Abstract
Phospholipase D (PLD) catalyzes the hydrolysis of phosphatidylcholine to generate the lipid second messenger, phosphatidic acid. PLD is localized in most cellular organelles, where it is likely to play different roles in signal transduction. PLD1 is primarily localized in vesicular structures such as endosomes, lysosomes and autophagosomes. However, the factors defining its localization are less clear. In this study, we found that four hydrophobic residues present in the N-terminal HKD catalytic motif of PLD1, which is involved in intramolecular association, are responsible for vesicular localization. Site-directed mutagenesis of the residues dramatically disrupted vesicular localization of PLD1. Interestingly, the hydrophobic residues of PLD1 are also involved in the interruption of its nuclear localization. Mutation of the residues increased the association of PLD1 with importin-β, which is known to mediate nuclear importation, and induced the localization of PLD1 from vesicles into the nucleus. Taken together, these data suggest that the hydrophobic amino acids involved in the interdomain association of PLD1 are required for vesicular localization and disturbance of its nuclear localization.
Keywords: cytoplasmic vesicles, nuclear localization signals, phospholipase D1
Introduction
Phospholipase D (PLD) catalyzes the hydrolysis of phosphatidylcholine (PC) to generate the lipid second messenger, phosphatidic acid (PA), and choline. PLD and PA have been implicated in a wide variety of cellular processes including cell proliferation, survival, vesicle transport, cytoskeletal organization and receptor signaling (Singer et al., 1997; Freyberg et al., 2003). PLD isozymes contain two highly-conserved copies of the HKD motif (HXK(X)4D), which is essential for catalysis (Colley et al., 1997b). Two distinct isoforms of mammalian PLD, PLD1 and PLD2, have been identified (Hammond et al., 1995; Colley et al., 1997b). The activation of PLD plays an important role in regulation of cell function and cell fate via extracellular signal molecules. The factors regulating the activity of PLD have been well characterized, but those defining its cellular localization are nearly unknown. PLD is localized in most, if not all, cellular organelles (plasma membrane, nucleus, endoplasmic reticulum, Golgi apparatus, transport/secretory vesicles), where it likely plays different roles in signal transduction (Liscovitch et al., 1999). We recently identified the nuclear localization sequence (NLS) of PLD1, and found that itsmutation abolished its nuclear import (Jang and Min, 2011). We alsoreported that caspase-mediated cleavage of PLD1 generates the N-terminal fragment (NF-PLD1) and C-terminal fragment (CF-PLD1) (Jang et al., 2008). Moreover, we demonstrated that CF-PLD1, but not NF-PLD1, is exclusively imported into the nucleus via its functional NLS, whereas only some portions of intact PLD1 were localized into the nucleus. The NLS of intact PLD1 or CF-PLD1 is required for interaction with importin-β, which is known to mediate nuclear import. The amount of intact PLD1 or CF-PLD1 translocated into nucleus is correlated with its binding affinity with importin-β (Jang and Min, 2011). Moreover, we have reported that the association ofcaspase-mediated cleavage fragments of PLD1 through hydrophobic residues within the catalytic motif inhibited nuclear localization of CF-PLD1, and that two catalytic motif and nuclear localization sequences regulated nucleocytoplasmic shuttling of PLD1 (Jang and Min, 2012). Although the conserved amino acids in the HKD motifs may have dual roles in both the catalytic reaction and the interdomain association, it is unknown whether the hydrophobic amino acids present in HKD motif affect localization of intact PLD1. Thus, the role of the hydrophobic amino acids in the localization of intact PLD1 should be identified. Furthermore, the molecular mechanisms that regulate shuttling between vesicular and nuclear localization of PLD1 remain unknown.
In the present study, we show for the first time that the hydrophobic residues of the HKD catalytic motif present in the N-terminal region of PLD1 are required for targetingPLD1 for vesicular localization and interruption of its nuclear localization. Specifically, we demonstrate that the hydrophobic amino acids involved in the interdomain association of PLD1 play an important role inregulation of shuttling PLD1 fromvesicular organelles into the nucleus.
