Abstract
Subcortical vascular dementia or cerebral small vessel disease is a common cause of disability in the elderly. On magnetic resonance imaging the disease is manifested as white matter lesions, lacunes and microbleeds. Its etiology is complex, with age and hypertension as established risk factors. The heritability of white matter lesions is constantly high over different populations. Linkage studies identified several loci for these lesions however no genes responsible for the linkage signals had been identified so far. Results from genetic association studies using the candidate gene approach support the role of APOE, the renin–angiotensin system, as well as the Notch3 signaling pathway in the development of subcortical vascular dementia. The recent genomegenome wide association study on white matter lesions identified a novel locus on chromosome 17q25 harboring several genes such as TRIM65 and TRIM47 which pinpoints to possible novel mechanisms leading to these lesions.
Keywords: Subcortical vascular dementia cerebral small vessel disease, Genetics, White matter lesions, Lacunes, Microbleeds, Risk factors
Highlights
► We review the genetics of MRI correlates of subcortical vascular dementia (sVaD). ► MRI correlates of sVaD includes white matter lesions, lacunes and microbleeds. ► White matter lesions are highly heritable. ► SNPs in APOE, NOTCH3 and the renin-angiotensin system are associated with sVaD. ► The first gemone wide association study identifies novel genes on chromosome 17.
1. Introduction
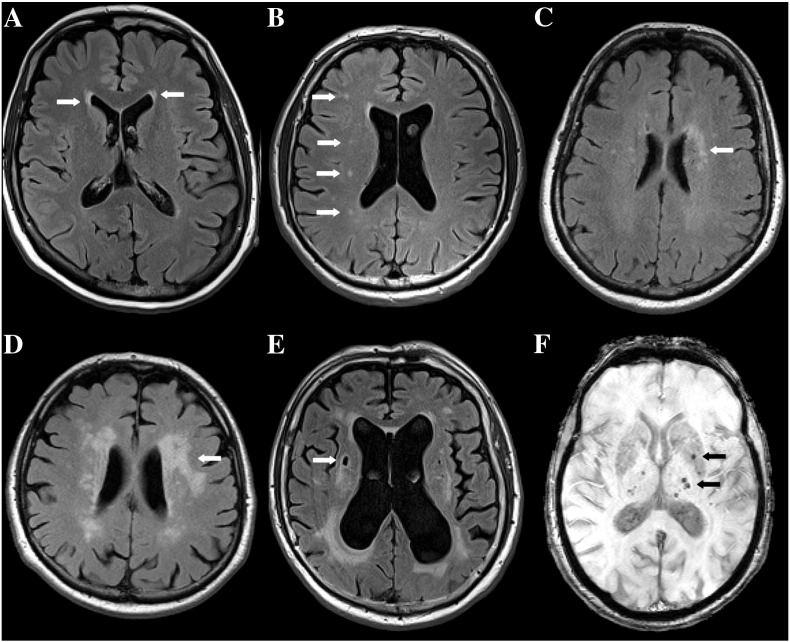
Subcortical vascular dementia (VaD) results from cerebral small vessel disease leading to ischemic and hemorrhagic brain tissue damage that can be observed on magnetic resonance imaging (MRI). Such brain lesions are white matter lesions (WML), lacunes and microbleeds (MB). WML are seen on T2/FLAIR weighted MRI as areas of high signal intensity of variable extent. (Fig. 1A–D). Mild periventricular lines and caps as well as punctate deep lesions are of non-ischemic origin, while early confluent and confluent lesions represent ischemic damage ranging from incomplete to complete tissue loss (Fazekas et al., 1993). The prevalence of any WML ranges between 40 and 95%, depending on the study population (van Dijk et al., 2002). Conventional risk factors such as age and hypertension explain only a small proportion of the occurrence of WML (O'Brien et al., 2003). Lacunes are observed on T2/FLAIR weighted MR images as 3 to 15 mm abnormalities with CSF-equivalent signal intensity (Koch et al., 2011) (Fig. 1E). Their frequency ranges between 6 and 20% (van Dijk et al., 2002). The third lesion type is MB seen as hypointense or black spots on susceptibility or T2* weighted MR images due to paramagnetic effects of hemosiderin deposits at the location of the bleeding (Greenberg et al., 2009) (Fig. 1F). In the Rotterdam Scan Study, prevalence of MB increased strongly with age, ranging from 6.5 to 35.7%.
