Abstract
Brain tumors are typically comprised of morphologically diverse cells that express a variety of neural lineage markers. Only a relatively small fraction of cells in the tumor with stem cell properties, termed brain tumor initiating cells (BTICs), possess an ability to differentiate along multiple lineages, self-renew, and initiate tumors in vivo. We applied culture conditions originally used for normal neural stem cells (NSCs) to a variety of human brain tumors and found that this culture method specifically selects for stem-like populations. Serum-free medium (NSC) allows for the maintenance of an undifferentiated stem cell state, and the addition of bFGF and EGF allows for the proliferation of multi-potent, self-renewing, and expandable tumorspheres.
To further characterize each tumor's BTIC population, we evaluate cell surface markers by flow cytometry. We may also sort populations of interest for more specific characterization. Self-renewal assays are performed on single BTICs sorted into 96 well plates; the formation of tumorspheres following incubation at 37 °C indicates the presence of a stem or progenitor cell. Multiple cell numbers of a particular population can also be sorted in different wells for limiting dilution analysis, to analyze self-renewal capacity. We can also study differential gene expression within a particular cell population by using single cell RT-PCR.
The following protocols describe our procedures for the dissociation and culturing of primary human samples to enrich for BTIC populations, as well as the dissociation of tumorspheres. Also included are protocols for staining for flow cytometry analysis or sorting, self-renewal assays, and single cell RT-PCR.
Keywords: Cancer Biology, Issue 67, Stem Cell Biology, Medicine, Cellular Biology, Molecular Biology, BTIC (brain tumor initiating cells), tumorspheres, self-renewal, flow cytometry, single cell RT-PCR
Introduction
Brain tumors are among the most aggressive and heterogeneous cancers known in humans. Although their earlier detection and diagnosis have been facilitated by modern neuro-imaging technology, we still lack curative therapies for many brain tumors, particularly for diffuse, invasive ones or those located deep in the brain.
Brain tumors represent the leading cause of cancer mortality in children due to their highly aggressive and often incurable nature. Glioblastoma (GBM), the most common primary brain tumor in adults, is one of the most aggressive human cancers, feared for its uniformly fatal prognosis1. This highly malignant astrocytic tumor (WHO Grade 4) usually occurs in the cerebral hemispheres of adults, and can also occur in young children and infants. Its growth is rapid and infiltrative, and diagnostic pathological features include nuclear pleomorphism, microvascular proliferation, and necrosis2,3. For adults with newly diagnosed GBM, median survival rarely extends beyond 12 months1, with generally poor responses to all therapeutic modalities. We noted that there are many functional and genetic similarities shared by somatic stem cells and cancer cells, and that the molecular pathways that regulate normal brain development are often dysregulated in cancer. In applying stem cell biology paradigms to the study of brain tumors, we were the first researchers to prospectively identify and purify a subpopulation of cells from human GBMs which exhibited the stem cell properties of proliferation, self-renewal, and differentiation in vitro4 and in vivo5. We applied culture conditions and assays originally used to characterize normal neural stem cells (NSCs) in vitro 6,7 to multiple pediatric and adult brain tumors, and enriched for these stem-like cells by cell sorting for the neural progenitor cell surface marker CD1338,9. The CD133+ brain tumor fraction contained cells that had a much higher frequency of tumor initiation than the CD133- fraction in NOD-SCID mouse brains5,10. This formally established that only a rare subset of brain tumor cells with stem cell properties are tumor-initiating, earning them the name "brain tumor initiating cells" or "BTICs". The novel identification of BTICs provides new insights into human brain tumorigenesis, giving strong support for the cancer stem cell hypothesis10-13 as the basis for many solid tumors, and establishes a novel cellular target for more effective cancer therapies14-20. Therapies that focus on killing the bulk of the tumor may miss the rare stem-like fraction, allowing the tumor to continue to grow. Therapies that focus on killing the cancer stem cell may provide better treatment and prognosis for patients with brain tumors.