Results
PLD1 is localized in vesicles
Numerous reports based on overexpression have described localization of PLDisoforms to separate regions of the cell. Specifically, PLD1 is localized to endosomal vesicles, the Golgi apparatus and perinuclear vesicles (Colley et al., 1997a; Toda et al., 1999; Lucocq et al., 2001), whilePLD2 is localized to the plasma membrane (Colley et al., 1997a; Lucocq et al., 2001; O'Luanaigh et al., 2002). However, localization of the isoforms is clearly complex. In some cells, PLD1 preferentially localizes to the plasma membrane (Vitale et al., 2001), and it generally cycles between perinuclear regions and the plasma membrane (Brown et al., 1998; Emoto et al., 2000; Du et al., 2003). Similarly, internalization of PLD2 subsequent to serum (Colley et al., 1997a) or insulin (Rizzo et al., 1999) and stimulation and its localization to early endosomes when one of its several membrane targeting motifs is altered (Sciorra et al., 2002) suggests that it alsocycles regularly from the plasma membrane through subcellular membrane vesicle compartments. We examined the localization of PLD1 using ectopically expressed GFP (green fluorescence protein)-PLD1 in HEK293 (human embryonic kidney 293). Fluorescence microscopy revealed that PLD1 primarily localized in punctate structures such as vesicles (Figure 1A). To further confirm the vesicular localizationof PLD1, we fractionated GFP-PLD1-transfected cells into the cytosol and vesicles. The fractions were then immunoblotted with antibody to PLD, which recognizes both PLD1 and PLD2. As shown in Figure 1B, both endogenous PLD1 and ectopically expressed GFP-PLD1 were present in the vesicular fraction. The purity of the cytosolic and vesicular fraction was confirmed by immunoblotting with antibody to the cytosolic marker, α-tubulin. Taken together, thesedata suggest that PLD1 is primarilylocalized in the vesicular organelle.
Figure 1.
PLD1 is localized in vesicles. (A) HEK293 cells were transfected with GFP-PLD1 and then observed by fluorescence microscopy. (B) HEK293 cells were transfected with GFP-PLD1, and the cells were then fractionated into cytosol (C) and vesicles (V) and analyzed by immunoblot analysis using antibody to PLD1 or α-tubulin. These data are representative of results obtained from three experiments.
PLD1 is localized in the late endosome, lysosome and autophagosome
We further examinedwhether PLD1 was localized in various vesicular structures using resident marker proteinsof some vesicles. RFP (red fluorescence protein)-PLD1 were cotransfected with GFP-Rab5 (early endosome marker), GFP-Rab7 (late endosome marker), GFP-Lamp1 (lysosomal-associated membrane protein;lysosome marker)or GFP-LC3 (microtubule associated protein light chain 3; autophagosome marker) in HEK293 cells. We investigated the distribution of RFP-PLD1 with early/late endosome marker (GFP-Rab5/Rab7), lysosome marker (GFP-Lamp1) and autophagosomemarker (GFP-LC3). As shown in Figure 2, RFP-PLD1 co-localized with GFP-Rab7, GFP-Lamp1 and GFP-LC3, but not GFP-Rab5. These findings are consistent with vesicular localization of PLD1 and indicate that intracellular distribution of PLD1 may be related to late endosome-lysosome trafficking or autophagosomal processing.
Figure 2.
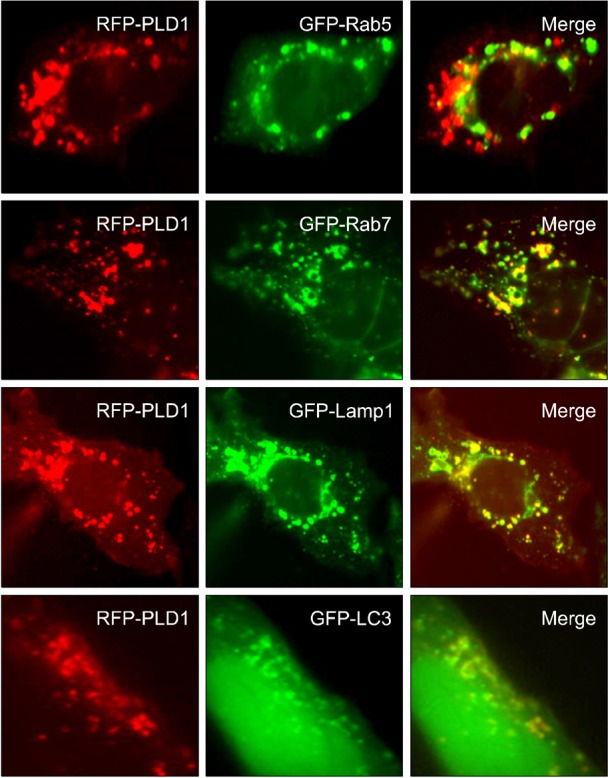
PLD1 is localized in the late endosome, lysosome and autophagosome. HEK293 cells were cotransfected with RFP-PLD1, and the indicated GFP constructs and fluorescent signals were then observed by fluorescence microscopy. These data are representative of results obtained from three experiments.
