Abstract
By virtue of being the product of the genetic admixture of three ancestral roots: Europeans, Africans, and Amerindians, the present-day Brazilian population displays very high levels of genomic diversity, which have important pharmacogenetic/-genomic (PGx) implications. Recognition of this fact has prompted the creation of the Brazilian Pharmacogenomics Network (Refargen), a nationwide consortium of research groups, with the mission to provide leadership in PGx research and education in Brazil, with a population heath impact. Here, we present original data and review published results from a Refargen comprehensive study of the distribution of PGx polymorphisms in a representative cohort of the Brazilian people, comprising 1,034 healthy, unrelated adults, self-identified as white, brown, or black, according to the Color categories adopted by the Brazilian Census. Multinomial log-linear regression analysis was applied to infer the statistical association between allele, genotype, and haplotype distributions among Brazilians (response variables) and self-reported Color, geographical region, and biogeographical ancestry (explanatory variables), whereas Wright’s FST statistics was used to assess the extent of PGx divergence among different strata of the Brazilian population. Major PGx implications of these findings are: first, extrapolation of data from relatively well-defined ethnic groups is clearly not applicable to the majority of Brazilians; second, the frequency distribution of polymorphisms in several pharmacogenes of clinical relevance (e.g., ABCB1, CYP3A5, CYP2C9, VKORC) varies continuously among Brazilians and is not captured by race/Color self-identification; third, the intrinsic heterogeneity of the Brazilian population must be acknowledged in the design and interpretation of PGx studies in order to avoid spurious conclusions based on improper matching of study cohorts.
Keywords: biogeographical ancestry, Brazilian pharmacogenomic network, FST statistics, pharmacogenomic diversity, population admixture, refargen
Introduction
The present-day Brazilian population, in excess of 190 million people, is highly heterogeneous and admixed, as result of five centuries of mating between native Amerindians, Europeans, and sub-Saharan Africans. This fact renders inappropriate extrapolation of pharmacogenetic/-genomic (PGx) data derived from well-defined ethnic groups to the majority of Brazilians. Recognition of this fact has prompted the creation of the Brazilian Pharmacogenomics Network or Refargen (Suarez-Kurtz, 2004), a nationwide consortium of research groups, mostly from academia1. In consonance with its mission to provide leadership in PGx research and education in Brazil, with impact on population heath (Suarez-Kurtz, 2009), Refargen has recently concluded a comprehensive study of the distribution of PGx polymorphisms among Brazilians. In this article, we will present original data and review previously published results (Suarez-Kurtz et al., 2010, 2012a,b,c; Pena et al., 2011; Sortica et al., 2012) from the Refargen study and discuss the PGx implications of the findings for Brazilians and possibly other admixed populations of the Americas.
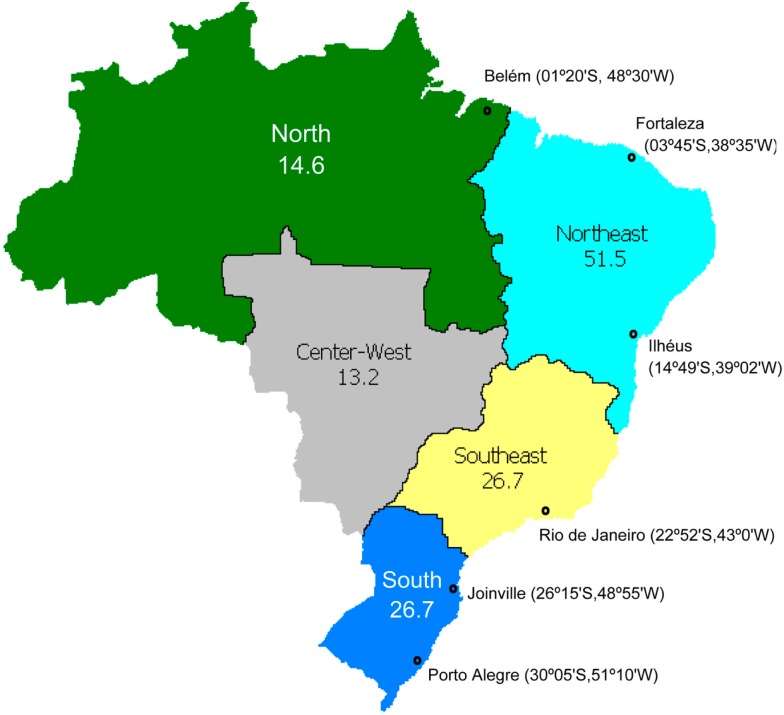
The study cohort consisted of 1,034 healthy, unrelated adults recruited in the North, Northeast, Southeast, and South regions of Brazil (Figure 1). Each individual signed a written informed consent and was asked to self-identify according to the classification scheme adopted by the Brazilian Census2, which relies on self-perception of skin color. Accordingly, the subjects were distributed into three groups: branco (White, n = 342), pardo (Brown, n = 350), and preto (Black, n = 342). The term Color is capitalized throughout the text, to call attention to its special meaning in the context of the Brazilian Census classification. This cohort is considered representative of the present-day Brazilian population since 99% of Brazilians self-identify in one of the three Color categories, and 93% live in one of the four regions, included in the study3. Individuals from the Center-West region (7% of the Brazilian population) and those classified as “Yellow” (meaning Asian descendants, 0.7%) or Amerindian (0.3%) were not included in the study. We genotyped 44 loci in 12 pharmacogenes (Table 1) which modulate drug metabolism (CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A5, COMT, and TPMT), transport (ABCB1, SLCO1B1, and SLCO1B3) and effect (VKORC1). Pharmacogenomics Knowledge Base (PharmGKB4) lists all these genes, except SCLO1B3, as “Important PGx genes (VIP)” and two thirds of the 44 polymorphisms investigated as “Important Variants.”