In order to study BTIC populations, we have refined our culture protocols to specifically select for cell populations within human brain tumors that possess stem cell properties. Serum-free, neural stem cell (NSC) medium allows for the maintenance of an undifferentiated stem cell state, and the addition of basic fibroblast growth factor (bFGF), epidermal growth factor (EGF), and leukemia inhibitory factor (LIF) allows for the proliferation of multi-potent, self-renewing, and expandable human tumorspheres. Here, we describe the methods involved in processing of primary brain tumors and culturing them in NSC medium to enrich for BTIC populations. We have called our experimental model system "BTIC patient isolates" to emphasize the fact that these cells are only minimally cultured under stem cell conditions to select for stem cell populations. Subsequent immunolabelling of BTIC populations for key stem cell markers such as CD133 and CD15 and flow cytometry analysis is also described. We then discuss the limiting dilution analysis, which aids in studying the self-renewal potential of BTICs. Finally, we explore the gene expression analysis of these rare cells by sorting single cells onto AmpliGrid slides and performing single cell RT-PCR. These techniques are also applicable to other brain tumors such as medulloblastoma, ependymoma and pediatric gliomas.
Protocol
1. Culture of Brain Tumor Tissue
Add 200 μl thawed Liberase (Roche Applied Science) to 15 ml of artificial CSF (aCSF- see Table 1) and place into 37 °C water bath. Liberase TM is a mix of proteolytic enzymes used to dissociate primary tissue samples, as well as cultured tumorspheres. Unlike Trypsin-EDTA, the Liberase method preserves the surface antigen CD133. For a tissue sample of about 0.5 cm3, we use 200 μl of Liberase. If the tissue is smaller, we use 100 ul.
Bring ammonium chloride solution (Stem cell technologies) to room temperature. Ammonium Chloride solution gently lyses red blood cells with minimal effect on other cells. It does not contain a fixative.
In sterile biological safety cabinet, add 5 ml of aCSF to specimen container, swirl to rinse tissue, then pipette off. This step helps to remove red blood cells (RBC).
Transfer brain tumor tissue to a sterile 100 mm Petri dish.
Using fine scissors or scalpels and forceps, disaggregate tissue to slurry consistency.
Collect sample using a 10 ml regular pipette or forceps and transfer fragments into the tube containing pre-warmed aCSF with Liberase.
Place on incubator-shaker (30 rpm) and set to 37 °C, for 15 min.
Filter the tissue lysate through 70 μm cell strainer into a 50 ml Falcon tube.
Spin the filtrate down at 280 x g for 5 min.
Remove supernatant carefully and evaluate size and colour of the resulting cell pellet: pellets which are pink or red indicate increasing numbers of red blood cells.
Resuspend pellet in 1 ml PBS.
Add an appropriate amount of ammonium chloride solution (4-12 ml) based on pellet size and red cell contamination (the ammonium chloride solution is very gentle and increased amounts are not harmful to cells other than red cells).
Incubate at room temperature for 5 min.
Spin cells down at 280 x g for 5 min.
Wash once with 10 ml of sterile PBS.
Resuspend in 5 ml NSC complete medium (Table 2) and transfer to an ultra-low binding 60 mm tissue culture plate (Corning). We use ultra-low binding culture plates with covalently bonded hydrogel surfaces that are hydrophilic and neutrally charged, to minimize cell attachment-mediated differentiation.
For the initial days in culture, do not change the media: top-up only with 1-2 ml media as needed, then continue to observe culture and change media when the color of the media becomes slightly yellow.
2. Tumorsphere Dissociation for Flow Cytometry
Evaluate tumorspheres under microscope: if the sphere size is >100 μm, dissociation is recommended as larger spheres may become necrotic in the center.
Transfer culture to 15 ml conical tube.
Add 2-3 ml sterile PBS to rinse plate thoroughly and add to conical tube.
Centrifuge at 280 x g for 5 min.
Remove supernatant and resuspend in 1- 2 ml sterile PBS.
Add 10 μl Liberase.
Incubate in 37 °C water bath for 3 min. Remove and visually evaluate suspension, if multiple clumps seen, triturate gently using a 1,000 μl pipette tip.
If clumping persists, continue incubation for an additional 1-2 min.
Wash the cells by adding 5 ml sterile PBS and centrifuging at 280 x g for 5 min.
Remove supernatant and resuspend in 500-1,000 μl sterile PBS+2 mM EDTA.
Assess cell number and viability using trypan blue.
Adjust cell count to 1 million/ml in PBS+2 mM EDTA.
3. Surface Staining
Transfer 100 μl of 1x106/ml cell suspension per test to 12x75 mm flow tubes.
Add appropriate amount of antibodies. Matched Isotype controls should be used for each antibody (see Table 3).
Incubate for 30 min on ice.
Add 2 ml PBS+2 mM EDTA to each flow tube.