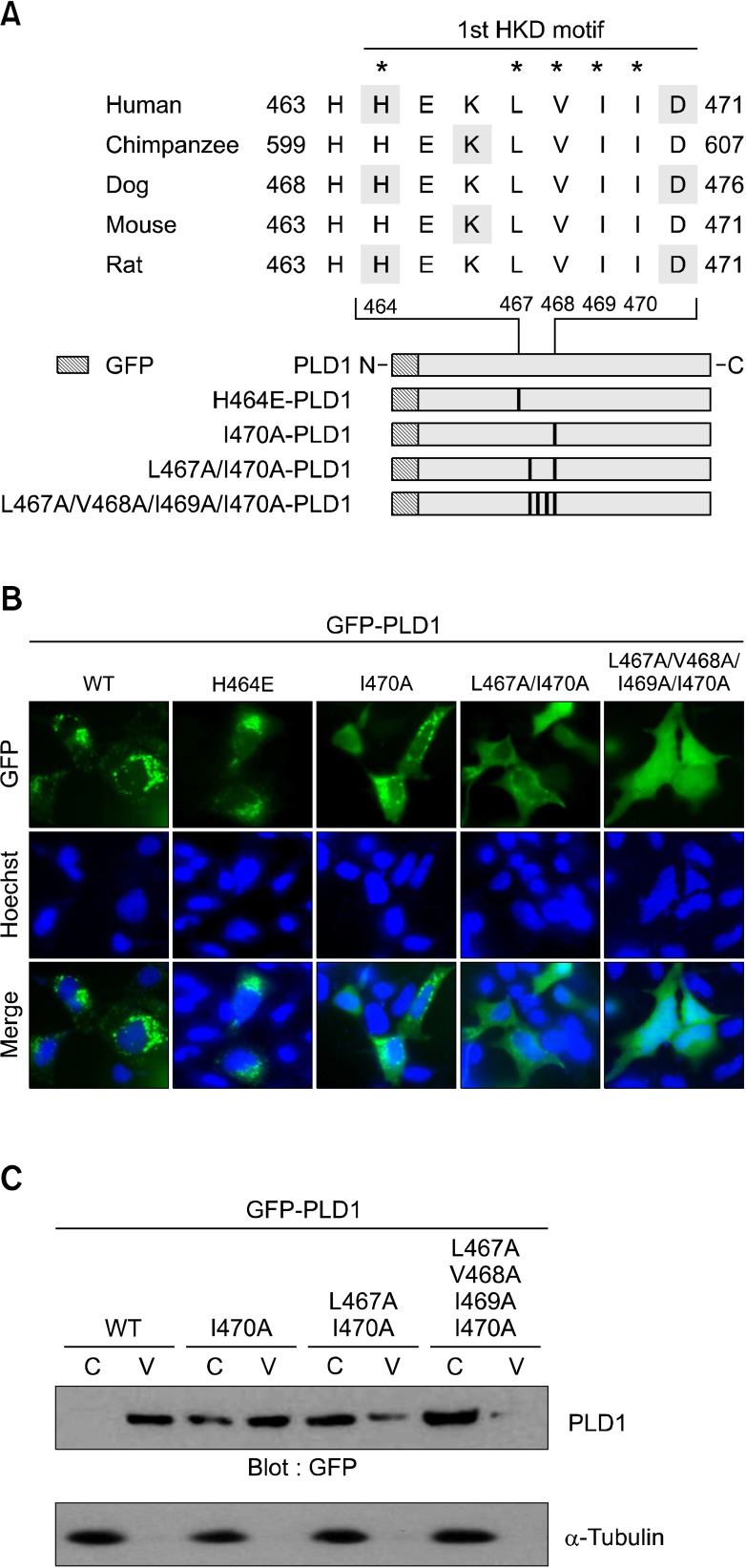
The hydrophobic amino acids involved in the interdomain association of PLD1 are required for its vesicular localization
We recently reported the importance of conserved hydrophobic amino acids within the HKD catalytic motif for intermolecular association between NF-PLD1 and endogenous intact PLD1 (Jang et al., 2008b). The integrity of the two HKD domains is essential for enzymatic activity. The hydrophobic residues of PLD1 involved in the intermolecular association are also required for both its enzymatic activity and anti-apoptotic function (Jang and Min, 2012). The hydrophobic residues (L467, V468, I469 and I470) in the HKD motif sequences are highly conserved in mammalian PLD1 from different species (Figure 3A). To examine the involvement of residues in the localization of PLD1, we generated GFP-PLD1 mutant constructs of the hydrophobic amino acids (Figure 3A). As shown in Figure 3B, wild type PLD1 shows punctate and cytosolic vesicular distribution, but not nuclear localization. Interestingly, as the number of mutated amino acids increased, the punctate form of PLD1 localized in vesicular structure gradually disappeared and its nuclear localization increased. The four hydrophobicamino acid mutation (L467A/V468A/I469A/I470A) showed the greatest increase in localization of PLD1 into the nucleus (Figure 3B). Mutation of H463, an amino acid critical for enzymatic activity, did not affect localization of PLD1. To further examine the localization pattern of these proteins, HEK293 cells were transfected with the mutant constructs and the lysates were fractionated into the cytosol and vesicles, after which they were subjected to immunoblot analysis. As shown in Figure 3C, intact wild type PLD1 was only detected inthevesicular fraction, whereas mutation of the hydrophobic amino acid decreased the amount of PLD1 localized in vesicles. The four hydrophobic amino acid mutant of PLD1 (L467A/V468A/I469A/I470A) was nearly completely absent from the vesicles (Figure 3C). Taken together, these results suggest that the four hydrophobic residues involved in the interdomain association of PLD1 are also required for its vesicular localization.