Figure 1.
Map of Brazil, showing its five geographical regions, their populations (in millions), and cities where individuals were recruited for the Refargen study, reviewed in this article.
Table 1.
Distribution of pharmacogenetic polymorphisms among Brazilians.
| Gene | Polymorphism | Id # | Effect | Minor allele frequency |
Chi square |
FST |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | White | Brown | Black | P value | White vs. Black | White vs. Brown | Brown vs. Black | ||||
| ABCB1 | 1267C > T | rs1128503 | G412G | 0.380 | 0.412 | 0.362 | 0.312 | 0.18 | 0.011 | 0.003 | 0.003 |
| 2677G > T/A | rs2032582 | S193A/T | 0.370 | 0.417 | 0.343 | 0.221 | 0.0006 | 0.044 | 0.006 | 0.019 | |
| 3435C > T | rs1045642 | I1145I | 0.427 | 0.458 | 0.411 | 0.317 | 0.037 | 0.021 | 0.002 | 0.009 | |
| COMT | 472G > A | rs4680 | V158M | 0.408 | 0.437 | 0.374 | 0.411 | 0.52 | 0.001 | 0.004 | 0.001 |
| CYP2B6 | 64C > T | rs8192709 | R22C | 0.081 | 0.067 | 0.099 | 0.078 | 0.60 | 0.001 | 0.003 | 0.001 |
| 785A > G | rs2279343 | K262R | 0.303 | 0.295 | 0.300 | 0.374 | 0.22 | 0.007 | 0.000 | 0.006 | |
| 1459C > T | rs3211371 | R487C | 0.172 | 0.213 | 0.135 | 0.127 | 0.068 | 0.013 | 0.011 | 0.000 | |
| 516G > T | rs3745274 | Q172H | 0.369 | 0.393 | 0.325 | 0.458 | 0.055 | 0.004 | 0.005 | 0.019 | |
| CYP2C8 | 805A > T | rs11572103 | I269F | 0.047 | 0.026 | 0.060 | 0.106 | 0.012 | 0.026 | 0.007 | 0.007 |
| 416G > A | rs11572080 | R139K | 0.098 | 0.125 | 0.075 | 0.058 | 0.08 | 0.014 | 0.007 | 0.001 | |
| 792C > G | rs1058930 | I264M | 0.038 | 0.040 | 0.039 | 0.020 | 0.54 | 0.004 | 0.000 | 0.003 | |
| CYP2C9 | 430C > T | rs1799853 | R144C | 0.108 | 0.137 | 0.082 | 0.066 | 0.08 | 0.004 | 0.001 | 0.007 |
| 1075A > C | rs1057910 | I359L | 0.052 | 0.049 | 0.059 | 0.026 | 0.26 | 0.014 | 0.008 | 0.001 | |
| 1080C > G | rs28371686 | D360E | 0.005 | 0.004 | 0.005 | 0.009 | 1.0 | 0.001 | 0.000 | 0.000 | |
| 1003C > T | rs28371685 | R335W | 0.008 | 0.010 | 0.005 | 0.009 | 0.78 | 0.000 | 0.001 | 0.000 | |
| CYP2C19 | 681G > A | rs4244285 | Splicing defect | 0.115 | 0.106 | 0.120 | 0.144 | 0.58 | 0.003 | 0.001 | 0.001 |
| 636G > A | rs4986893 | W212X | 0.000 | 0.000 | 0.000 | 0.003 | 1.0 | 0.000 | 0.000 | 0.000 | |
| −806C > T | rs12248560 | Increased transcription | 0.169 | 0.168 | 0.167 | 0.175 | 0.98 | 0.000 | 0.000 | 0.000 | |
| CYP2D6 | −1584C > G | rs1080985 | Promoter region | 0.218 | 0.235 | 0.207 | 0.178 | 0.64 | 0.003 | 0.003 | 0.000 |
| 31G > A | rs769258 | V11M | 0.035 | 0.043 | 0.027 | 0.023 | 0.60 | 0.002 | 0.002 | 0.000 | |
| 100C > T | rs1065852 | P34S | 0.136 | 0.150 | 0.124 | 0.115 | 0.79 | 0.003 | 0.002 | 0.000 | |
| 1023C > T | rs28371706 | T107I | 0.044 | 0.021 | 0.057 | 0.111 | 0.08 | 0.025 | 0.011 | 0.003 | |
| 1661G > C | rs1058164 | V136V | 0.456 | 0.461 | 0.445 | 0.482 | 0.89 | 0.000 | 0.000 | 0.001 | |
| 1846G > A | rs3892097 | Splicing defect | 0.117 | 0.134 | 0.105 | 0.080 | 0.61 | 0.003 | 0.004 | 0.000 | |
| 2549A > del | rs35742686 | Frame shift | 0.009 | 0.011 | 0.007 | 0.006 | 0.60 | 0.000 | 0.000 | 0.000 | |
| 2615-2617delAAG | rs5030656 | K281del | 0.013 | 0.014 | 0.011 | 0.013 | 1.0 | 0.001 | 0.002 | 0.001 | |
| 2850C > T | rs16947 | R296C | 0.390 | 0.352 | 0.417 | 0.480 | 0.23 | 0.016 | 0.003 | 0.005 | |
| 3183G > A | rs59421388 | V287M | 0.026 | 0.011 | 0.038 | 0.056 | 0.25 | 0.021 | 0.011 | 0.002 | |
| 4180G > C | rs1135840 | S486T | 0.531 | 0.513 | 0.541 | 0.582 | 0.74 | 0.004 | 0.000 | 0.002 | |
| CYP3A5 | 6986A > G | rs776746 | Frame shift | 0.698 | 0.785 | 0.627 | 0.541 | <0.0001 | 0.067 | 0.030 | 0.007 |
| 14690G > A | rs10264272 | Splicing defect | 0.027 | 0.004 | 0.039 | 0.105 | 0.0001 | 0.049 | 0.014 | 0.017 | |
