room temperature for 1 h, reacted with primary antibodies in 5% normal goat serum at room temperature for 2 h, and subsequently incubated with Alexa Fluor® conjugated secondary antibodies in 5% normal goat serum at room temperature for 1 h. Note that between each individual step, the cells were always rinsed with D-PBS 3 times, 5 min each. Finally, the cells were stained with Hoechst 33342 solution. The samples were examined under an Axiovert 200 fluorescence microscope (Zeiss, Jena, Germany) equipped with Adobe-Photohop® or ApoTOME AxioVision Rel 4.6 (Zeiss) acquisition systems. All images used for quantitative analysis were obtained under unsaturated exposure conditions and analyzed by the ImageJ program (National Institutes of Health, Bethesda, MD).
Teratoma formation assays
WA01 (H1) cells under NCM conditions (passage number: 13) were grown on 5% Matrigel (BD Biosciences) and shipped to Applied StemCell Inc. (Menlo Park, CA) for teratoma formation assays. The cells were dissociated with dispase and triturated as small clumps and injected into kidney capsule and testis sites (~2×106 cells per site) of three 6-week-old male mice (strain: Fox Chase SCID®-beige from Charles River Laboratories, Wilmington, MA). Cells from a human iPSC line were injected into the kidney capsule and testis sites of one mouse as control. Tumor formation was observed in the three mice and harvested at day 40 post injection. The tumor samples at the 6 sites were fixed in 10% formalin overnight, embedded in paraffin, cut into 5-μm serial sections, stained with hematoxylin and eosin (H&E), and photographed for histological analyses.
Full methods and associated references regarding hESC culture on MEF feeder layers, Matrigel-mediated NCM, flow cytometry, cDNA microarray, array-based CGH, differentiation, plasmid DNA transfection, lentiviral transduction, and 96-well based high-throughput assays can be found in Supplemental Information and the website of the NIH Stem Cell Unit (http://stemcells.nih.gov/research/nihresearch/scunit/).
Results and discussion
Growth control under the non-colony type monolayer (NCM) culture
Adaptation to the NCM method proceeds through four phases that include high-density single-cell plating, multicellular association, monolayer fusion, and cellular condensation (Fig. 1, Supplemental Figs. 1 and 2). We found that to maintain a healthy undifferentiated culture, it was essential to plate the cells at high density (1.4 to 2.1 ×106 per well in a 6-well plate) and passage strictly upon reaching confluence, usually at day 3 or 4 (Fig. 1A). Although this NCM method was initially developed using MEF-conditioned media on 7.5% Matrigel-coated plastic-ware, it was readily adapted to a defined medium, mTeSR™1, on 2.5% Matrigel-coated plasticware (Fig. 1, Table 1, Supplemental Figs. 1 and 2). Eleven hESC and two induced pluripotent stem cell (iPSC) lines were adapted to NCM under this growth condition and showed similar growth patterns (Table 1).
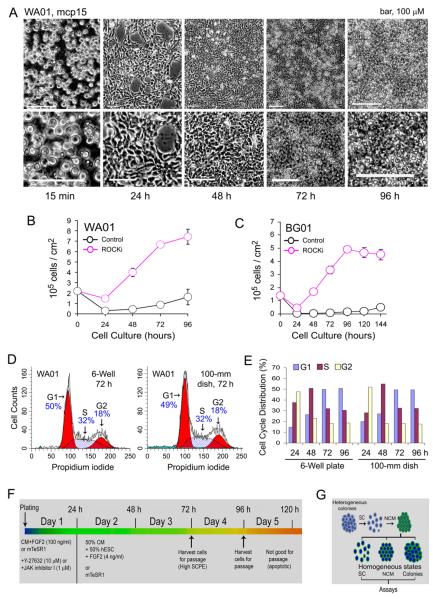
Figure 1.
Control of hESC growth rates by non-colony type monolayer (NCM) culture. (A) Representative phase images of the NCM culture of WA01 cells (passage number 15 or mcp15) at different time points after plating on 2.5% BD Matrigel with the use of 10 μM ROCK inhibitor Y-27632 (ROCKi). Phase images in the lower panel of Figure 1A are regionally enlarged views of the images in the upper panel. (B and C) Growth curves of both WA01 and BG01 cell lines were determined in 2.5% BD Matrigel in the presence or absence of 10 μM Y-27632 (ROCKi) for the initial 24-hour single-cell plating. (D and E) Cell cycle analysis was performed in WA01 cells grown under NCM conditions in both 6-well plates and 100-mm culture dishes (1.3×107 cells per dish) at different time points. Representative histograms are shown. (F) A schema of the NCM culture. (G) A flow chart for the generation of homogeneous hESC conditions (e.g., SC, single cells; NCM; and the NCM-derived colonies) via the NCM intermediate step. Abbreviations: SCPE, single-cell plating efficiency.
Table 1.