Fig. 1.
Different lesion types commonly found on MRI scans of elderly people (arrows): (A–E) show Fluid Attenuated Inversion Recovery (FLAIR) sequences where white matter lesions (WML) appear as areas of high signal intensity while cerebrospinal fluid (CSF) signal is suppressed. (A) So-called “caps” located symmetrically around the anterior horns of both lateral ventricles. (B) Punctate WML in the centrum semiovale (CS) of the right hemisphere. (C) Early confluent and (D) confluent WML in the left CS. (E) Lacune seen as CSF-isointense lesion surrounded by a small hyperintense rim in the right CS. (F) shows a T2* weighted MRI sequence that is sensitive to susceptibility effects of hemosiderin. Spots of low signal intensity correspond to microbleeds in the left basal ganglia and thalamus.
2. Heritability of cerebral small vessel disease
Although several monogenic forms of cerebral small vessel disease with early onset have been described, the majority of patients with subcortical VaD represents sporadic cases with enhanced aggregation in families but without a clear Mendelian pattern of inheritance. The heritability of a trait gives the proportion of the total phenotypic variance (Vp) in the population which can be explained by genetic differences among individuals (Vg). Importantly, the heritability index is a quotient (h2 = Vg/Vp) and its estimate is high, when there is a strong phenotypic variability due to genetic factors, as well as when there is low phenotypic variability due to environmental factors in a population. Therefore heritability indices strictly apply to the population they have been estimated for. Heritability estimates for these age-related forms have been reported for WML in elderly male twins (Carmelli et al., 1998) in a large population based sample of middle aged–elderly sibships (Atwood et al., 2004), within non-Hispanic white and black hypertensive siblings (Turner et al., 2004), and in siblings discordant for Alzheimer disease (Cuenco et al., 2008). In spite of major differences in study design, heritability estimates were comparable among these studies, and were within the range of 50–80%. The heritability of WML may however be dependent on gender and age as reported by the Framingham study with young women having the highest estimates (h2 = 0.9 (Atwood et al., 2004). Bivariate heritability analyses explore the genetic correlation (rg) between traits, which gives the proportion of variance that two traits share due to common genetic factors. The GENOA study showed significant genetic correlations (rg) between WML and mean arterial pressure both in whites (rg = 0.61) and in blacks (rg = 0.40) and with pulse pressure in blacks (0.29; P < .001). This indicates common genetic components between WML and blood pressure (Turner et al., 2004). Bivariate heritability analyses also suggested common genetic influences on WML volume and the volume of specific brain regions such as frontal (rg = 0.30) and parietal lobe (rg = − 0.39) (DeStefano et al., 2009). Heritability estimates for lacunes and for MB are not available so far.
In order to identify genetic variants contributing to subcortical VaD both association and linkage studies have been performed. These studies mainly focused on WML, but some candidate gene based association studies also investigated lacunes and MB.
3. Linkage studies
So far significant logarithm of the odds (LOD) scores for WML have been found at 4 cM on chromosome 4 (DeStefano et al., 2006; Seshadri et al., 2007), in healthy elderly white subjects, and at chromosome 1q24 in hypertensive sibships (Turner et al., 2009). Suggestive LOD scores by these studies have been reported at 95 cM on chromosome 17 (LOD = 1.78) (DeStefano et al., 2006), at 118 cM on chromosome 11, and at 13 cM on chromosome 21 in white, and at 36 cM on chromosome 22 as well as at 58 cM on chromosome 21 in black subjects (Turner et al., 2009). Bivariate linkage studies identified significant (chromosome 1q24) and suggestive (chromosome 1q42, 10q22–q26, 15q26) shared loci for WML volumes and blood pressure measurements, such as pulse pressure and systolic blood pressure, indicating the presence of genes with pleiotropic effects at these regions (Kochunov et al., 2010; Turner et al., 2009).