| 23132insT | rs413003343 | Frame shift | 0.024 | 0.013 | 0.027 | 0.079 | 0.003 | 0.025 | 0.003 | 0.013 | |
| SLCO1B1 | 388A > G | rs2306283 | N130D | 0.553 | 0.498 | 0.601 | 0.635 | 0.036 | 0.019 | 0.011 | 0.001 |
| 463C > A | rs11045819 | P155T | 0.118 | 0.118 | 0.122 | 0.097 | 0.77 | 0.001 | 0.000 | 0.002 | |
| 521T > C | rs4149056 | V174A | 0.135 | 0.135 | 0.143 | 0.089 | 0.24 | 0.005 | 0.000 | 0.007 | |
| SLCO1B3 | 334T > G | rs4149117 | S112A | 0.741 | 0.799 | 0.702 | 0.592 | 0.0003 | 0.051 | 0.013 | 0.013 |
| 699G > A | rs7311358 | M233I | 0.741 | 0.799 | 0.702 | 0.592 | 0.0003 | 0.051 | 0.013 | 0.013 | |
| TMPT | 238G > C | rs1800462 | A80P | 0.008 | 0.001 | 0.014 | 0.014 | 0.36 | 0.006 | 0.006 | 0.000 |
| 460G > A | rs1800460 | A154T | 0.010 | 0.009 | 0.011 | 0.010 | 0.82 | 0.000 | 0.000 | 0.000 | |
| 719A > G | rs1142345 | Y240C | 0.026 | 0.017 | 0.037 | 0.017 | 0.46 | 0.000 | 0.004 | 0.004 | |
| VKORC1 | 3673G > A | rs9923231 | Reduced transcription | 0.333 | 0.371 | 0.306 | 0.238 | 0.038 | 0.021 | 0.005 | 0.006 |
| 5808C > T | rs2884737 | Intronic | 0.193 | 0.255 | 0.135 | 0.128 | 0.003 | 0.026 | 0.023 | 0.000 | |
| 6853G > C | rs8050894 | Intronic | 0.392 | 0.404 | 0.386 | 0.357 | 0.65 | 0.002 | 0.000 | 0.001 | |
| 9104G > A | rs7294 | 3-UTP | 0.375 | 0.376 | 0.372 | 0.379 | 0.65 | 0.000 | 0.000 | 0.000 | |
Bold numbers in the column “chi square” indicate statistically significant P values; bold numbers in the “White vs. Black” column indicate moderate pharmacogenetic divergence.
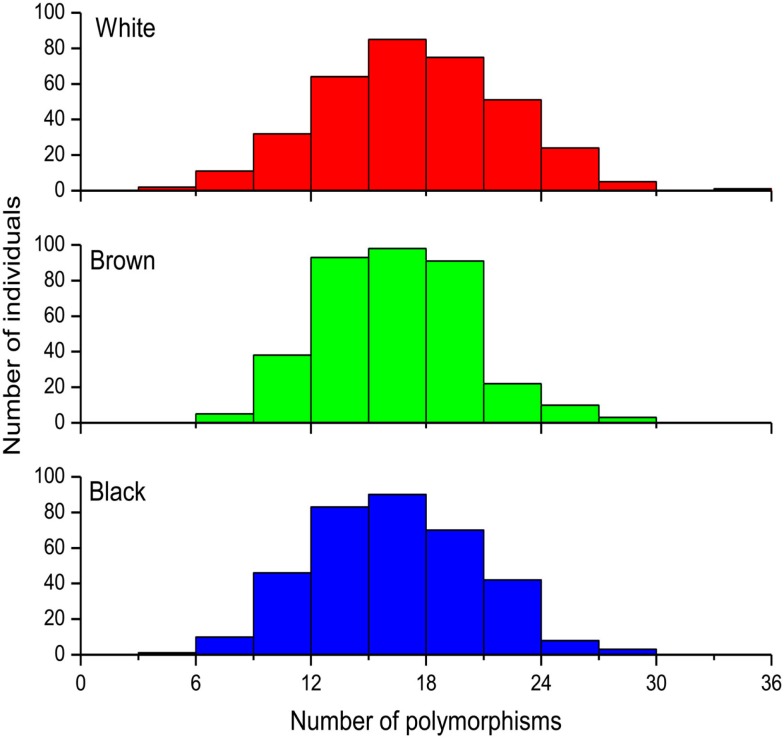
We will initially present data for the overall cohort and for each Color group within this cohort. Figure 2 shows frequency histograms of the total number of minor alleles identified in each individual. No statistically significant difference (Kruskal–Wallis test p = 0.92) was detected across the three Color groups, the median (interquartile range) number of polymorphisms being 17 (14–20), 16 (13–18), and 16 (13–19) in White, Brown, and Black individuals. This adds to 18.9% of the total number of alleles genotyped at the 44 loci in the overall cohort. However, the allele frequency at 11 of these loci differed significantly (chi square p < 0.05) across the Color groups. The pharmacogenes affected were ABCB1 (2 SNPs), CYP2C8 (1) CYP3A5 (3), NAT2 (3), SLCO1B1 (1), SCLO1B3 (2 SNPs, which are in complete LD) and VKORC1 (2). We applied the Wright’s FST statistics (Wright, 1951) to estimate the extent of PGx divergence among the three Color strata, and observed mean FST values of 0.005 (SD 0.006), 0.013 (0.017), and 0.004 (0.005) for pair-wise comparisons of White vs. Brown, White vs. Black, and Brown vs. Black, respectively (Suarez-Kurtz et al., 2012b). According to Wright’s qualitative guidelines (Wright, 1978), FST values lower than 0.05 denote low genetic diversity, whereas values between 0.05 and 0.15 indicate moderate diversity. As shown in Table 1, only three SNPs, namely CYP3A5*3 and the linked SLCO1B3 334T > C and 699G > A transitions exceeded, and two other SNPs (ABCB1 2677G > nonG and CYP3A5*6) approached, the FST threshold for moderate genetic divergence in White vs. Black Brazilians in the entire cohort. Not surprisingly, these were the SNPs with the smallest p values for the Kruskal–Wallis analyses of frequency distribution in the overall cohort (<0.0001–0.0006, Table 1). Taken together, the FST analyses in the overall cohort suggest low PGx divergence at all loci interrogated in self-identified Brown vs. White or Black individuals, whereas moderate divergence was observed at three, and possibly five loci (out of the 44 investigated) in pair-wise comparisons of White vs. Black Brazilians.