Non-colony type monolayer culture (NCM) of hESC and iPSC lines.a
| Cell | Resource | pb | mcpc | Substrate | Medium | Inhibitord | Karyotypee |
|---|---|---|---|---|---|---|---|
| BG01 | BresaGenf | 67 | 10 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| ES01 | ESCI | 74 | 9 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Trisomy 20, 14% |
| ES04 | ESCI | 63 | 9 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| WA01 | WiCell | 35 | 10 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| WA07 | WiCell | 35 | 12 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| WA09 | WiCell | 33 | 11 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Elevated polyploidy |
| WA09 | WiCell | 33 | 12 | BD Purecoat/FN | X-KSR/TeSR2 | Y-27632 | Elevated polyploidy |
| WA13 | WiCell | 27 | 10 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| WA14 | WiCell | 30 | 10 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| WA14 | WiCell | 30 | 11 | Matrigel (2.5%) | TeSR2 | Y-27632 | Normal |
| WA14 | WiCell | 30 | 30 | Matrigel (2.5%) | TeSR2 | Y-27632 | Normal |
| WA14 | WiCell | 30 | 11 | BD Purecoat/FN | X-KSR/TeSR2 | Y-27632 | Normal |
| WA14 | WiCell | 30 | 7 | BD Purecoat/FN | XGFS/TeSR2 | Y-27632 | NDg |
| TE03 | Technion | 59 | 6 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| UC06 | UCSF | 67 | 12 | Matrigel (2.5%) | mTeSR1 | JAKi | Normal |
| UC06 | UCSF | 67 | 7 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| SA01 | CellArtis AB | 22 | 3 | BD Purecoat | MEF-CM | Y-27632 | ND |
| BC1 | Chen Gh | 64 | 10 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
| SCU-i10 | NIH SCU | ND | 13 | Matrigel (2.5%) | mTeSR1 | Y-27632 | Normal |
Notes and abbreviations:
All hESC cell lines described above were initially cultured on MEF-feeder layers as documented on the website of the NIH Stem Cell Unit (SCU). BG01 cells (p70) were adapted to NCM conditions in the presence of 50% MEF-conditioned medium and 50% regular hESC medium (containing 4 ng/ml FGF2). Additionally, 10 hESC cell lines were cultured under NCM conditions in mTesR medium (StemCell Technologies) or 2% XGFS/TeSR2 or 10% X-KSR/TeSR2 as indicated.
Passage numbers, designated for hESC lines grown on MEF-feeder layer.
Passage numbers, designated for the NCM culture of hESCs grown on Matrigel.
The chemical molecules, Y27632 (10 μM) and JAK inhibitor I (JAKi, 1 μM), were used for enhancing single-cell plating efficiency within the initial 24 h after plating the cells. The inhibitors were then removed and cells were grown in the media described above.
Karyotype: elevated percentages of polyploidy were found in WA09 (H9) cells.
Abbreviations are: BresaGen: BresaGen, Inc., Athens, GA; ESCI, ES Cell International; FN, fibronectin; Technion: Technion-Israel Institute of Technology, Haifa, Israel; UCSF: University of California, San Francisco, CA; and WiCell: Wisconsin Alumni Research Foundation (WARF) (WiCell Research Institute), Madison, WI.
ND, not determined.
Provided by Dr. Guokai Chen (NIH).
In general, cells with higher single-cell plating efficiency exhibit an exponential growth pattern between 24 and 72 h (Figs. 1B and C), followed by a delayed growth period and a stage with decreased cell numbers (Fig. 1C). Under these optimized growth conditions, hESCs exhibit a cell cycle profile (Figs. 1D and E, Supplemental Fig. 1) that corresponds to different phases on the growth curves (Figs. 1B and C). Moreover, the mode of cell cycle distribution remains consistent when WA01 cells were grown in a scaled-up condition (i.e., plating 2.1×107 hESCs per 100-mm culture dish) (Figs. 1D and E). Consequently, by selecting specific time windows after plating, we could precisely select a population of cells with specific growth rates, cell density, and cell cycle.
Human ES cells under the NCM culture have normal karyotypes
To exclude the existence of potential chromosomal or genomic abnormalities in hESCs under NCM, we performed cytogenetic karyotyping, fluorescence in situ hybridization (FISH), and array-based comparative genomic hybridization (aCGH) (Fig. 2; Supplemental Fig. 3, A-C, Table 1). Karyotypes were reported normal at passage 10 in WA01 cells (Fig. 2A) and at passage 33 in WA14 cells (Fig. 2B). Similarly, BG01 cells were also reported karyotypically normal (Supplemental Fig. 3A) and showed no gross abnormalities by aCGH up to 20 passages (Supplemental Fig. 3C). In addition, no trisomy of chromosomes 17 or 12, the most common abnormalities arising in hESCs in culture, was found by FISH in BG01 cells (Supplemental Fig. 3B). We performed karyotype analysis in 8 additional hESC and 2 iPSC lines under NCM conditions (Table 1). Although the majority of cell lines showed normal karyotypes, one line, ES01, showed 14% of trisomy 20 at passage 9 (Table 1). Elevated polyploidy was also found in WA09 cells. In these cases, it is not clear whether these genomic abnormalities were due to the consequences of NCM or pre-existing alterations under the colony-type culture of the same cells on MEF feeder layers. Nonetheless, these data suggest that we can grow hESCs under NCM without disturbing chromosomal stability.