4. Association studies
Association studies aim to identify genetic variants explaining phenotypic variations in a trait or in susceptibility to a disease in the population. The most frequently investigated genetic variants are the single nucleotide polymorphisms (SNPs), which are bi-allelic variants in the human genome involving a nucleotide exchange. The phenotypes investigated by association studies are either qualitative or quantitative. In case of dichotomized outcomes the effect size estimates of the variant is given as the odds ratio (OR) in those carrying the variant allele compared to non-carriers. If an observed genetic association is true, then it implies that either the investigated variant is causal (direct association) or there is a causal variant in linkage disequilibrium with the investigated one in its neighborhood (indirect association). Importantly, observed associations in one sample must be replicated in independent samples in order to reduce the likelihood of false positive findings, which had discredited association studies in early days. Association studies performed according to published guidelines ensure high quality and have substantially contributed to our understanding of subcortical VaD. During the last two decades numerous association studies using the candidate gene approach have focused on pathways hypothesized to be involved in the pathogenesis of cerebral small vessel disease. According to a recent meta-analyses 46 candidate gene studies investigating variants in 19 genes have been performed (Paternoster et al., 2009). Markus (2008) reviewed studies investigating genes involved in vascular dysfunction in cerebral small vessel disease such as EDN1, MHTFR, and NOS3 (GenBank ID 1906, 4524, 4846). Due to small sample size of the studies and lack of replications, there is little statistical evidence presently that these genes harbor causal variants for subcortical VaD. Recently hypothesis free genome wide approaches became affordable and identified novel genetic variants in pathways not being suspected to be involved in the development of these lesions previously. Here we focus on those candidate genes which are strongly supported by replication studies or functional data and discuss the findings of the first genome wide association (GWA) study (Table 1).
Table 1.
Summary of association studies on white matter lesions, lacunes and microbleeds discussed in the text.
| Phenotype | Gene/ref seq Gene ID | SNP rs number | Risk allele | OR/p value | Sample | Reference |
|---|---|---|---|---|---|---|
| WML burden | ACE (1636) | I/D | D/D | OR = 1.95, 95% CI = 1.09–3.48 | Meta-analysis of 46 studies | Paternoster et al., 2009 |
| WML presence | AGT (183) | Promoter haplotype | B/B or B +/A − | OR = 8.0 P = 0.003 | Cohort study in elderly Caucasian | Schmidt et al., 2001 |
| WML burden | M235T/rs699 | 235T | NS | Meta-analysis of 6 studies | Paternoster et al., 2009 | |
| WML burden | M235T/rs699 | 235T | P = 0.01 | Meta‐analyses of GWAS in elderly Caucasian | Fornage et al., 2011 | |
| WML progression | AGTR1 (185) | A1166C/rs5186 | 1166C | P = 0.0002 in males | Depressed and non-depressed elderly Caucasian | Taylor et al., 2010 |
| WML progression | AGTR2 (11609) | C3123A | 3123A | P = 0.014 in males | Depressed and non-depressed elderly Caucasian | Taylor et al., 2010 |
| WML burden | APOE (348) | ε2/ε3/ε4 | ε4 carrier | P = 0.016 in hypertensives. | Cohort study elderly Caucasian | Leeuw et al., 2004 |
| WML burden | ε2/ε3/ε4 | ε4 carrier | P = 0.003 | Cohort study elderly Caucasian | Godin et al., 2009 | |
| WML and lacunes presence | ε2/ε3/ε4 | ε2/ε3 genotype | OR 3.0. | Cohort study elderly Caucasian | Schmidt et al., 1997 | |
| WML presence | ε2/ε3/ε4 | ε4 carrier | NS | Meta-analysis of 24 studies | Paternoster et al., 2009 | |
| Lacunes | APOE (348) | ε2/ε3/ε4 | ε4 +/ε4 + | OR 4.7 p = 0.04 | Genetic isolate, elderly hypertensives | Schuur et al., 2011 |
| Microbleeds | APOE (348) | ε2/ε3/ε4 | ε4 carrier | OR = 1.35, 95% CI = 1.10–1.65 | Cohort study elderly Caucasian | Poels et al., 2010 |
| WML presence | NOTCH3 (4854) | rs10404382 | 1036+846G>T | OR 1.8 p = 0.02; OR 3,2 p = 0.002 in hypertensives |
Cohort study elderly Caucasian | Schmidt et al., 2011 |
| WML progression | rs10404382 | 1036+846G>T | β = 0.087 p = 0.05; β = 0.136 p = 0.013 in hypertensives; |
|||
| Microbleeds | SORL1 (6653) | rs2282649. | 5239+73TT | OR 6.87 p = 0.005 | Genetic isolate, elderly hypertensives | Schuur et al., 2011 |
| WML burden | TRIM 47 (91107) | rs3744017 | A | p = 7.3 × 10− 9 | Meta‐analyses of GWAS in elderly Caucasian | Fornage et al., 2011 |
| WML burden | TRIM 65 (201292) | rs3744028 | C | p = 4.0 × 10− 9 | Meta‐analyses of GWAS in elderly Caucasian | Fornage et al., 2011 |
| WML burden | WBP2 (23558) | rs11869977 | G | p = 5.7 × 10− 9 | Meta‐analyses of GWAS in elderly Caucasian | Fornage et al., 2011 |
WML = white matter lesions; OR = odds ratio; 95% CI = 95% confidence interval; β = regression coefficient; GWAS = genome-wide association study, NS = non significant.