Figure 2.
Frequency histograms of the distribution of 44 PGx polymorphisms among self-identified (White n = 342), Brown (n = 352), and Black Brazilians (n = 342). The data represent the total number of minor alleles at the 44 loci, in each individual.
Distribution of Pharmacogenetic Polymorphisms among Brazilians According to Color Categories and Geographical Regions
With an area of 8,511,960 Km2, Brazil is a country of continental size (the fifth largest in the world) and different regions have diverse population histories. For instance, the North had a large influence of the Amerindian root, the Northeast had a history of strong African presence due to slavery and the South was mostly settled by European immigrants (Pena et al., 2011). We have applied multinomial log-linear regression analyses (Suarez-Kurtz et al., 2010, 2012c; Sortica et al., 2012) to infer the statistical association between allele, genotype, and haplotype distributions among Brazilians (response variables) and self-reported Color and geographical region (explanatory variables). This procedure obviates the need for correction for multiple comparisons, because the main effects and interaction terms are tested simultaneously within each regression context. Table 2 illustrates results from this exercise, applied to selected genes affecting drug metabolism (CYP2C8, CYP2C9, and CYP2C19), transport (ABCB1 and SLCO1B1) and response (VKORC1). Color per se associates significantly with the frequency distribution of CYP2C8 and CYP2C9 variant alleles, ABCB1 and SLCO1B1 haplotypes, and VKOC1 3673G > A alleles and genotypes; no association is observed with respect to the CYP2C19 polymorphisms. Color in combination with geographical region is significantly associated with distribution of CYP2C8 and CYP2C9 alleles, ABCB1 and SLCO1B1 haplotypes, whereas geographical region per se associates with CYP2C8 and CYP2C9 allele frequency.
Table 2.
Multinominal log-linear analyses of the distribution of pharmacogenetic polymorphisms alleles among Brazilians according to self-reported color and geographical region.
| Gene | Alleles | Explanatory variablesa |
||
|---|---|---|---|---|
| Color | Geographical region | Color: geographical region | ||
| CYP2C8 | *2, *3, *4 | <0.0001 | 0.71 | 0.04 |
| CYP2C9 | *2, *3, *5, *11 | <0.0001 | 0.23 | 0.01 |
| CYP2C19 | *2, *3, *17 | 0.60 | 0.11 | 0.33 |
| ABCB1 | haplotypesb | <0.001 | 0.001 | 0.013 |
| SLCO1B1 | haplotypesc | <0.001 | 0.001 | 0.003 |
| VKORC1 | 3673A allele | 0.0004 | 0.07 | 0.11 |
| 3673A genotype | 0.002 | 0.18 | 0.19 | |
ap values associated to the “main effects” (Color and geographical region) and their “interaction.”
bHaplotypes comprising the 1236C > T, 2677G > nonG, and 3435C > T loci.
cHaplotypes comprising the 388A > G, 463C > A, and 521T > C loci.