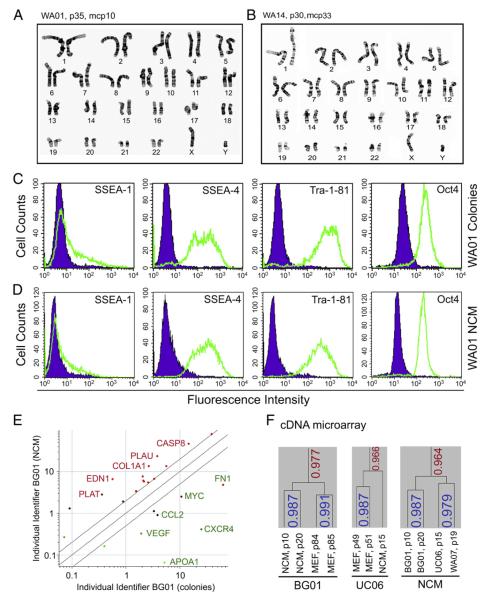
Figure 2.
Characterization of hESCs under NCM conditions. (A) Karyotyping of WA01 cells cultured on MEFs for 35 passages (p35) and then grown under NCM conditions for 10 passages (i.e., mcp10). (B) Karyotype analysis of WA14 cells grown on MEFs for 30 passages (p30) and under NCM for 33 passages (mcp33). (C and D) Flow cytometric analysis of hESC marker expression in WA01 cells at mcp15 and WA01 cells grown on MEFs. (E) Two-way microarray analysis of BG01 cells grown as colonies and as NCM. Two-fold changes in mRNA levels (P<0.05) after normalization are shown. Partial up-regulated (red) and down-regulated (green) genes are indicated. (F) Hierarchical cluster analysis of mRNA expression in hESCs under NCM conditions and colony-type culture on MEFs.
Expression of hESC markers in hESCs with NCM
With respect to hESC markers, NCM not only retained the expression of all examined hESC markers, but also reduced SSEA-4 heterogeneity observed in both WA01 (Figs. 2C and D) and BG01 cells grown on MEFs by flow cytometric analysis (Supplemental Fig. 3D). Interestingly, there was an increase in the fluorescence intensity of Oct-4 in both WA01 and BG01 cells under NCM compared with their control (Figs. 2C and D; Supplemental Fig. 3D). A similar Oct-4 expression pattern was also found in NCM of UC06 cells (data not shown). In addition, immunostaining indicates that NANOG and Oct-4 are coexpressed in both WA01 and WA09 hESC lines under NCM conditions (Supplemental Fig. 4).
Genome-wide mRNA expression in hESCs with NCM
We continued to characterize hESCs under NCM in terms of gene expression profile using cDNA microarray analysis (Figs. 2E and F). Two-way microarray analysis in a representative hESC line (i.e., BG01) revealed that the mRNA profile under NCM was comparable to that of BG01 colonies grown on MEFs (Fig. 2E). Only limited numbers of genes (n = 25) were found to be expressed at significantly different levels (P<0.05) between the NCM and colony-type culture on MEFs (Fig. 2E, Supplemental Table 1). Similar results from the two-way microarray analysis were also found in WA07 cells. Both types of culture shared 97.7% similarity in BG01 cells and 96.6% similarity in UC06 cells based on their mRNA expression patterns (Fig. 2F). Interestingly, hESC lines BG01, WA07, and UC06 under NCM conditions had an overall 96.4% similarity (Fig. 2F). These data suggest that hESCs under both conditions exhibit similar mRNA expression patterns.
Dynamic changes in cell cycle are not associated with differentiation
We should point out that the dynamic change of cell cycle parameters (e.g., elevated G1 fractions at 72 and 96 h) is an important property of hESCs under NCM conditions. Flow cytometric analysis, cDNA microarray, and immunofluorescence were performed in hESCs under the NCM culture at 72 and 96 h. Our results indicate that hESCs (e.g., WA01 cells) with an elevated G1 fraction still retained the expression of pluripotent markers and had decreased the expression of the differentiation marker SSEA-1 compared with hESC colonies (Figs. 2C and D). Interestingly, the intensity of the pluripotent marker Oct4 was increased in BG01 cells at this stage under NCM (Supplemental Fig. 3D). Two-way gene expression analyses also indicate that there is no increase in expression of genes or gene clusters that are commonly associated with differentiation pathways (Figs. 2E and F). In addition, we observed that the increase in G1 fraction under NCM was reduced 24 h after passaging the cells, indicating that it is a transient, not progressive, state. Taken together, these data suggest that the increase in G1 fraction at 72 and 96 h reflects only the change of growth rates that correspond to the growth curves, and is not associated with differentiation.