Figure 3.
The hydrophobic amino acids involved in the interdomain association of PLD1 are required for its vesicular localization. (A) Alignment of the HKD motifs in mammalian PLD1 from different species and schematic representation of mutations generated in the GFP-PLD1. (B) HEK 293 cells were transfected with the indicated GFP-PLD1 constructs and its localization was observed using fluorescence microscopy. WT, wild type (C) HEK 293 cells were transfected with the indicated GFP-PLD1 constructs and the lysates were fractionated into cytosol (C) and vesicles (V), after which the samples weresubjected to immunoblotanalysis with the indicated antibodies. These data are representative of results obtained from three experiments.
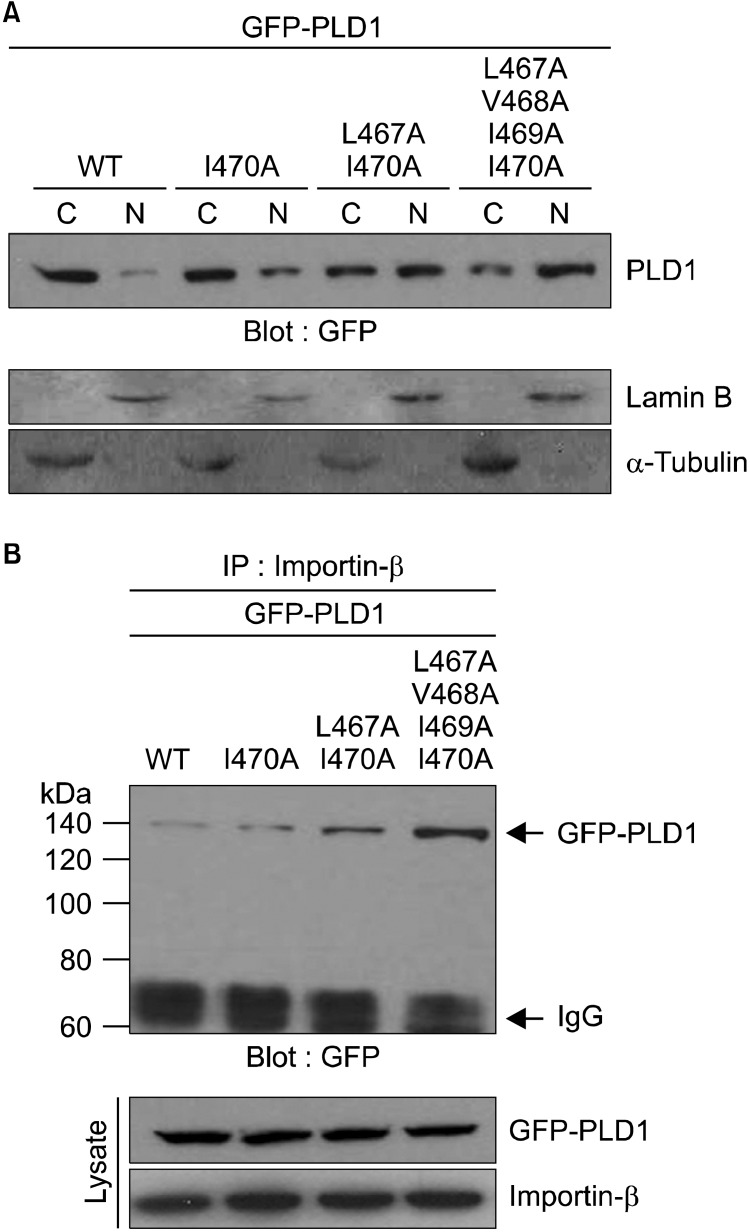
The hydrophobic residues responsible for vesicular localization of PLD1 interrupt its nuclear localization
We recently reported that intermolecular association of NF- and CF-PLD1 through the hydrophobic amino acids present in the HKD motif results in suppression of nuclear translocation of CF-PLD1. We also found that PLD1 was primarily detected in the cytoplasm, but that a minor portion was also detectedin the nucleus of HEK293 cells, even though PLD1 contains NLS in its C-terminal half region (Jang and Min, 2012). It has also been suggested that interaction of two HKD motifs prevents exposure of NLS to NLS machinery, such as importin-β, and that HKD and NLS motifs regulate nucleocytoplasmic shuttling of PLD1 (Jang and Min, 2012). Thus, we attempted to examine whether the hydrophobic residues in the HKD motif of PLD1 also affect its nuclear localization. To accomplish this, HEK293 cells were transfected with wild type or hydrophobic mutant of full length PLD1. As shown in Figure 4A, wild type PLD1 was primarily detected in the cytosolic fraction, whereas translocation of the mutant PLD1 from cytosol to the nuclear fraction increased as the number of mutated residues increased. The four hydrophobic amino acid mutation (L467A/V468A/I469A/I470A) showed the greatest increase in localization of PLD1 in the nuclear fraction. These results are consistent with those obtained from fluorescence microscopy (Figure 3B). We also examined whether the extent of PLD1 localization in the nuclear fraction is involved in the interaction with importin-β. Wild type PLD1 was slightly associated with importin-β, whereas the hydrophobic mutants of PLD1 showed a stronger interaction with importin-β than with wild type PLD1 (Figure 4B). L467A/V468A/I469A/I470A mutant of PLD1 showed the strongest interaction with importin-β. Taken together, these data suggest that the hydrophobic residues responsible for vesicular localization of PLD1 interrupt its nuclear localization. Overall, our results demonstrate that the hydrophobic amino acids involved in the interdomain association of PLD1 play an important role inregulation of shuttling of PLD1 fromvesicular organelles into the nucleus.
Figure 4.
