Abstract
Hydroperoxide lyase (HPL) cleaves lipid hydroperoxides to produce volatile flavor molecules and also potential signal molecules. We have characterized a gene from Arabidopsis that is homologous to a recently cloned HPL from green pepper (Capsicum annuum). The deduced protein sequence indicates that this gene encodes a cytochrome P-450 with a structure similar to that of allene oxide synthase. The gene was cloned into an expression vector and expressed in Escherichia coli to demonstrate HPL activity. Significant HPL activity was evident when 13S-hydroperoxy-9(Z),11(E),15(Z)-octadecatrienoic acid was used as the substrate, whereas activity with 13S-hydroperoxy-9(Z),11(E)-octadecadienoic acid was approximately 10-fold lower. Analysis of headspace volatiles by gas chromatography-mass spectrometry, after addition of the substrate to E. coli extracts expressing the protein, confirmed enzyme-activity data, since cis-3-hexenal was produced by the enzymatic activity of the encoded protein, whereas hexanal production was limited. Molecular characterization of this gene indicates that it is expressed at high levels in floral tissue and is wound inducible but, unlike allene oxide synthase, it is not induced by treatment with methyl jasmonate.
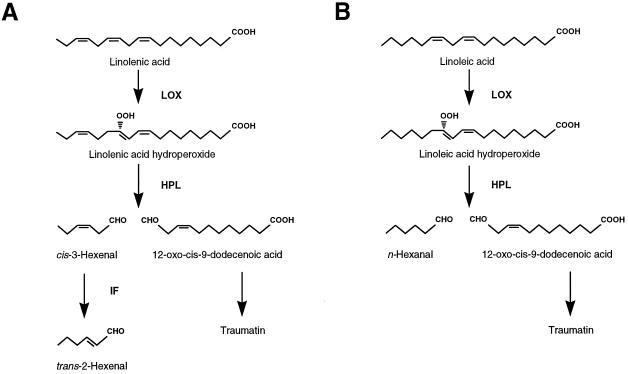
In green plant tissue HPL cleaves C18-lipid hydroperoxides to form a C6-aldehyde and a 12-carbon oxoacid (Hatanaka, 1993). The C12 product of HPL leads to the formation of traumatin, which is implicated in wound signaling (Zimmerman and Coudron, 1979). The C6-aldehyde products of the HPL reaction depend on the substrate; cis-3-hexenal is formed from HPOT and hexanal is formed from HPOD. The lipid hydroperoxide substrates were generated from C18 free fatty acids by the enzymatic activity of lipoxygenase (see Fig. 1). HPL activity has been found in a variety of plants and is thought to be associated with the chloroplast envelope (Blée and Joyard, 1996), developmentally regulated (Vick and Zimmerman, 1976; Gardner et al., 1991; Riley et al., 1996), and discernible as two distinct activities in tea leaves (Matsui et al., 1991).
Figure 1.
Biochemical pathway illustrating the enzymatic activity of HPL. HPL cleaves either HPOT (A) or HPOD (B), forming 12-oxo-trans-10-dodecenoic acid and cis-3-hexenal or hexanal, respectively. An isomerization factor (IF) interconverts cis-3-hexenal to trans-2-hexenal in vivo.
Although the central lipoxygenase pathway is present in animal systems, and the formation of prostaglandins and leukotrienes in animals is analogous to the formation of jasmonates in plant tissue (Anderson, 1989), there is no pathway in animals analogous to the HPL branch pathway. The C6 compounds produced by HPL are released rapidly from disrupted plant tissue and form the basis for the “green note” flavor characteristic of plant tissue. The green note flavor is an important determinant of fresh fruit and vegetable quality, and C6 volatiles are widely used as a prepared food additive (Hatanaka, 1993). C6 volatiles produced from this pathway also have antimicrobial properties, suggesting that they may play a protective role in plant defense (Croft et al., 1993). They may also have an application as antimicrobial fumigants in postharvest storage (Archbold et al., 1997). In addition, C6 volatiles of the HPL pathway induce phytoalexin accumulation (Zeringue, 1992) and inhibit seed germination (Gardner et al., 1990), suggesting that they may also play a signaling role in plants.