Pluripotency assays of hESCs under NCM culture conditions
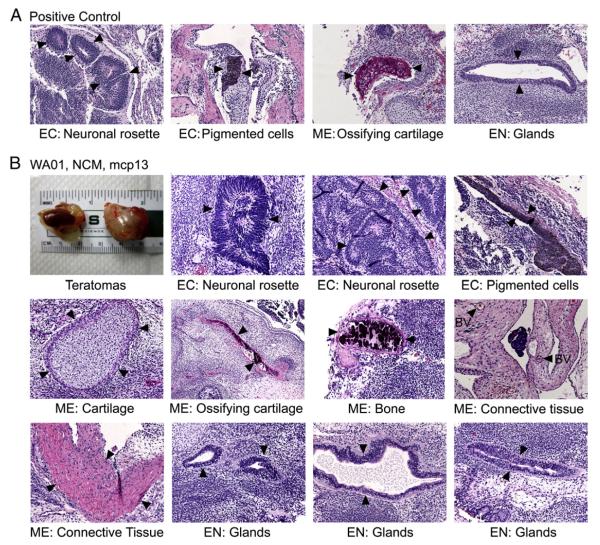
We used teratoma assays to define the pluripotent state of WA01 cells under NCM conditions (Fig. 3). We found that three kidney and three testis tumors were composed of scattered and differentiated cells and a large population of undifferentiated neoplastic cells (Fig. 3). Organized structures representative of the three germ layers (i.e., ectoderm, mesoderm, and endoderm) were clearly identified by histological analyses. Such structures include neuronal rosettes, pigmented cells, cartilage, ossifying cartilage, bony tissues, and different glandular structures (Fig. 3B). These data suggest that hESCs such as WA01 cells under the NCM culture retain pluripotency.
Figure 3.
Teratoma formation assays. (A) Teratoma formation control from an iPSC line. (B) Teratoma formation assays in WA01 cells under NCM conditions (passage number 13, designated as mcp13). Cells were injected into the kidney capsule and testis sites of three mice. Tumor samples at day 40 after injection were fixed and stained with hematoxylin and eosin (H&E), and photographed for histological analyses. Variously differentiated tissues that represent the three germ layers are shown and indicated by arrowheads. Abbreviations: BV, blood vessels; CT, connective tissues; EC, ectoderm; EN, endoderm; ME, mesoderm.
In vitro differentiation potentials of hESCs under NCM culture conditions
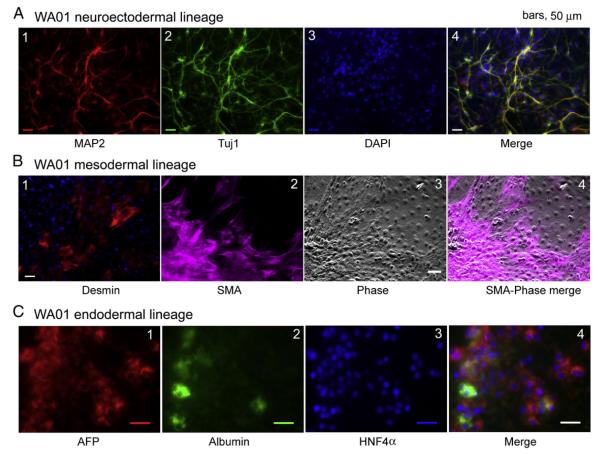
We examined the potentials of hESCs under NCM conditions to differentiate into cell types representative of the three germlayers in vitro. We converted WA01 cells to neurons (ectoderm lineage) using a recently described protocol (Kozhich et al., 2012). This is demonstrated by the neuronal morphology of the differentiated cells and by positive immunostaining with specific antibodies to the neural stem cell marker, nestin, and the neuronal markers MAP2 and β-tubulin III (Tuj1) (Fig. 4A). Similar results were also shown in UC06 and in the iPSC line BC1 under the NCM culture (Supplemental Figs. 5A and 6A). No glial fibrillary acidic protein (GFAP), an intermediate filament protein specifically associated with astrocytes, was found among derived neurons (Supplemental Fig. 5A). These data suggest that hESCs and some iPSCs under NCM retain the potential to differentiate into neuroectodermal precursors and neurons.
Figure 4.