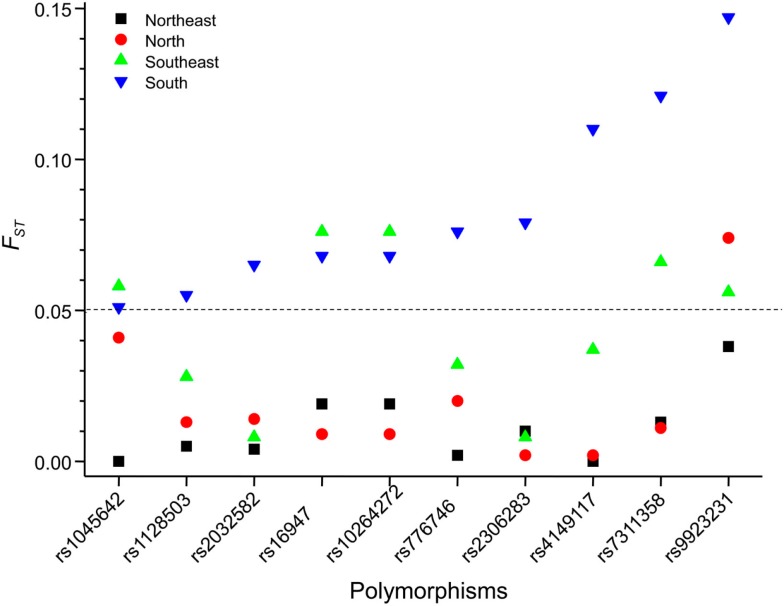
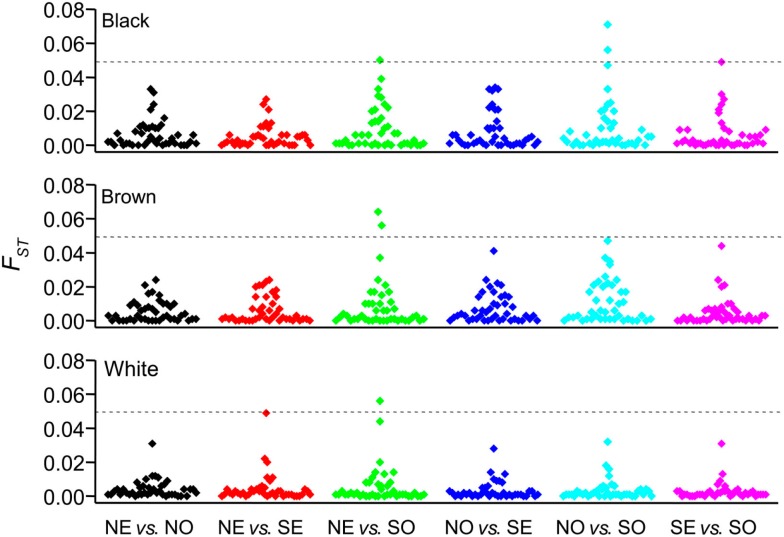
We explored further the PGx heterogeneity among Brazilians by the FST statistics. First, we performed pair-wise comparisons between Color groups within each geographical region, and detected significant differences in the distribution of FST values for White vs. Brown (P < 0.0001, ANOVA) and White vs. Black (P < 0.0001), but not Brown vs. Black individuals, across regions (Suarez-Kurtz et al., 2012b). This implies that the extent of pharmacogenetic divergence between Whites and Non-Whites (i.e., Black and Brown individuals) varies significantly among regions. The data presented in Figure 3 supports this interpretation: we show that 10 selected polymorphisms in ABCB1, CYP2D6, CYP3A5, SLCO1B1, SCLO1B3, and VKORC1 display moderate divergence between Whites and Blacks in the South, compared to five, one, and zero in the Southeast, North, and Northeast, respectively. In a second exercise, we compared FST values for each Color between regions and present the results in Figure 4. Of the 792 (44 polymorphisms × six pair-wise regions × three Color groups) comparisons, only three SNPs among Black, one among Brown, and one among White individuals exceeded the threshold (FST = 0.05) for moderate PGx divergence. Taken together, these FST results extend the conclusions of the multinomial analyses described above, that the distribution of PGx polymorphisms among Brazilians is influenced by self-reported Color, geographical region, and the interaction of these two variables. Collectively, these data reflect the notorious heterogeneity of the Brazilian population and highlight the inappropriateness of ascribing PGx polymorphisms’ frequencies for “Brazilians” based on data from one or more Color strata recruited at a given region (or city).
Figure 3.
Allele-specific FST values for 10 PGx polymorphisms (x-axis) in White vs. Black Brazilians recruited at the North, Northeast, Southeast, and South regions. The dashed line shows the threshold FST values (0.05) for moderate genetic divergence. The genes and loci for each polymorphism are presented in Table 1.
Figure 4.
Allele-specific FST values (y-axis) for pair-wise comparisons between geographical regions (x-axis). Data are presented separately for self-identified Black (top panel), Brown (middle panel), and White (bottom panel) individuals. Each symbol correspond to one of the 44 polymorphisms listed in Table 1. The dashed line shows the threshold FST values (0.05) for moderate genetic divergence. NO, North; NE, Northeast; SE, Southeast; SO, South.