The hydrophobic residues responsible for vesicular localization of PLD1 interrupt its nuclear localization. (A) HEK 293 cells were transfected with the indicated GFP-PLD1 constructs and the lysates were fractionated into the cytosol (C) and nuclei (N), after which the samples weresubjected to immunoblotanalysis with the indicated antibodies. (B) HEK293 cells were transfected with the indicated GFP-PLD1 constructs, and the lysates were then immunoprecipitated with antibody to importin-β, after which the samples were subjected toimmunoblot analysis-with antibody to GFP. The level of GFP-PLD1 and importin-β was determined by immunoblottingusing the indicated antibody. These data are representative of results obtained from three experiments.
Discussion
To the best of our knowledge, this is the first study to demonstratethat the hydrophobic amino acids involved in the interdomain association of PLD1 are required for vesicular localization and interruption of its nuclear localization. Membrane compartments in eukaryotic cells are in constant dynamic flux, with-transport vesicles emerging from donor compartments and being targeted specifically to acceptor compartments, where they deliver cargo through membrane fusion. PLD generates PA, a multifunctional lipid that has been proposed to alter membrane curvature, serve as a protein attachment site, activate selected enzymes or serve as starting material for the production of additional signaling lipids, particularly in the context of membrane vesicle trafficking and cytoskeletal dynamics (Vitale et al., 2001). PLD is a key facilitator of multiple types of membrane vesicle trafficking events. In resting cells, PLD1 is primarily localized to endosomal vesicles, lysosomes, and Golgi and perinuclear vesicles in multiple cell types. PLD2 has been shown to primarily localize to the plasma membrane. It has also been reported that some factors may be determinants of membrane localization. Specifically, the N-terminal pleckstrin homology domain of PLD1 (Hodgkin et al., 2000) and a central domain of PLD1 (Sciorra et al., 1999) have both been shown to bind phosphatidylinositol polyphosphates, and fatty acylation of cysteine residues within the N-terminus of the protein (Sugars et al., 1999) may also influence membrane association. However, the translocation of PLD from one compartment to other organelles is not well understood. Although collapse of the Golgi apparatus in response to brefeldin A has been reported to alter PLD1 localization to the endoplasmic reticulum and significantly enhance its nuclear translocation (Freyberg et al., 2001), we did not observe any nuclear translocation of PLD1 in response to treatment of HEK293 cells with brefeldin A. These discrepancies might be attributable to differences in cell types and/or agonist-activated signaling pathways. The significance of the translocation of PLD isoform from the Golgi apparatus to the nucleus is currently unclear, and the factors defining its localization are even less clear. In the present study, we found that PLD1 was primarily localized in the vesicles using fractionation, and that four hydrophobic residues present in the N-terminal HKDcatalytic motif of PLD1, which are involved in the intramolecular association, are responsible for its vesicular localization. Fluorescence microscopy and fractionation revealed that mutation of all four hydrophobic amino acids dramatically disrupted vesicular localization of PLD1 and increased its nuclear localization. We previously identified a functional and highly conserved NLS required for nuclear import of PLD1 (Jang and Min, 2011); however, we have been unable to identify conditions (e.g. growth factors, apoptosis, normal