Centrifuge at 200 x g for 4 min, decant and blot.
Resuspend pellet in 300 μl PBS+2 mM EDTA.
Add 10 μl 7AAD viability dye to each tube and incubate for at least 15 min on ice.
Analyze by flow cytometry.
4. Flow Cytometry Acquisition and Analysis
The specifics of acquisition and analysis of flow data are instrument-dependent. Representative negative samples are run and instrument settings established for Forward (size) and Side (granularity) scatter. This scatter pattern allows the end-user to view all cells in the sample, including debris. A region is usually drawn around the cells of interest. (Figure 4a) Settings are then established for all fluorescence detectors required to position the negative cells within the first decade of a fluorescence intensity plot. When more than one dye or fluorochrome is used, single stained controls must be run to establish color compensation values (subtraction of interfering fluorescence emissions). When running live cells, a viability dye such as 7-AAD (7-Amino-actinomycin D - excitation 488/emission 655) is used and a second region drawn to exclude dead cells (Figure 4b). Both of these regions are applied to any further analysis of the samples.
5. Self-renewal Assay
The key assay that is greatly facilitated by clonal sphere growth is the in vitro test of self-renewal capacity, the limiting dilution assay. Tumorspheres are dissociated and distributed at dilutions down to a single cell per well, and the rate of subsequent sphere formation over 7 days in culture is calculated. On day 7, the percentage of wells not containing spheres for each cell plating density (F0) is calculated and plotted against the number of cells per well (x). The number of cells required to form at least one tumorsphere in every well is determined from the point at which the line crosses the 0.37 level (F0 = e-x). In a Poisson distribution of cells, F0 = 0.37 corresponds to the dilution of one cell per well, so that this calculation (the 37% intersect) reflects the frequency of clonogenic stem cells in the whole cell population (Figure 5).
6. Single Cell RT-PCR
Single cells from different populations are sorted by Beckman Coulter MoFlo XDP on reaction sites on an AmpliGrid slide (Beckman Coulter cat#AG480F). Single cell RT-PCR is performed using AmpliGrid Single One Step RT-PCR system (Beckman Coulter cat#OAX04515) according to manufacturer's specifications. Briefly, a single cell is deposited into each reaction site of an AmpliGrid slide; the presence of a single cell in each reaction site confirmed by microscopy. Reverse transcription (RT) is performed immediately after sorting to prevent the sample from being compromised. The RT master mix is prepared fresh, according to kit instructions (Table 4). We used 0.03 μl of 25 μM random primers (Promega) per reaction; 1 μl of master mix is added to the centre of each reaction site. This is immediately coated with 5 μl of sealing solution (Beckman Coulter cat#OAX04503). Using an AmpliSpeed slide cycler (Beckman Coulter; cat#OAX04101), slides are incubated at 25 °C for 10 min, 42 °C for 10 min, and 55 °C for 45 min. After reverse transcription, 3 μl of DNase/RNase-free water is added to each reaction site, and the entire mixture transferred to a PCR tube (1 tube/site). In a PCR plate, a reaction volume of 10 μl is used for quantitative PCR: 8 μl qPCR master mix (5 μl Sybr Green (Quanta), 1 μl water, 1 μl of 10 μM forward and reverse primer mix) plus 2 μl of the RT mixture. For real-time quantification, samples are run on a BioRad cycler using the following program: 95 °C for 10 min, 45 cycles of 95 °C for 30 sec, 60 °C for 60 sec, 72 °C for 60 sec, followed by 10 min at 72 °C and melting curve analysis. Ct values were determined using Opticon Monitor 3 (BioRad). Three genes can be assessed per cell, including GAPDH as a housekeeping gene. Gene expression is normalized to GAPDH expression, according to 2-ΔCt. At least a dozen single cells of the same population from the same sort can be used as biological replicates to compensate for lack of technical replicates.
7. Representative Results
Figure 1 shows a Magnetic Resonance Imaging (MRI) scan of a representative patient with a GBM. Brain tumor samples are obtained from consenting patients immediately after surgery, as approved by the Hamilton Health Sciences/McMaster Health Sciences Research Ethics Board. A part of each specimen is given to the neuropathologist for routine clinical diagnosis. The remaining sample is processed as shown in Figure 2. The tumor cells, once plated in complete neural stem cell media with growth factors, form tumorspheres as shown in Figure 3.
The tumor cells are labeled with neural surface stem cell markers CD133 and CD15 and analyzed by flow cytometry as shown in Figure 4.