Human ES cells under NCM culture maintain the potential to differentiate to multiple cellular fates in vitro. (A) Neuronal differentiation of WA01 cells: differentiated cells were examined with specific antibodies against MAP2 and β-tubulin III (Tuj1). The cells were co-stained with 4′,6-diamidino-2-phenylindole (DAPI). The images were merged. (B) Mesodermal differentiation of WA01 cells: differentiated cells were stained with specific antibodies against desmin and smooth muscle actin (SMA). (C) Endodermal and hepatic differentiation of WA01 cells: expression patterns of the hepatic markers α-fetoprotein (AFP), human albumin, and HNF4α by immunostaining the cells with specific antibodies.
We also examined the ability of the NCM-based growth of hESCs to differentiate toward mesodermal fates. With a spontaneous differentiation protocol in the presence of 20% fetal bovine serum (FBS), WA01 cells were readily directed into multiple cell types that included desmin immunopositive cells and cells that demonstrated smooth muscle actin (SMA) immunoreactivity (Fig. 4B). Furthermore, under this spontaneous differentiation condition, both UC06 and BC1 cells also generated patches of synchronously contracting cells indicative of cardiomyocytes (Supplemental Fig. 6B, Supplemental Movies 1 and 2). Hence, our data indicate that NCM sustains the potential of hESCs or iPSCs to differentiate toward mesodermal fates.
Finally, we tested the ability of WA01 cells under NCM conditions to differentiate into endodermal fates (Fig. 4C). These differentiated cells expressed early (i.e. α-fetoprotein or AFP) and intermediate markers of hepatic fates such as albumin and hepatocyte nuclear factor 4α (HNF4α) (Fig. 4C). The capacity of hESCs under NCM conditions to differentiate toward hepatic lineages was also demonstrated in both WA14 and BC1 cells (Supplemental Figs. 5B and 6C). Co-expression of α-fetoprotein with both cytokeratin 8 and albumin was also evident in WA14 differentiated cells (Supplemental Fig. 5B). Taken together, our data indicate that the NCM culture supports the growth of hESCs, retains the pluripotent state, and sustains the potential to differentiate into the three germ layers.
Non-colony type monolayer culture is a novel method for controlling hESC growth
Various single-cell passaging methods have been documented in the literature in which cells are enzymatically dissociated to single cells or to 2-5 cell groups during passage (Bajpai et al., 2008; Chan et al., 2008; Costa et al., 2008; Damoiseaux et al., 2009; Ellerstrom et al., 2007; Emre et al., 2010; Hanna et al., 2010; Hockemeyer et al., 2009; Kibschull et al., 2011; Krawetz et al., 2009; Li et al., 2009; Mei et al., 2010; Saha et al., 2011; Tsutsui et al., 2011; Ungrin et al., 2008; Xu et al., 2010). In contrast to our method, all of above culture methods employed colony-type cultures. For example, Wu and colleagues used dissociated cells for analysis of the effects of small-molecule cocktails that support the maintenance of hESCs (Tsutsui et al., 2011). In their report, the cells had 20 passages under the dissociated condition, but these cells were grown and assayed as colonies (Tsutsui et al., 2011). Thus, the fundamental difference between our NCM culture and previous cultures using single-cell passaging is that their final products of hESC culture are colonies. These colony-type cultures have been shown to associate with heterogeneous cellular states and the occurrence of karyotypically abnormal hESCs (Baker et al., 2007; Draper et al., 2004; Lefort et al., 2008; Maitra et al., 2005; Spits et al., 2008). Thus, no independent culture systems based on NCM have been previously developed and characterized.
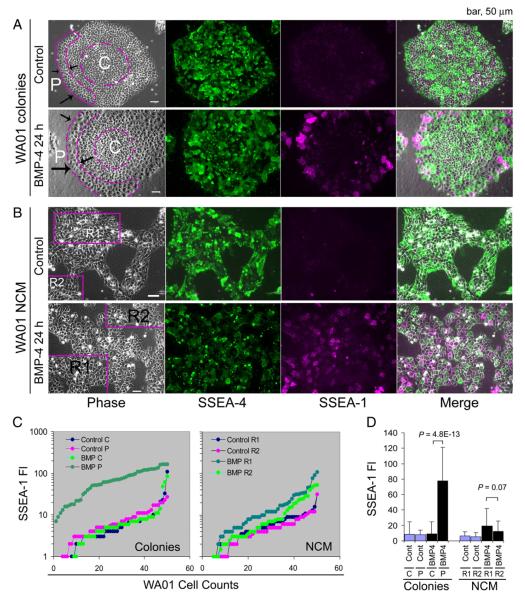
Moreover, the colony-type culture generates heterogeneous hESCs regardless of the size of cell clumps plated, even in those colonies grown from single cells. We found that hESCs at different zones of a colony have differential responses to signal molecules. For example, the peripheral cells in a WA01 colony are 8.6-fold more sensitive to BMP4-induced differentiation (as manifested by elevated SSEA-1 expression) than the cells at the center of the colonies (P=4.8×10−13; Figs. 5A, C,D). In contrast, cells under NCM conditions showed a more homogeneous response to BMP-4 signaling (i.e., variances ranging from 184 to 512) than those WA01 cells in the colonies (variances ranging from 231 to 2156) (Figs. 5B-D).