4.1. APOE
APOE (GenBank ID 348) is the only gene, which had been investigated in all three MRI correlates of subcortical VaD. The APOE E4 allele was reported to be associated both with the extent and the progression of WML (Godin et al., 2009; Leeuw et al., 2004). Contrarily, the Austrian Stroke Prevention Study found a higher prevalence of WML and lacunar infarcts in carriers of the E2 allele (Schmidt et al., 1997). The effect of APOE variants on WML may depend on vascular risk factors such as the presence hypertension (Leeuw et al., 2004). Homozygosity for the E4 allele was also significantly associated with the presence of lacunes in an isolated population (Schuur et al., 2011). In the Rotterdam study, the presence of APOE E4 allele enhanced the risk for cerebral MB (Poels et al., 2010), but this finding is controversial (Lemmens et al., 2007).
4.2. The renin–angiotensin system (RAS)
Genes coding for the RAS are excellent candidates for cerebral small vessel disease as RAS is a major regulator of systemic blood pressure, and of cerebral blood flow. Plasma angiotensinogen (AGT, GenBank ID 183) synthesized by the liver is processed to angiotensin II (Ang II) by the serial action of renin and angiotensin-converting enzyme (ACE GenBank ID 1636). Ang II exerts its effect through the angiotensin receptors (AGTR GenBank ID 185). The association of ACE I/D polymorphism with WML has been investigated in nine studies including 2396 individuals and the DD genotype remained a significant predictor of WML (OR = 1,95; 95% CI 1,09:3,48) upon meta-analysis (Paternoster et al., 2009). The most frequently investigated single nucleotide polymorphism (SNP) in the AGT gene is the M235T. Meta-analyses of 6 candidate gene studies including 2702 individuals showed no effect of this SNP on WML (Paternoster et al., 2009). In contrast, linkage studies repeatedly implicated the AGT locus (1q42) (Kochunov et al., 2010; Turner et al., 2009) and the recent GWA study had replicated the association between the 235T allele and WML (Fornage et al., 2011). It is likely that the 235T SNP by itself is not causal, and functional variants in linkage disequilibrium are responsible for these findings. One such variant might be a haplotype (-6:a, -20:c, -153:g, -218:g, positions relative to transcriptional start site) at the AGT promoter. This haplotype was first described by the Austrian Stroke Prevention Study and was significantly related to the presence of severe WML (Schmidt et al., 2001). The association was independent of hypertension and the haplotype enhanced the basal transcriptional activity of the AGT promoter in astrocytes but not in hepatocytes suggesting that the association is mediated by an altered activity of the cerebral and not the systemic RAS (Schmidt et al., 2004). Recently, a longitudinal study reported that progression rate of WML in elderly men is influenced by polymorphisms in the AGTR1 and AGTR2 genes. Homozygotes for the 1166A allele at AGTR1 showed less change over time than 1166C carriers (Taylor et al., 2010).