Single cells from different populations for e.g., CD15+/CD133+, CD15+/CD133-, CD15-/CD133+, CD15-/CD133- are then sorted into 96 well plates (Figure 5a) or into AmpliGrid slides (Figure 6a).
In the 96 well plate, single cells which possess self-renewal capacity (e.g. CD133+ cells), (Figure 5b), can form tumorspheres (Figure 5c) following incubation. A self-renewal graph can be plotted (Figure 5d) by counting the number of spheres formed per well. Single cells sorted into the reaction site of the Ampligrid slide (Figure 6a). The RNA extracted from single cells can be reverse transcribed using the Advalytix AmpliSpeed (Figure 6b). The cDNA obtained is used to perform Real time RT-PCR (Figure 6c).
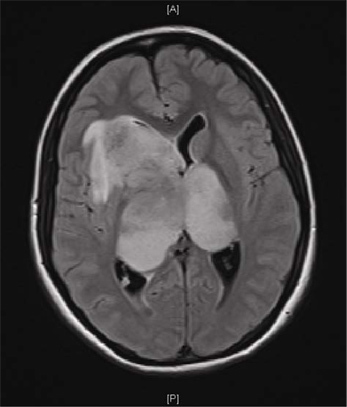 Figure 1. Magnetic resonance image (MRI) of a patient GBM. MRI scan of the brain of a 16-year old girl with a month-long history of headache and a one week history of vomiting, lethargy and visual blurring. Axial FLAIR sequence shows a large bi-hemispheric ("butterfly") GBM.
Figure 1. Magnetic resonance image (MRI) of a patient GBM. MRI scan of the brain of a 16-year old girl with a month-long history of headache and a one week history of vomiting, lethargy and visual blurring. Axial FLAIR sequence shows a large bi-hemispheric ("butterfly") GBM.
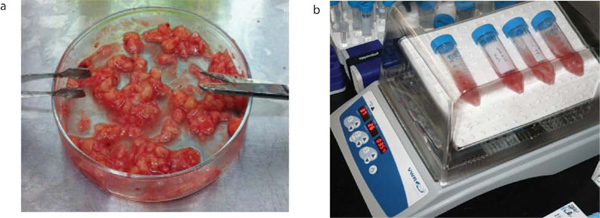 Figure 2. Brain tumor processing. Brain tumor samples are obtained from consenting patients immediately after surgery. They are mechanically dissociated as shown (a) and are then enzymatically dissociated in aCSF with Liberase by incubating in a 30 rpm rocking incubator at 37 °C for 15 min (b).
Figure 2. Brain tumor processing. Brain tumor samples are obtained from consenting patients immediately after surgery. They are mechanically dissociated as shown (a) and are then enzymatically dissociated in aCSF with Liberase by incubating in a 30 rpm rocking incubator at 37 °C for 15 min (b).
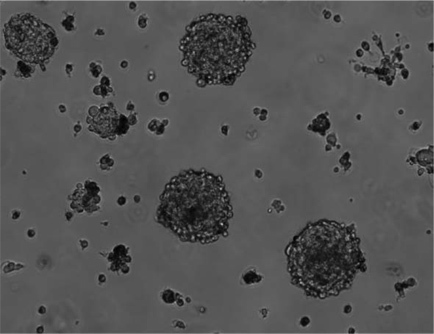 Figure 3. BTIC populations form tumorspheres in culture. Dissociated brain tumor cells plated in complete neural stem cell media with growth factors form tumorspheres.
Figure 3. BTIC populations form tumorspheres in culture. Dissociated brain tumor cells plated in complete neural stem cell media with growth factors form tumorspheres.
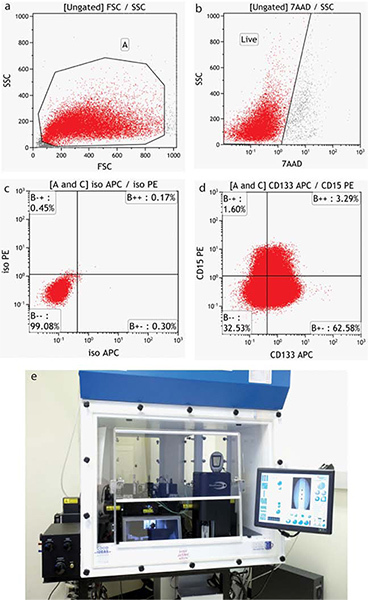 Figure 4. Flow cytometric analysis of BTIC populations. (a) Forward versus side scatter properties provide an image of all cells, including debris (b) Cells which stain with the viability dye 7-AAD are excluded from analysis. (c) The position of the statistical quadrants is determined using appropriate Isotype controls (d) Tumor cells stained with surface markers CD15-PE and CD133-APC. (d) Moflo XDP cell sorter. Click here to view larger figure.