versus transformed phenotype) that might alter the relative distribution of endogenous PLD1 between the cytosolic and nuclear compartments. Although we have disclosed the importance of NLS and its interaction with β-importin in the nuclear localization of PLD1, further study for ligand or condition-induced nuclear translocation of endogenous PLD1 will strengthen the molecular mechanism of nucleocytoplasmic shuttling of PLD1 under physiological conditions. We recently reported that caspase-mediated cleavage of PLD1 during apoptosis results in production of NF-PLD1 and CF-PLD1 (Jang et al., 2008). Interestingly, CF-PLD1 containing the functional NLS, but not NF-PLD1 lacking the NLS, is exclusively imported into the nucleus, whereas full length PLD1 is partially imported into the nucleus. The extent of nuclear localization of CF-PLD1 and full length PLD1 is different, even though these two proteins have the same NLS motif. We also found that intermolecular association between the two fragmentsof PLD1 through hydrophobic residues within the catalytic motif inhibited nuclear localization of CF-PLD1 (Jang and Min, 2012). Thus, it is suggested that the hydrophobic amino acids in the catalytic motif and NLS regulate nuclocytoplasmic shuttling of intact PLD1. Furthermore, mutation of four hydrophobic residues of full length PLD1 showed a stronger interaction with β-importin than that of wild type orone or two residue mutants of PLD1. Thus, it is possible that PLD1 harboring the four mutated hydrophobic residues may be more easily exposed to NLS machinery such as importin-β than wild type PLD1 and thus, largely localized in the nucleus. In summary, the hydrophobic amino acids involved in the interdomain association of PLD1 play an important role inregulation of shuttling of PLD1 from vesicular organelles into the nucleus through vesicular localization and disturbance of nuclear localization. Further investigation of signal-dependent translocation of endogenous PLD1 from one compartment to other organellesshould help elucidate the role of PLD1 in the compartment.
Methods
Cell culture and transient transfection
HEK293 cells were cultured at 37℃ in DMEM (Life Technologies, CA) containing 10% fetal bovine serum and 1% antibiotic-antimycotic. Cells were grown to 50% to 60% confluence for transient transfection using LipofectAMINE Plus (Life Technologies, CA) according to the manufacturer's instructions.
Construction of plasmids and site directed mutagenesis
Cloning of GFP-tagged full length (FL)-PLD1 has been previously described (Jang et al., 2008). The catalytic active site (H464E) and interdomain-association sites (I470A, I467A/I470A and I467A/V468A/I469A/I470A) of PLD1 were generated by PCR site-directed mutagenesis using a Quick Change Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA) according to the manufacturer's instructions. Mutation was then confirmed by DNA sequencing. Primers for generation of the H464E, I467A and I470A have been previously described (Jang et al., 2008). The primers used for generation of the I468A/I469A were as follows:5'-Forward; CACCATGAGAAGGCTGCCGCCGCTGACCAATCGGTGGCC, 3'-Reverse;CCGGTGGCTAACCAGTCGCCGCCGTCGGAAGAGTACCAC. Mutagenesis for generation of I467A/I470A and I467A/V468A/I469A/I470A was performed using I470A-FL-PLD1 and I467A/I470A-FL-PLD1 as a template.