4.3. NOTCH3
CADASIL is caused by mutations in the NOTCH3 (GenBank ID 4854) gene (Joutel et al., 1996), which plays a central role in the functional and structural integrity of small arteries (Domenga et al., 2004). Recently sequencing of the NOTCH3 gene in the Austrian Stroke Prevention Study showed that this gene is highly variable in the elderly with both common and rare SNPs spreading over the gene (Schmidt et al., 2011). Common SNPs (rs1043994, rs10404382, rs10423702, rs1043997) were significantly associated with the presence and the progression of WML. The association was confined to hypertensives and was replicated in the Cohorts for Heart and Ageing Research in Genomic Epidemiology Consortium (CHARGE) in an independent sample of 4773 stroke-free hypertensive elderly individuals of European descent. Beside the common SNPs there were 6 non-synonymous rare SNPs (H170R, P496L, V1183M, L1518M, D1823N and V1952M) in this study, which were detected only in subjects with severe WML and which were predicted to be functional by protein modeling tools. Importantly none of these SNPs affected a cysteine residue and none of the subjects carrying these mutations had MRI signs typical of CADASIL. The results on NOTCH3 resembles other studies on complex traits, such as dyslipidemia where genes carrying common variants associated with modest effect in the population also carry rare variants with large effects segregating in families.
4.4. Genome wide association study
The first GWA study on WML was performed within the CHARGE consortium (Fornage et al., 2011), including 9361 elderly individuals of European descent from 7 community-based cohort studies: the Aging Gene–Environment Susceptibility-Reykjavik Study, the Atherosclerosis Risk in Communities Study, the Austrian Stroke Prevention Study, the Cardiovascular Health Study, the Framingham Heart Study, and 2 cohorts from the Rotterdam Study. Genome wide significant association was identified for 6 SNPs on chromosome 17q25. The findings had been replicated in an independent sample of 1607 AGES-Reykjavik participants and in 1417 elderly white participants from the 3C-Dijon Study in the original report (Fornage et al., 2011) and in 1677 persons of the Rotterdam Study III (Verhaaren et al., 2011). In the ~ 100 kb long region on chromosome 17 there are several genes with diverse functions such as the 2 tripartite motif-containing genes (TRIM65 and TRIM47 GenBank ID 201292, 91107) the WW domain binding protein 2 gene (WBP2 GenBank ID 23558), the mitochondrial ribosomal protein L38 gene (MRPL38 GenBank ID 64978), the Fas-binding factor 1 gene (FBF1 GenBank ID 217335), the acyl-coenzyme A oxidase 1 gene (ACOX1 GenBank ID 51) and the C-Elegans homolog (UNC13D GenBank ID 201294) gene which may account for the association. The study also reported 3 additional SNPs with highly suggestive p-values of < 5 ∗ 10− 5 which were located within genes coding for the polyamine-modulated factor 1 gene (PMF1 GenBank ID11243), the collagen type XXV alpha gene (COL25A1 GenBank ID 84570), and the methylenetetrahydrofolate dehydrogenase 1 gene (MTHFD1 GenBank ID 4522).
5. Summary
The last decade of research on the genetics of cerebral small vessel disease showed that this phenotype is highly heritable over different populations and subgroups. Candidate gene studies provided strong evidence for at least three pathways involved, namely the RAS, the NOTCH3 signaling pathway and pathways related to APOE. These pathways play an important role in the normal functioning of the neurovascular unit by ensuring dynamic communication between astrocytes, vascular smooth muscle cells, pericytes and endothelial cells. It is conceivable that mutations within these genes interfere with normal cellular communications and lead to a dysfunction of the neurovascular unit, which in turn leads to impaired vasoregulation, cerebral autoregulation, and blood brain barrier permeability. This may ultimately result in ischemic parenchymal damage with myelin rarefaction as histopathologically observed in areas of WML. The recent GWA study was successful in identifying common variants at chromosome 17, and pinpointed to possible novel mechanisms leading to WML. Although association studies had been successful in identifying many common genetic variants, these variants have small effect sizes, odds ratios are typically below 2, and are therefore not useful in clinical settings. Common variants identified so far also explain only a small proportion of the heritability. The same is true for genetic variants that have been identified for other dementia types including Alzheimer's disease. Other than in dementia of the Alzheimer type in which many of the identified susceptibility genes and even of loci identified in GWA studies have been replicated by multiple studies with large sample sizes (Paulson and Igo, 2011), the genetic architecture of vascular dementia and its subtypes is less well explored. Many of the reported genetic associations seen with vascular dementia-related phenotypes rely on small to mid-sized single studies and replication will be mandatory in order to judge on the robustness of these findings.