Figure 5.
Cellular responses of hESCs to BMP-4 signaling. (A) WA01 cells were grown as colonies on 2.5% BD Matrigel in mTeSR™1 medium and treated with or without 100 ng/ml of BMP-4 for 24 h. (B) WA01 cells were cultured under NCM conditions and treated with or without 100 ng/ml of BMP-4 for 24 h. Cells were fixed and immunostained with antibodies against the surface markers SSEA-1 and SSEA-4. (C) Quantitative analysis of SSEA-1 expression in cells located at central (C) and peripheral (P) regions of the WA01 colonies and in WA01 cells from different regions (i.e., R1 and R2) under NCM conditions by the ImageJ program. Initially, fifty WA01 cells were randomly assigned from the indicated regions of interest (ROI) in the phase images. SSEA-1 fluorescence intensity (FI) was then measured at the single pixel level at the plasma membrane region of each cell. SSEA-1 fluorescence intensity values were sorted and presented as line graphs in (C) to visualize the heterogeneous signaling responses. (D) Statistical analysis of the mean (columns) and standard deviation (bars) of SSEA-1 fluorescence intensity among the 50 cells presented in (C). P values were derived from an unpaired and two-sided student t-test. Abbreviations: BMP, BMP-4; Cont, BMP-4 untreated control.
Thus, several immediate advantages of the NCM method are to homogenize heterogeneous cellular states in hESC colonies and to establish standards to assess hESC biology under various multicellular associations (Fig. 1G). Since the production and quality of hESCs from colony-type cultures in vitro are not always consistent, the NCM culture provides an efficient way to eliminate such inconsistency by precisely regulating the growth rate and cell numbers in a density- and time-dependent manner (Fig. 1).
Other applications of the NCM culture
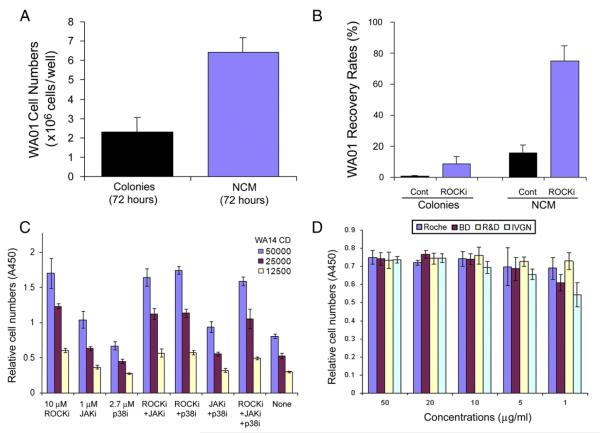
There are many other advantages to applying NCM. We have shown that NCM increases WA01 cell production by approximately 2.8-fold when compared with the colony-type culture of the same cells on MEFs (Fig. 6A), and significantly enhances recovery rate of WA01 cells (i.e., 75%) from frozen single-cell suspensions with the use of 10 μM ROCKi (Fig. 6B). In contrast, the recovery rate of frozen clumps of WA01 cells, grown as colonies on MEFs, was approximately 8.7% with the use of ROCKi and 0.7% without ROCKi. Similar results were also found in UC06 cells (Supplemental Fig. 7).
Figure 6.
Applications of NCM culture. (A) Comparison of cell yields under NCM to conventional colony-type culture of hESCs (e.g., WA01 cells) on MEF feeder layers. (B) Recovery rate of WA01 cells: WA01 cells were cultured under ncm conditions, frozen in liquid nitrogen, and thawed in the presence or absence of 10 μM ROCKi. WA01 colonies, grown on MEFs, were used for comparison. The cell numbers were counted 24 h after thawing. (C) High throughput assay of growth conditions for hESCs in a 96-well format: to demonstrate the ease with which the NCM method may be used in high throughput applications, we performed single-cell plating efficiency assays of WA14 hESCs under NCM conditions with various small-molecules and their combinations. (D) Cell survival assays: to test different sources (i.e., purchased from Roche, R&D Systems, Invitrogen, and BD Biosciences; abbreviated as Roche, R&D, IVGN, and BD respectively) and concentrations of fibronectin on plating efficiency using a 96-well format: approximately 31,000 WA14 cells were plated in 10% KSR-X/TeSR2 on BD Purecoat (BD Biosciences), additionally coated with a range of concentrations of human fibronectin from different vendors. After 24 h, the cells were subjected to the CCK-8 survival assay (Dojino Molecular Technologies, Rockville, MD) by measuring the absorbance at 450 nm (A450). Abbreviations: Cont, drug untreated control; JAKi, JAK inhibitor I; p38i, the p38MAPK inhibitor SB203580; ROCKi, the ROCK inhibitor Y27632; WA14 CD, WA14 cell density.