Total vesicle preparation by flotation centrifugation
The procedure for vesicle preparation has been described previously (Lee et al., 2005). Briefly, buffer (10 mM HEPES, pH 7.2, 10 mM KCl, 1 mM EGTA, 250 mM Sucrose) with protease inhibitor cocktail was used for cell harvest. Cells were disrupted using either an N2 cell disruption bomb (Parr Instrument, Moline, IL) or a Dounce homogenizer. The samples were then centrifuged at 1000 g, after which the supernatant was mixed with iodixanol (OPTI-PREP reagent) to obtain a final concentration of 35%. The mixture was then layered under 30% and 5% step gradient layers and centrifuged at 200,000 g for 2 h, after which total vesicles were collected at the interphase between 5% and 30%, and the cytosolic fraction was collected from the 35% bottom fraction.
Western blotting and immunoprecipitation
The cell lysates were analyzed for immunoblot or immunoprecipitation as previously described (Ahn et al., 2003) using purchased anti-α-tubulin (Sigma, MO), anti-lamin B (Santa Cruz, CA), anti-importin-β (Santa Cruz, CA), and anti-GFP (Santa Cruz, CA) antibody. In addition, rabbit polyclonal anti-PLD1 antibody that recognizes the C-terminal region of PLD1 was generated as previously described (Min et al., 2001).
Fluorescence microscopy
Cells were transfected with various constructs of PLD1 and incubated with 10 µg/ml Hoechst (Molecular Probe, CA) for 20 min. Cells were then visualized, and images were collected by fluorescence microscopy (Axiovert 200M, Zeiss, Germany).
Acknowledgements
This work was supported by a 2-yr Research Grant from Pusan National University.
Abbreviations
- CF-PLD1
C-terminal fragment-PLD1
- HEK293
human embryonic kidney 293
- Lamp1
lysosomal-associated membrane protein 1
- LC3
microtubule associated protein light chain 3
- NF-PLD1
N-terminal fragment-PLD1
- NLS
nuclear localization sequence
- PA
phosphatidic acid
- PLD1
phospholipase D1
- RFP
red fluorescence protein
References
- 1.Ahn BH, Kim SY, Kim EH, Choi KS, Kwon TK, Lee YH, Chang JS, Kim MS, Jo YH, Min DS. Transmodulation between phospholipase D and c-Src enhances cell proliferation. Mol Cell Biol. 2003;23:3103–3115. doi: 10.1128/MCB.23.9.3103-3115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown FD, Thompson N, Saqib KM, Clark JM, Powner D, Thompson NT, Solari R, Wakelam MJ. Phospholipase D1 localises to secretory granules and lysosomes and is plasma-membrane translocated on cellular stimulation. Curr Biol. 1998;8:835–838. doi: 10.1016/s0960-9822(98)70326-4. [DOI] [PubMed] [Google Scholar]
- 3.Colley WC, Altshuller YM, Sue-Ling CK, Copeland NG, Gilbert DJ, Jenkins NA, Branch KD, Tsirka SE, Bollag RJ, Bollag WB, Frohman MA. Cloning and expression analysis of murine phospholipase D1. Biochem J. 1997a;326(Pt 3):745–753. doi: 10.1042/bj3260745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997b;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- 5.Du G, Altshuller YM, Vitale N, Huang P, Chasserot-Golaz S, Morris AJ, Bader MF, Frohman MA. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J Cell Biol. 2003;162:305–315. doi: 10.1083/jcb.200302033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emoto M, Klarlund JK, Waters SB, Hu V, Buxton JM, Chawla A, Czech MP. A role for phospholipase D in GLUT4 glucose transporter translocation. J Biol Chem. 2000;275:7144–7151. doi: 10.1074/jbc.275.10.7144. [DOI] [PubMed] [Google Scholar]
- 7.Freyberg Z, Sweeney D, Siddhanta A, Bourgoin S, Frohman M, Shields D. Intracellular localization of phospholipase D1 in mammalian cells. Mol Biol Cell. 2001;12:943–955. doi: 10.1091/mbc.12.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freyberg Z, Siddhanta A, Shields D. "Slip, sliding away": phospholipase D and the Golgi apparatus. Trends Cell Biol. 2003;13:540–546. doi: 10.1016/j.tcb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, Morris AJ, Frohman MA. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- 10.Hodgkin MN, Masson MR, Powner D, Saqib KM, Ponting CP, Wakelam MJ. Phospholipase D regulation and localisation is dependent upon a phosphatidylinositol 4,5-biphosphate-specific PH domain. Curr Biol. 2000;10:43–46. doi: 10.1016/s0960-9822(99)00264-x. [DOI] [PubMed] [Google Scholar]