The role that HPL plays in flavor formation as well as in plant defense indicates that this enzyme is an important biotechnology target. In this paper we present the biochemical and molecular characterization of an Arabidopsis clone that, when expressed in Escherichia coli, yields a product possessing HPL activity.
MATERIALS AND METHODS
HPL Characterization and Sequencing
Using the BLASTX algorithm (Altschul et al., 1990), we found an EST clone (EST designation 94J16T7) that has a high degree of homology with the published HPL sequence for bell pepper (Capsicum annuum) (Matsui et al., 1996). Because there is variability in the length of the N-terminal transit peptide of chloroplast-targeted proteins, we used 5′ RACE to ensure that a full-length sequence was obtained. 5′ RACE was performed as described by Goring et al. (1992). Total RNA isolated from leaf tissue was tailed at the 5′ end with terminal transferase and dATP. An adapter primer (adapter-dT17) and a gene-specific primer (5′-AGA-TGG-CTA-AAA-GAC-TTG-ACG-TCA-AGA-3′) were used for the first set of PCR reactions. PCR reaction products were separated on an agarose gel, and plugs were taken from the gel in the expected size range (300–400 bp). A second round of PCR reactions was performed using the agarose plugs as a source of template, an adapter primer (Goring et al., 1992), and a second gene-specific primer (5′TGA-CGT-CAA-GAA-CGG-CGA-CGA-TGT-3′). After gel purification, PCR products of the expected size were cloned into pGEM-T (Promega Biotech). Four of the 5′ RACE clones from two different sources of RNA had identical sequences, but varied slightly in length. The sequence of the longest 5′ RACE product is presented.
Expression of Fusion Proteins in Escherichia coli
To demonstrate HPL activity, a fusion protein was produced in E. coli using the pGEX system (Pharmacia). Preliminary experiments demonstrated that the full-length clone had no HPL activity, and so the putative transit peptide was removed using a PCR strategy. An oligonucleotide was designed (5′-TCA-CAG-CTT-CCC-CTC-CGT-ACA-ATG-3′) so that the corresponding HPL protein sequence started at amino acid 30 (Ser). A PCR reaction product was obtained using this primer and one located downstream of an internal DraII site (5′-CGC-AGA-GGA-AAC-TGA-AGA-TGC-AAC-3′, corresponding to nucleotides 650–672) that was cloned into pGEM-T and sequenced. This clone was digested with a pGEM-T polylinker restriction enzyme (PstI) and DraII, and the fragment was purified and ligated to the remainder of the full-length HPL insert cut at the polylinker PstI site and the internal DraII site, effectively replacing the 5′ end of the gene with the truncated version. This truncated and reassembled clone was fused in-frame to the GST moiety of the expression plasmid pGEX-5X-2 (Pharmacia). The protocols used for protein expression and purification were as specified by the manufacturer. Control protein consisted of the GST protein expressed and purified in the same manner. Protein quantity was determined by the method of Bradford (1976).
Growth Conditions, Wounding, and Treatment with MeJA
Arabidopsis (ecotype Columbia) was soil grown in controlled-environment growth chambers with a 16-h/8-h day/night regime. Temperature was maintained at 23°C for both cycles. After approximately 3 weeks of growth, Arabidopsis plants were treated with MeJA or wounded, and RNA was isolated by the guanidinium isothiocyanate procedure (Ausubel et al., 1987). MeJA treatment consisted of placing the potted Arabidopsis plants into a sealable 1-L canning jar for 24 h to allow for acclimation before treatment with either 10 μL of methanol (negative control) or 10 μL of 0.1 m MeJA (97% purity; Firmenich, Geneva, Switzerland). Three samples were collected: 4 h of exposure to methanol, 4 h of exposure to MeJA, and 24 h of exposure to MeJA. After treatments, plants were removed and leaf tissue was extracted for RNA. For wound induction, leaves of 4-week-old plants were sliced with a razor blade five times across the mid-vein and placed in a high-humidity growth chamber for 15, 30, or 60 min before wounded leaf tissue was removed for RNA extraction.
Quantification of Transcript Levels by Northern-Blot Analysis and RT-PCR
Total RNA was isolated by the guanidinium isothiocyanate method (Ausubel et al., 1987). For northern-blot analysis, equal quantities of total RNA (10 μg) were separated on a formaldehyde denaturing gel, transferred to nylon membranes, and hybridized with radiolabeled gene-specific probes according to the method of Church and Gilbert (1984).