Figure 4. Flow cytometric analysis of BTIC populations. (a) Forward versus side scatter properties provide an image of all cells, including debris (b) Cells which stain with the viability dye 7-AAD are excluded from analysis. (c) The position of the statistical quadrants is determined using appropriate Isotype controls (d) Tumor cells stained with surface markers CD15-PE and CD133-APC. (d) Moflo XDP cell sorter. Click here to view larger figure.
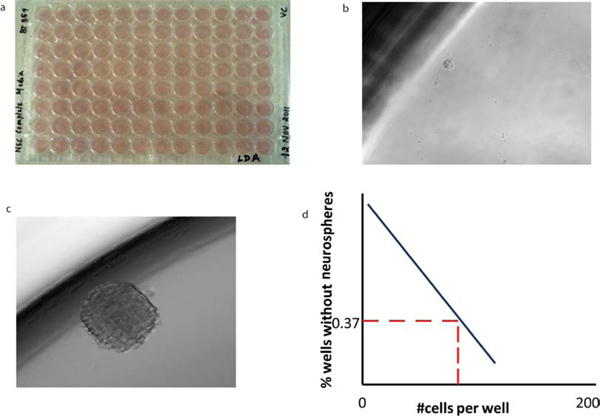 Figure 5. Limiting dilution analysis of BTICs reveals their self-renewal capacity. (a) Cells of varying dilutions are sorted into 96 wells containing 200 μl of NSC to (b) a single cell per well. (c) Following a 7 day incubation period, a tumorsphere is formed. The morphology of secondary tumorspheres is identical to that of primary tumorspheres. (d) The mean x-intercept values can be calculated from limiting dilution analysis for each tumor subtype to reveal the number of cells required to form at least one tumorsphere per well.
Figure 5. Limiting dilution analysis of BTICs reveals their self-renewal capacity. (a) Cells of varying dilutions are sorted into 96 wells containing 200 μl of NSC to (b) a single cell per well. (c) Following a 7 day incubation period, a tumorsphere is formed. The morphology of secondary tumorspheres is identical to that of primary tumorspheres. (d) The mean x-intercept values can be calculated from limiting dilution analysis for each tumor subtype to reveal the number of cells required to form at least one tumorsphere per well.
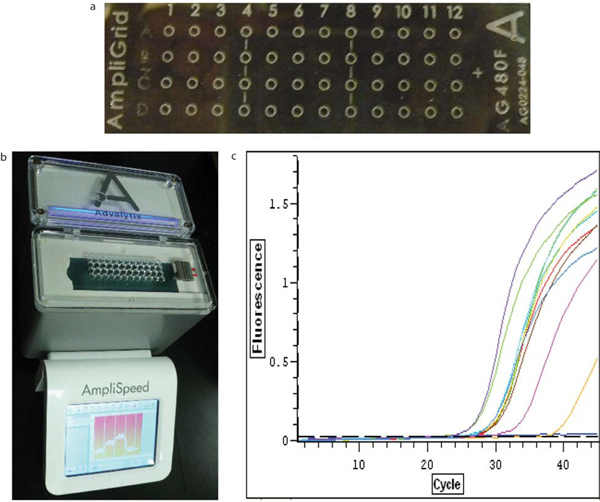 Figure 6. Gene expression analysis of a single BTIC cell is possible using single cell RT-PCR technology. (a) Single cells from a BTIC population can be sorted onto AmpliGrid slides. (b) cDNA synthesis is performed on single cells on AmpliGrid slides using the Advalytix AmpliSpeed. (c) Quantitative Realtime PCR is performed on the cDNA synthesized from single BTIC cells.