With respect to genetic manipulation, hESCs under colony-type culture on feeder layers had low transfection efficiencies and a greater variability (i.e., transfection efficiency ranging from 3 to 35%) (Braam et al., 2008; Liew et al., 2007). We also found no detectable transduced signals in BG01 cells under the colony-type culture condition (Supplemental Fig. 8B, upper panel). However, approximately 75% to 77% transduction efficiencies were achieved in BG01 cells under the NCM condition (Supplemental Fig. 8B, middle and lower panels). These transduction efficiencies are comparable to previously reported efficiencies (~90%) mediated by lentivirus (Braam et al., 2008). Thus, the NCM culture represents a simple, robust, and scalable system that may be particularly suitable for genetic manipulation of hESCs (Supplemental Fig. 8).
Finally, we also provide high-throughput proof-of-principle experiments in a 96-well format in which we screened small molecules that affect cell survival individually and in combination (Fig. 6C). We examined the effects of different sources and concentrations of fibronectin on hESC plating efficiency in a xeno-free culture system using the96-well format (Fig. 6D). These data suggest a possible use of this method in high-throughput assays for pharmacological drug screening and drug discovery.
Conclusions
We describe a NCM method for culturing hESCs with defined feeder-free conditions. Human ES cells under these conditions are cytogenetically stable. This novel culture system is simple, robust, and scalable, making it ideal for the efficient production of a large amount of homogeneously undifferentiated hESCs for regenerative medicine and high-throughput drug discovery.
Supplementary Material
Acknowledgments
The NIH Stem Cell Unit is supported by NIH funds. We thank Dr. Joshua Chenoweth for the discussion, Dr. Peter Andrews for providing antibodies described in Supplemental information, Dr. Guokai Chen for the BC1 line, and Dr. Tianmin Ivy Zhang and Dr. Qi Zheng (Applied StemCell Inc.) for conducting the teratoma assays.
Abbreviations
- hESCs
human embryonic stem cells
- NCM
non-colony type monolayer.
Footnotes
Disclosure of potential conflicts of interest
The authors indicate no potential conflict of interest.
Supplementary materials related to this article can be found online at http://dx.doi.org/10.1016/j.scr.2012.06.003.
References
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- Bajpai R, Lesperance J, Kim M, Terskikh AV. Efficient propagation of single cells accutase-dissociated human embryonic stem cells. Mol. Reprod. Dev. 2008;75:818–827. doi: 10.1002/mrd.20809. [DOI] [PubMed] [Google Scholar]
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Braam SR, Denning C, van den Brink S, Kats P, Hochstenbach R, Passier R, Mummery CL. Improved genetic manipulation of human embryonic stem cells. Nat. Methods. 2008;5:389–392. doi: 10.1038/nmeth.1200. [DOI] [PubMed] [Google Scholar]
- Chan EM, Yates F, Boyer LF, Schlaeger TM, Daley GQ. Enhanced plating efficiency of trypsin-adapted human embryonic stem cells is reversible and independent of trisomy 12/17. Cloning Stem Cells. 2008;10:107–118. doi: 10.1089/clo.2007.0064. [DOI] [PubMed] [Google Scholar]
- Chen G, Hou Z, Gulbranson DR, Thomson JA. Actinmyosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240–248. doi: 10.1016/j.stem.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J, Cheng L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Sourris K, Hatzistavrou T, Elefanty AG, Stanley EG. Expansion of human embryonic stem cells in vitro. Curr. Protoc. Stem Cell Biol. 2008 doi: 10.1002/9780470151808.sc01c01s5. Chapter 1: Unit 1C.1.1-1C.1.7. [DOI] [PubMed] [Google Scholar]
- Damoiseaux R, Sherman SP, Alva JA, Peterson C, Pyle AD. Integrated chemical genomics reveals modifiers of survival in human embryonic stem cells. Stem Cells. 2009;27:533–542. doi: 10.1634/stemcells.2008-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat. Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- Ellerstrom C, Strehl R, Noaksson K, Hyllner J, Semb H. Facilitated expansion of human embryonic stem cells by single-cell enzymatic dissociation. Stem Cells. 2007;25:1690–1696. doi: 10.1634/stemcells.2006-0607. [DOI] [PubMed] [Google Scholar]