RT-PCR was used to detect mRNA quantity for HPL, AOS, and the constitutive control β-ATPase after wounding and MeJA treatments. Ten micrograms of total RNA was treated with DNase I (GIBCO-BRL) according to the manufacturer's instructions, and first-strand cDNA was synthesized using Moloney murine leukemia virus RT (GIBCO-BRL) and 0.1 unit of random hexanucleotides (Pharmacia Biotech) according to the method of Ausubel et al. (1987). After the RT reaction and heat inactivation of Moloney murine leukemia virus RT, one-tenth of the reaction was used for PCR amplification using gene-specific primers for Arabidopsis HPL, AOS, and β-ATPase. Primers for PCR were chosen so that genomic DNA amplification products would include an intron, thus allowing a distinction between DNA and cDNA amplification products based on size. Arabidopsis HPL gene-specific primers were 5′-GCT-CAA-AAG-ATG-TTG-TTG-AGA-ACG-3′ and 5′-CGC-AGA-GGA-AAC-TGA-AGA-TGC-AAC-3′, corresponding to nucleotides 54 to 77 and 648 to 672 of the Arabidopsis HPL sequence, respectively. A 302-bp AOS fragment was amplified using the primers 5′-CTT-TTC-ACC-GGT-ACT-TAC-ATG-CCG-3′ and 5′-GAG-CTT-GTA-TCT-GCG-GGA-TTC-GTC-3′, corresponding to bases 447 to 470 and 723 to 746, respectively (Laudert et al., 1996).
β-ATPase gene-specific primers were designed based on conserved regions of the tobacco sequence (Boutry and Chua, 1985) and an Arabidopsis partial EST sequence (G3 h9T7). The upstream primer was 5′-TGC-TCG-TGC-CCG-TGT-TGG-ACT-3′ and the downstream primer was 5′-CTT-TCT-GCA-CAC-CAC-GAG-CAG-3′, corresponding to nucleotides 2798 to 2806 and 3409 to 3430 of the published tobacco sequence (Boutry and Chua, 1985), respectively. Reactions were run through a thermal cycler (model 9600, Perkin-Elmer) for 20 cycles (40 s at 94°C, 40 s at 55°C, and 40 s at 72°C). PCR amplification conditions were optimized for linearity of template quantity. To detect PCR products, 10% of the reaction volume was separated on a 1.2% agarose gel and transferred to a nylon membrane in 0.4 n NaOH. Blots were prehybridized, hybridized with radiolabeled probe, and washed according to the method of Church and Gilbert (1984). Hybridization and washes were performed at 65°C.
HPL Enzyme Assay and Volatile Measurements
HPL activities of the crude bacterial lysate, affinity-purified GST-94J16 fusion protein, and pure GST protein were determined spectrophotometrically using the coupled-enzyme assay described by Vick (1991), which measures the oxidation of reduced nicotinamide adenine dinucleotide. The activity is expressed as the decrease in A340 min−1 mg−1 protein.
Volatile emissions from samples were analyzed and quantified by GC-MS after purge trapping. Equimolar concentrations of HPOT or HPOD in 5-mL reaction volumes were incubated with 40 g of affinity-purified fusion protein or GST for 15 min. After this period the reaction was stopped by placing the samples on ice, and the volatiles produced were contained in the solution by the addition of 5 mL of saturated calcium chloride. The entire mixture was then added to the purge vessel of a Dynatherm thermal stripper (model 1000, Supelco, Inc., Bellefonte, PA), which was preheated for 5 min at 50°C before being purged with nitrogen for 10 min at a flow rate of 120 mL min−1. The volatiles emitted were entrained on a Carbotrap 100 thermal desorption tube (Supelco) and released by heating to 300°C for 5 min in a Dynatherm model 860 thermal desorption unit (model 860, Supelco), and individual compounds were separated and identified using a gas chromatograph (model 5890, Hewlett-Packard) fitted with a mass selective detector (model 5970, Hewlett-Packard). A Supelco wax 10 capillary column (30 m × 0.25 mm i.d., 0.25-mm film thickness) was used for volatile separation, and the internal mass spectra library of the detector, together with authentic external standards, were used to identify resolved peaks of interest.