Impact of Biogeographical Ancestry on the Distribution of Pharmacogenetic Polymorphisms among Brazilians
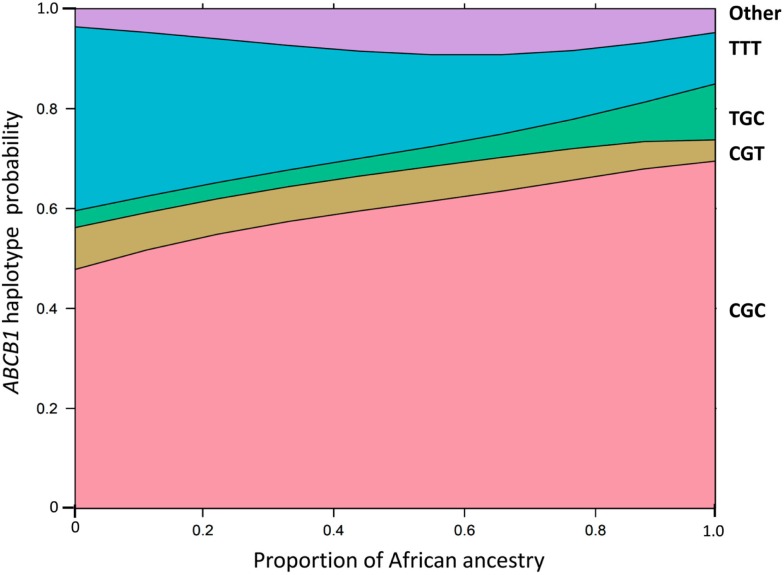
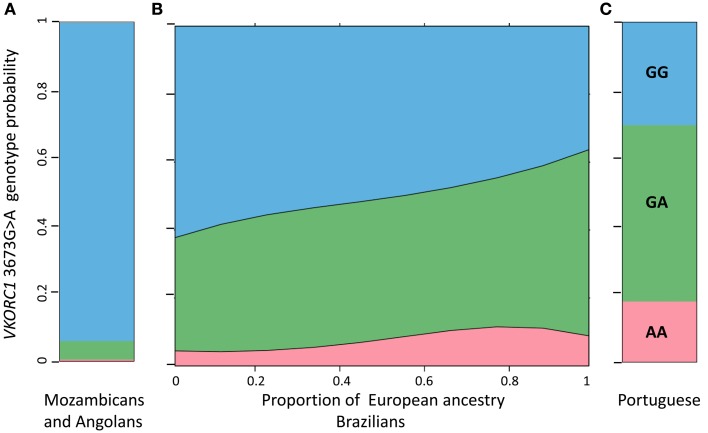
These analyses were based on the individual proportions of European, African, and Amerindian ancestry, estimated using a panel of short insertion/deletion polymorphisms, validated as ancestry-informative markers (Bastos-Rodrigues et al., 2006), and the STRUCTURE clustering software (Pritchard et al., 2000). These data, available for 965 subjects confirmed that the vast majority of Brazilians, irrespective of self-reported Color, share European and African ancestries in variable proportions, and a sizable number of individuals display also distinct Amerindian ancestry (Suarez-Kurtz and Pena, 2006, 2007; Suarez-Kurtz et al., 2010; Pena et al., 2011). The average proportions of European ancestry decrease progressively from self-reported White (mean 0.80, SD 0.21, n = 325), to Brown (0.62, 0.29, 322) and then to Black individuals (0.46, 0.20, 318), and the opposite trend is observed with respect to African ancestry, which averaged 0.10 (SD 0.14) in White, 0.25 (0.26) in Brown, and 0.42 (0.29) in Black persons. However, the individual proportions of European and African ancestry varies widely, and most importantly, as a continuum within each of these three Color categories, whereas the individual proportion of Amerindian ancestry remains relatively constant across the three groups, ranging from 0.10 to 0.13. To describe the association between PGx polymorphisms and the estimated individual biogeographical ancestry we fitted non-linear logistic regression modeling using maximum likelihood estimation. A consistent finding in these analyses (Suarez-Kurtz et al., 2007a,b, 2010, 2012c; Estrela et al., 2008; Vargens et al., 2008) is that the frequency distribution of PGx polymorphisms among Brazilians is best fit by continuous functions of the individual proportions of African and European ancestry. This is illustrated in Figures 5 and 6. In Figure 5 we show that the probability of having the wild-type (C/G/C) and the T/G/C ABCB1 haplotypes increases continuously with the increase in African ancestry, whereas the opposite trend is observed for the T/nonG/T haplotype. Figure 6 shows that the odds of having the heterozygous, and to a lesser extent, the homozygous variant genotype at the VKORC1 3673G > A locus increase progressively as the individual proportion of European ancestry increases. For comparison, we also display in Figure 6 the distribution of VKORC1 3673G > A genotypes among Portuguese, by far the most important source of European migrants from Brazil, and individuals from Angola and Mozambique, two former Portuguese colonies in Africa, and origin of enslaved Africans brought to Brazil.
Figure 5.
Effect display for the distribution of ABCB1 haplotypes in the logit model fit to the data for African ancestry in 965 Brazilians. The haplotypes comprising the 1236C > T, 2677G > nonG, and 3435C > T SNPs are shown at the right of the plot. The individual proportion of African ancestry is shown in the x-axis. The y-axis is labeled on the probability scale. The plot was generated as described by Venables and Ripley (2002) and implemented as function “multinom” available in the R package “nnet.” Data from Sortica et al. (2012).
Figure 6.
Effect display for the distribution of VKORC1 3673G > A genotypes in the logit model fit to the data for African ancestry in 965 Brazilians (B). For comparison the frequency of each genotype in a cohort of Angolans and Mozambicans [n = 216, (A)] and in a Portuguese cohort [n = 89, (C)] are also shown. The individual proportion of African ancestry in Brazilians is shown in the x-axis. The y-axis represents the genotype probability for Brazilians and the observed genotype frequency for the African and Portuguese cohorts. Data from Suarez-Kurtz et al. (2010). The plot for Brazilians was generated as described in Figure 5.