Figure 6. Gene expression analysis of a single BTIC cell is possible using single cell RT-PCR technology. (a) Single cells from a BTIC population can be sorted onto AmpliGrid slides. (b) cDNA synthesis is performed on single cells on AmpliGrid slides using the Advalytix AmpliSpeed. (c) Quantitative Realtime PCR is performed on the cDNA synthesized from single BTIC cells.
| 2M NaCl | 62 ml |
| 1M KCl | 5 ml |
| 1M MgCl2 | 3.2 ml |
| 155 mM NaHCO3 | 169 ml |
| 1M Glucose | 10 ml |
| 108 mM CaCl2 | 0.9256 ml |
| Purelab Ultra H20 | 749.84 ml |
Table 1. Artificial Cerebrospinal Fluid (aCSF) - 1,000 ml.
| Stock Basal media | 500 ml |
| 1:1 DMEM:F12 | 480 ml |
| N2 supplement | 5 ml |
| 1M HEPES | 5 ml |
| Glucose | 3.0 g |
| N-acetylcysteine (60 μg/ml) | 1 ml |
| Neural survival factor-1 (NSF-1) | 10 ml |
Table 2. Neural stem cell media.
NSC complete media (made fresh prior to use): NSC basal media + 20 ng/ml EGF (epidermal growth factor) + 20 ng/ml bFGF (basic fibroblast growth factor) + 10 ng/ml LIF (leukemia inhibitory factor)* + 10 μl/ml antibiotic-antimycotic
*Leukemia inhibitory factor (LIF): This reagent from Millipore contains an interleukin 6 class cytokine, a protein which suppresses spontaneous differentiation of stem cells promoting longer term maintenance of stem cell cultures.
| Antibodies | Supplier/CAT# | μl/test (in 100 μl) |
| CD133/2 APC (293C3) | Miltenyi Biotec/130-090-854 | 5 |
| IgG2b APC (Isotype control) | Miltenyi Biotec/130-092-217 | 5 |
| CD15PE | Beckman Coulter/IM1954U | 5 |
| IgG2a PE (Isotype control) | Beckman Coulter/A09141 | 5 |
| 7AAD | Beckman Coulter/A07704 | 10 |
Table 3. Flow antibodies.
| Component | Volume (1 reaction) | Volume ( 48 reaction) |
| 2x Single Cell RT reaction Buffer | 0.50 μl | 30.0 μl |
| RNase Inhibitor (10 U/μl) | 0.02 μl | 1.2 μl |
| 5X Single Cell RT enhancer | 0.15 μl | 9.00 μl |
| Single Cell RT Enzyme Mix | 0.04 μl | 2.4 μl |
| Advablue (OAX04227) | 0.1 μl | 6.00 μl |
| Nuclease free water | make up to 1 μl | make up to 60 μl |
Table 4. Composition of RT master mix for Single Cell RTPCR (cat# OAX04515).
Discussion
The cancer stem cell hypothesis10, based on work in leukemia21, breast cancer11 and brain cancer 4,5, suggests that only a relatively small fraction of cells in the tumor, termed cancer stem cells, possess an ability to extensively proliferate and self-renew. Most of the tumor cells lose the ability to proliferate and self-renew as they differentiate into cells that become the phenotypic signature of the tumor. Finding the key cells in the brain tumor population that are able to maintain the tumor will give insight into the mechanism of brain tumorigenesis and will allow us to trace back to the cell of origin.
Stem cells are functionally defined as self-renewing cells that exhibit multi-lineage differentiation22-24. Primary brain tumor tissue is grown under culture conditions that favor stem cell growth, previously established for isolation of neural stem cells as tumorspheres25. Neural stem cell media (NSC) allows for the maintenance of an undifferentiated stem cell state, and the addition of bFGF and EGF induces the proliferation of multi-potent, self-renewing and expandable stem cells6,26. Regardless of pathological subtype, within 24 to 48 hours of primary culture, most brain tumors yield a minority fraction of cells that demonstrate growth into clonally derived neurosphere-like clusters, or tumorspheres. Thus we are able to enrich for BTIC populations from patient brain tumor tissues.
The advent of multi-parameter fluorescent activated cell sorting27 and monoclonal antibody technology28 has allowed for the purification of cancer stem cells based on cell surface markers. By using stem cell markers such as CD133 and CD15, we have been able to prospectively sort for specific stem cell-like populations and to study their self-renewal capacity by performing limiting dilution assays. A caveat of self-renewal assays was introduced by a recent study showing that in vitro self-renewal does not always correlate with tumor formation in mouse models 29. However our recent work has shown that in vitro self-renewal assays do correlate with patient survival, rendering these assays valuable to the ongoing study of BTIC biology30 .