RESULTS AND DISCUSSION
Gene Isolation and Sequence Analysis
A partial sequence from the Arabidopsis EST database had significant homology to an HPL isolated from bell pepper, and the corresponding clone (94J16) was sequenced in its entirety. To ensure that the full-length clone was obtained, 5′ RACE was used to gain additional sequence upstream from where the EST sequence ended. Several RACE products were sequenced and, although they differed slightly in length, they all had the same sequence upstream from the 5′ end of the EST (Fig. 2). In the RACE sequences, an in-frame stop codon was present at nucleotides 51 to 53 upstream of an AUG codon (nucleotides 63–65), suggesting that this AUG is the start codon. Significant homology between bell pepper HPL and this clone began at residue 31 (Leu-31), suggesting that the protein sequence before Leu-31 functions as a transit peptide for chloroplast targeting. The deduced protein sequence up to Ser-29 has structural features of a chloroplast transit peptide, including an enrichment of Ser and the absence of Asp, Glu, and Tyr residues (Von Heijne et al., 1989). The structure of this putative transit peptide is similar to those for proteins previously demonstrated to be targeted to the chloroplast envelope (K. Ko, Queens University, Kingston, Ontario, Canada, personal communication). Thus, the structural features of the deduced transit peptide sequence are consistent with enzyme-activity data, which localize HPL to the chloroplast envelope (Blée and Joyard, 1996).
Figure 2.
Nucleotide and deduced protein sequence of Arabidopsis HPL. The arrowhead denotes the start of the sequence used in protein-expression studies. Cyt P-450 domains A to D have lines above the protein sequence. An internal EcoRI site is boxed.
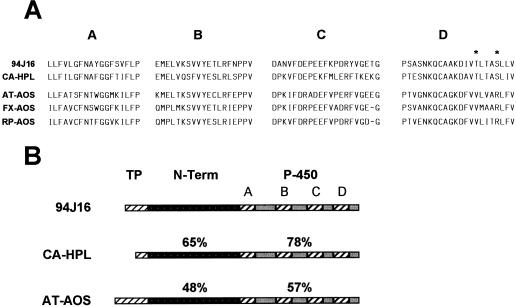
At the C terminus of the protein, there are four domains characteristic of Cyt P-450. Figure 3A compares the Cyt P-450 domains of the 94J16 clone with related sequences, including green pepper HPL (CA-HPL; Matsui et al., 1996), Arabidopsis AOS (AT-AOS; Laudert et al., 1996), flax AOS (FX-AOS, Song et al., 1993), and rubber particle protein (RP-AOS, Pan et al., 1995). There is a striking homology between these sequences relative to other plant Cyt P-450 proteins, suggesting that they are related enzymes. The homology in the Cyt-P-450 domains is greatest between the 94J16 clone and bell pepper HPL, however, and there are two conserved residues in the AOS protein sequences that are replaced by nonconservative residues in the HPL sequences. Specifically, in domain D, Val and Arg are present in all of the AOS-related sequences, but are replaced by Thr-342 and Ser-346 in the aligned HPL sequences. Sequence information from additional HPL genes will determine if these are conserved residues that are diagnostic for HPL. HPL may exist as a small gene family in Arabidopsis, since Southern-blot analysis at moderate stringency (50°C hybridization temperature) using the 5′ end of the 94J16 sequence (up to the EcoRI site) revealed a weak hybridization signal in addition to the primary signal from the 94J16 probe (data not shown).
Figure 3.
Protein-sequence comparison between Arabidopsis HPL (94J16) and related proteins. A, Comparison of the four Cyt P-450 domains (A–D) of Arabidopsis HPL (94J16) with those of green pepper HPL (CA-HPL), as well as those of AOS from Arabidopsis (AT-AOS), flax (FX-AOS), and rubber plant (RP-AOS). Asterisks in Cyt P-450 domain D denote residues conserved in HPL sequences that are dissimilar from corresponding residues in published AOS sequences. B, Structural comparison of the full-length sequences of 94J16, green pepper HPL (CA-HPL), and Arabidopsis AOS (AT-AOS). The figure divides the protein sequence into three regions: TP, putative transit peptide; N-Term, N-terminal region; and P-450, Cyt P-450 region, indicating the four Cyt P-450 domains (A–D). Percentage figures above the individual regions indicate the degree of similarity at the protein level between 94J16 and CA-HPL or AT-AOS.