- 11.Jang YH, Namkoong S, Kim YM, Lee SJ, Park BJ, Min DS. Cleavage of phospholipase D1 by caspase promotes apoptosis via modulation of the p53-dependent cell death pathway. Cell Death Differ. 2008;15:1782–1793. doi: 10.1038/cdd.2008.111. [DOI] [PubMed] [Google Scholar]
- 12.Jang YH, Min DS. Nuclear localization of phospholipase D1 mediates the activation of nuclear protein kinase C (alpha) and extracellular signal-regulated kinase signaling pathways. J Biol Chem. 2011;286:4680–4689. doi: 10.1074/jbc.M110.162602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang YH, Min DS. Intermolecular association between caspase-mediated cleavage fragments of phospholipase D1 protects against apoptosis. Int J Biochem Cell Biol. 2012;44:358–365. doi: 10.1016/j.biocel.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Lee H, Patel S, Lee S. Intravesicular localization and exocytosis of alpha-synuclein and its aggregates. J Neurosci. 2005;25:6016–6024. doi: 10.1523/JNEUROSCI.0692-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liscovitch M, Czarny M, Fiucci G, Lavie Y, Tang X. Localization and possible functions of phospholipase D isozymes. Biochim Biophys Acta. 1999;1439:245–263. doi: 10.1016/s1388-1981(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 16.Lucocq J, Manifava M, Bi K, Roth MG, Ktistakis NT. Immunolocalisation of phospholipase D1 on tubular vesicular membranes of endocytic and secretory origin. Eur J Cell Biol. 2001;80:508–520. doi: 10.1078/0171-9335-00186. [DOI] [PubMed] [Google Scholar]
- 17.Min DS, Ahn BH, Rhie DJ, Yoon SH, Hahn SJ, Kim MS, Jo YH. Expression and regulation of phospholipase D during neuronal differentiation of PC12 cells. Neuropharmacology. 2001;41:384–391. doi: 10.1016/s0028-3908(01)00070-3. [DOI] [PubMed] [Google Scholar]
- 18.O'Luanaigh N, Pardo R, Fensome A, Allen-Baume V, Jones D, Holt MR, Cockcroft S. Continual production of phosphatidic acid by phospholipase D is essential for antigen-stimulated membrane ruffling in cultured mast cells. Mol Biol Cell. 2002;13:3730–3746. doi: 10.1091/mbc.E02-04-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzo MA, Shome K, Vasudevan C, Stolz DB, Sung TC, Frohman MA, Watkins SC, Romero G. Phospholipase D and its product, phosphatidic acid, mediate agonist-dependent raf-1 translocation to the plasma membrane and the activation of the mitogen-activated protein kinase pathway. J Biol Chem. 1999;274:1131–1139. doi: 10.1074/jbc.274.2.1131. [DOI] [PubMed] [Google Scholar]
- 20.Sciorra VA, Rudge SA, Prestwich GD, Frohman MA, Engebrecht J, Morris AJ. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 1999;18:5911–5921. doi: 10.1093/emboj/18.21.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sciorra VA, Rudge SA, Wang J, McLaughlin S, Engebrecht J, Morris AJ. Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J Cell Biol. 2002;159:1039–1049. doi: 10.1083/jcb.200205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer WD, Brown HA, Sternweis PC. Regulation of eukaryotic phosphatidylinositol-specific phospholipase C and phospholipase D. Annu Rev Biochem. 1997;66:475–509. doi: 10.1146/annurev.biochem.66.1.475. [DOI] [PubMed] [Google Scholar]
- 23.Sugars JM, Cellek S, Manifava M, Coadwell J, Ktistakis NT. Fatty acylation of phospholipase D1 on cysteine residues 240 and 241 determines localization on intracellular membranes. J Biol Chem. 1999;274:30023–30027. doi: 10.1074/jbc.274.42.30023. [DOI] [PubMed] [Google Scholar]
- 24.Toda K, Nogami M, Murakami K, Kanaho Y, Nakayama K. Colocalization of phospholipase D1 and GTP-binding-defective mutant of ADP-ribosylation factor 6 to endosomes and lysosomes. FEBS Lett. 1999;442:221–225. doi: 10.1016/s0014-5793(98)01646-9. [DOI] [PubMed] [Google Scholar]
- 25.Vitale N, Caumont AS, Chasserot-Golaz S, Du G, Wu S, Sciorra VA, Morris AJ, Frohman MA, Bader MF. Phospholipase D1: a key factor for the exocytotic machinery in neuroendocrine cells. EMBO J. 2001;20:2424–2434. doi: 10.1093/emboj/20.10.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]