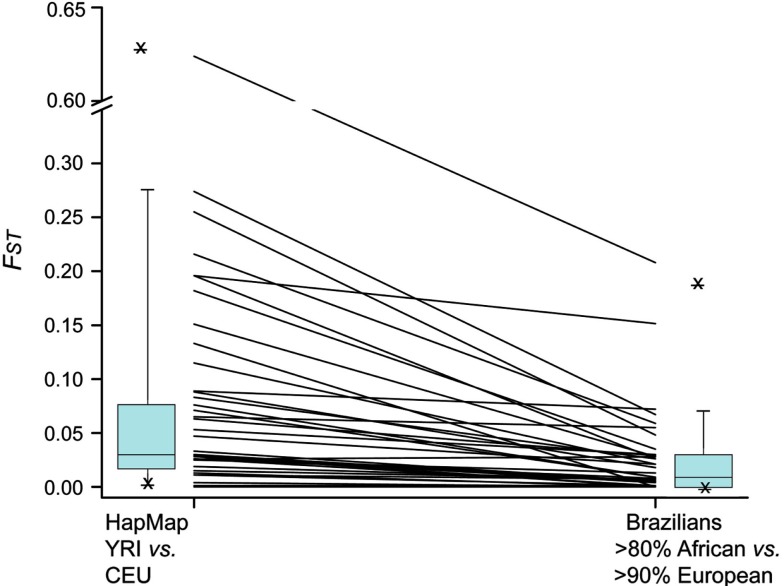
Considering that the European and African components together account for 89% of the diversity in individual genetic ancestry in the Refargen cohort (Pena et al., 2011), it might be anticipated that: (a) the greater the difference in frequency of a given polymorphism between Europeans and sub-Saharan Africans, the wider the range of frequency variation among Brazilians; (b) the range of variation among Brazilians will be smaller than the difference in frequency between Europeans and Africans, because of the admixture of these ancestral roots in Brazilians. We have previously verified both these predictions for polymorphisms in VKORC1 (Suarez-Kurtz et al., 2010) and within the CYP2C cluster (Suarez-Kurtz et al., 2012c). We applied the FST statistics to examine these predictions in 38 polymorphisms which were genotyped in the Refargen cohort and also in the HapMap project. In Figure 7 we shown the pair-wise FST values for each polymorphism in HapMap CEU vs. YRI cohorts – taken as proxies of the European and sub-Saharan African ancestral roots of Brazilians, respectively – and Brazilians with >90% European ancestry vs. Brazilians with >80% African ancestry. The attenuation of pharmacogenetic divergence between the Brazilian groups compared to the HapMap populations is evident.
Figure 7.
Allele-specific FST values for 38 PGx polymorphisms in HapMap CEU vs. YRI and in Brazilians with >90% European ancestry vs. Brazilians with >80% African ancestry. The lines connect the FST values for each polymorphism in the two data sets and the box plots at the left and right summarize the ensemble of the data for each set.
Concluding Remarks and Perspectives
The kaleidoscopic diversity of the admixed Brazilian population, with tri-hybrid biogeographical ancestry in Europe, Africa, and America adds complexity to, but also creates advantages for PGx research. Advantages include the opportunity to explore PGx associations in individuals with heterogeneous genetic ancestry under similar environmental and socio-economical conditions, and to gather information on peoples that are excluded or under-represented in clinical drug trials, such as sub-Saharan Africans and Native Americans. A major challenge to PGx studies in Brazil is population stratification, which if not controlled for, will confound the outcomes of PGx association studies. Our studies describe ways to control for this caveat, by combining ancestry-informative markers and appropriate statistical approaches. A distinct message that emerges from these studies is that race/color categorization does not capture the distribution of PGx polymorphisms among Brazilians, which is best modeled by continuous functions of the individual proportions of European and African ancestry, irrespective of self-identified Color (Suarez-Kurtz, 2010). Recognition of this fact is important in the design and interpretation of PGx clinical trials in Brazilians but does not imply that PGx-informed drug prescription requires investigation of individual ancestry. Rather, individual genotyping should be directed to PGx polymorphisms of proven clinical utility for the specific medical condition being treated, irrespective of biogeographical ancestry.
Drug assessment and regulatory processes in Brazil are carried out by the National Health Surveillance Agency, ANVISA, an independently administered, financially autonomous agency, managed by a Collegiate Board of Directors5. ANVISA has the mandate to grant, and withdraw, product registration permits within its areas of activity, which comprise medicines for human use. Registration of new medicines do not require, that clinical trials be carried out in the Brazilian population, and evaluation of the medicine’s efficacy and toxicity is based mainly, if not exclusively, on foreign data. Despite the increasing enrolment of non–Caucasian subjects in global drug development programs, most data submitted to ANVISA derive from white Europeans and North Americans. We have recently shown that there is little pharmacogenetic divergence between the HapMap CEU cohort of European extraction and White Brazilians, such that only CYP3A5*3 among 44 polymorphisms exceeded the FST threshold for moderate divergence. By contrast, FST analyses revealed very large divergence between CEU and Black Brazilians for CYP3A5*3 and moderate divergence for eight other polymorphisms, including another CYP3A5 SNP (CYP3A5*6) and SNPs in the ABCB1, SLCO1B3, and SLCO1B1 genes. These findings represent a caveat against extrapolation of PGx data from European-derived (“Caucasian”) cohorts to the ensemble of Brazilians.
Admixture is common in all developing nations in the American continent, although the relative contributions of the three major ancestral roots – native American, European, and sub-Saharan African – vary among these nations, as well as among ethnic groups and geographical regions within a given country. Hence, extrapolation of conclusions drawn from PGx studies in Brazilians to other admixed Latin American populations must take into account the specific patterns of population structure and diversity across the Americas. Therapeutic drugs are usually developed and investigated for their safety and efficacy in geographical and ethnical populations that do not encompass the diversity of Latin American peoples. Drivers and barriers to the adoption of PGx in developing countries, and specific ways in which these countries could benefit from PGx-based drug therapy deserve greater attention from academic and industrial scientists, prescribers, and legislators in developing nations across the Americas. This goal is not likely to be achieved simply by mandates to include subjects from ethnic minorities in clinical drug trials, especially when these groups are labeled by phenotypes which do not accurately reflect genetic ancestry (Suarez-Kurtz, 2005, 2010; Suarez-Kurtz and Pena, 2006, 2007).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research was supported by a grant from Financiadora de Estudos e Projetos (FINEP 01.08.01230.00). Guilherme Suarez-Kurtz and Claudio José Struchiner are supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). The authors acknowledge the contribution of Refargen researchers for providing blood and/or DNA samples and genotyping pharmacogenetic polymorphisms.