Working with primary human tissue presents many technical challenges. Tissue processing should be performed very gently to preserve the viability of isolated cells. The amount of Liberase used and time of incubation is critical. Flow sorting of cells can be optimised to increase post-sort viability by using different nozzles and pressures. We find that our cells prefer to be sorted through a 100μm nozzle at 30 psi of sheath pressure. Sorting onto Ampligrid slide reactive sites may be somewhat difficult: it is important to confirm the presence of the deposited cell by microscopy.
Tumor heterogeneity represents one of the biggest challenges in the ongoing search for new cancer biomarkers and targeted therapies 31. Brain tumors are typically comprised of morphologically diverse cells that express a variety of neural lineage markers. Study of brain tumors by traditional histopathology has only yielded a limited amount of knowledge of the clinical behaviour of the tumor. Although major advances have been made in the understanding of the molecular genetic alterations of some brain tumors3,32-34 , particularly medulloblastomas and malignant gliomas, and some of these identified alterations are now beginning to guide treatment, it is not clear whether all tumor cells are equivalent in their ability to maintain tumor growth. Analysis of key genes involved in signaling pathways in brain tumor populations is critical to further our understanding of aberrant mechanisms leading to brain tumor formation. Even in a seemingly homogenous tissue, cells prove to be unique in their potential abilities. Analysis of transcript and protein expression levels in single cells reveal that there is a large cell-cell variation both in the resting state and when exposed to stimuli35-39. Therefore, analyzing the whole population of cells will not reveal the behaviour of a single cell. qRT-PCR is considered to be the gold standard for mRNA quantification40-41. In this study, we demonstrate prospective sorting of individual cells from distinct populations on AmpliGrid slides. Using single cell RT-PCR, we are able to perform gene expression analysis on a single cell from a heterogeneous population, thus evaluating the biological significance of a selected cell within a mixed population.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was funded by the Ontario Institute of Cancer Research (OICR), the Terry Fox Foundation and the American Association of Neurological Surgeons.
References
- Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma. 2010;10:319. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R, Scott MP. The developmental biology of brain tumors. Annu. Rev. Neurosci. 2001;24:385. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- Singh SK. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821. [PubMed] [Google Scholar]
- Singh SK. Identification of human brain tumour initiating cells. Nature. 2004;432:396. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev. Biol. 1996;175:1. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- Tropepe V. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev. Biol. 1999;208:166. doi: 10.1006/dbio.1998.9192. [DOI] [PubMed] [Google Scholar]
- Yin AH. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002. [PubMed] [Google Scholar]
- Yu Y, Flint A, Dvorin EL, Bischoff J. AC133-2, a novel isoform of human AC133 stem cell antigen. J. Biol. Chem. 2002;277:20711. doi: 10.1074/jbc.M202349200. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3983. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Matsui W. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103:2332. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Bao S. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- Beier D. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68:5706. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG. morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Rich JN, Bao S. Chemotherapy and cancer stem cells. Cell Stem Cell. 2007;1:353. doi: 10.1016/j.stem.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Lapidot T. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Segre JA. Stem cells: a new lease on life. Cell. 2000;100:143. doi: 10.1016/s0092-8674(00)81691-8. [DOI] [PubMed] [Google Scholar]
- Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J. Neurosci. 1992;12:4565. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett HR, Bonner WA, Barrett J, Herzenberg LA. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science. 1969;166:747. doi: 10.1126/science.166.3906.747. [DOI] [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Barrett LE. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21:11. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Venugopal C. Bmi1 marks intermediate precursors during differentiation of human brain tumor initiating cells. Stem Cell Res. 2012;8:141. doi: 10.1016/j.scr.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Gerlinger M. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson RJ. Medulloblastoma: signalling a change in treatment. Lancet. Oncol. 2004;5:209. doi: 10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Parada LF. The molecular and genetic basis of neurological tumours. Nat. Rev. Cancer. 2002;2:616. doi: 10.1038/nrc866. [DOI] [PubMed] [Google Scholar]
- Maher EA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Maheshri N, O'Shea EK. Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu. Rev. Biophys. Biomol. Struct. 2007;36:413. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- Raj A. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross IL, Browne CM, Hume DA. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol. Cell Biol. 1994;72:177. doi: 10.1038/icb.1994.26. [DOI] [PubMed] [Google Scholar]
- Kubista M. The real-time polymerase chain reaction. Mol. Aspects Med. 2006;27:95. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]