This missing heritability might be explained by the fact, that the association studies conducted were designed to test common SNPs with minor allele frequency of ≥ 5%. Other kinds of genetic variations such as rare or even individual SNPs, copy number variations and epigenetic modifications though maybe equally important, have not yet been targeted. An alternative to the “common disease common variants” hypotheses is the “common disease rare variant” hypothesis which supposes that rare variants (minor allele frequency ≤ 1–5%) with large effect size (OR > 2) segregating in families may substantially contribute to the heritability of common diseases. Linkage studies, which have the highest power to test this hypothesis, had detected significant loci for WML, but the causal variants responsible for the linkage signals have not yet been identified. With the advent of next generation sequencing methods and the comprehensive catalog of rare variants by the 1000 Genomes Project (http://www.1000genomes.org/page.php), sequencing these regions in large number of individuals became feasible, and we expect that rare causal variants with a large effect size will be identified in the future.
Acknowledgment
This work was supported by the Austrian Science Fond (FWF) grant number P20545-P05 to HS and P13180 to RS.
Section Editor: Christian Humpel
References
- Atwood L.D., Wolf P.A., Heard-Costa N.L., Massaro J.M., Beiser A., D'Agostino R.B., deCarli C. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke. 2004;35:1609–1613. doi: 10.1161/01.STR.0000129643.77045.10. [DOI] [PubMed] [Google Scholar]
- Carmelli D., DeCarli C., Swan G.E., Jack L.M., Reed T., Wolf P.A., Miller B.L. Evidence for genetic variance in white matter hyperintensity volume in normal elderly male twins. Stroke. 1998;29:1177–1181. doi: 10.1161/01.str.29.6.1177. [DOI] [PubMed] [Google Scholar]
- Cuenco K.T., Green R.C., Zhang J., Lunetta K., Erlich P.M., Cupples L.A., Farrer L.A., Decarli C. Magnetic resonance imaging traits in siblings discordant for Alzheimer disease. J. Neuroimaging. 2008;18:268–275. doi: 10.1111/j.1552-6569.2007.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano A.L., Atwood L.D., Massaro J.M., Heard-Costa N., Beiser A., Au R., Wold P.A., DeCarli C. Genome-wide scan for white matter hyperintensity the Framingham Heart Study. Stroke. 2006;37:77–81. doi: 10.1161/01.STR.0000196987.68770.b3. [DOI] [PubMed] [Google Scholar]
- DeStefano A.L., Seshadri S., Beiser A., Atwood L.D., Massaro J.M., Au R., Wolf P.A., DeCarli C. Bivariate heritability of total and regional brain volumes: the Framingham Study. Alzheimer Dis. Assoc. Disord. 2009;23:218–223. doi: 10.1097/WAD.0b013e31819cadd8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenga V., Fardoux P., Lacombe P., Monet M., Maciazek J., Krebs L.T., Klonjkowsko B., Berrou E., Mericskay M., Li Z., Tournier-Lasserve E., Gridley T., Joutel A. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas F., Kleinert R., Offenbacher H., Schmidt R., Kleinert G., Payer F., Radner H., Lechner H. Pathologic correlates of incidental MRI white matter signal hyperintensities. Neurology. 1993;43:1683–1689. doi: 10.1212/wnl.43.9.1683. [DOI] [PubMed] [Google Scholar]