Footnotes
References
- Bastos-Rodrigues L., Pimenta J. R., Pena S. D. J. (2006). The genetic structure of human populations studied through short insertion-deletion polymorphisms. Ann. Hum. Genet. 70, 658–665 10.1111/j.1469-1809.2006.00287.x [DOI] [PubMed] [Google Scholar]
- Estrela R. C., Ribeiro F. S., Carvalho R. S., Gregório S. P., Dias-Neto E., Struchiner C. J., et al. (2008). Distribution of ABCB1 polymorphisms among Brazilians, impact of population admixture. Pharmacogenomics 9, 267–276 10.2217/14622416.9.3.267 [DOI] [PubMed] [Google Scholar]
- Pena S. D. J., Di-Pietro G., Fuchshuber-Moraes M., Pasqualini-Genro J., Hutz M. H., Kehdi F., et al. (2011). The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS ONE 6, e17063. 10.1371/journal.pone.0017063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sortica V. de A., Ojopi E. B., Genro J. P., Callegari-Jacques S., Ribeiro-Dos-Santos A., de Moraes M. O., et al. (2012). Influence of genomic ancestry on the distribution of SLCO1B1, SLCO1B3 and ABCB1 gene polymorphisms among Brazilians. Basic Clin. Pharmacol. Toxicol. 110, 460–468 10.1111/j.1742-7843.2011.00838.x [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G. (2004). Pharmacogenomics in admixed populations, the Brazilian pharmacogenetics/pharmacogenomics network – REFARGEN. Pharmacogenomics J. 4, 347–438 10.1038/sj.tpj.6500287 [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G. (2005). Pharmacogenetics in admixed populations. Trends Pharmacol. Sci. 26, 196–201 10.1016/j.tips.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G. (2009). Editorial: pharmacogenomics and the genetic diversity of the Brazilian population. Cad. Saúde Pública 25, 1650–1651 10.1590/S0102-311X2009000800001 [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G. (2010). Pharmacogenetics in the Brazilian population. Front. Pharmacol. 1:118. 10.3389/fphar.2010.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Kurtz G., Amorim A., Damasceno A., Hutz M. H., Moraes M. O., Ojopi E. B., et al. (2010). VKORC1 polymorphisms in Brazilians, comparison with the Portuguese and Portuguese-speaking Africans and pharmacogenetic implications. Pharmacogenomics 11, 1257–1267 10.2217/pgs.10.89 [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G., Pena S. D. J. (2006). Pharmacogenomics in the Americas, impact of genetic admixture. Curr. Drug Targets 7, 1649–1658 10.2174/138945006779025392 [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G., Pena S. D. J. (2007). “Pharmacogenetic Studies in the Brazilian Population,” in Pharmacogenomics in Admixed Populations, ed. Suarez-Kurtz G. (Austin: Landes Biosciences; ), 75–98 [Google Scholar]
- Suarez-Kurtz G., Pena S. D. J., Hutz M. H. (2012a). Application of the FST statistics to explore pharmacogenomic diversity in the Brazilian population. Pharmacogenomics 13, 771–777 10.2217/pgs.12.128 [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G., Sortica V. A., Vargens D. D., Bruxel E. M., Petz-Erler M. L., Tsuneto L. T., et al. (2012b). Impact of population diversity on the prediction of 7-SNP NAT2 phenotypes using the tagSNP rs1495741 or paired SNPs. Pharmacogenet. Genomics 22, 305–309 10.1097/FPC.0b013e3283519c7c [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G., Genro J. P., Moraes M. O., Ojopi E. B., Pena S. D. J., Perini J. A., et al. (2012c). Global pharmacogenomics, impact of population diversity on the distribution of polymorphisms in the CYP2C cluster among Brazilians. Pharmacogenomics J. 12, 267–276 10.1038/tpj.2010.89 [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G., Vargens D. D., Struchiner C. J., Bastos-Rodrigues L., Pena S. D. J. (2007a). Self-reported skin color, genomic ancestry and the distribution of GST polymorphisms. Pharmacogenet. Genomics 17, 765–771 10.1097/FPC.0b013e3281c10e52 [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G., Perini J. A., Bastos-Rodrigues L., Pena S. D. J., Struchiner C. J. (2007b). Impact of population admixture on the distribution of the CYP3A5*3 polymorphism. Pharmacogenomics 8, 1299–1306 10.2217/14622416.8.10.1299 [DOI] [PubMed] [Google Scholar]
- Vargens D. D., Almendra L., Struchiner C. J., Suarez-Kurtz G. (2008). Distribution of the GNB3 825C>T polymorphism among Brazilians, impact of population structure. Eur. J. Clin. Pharmacol. 64, 253–256 10.1007/s00228-007-0413-2 [DOI] [PubMed] [Google Scholar]
- Venables W. N., Ripley B. D. (2002). Modern Applied Statistics with S, 4th Edn New York: Springer [Google Scholar]
- Wright S. (1951). The genetic structure of populations. Ann. Eugen. 15, 323–343 [DOI] [PubMed] [Google Scholar]
- Wright S. (1978). Variability Within and Among Natural Populations. Chicago, IL: University of Chicago [Google Scholar]