- Emre N, Vidal JG, Elia J, O’Connor ED, Paramban RI, Hefferan MP, Navarro R, Goldberg DS, Varki NM, Marsala M, Carson CT. The ROCK inhibitor Y-27632 improves recovery of human embryonic stem cells after fluorescence-activated cell sorting with multiple cell surface markers. PLoS One. 2010;5:e12148. doi: 10.1371/journal.pone.0012148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung O, Huo H, Daley GQ, Schlaeger TM. Clump passaging and expansion of human embryonic and induced pluripotent stem cells on mouse embryonic fibroblast feeder cells. Curr. Protoc. Stem Cell Biol. 2010 doi: 10.1002/9780470151808.sc01c10s14. Chapter 1: Unit 1C.10.1-1C.10.15. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibschull M, Mileikovsky M, Michael IP, Lye SJ, Nagy A. Human embryonic fibroblasts support single cell enzymatic expansion of human embryonic stem cells in xeno-free cultures. Stem Cell Res. 2011;6:70–82. doi: 10.1016/j.scr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat. Methods. 2010;7:989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimanskaya I, Chung Y, Meisner L, Johnson J, West MD, Lanza R. Human embryonic stem cells derived without feeder cells. Lancet. 2005;365:1636–1641. doi: 10.1016/S0140-6736(05)66473-2. [DOI] [PubMed] [Google Scholar]
- Kozhich OA, Hamilton RS, Mallon BS. Standardized generation and differentiation of neural precursor cells from human pluripotent stem cells. Stem Cell Rev. Rep. 2012 doi: 10.1007/s12015-012-9357-8. http://dx.doi.org/10.1007/s12015-012-9357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz RJ, Li X, Rancourt DE. Human embryonic stem cells: caught between a ROCK inhibitor and a hard place. Bioessays. 2009;31:336–343. doi: 10.1002/bies.200800157. [DOI] [PubMed] [Google Scholar]
- Lefort N, Feyeux M, Bas C, Feraud O, Bennaceur-Griscelli A, Tachdjian G, Peschanski M, Perrier AL. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat. Biotechnol. 2008;26:1364–1366. doi: 10.1038/nbt.1509. [DOI] [PubMed] [Google Scholar]
- Li X, Krawetz R, Liu S, Meng G, Rancourt DE. ROCK inhibitor improves survival of cryopreserved serum/feeder-free single human embryonic stem cells. Hum. Reprod. 2009;24:580–589. doi: 10.1093/humrep/den404. [DOI] [PubMed] [Google Scholar]
- Liew CG, Draper JS, Walsh J, Moore H, Andrews PW. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1521–1528. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, Brimble SN, et al. Genomic alterations in cultured human embryonic stem cells. Nat. Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Park KY, Chen KG, Hamilton RS, McKay RD. Toward xeno-free culture of human embryonic stem cells. Int. J. Biochem. Cell Biol. 2006;38:1063–1075. doi: 10.1016/j.biocel.2005.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, Cho SW, Mitalipova M, Pyzocha N, Rojas F, et al. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat. Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner LF, Johnson JA. Protocols for cytogenetic studies of human embryonic stem cells. Methods. 2008;45:133–141. doi: 10.1016/j.ymeth.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Melkoumian Z, Weber JL, Weber DM, Fadeev AG, Zhou Y, Dolley-Sonneville P, Yang J, Qiu L, Priest CA, Shogbon C, et al. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat. Biotechnol. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, Muguruma K, Nakano T, Suga H, Ueno M, et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- Pakzad M, Totonchi M, Taei A, Seifinejad A, Hassani SN, Baharvand H. Presence of a ROCK inhibitor in extracellular matrix supports more undifferentiated growth of feeder-free human embryonic and induced pluripotent stem cells upon passaging. Stem Cell Rev. 2010;6:96–107. doi: 10.1007/s12015-009-9103-z. [DOI] [PubMed] [Google Scholar]
- Rodin S, Domogatskaya A, Strom S, Hansson EM, Chien KR, Inzunza J, Hovatta O, Tryggvason K. Long-term selfrenewal of human pluripotent stem cells on human recombinant laminin-511. Nat. Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- Rosler ES, Fisk GJ, Ares X, Irving J, Miura T, Rao MS, Carpenter MK. Long-term culture of human embryonic stem cells in feeder-free conditions. Dev. Dyn. 2004;229:259–274. doi: 10.1002/dvdy.10430. [DOI] [PubMed] [Google Scholar]
- Saha K, Mei Y, Reisterer CM, Pyzocha NK, Yang J, Muffat J, Davies MC, Alexander MR, Langer R, Anderson DG, Jaenisch R. Surface-engineered substrates for improved human pluripotent stem cell culture under fully defined conditions. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18714–18719. doi: 10.1073/pnas.1114854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits C, Mateizel I, Geens M, Mertzanidou A, Staessen C, Vandeskelde Y, Van der Elst J, Liebaers I, Sermon K. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat. Biotechnol. 2008;26:1361–1363. doi: 10.1038/nbt.1510. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tsutsui H, Valamehr B, Hindoyan A, Qiao R, Ding X, Guo S, Witte ON, Liu X, Ho CM, Wu H. An optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cells. Nat. Commun. 2011;2:167. doi: 10.1038/ncomms1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Diaz LG, Nandivada H, Ding J, Nogueira-de-Souza NC, Krebsbach PH, O’Shea KS, Lahann J, Smith GD. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat. Biotechnol. 2010;28:581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Muguruma K, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, Carpenter MK. Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.