- Fornage M., Debette S., Bis J.C., Schmidt H., Ikram M.A., Dufouil C., Sigurdsson S., Lumley T., DeStefano A.L., Fazekas F., Vrooman H.A., Shibata D.K., Maillard P., Zijdenbos A., Smith A.V., Gudnason H., de Boer R., Cushman M., Mazoyer B., Heiss G., Vernooij M.W., Enzinger C., Glazer N.L., Beiser A., Knopman D.S., Cavalieri M., Niessen W.J., Harris T.B., Petrovic K., Lopez O.L., Au R., Lambert J.C., Hofman A., Gottesman R.F., Garcia M., Heckbert S.R., Atwood L.D., Catellier D.J., Uitterlinden A.G., Yang Q., Smith N.L., Aspelund T., Romero J.R., Rice K., Taylor K.D., Nalls M.A., Rotter J.I., Sharrett R., van Duijn C.M., Amouyel P., Wolf P.A., Gudnason V., van der Lugt A., Boerwinkle E., Psaty B.M., Seshadri S., Tzourio C., Breteler M.M., Mosley T.H., Schmidt R., Longstreth W.T., DeCarli C., Launer L.J. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann. Neurol. 2011;69:928–939. doi: 10.1002/ana.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin O., Tzourio C., Maillard P., Alpérovitch A., Mazoyer B., Dufouil C. Apolipoprotein E genotype is related to progression of white matter lesion load. Stroke. 2009;40:3186–3190. doi: 10.1161/STROKEAHA.109.555839. [DOI] [PubMed] [Google Scholar]
- Greenberg S.M., Vernooij M.W., Cordonnier C., Viswanathan A., Salman R.A.S., Warach S., Launer L.J., VanBuchem M.A., Breteler M.M. Cerebral microbleeds: a guide to detection and interpretation. Lancet Neurol. 2009;8:165–174. doi: 10.1016/S1474-4422(09)70013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A., Corpechot C., Ducros A., Vahedi K., Chabriat H., Mouton P., Alamowitch S., Domenga V., Cécillion M., Marechal E., Macizek J., Vayssiere C., Cruaud C., Cabanis E.A., Ruchoux M.M., Weissenbach J., Bach J.F., Bouser M.G. Notch3 mutations in CADASIL a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- Koch S., McClendon M.S., Bhatia R. Imaging evolution of acute lacunar infarction: leukoariosis or lacune? Neurology. 2011;77:1091–1095. doi: 10.1212/WNL.0b013e31822e1470. [DOI] [PubMed] [Google Scholar]
- Kochunov P., Glahn D., Lancaster J., Winkler A., Kent J.W., Olvera R.L., Cole S.A., Dyer T.D., Almasy L., Duggirala R., Fox P.T., Blangero J. Whole brain and regional hyperintense white matter volume and blood pressure: overlap of genetic loci produced by bivariate, whole-genome linkage analyses. Stroke. 2010;41:2137–2142. doi: 10.1161/STROKEAHA.110.590943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw F.E.De., Richard F., Groot J.C.D., Duijn C.M.V., Hofman A., Van Gijn J., Breteler M.M. Interaction between hypertension apoE and cerebral white matter lesions. Stroke. 2004;35:1057–1060. doi: 10.1161/01.STR.0000125859.71051.83. [DOI] [PubMed] [Google Scholar]
- Lemmens R., Görner A., Schrooten M., Thijs V. Association of apolipoprotein E epsilon2 with white matter disease but not with microbleeds. Stroke. 2007;38:1185–1188. doi: 10.1161/01.STR.0000259816.31370.44. [DOI] [PubMed] [Google Scholar]
- Markus H.S. Genes, endothelial function and cerebral small vessel disease in man. Exp. Physiol. 2008;93:121–127. doi: 10.1113/expphysiol.2007.038752. [DOI] [PubMed] [Google Scholar]
- O'Brien J.T., Erkinjuntti T., Reisberg B., Roman G., Sawada T., Pantoni L., Bowler J.V., Ballard C., DeCarli C., Gorelick P.B., Rockwood K., Burns A., Gauthier S., DeKosky S.T. Vascular cognitive impairment. Lancet Neurol. 2003;2:89–98. doi: 10.1016/s1474-4422(03)00305-3. [DOI] [PubMed] [Google Scholar]
- Paternoster L., Chen W., Sudlow C.L.M. Genetic determinants of white matter hyperintensities on brain scans: a systematic assessment of 19 candidate gene polymorphisms in 46 studies in 19,000 subjects. Stroke. 2009;40:2020–2026. doi: 10.1161/STROKEAHA.108.542050. [DOI] [PubMed] [Google Scholar]
- Paulson H.L., Igo I. Genetics of dementia. Semin. Neurol. 2011;31:449–460. doi: 10.1055/s-0031-1299784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poels M.M.F., Vernooij M.W., Ikram M.A., Hofman A., Krestin G.P., Lugt A.V.D., Breteler M.M.B. Prevalence and risk factors of cerebral microbleeds: an update of the Rotterdam scan study. Stroke. 2010;41(10 Suppl):S103–S106. doi: 10.1161/STROKEAHA.110.595181. [DOI] [PubMed] [Google Scholar]
- Schmidt R., Schmidt H., Fazekas F., Schumacher M., Niederkorn K., Kapeller P., Weinrauch V., Kostner G.M. Apolipoprotein E polymorphism and silent microangiopathy-related cerebral damage. Results of the Austrian Stroke Prevention Study. Stroke. 1997;28:951–956. doi: 10.1161/01.str.28.5.951. [DOI] [PubMed] [Google Scholar]
- Schmidt H., Fazekas F., Kostner G.M., Duijn C.M.V., Schmidt R. Angiotensinogen gene promoter haplotype and microangiopathy‐related cerebral damage: results of the Austrian Stroke Prevention Study. Stroke. 2001;32:405–412. doi: 10.1161/01.str.32.2.405. [DOI] [PubMed] [Google Scholar]
- Schmidt H., Aulchenko Y.S., Schweighofer N., Schmidt R., Frank S., Kostner G.M., Ott E., van Duijn C. Angiotensinogen promoter B‐haplotype associated with cerebral small vessel disease enhances basal transcriptional activity. Stroke. 2004;35:2592–2597. doi: 10.1161/01.STR.0000144646.96121.d2. [DOI] [PubMed] [Google Scholar]
- Schmidt H., Zeginigg M., Wiltgen M., Freudenberger P., Petrovic K., Cavalieri M., Gider P., Enzinger C., Fornage M., Debette S., Rotter J.I., Ikram M.A., Launer L.J., Schmidt R. Genetic variants of the NOTCH3 gene in the elderly and magnetic resonance imaging correlates of age-related cerebra small vessel disease. Brain. 2011;134:3384–3397. doi: 10.1093/brain/awr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuur M., Swieten J.C.V., Schol-Gelok S., Ikram M.A., Vernooij M.W., Liu F., Isaacs A., de Boer R., de Koning I., Niessen W.J., Vrooman H., Oostra B.A., van der Lugt A., Breteler M.M., van Dujin C.M. Genetic risk factors for cerebral small-vessel disease in hypertensive patients from a genetically isolated population. J. Neurol. Neurosurg. Psychiatry. 2011;82:41–44. doi: 10.1136/jnnp.2009.176362. [DOI] [PubMed] [Google Scholar]
- Seshadri S., DeStefano A.L., Au R., Massaro J.M., Beiser A.S., Kelly-Hayes M., Kase C.S., D'Agostino R.B., Sr., DeCarli C., Atwood L.D., Wolf P.A. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med. Genet. 2007;8(Suppl. 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W.D., Steffens D.C., Ashley-Koch A., Payne M.E., MacFall J.R., Potocky C.F., Krishnan K.R. Angiotensin receptor gene polymorphisms and 2-year change in hyperintense lesion volume in men. Mol. Psychiatry. 2010;15:816–822. doi: 10.1038/mp.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.T., Jack C.R., Fornage M., Mosley T.H., Boerwinkle E., Andrade M.D. Heritability of leukoaraiosis in hypertensive sibships. Hypertension. 2004;43:483–487. doi: 10.1161/01.HYP.0000112303.26158.92. [DOI] [PubMed] [Google Scholar]
- Turner S.T., Fornage M., Jack C.R., Mosley T.H., Knopman D.S., Kardia S.L.R., Boerwinkle E., de Andrade M. Genomic susceptibility Loci for brain atrophy ventricular volume and leukoaraiosis in hypertensive sibships. Arch. Neurol. 2009;66:847–857. doi: 10.1001/archneurol.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E.J.V., Prins N.D., Vermeer S.E., Koudstaal P.J., Breteler M.M.B. Frequency of white matter lesions and silent lacunar infarcts. J. Neural Transm. Suppl. 2002;62:25–39. doi: 10.1007/978-3-7091-6139-5_2. [DOI] [PubMed] [Google Scholar]
- Verhaaren B.F.J., Boer R.D., Vernooij M.W., Rivadeneira F., Uitterlinden A.G., Hofman A., Krestin G.P., van der Lugt A., Niessen W.J., Breteler M.M., Ikram M.A. Replication study of chr17q25 with cerebral white matter lesion volume. Stroke. 2011;42:3297–3299. doi: 10.1161/STROKEAHA.111.623090. [DOI] [PubMed] [Google Scholar]