There is significant homology between the 94J16 clone and AOS from Arabidopsis and other species throughout the protein sequence. Indeed, this clone has been identified as AOS in unpublished reports (Creelman and Mullet, 1997; European Union sequencing project, accession no. Z97339) and was first assumed by us to be another member of the AOS gene family in Arabidopsis. It is not surprising that HPL and AOS would have a similar protein structure, since they use the same substrate and are both associated with the chloroplast. However, close examination of the sequence revealed that the homology between 94J16 and Arabidopsis AOS was lower than is commonly seen for gene family members. For example, the similarity in amino acid sequence between 94J16 and Arabidopsis AOS was only 57% in the P-450 domains (Fig. 3B). This, coupled with the greater similarity of 94J16 to a recently published green pepper HPL sequence (Matsui et al., 1996), led us to explore the properties of this gene and its encoded protein more closely.
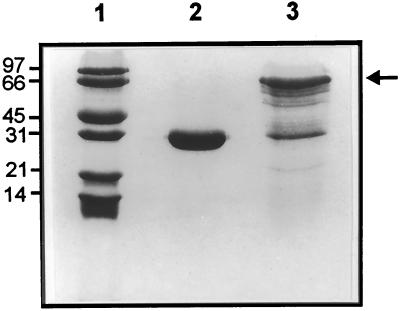
To determine if this clone encoded HPL, we produced the protein in E. coli and assayed for HPL enzyme activity and the production of the C6-aldehyde product. Because preliminary results indicated that the full-length protein had minimal activity, we removed the putative transit peptide in an attempt to increase specific activity, as has been demonstrated for other P-450 enzymes, including Arabidopsis AOS (Laudert et al., 1996). The pGEX prokaryotic protein expression system was used to produce the protein in E. coli as a N-terminal fusion with GST. Figure 4 shows that a full-length, soluble fusion protein of the expected size is produced in large quantity. A number of protein bands are detected by SDS-PAGE and Coomassie blue staining that are smaller than the full-length fusion protein, however (Fig. 4, lane 3), presumably as a result of protein degradation during purification or premature termination of translation.
Figure 4.
SDS-PAGE analysis of fusion proteins produced in E. coli. Protein Mr marker (lane 1), purified GST (lane 2), and purified GST-94J16 fusion protein (lane 3) were run on a 12.5% polyacrylamide gel and the bands visualized by staining with Coomassie blue. The arrow indicates the full-length GST-94J16 fusion protein.
HPL enzyme activity was demonstrated using a coupled-enzyme assay (Riley et al., 1996) modified from Vick (1991). Crude bacterial lysate containing the protein produced from 94J16 and affinity-purified protein were compared with a negative control consisting of equivalent quantities of GST protein. Table I demonstrates that this clone clearly possesses HPL enzyme activity. Activity for the 94J16 protein was very high when HPOT was used as the substrate in both the crude extract and purified protein, whereas activity was considerably lower (approximately 10-fold) when HPOD was used as the substrate. This suggests that the HPL encoded by 94J16 had some selectivity for a substrate. Matsui et al. (1991) purified two HPL enzyme fractions and biochemically characterized one fraction with approximately 10-fold higher activity for the HPOT substrate. Likewise, in soybean there appear to be tissue-specific HPL isozymes with differing affinities for 9- or 13-lipid hydroperoxides (Gardner et al., 1991). In Arabidopsis green leaf tissue, both hexanal (the product of HPOD cleavage) and the hexenals (the product of HPOT cleavage) are present (Avdiushko et al., 1995), indicating that HPOD and HPOT cleavage activity are present. These results collectively suggest that the 94J16 gene product accounts for the majority of the HPOT-cleaving activity and that another isozyme of HPL activity exists to account for HPOD cleavage. The existence of a related sequence, as shown by a second hybridizing signal in Southern blots under moderate stringency (see above), suggests that in Arabidopsis, two gene products account for total HPL activity.
Table I.
HPL activity of E. coli extracts containing GST protein or GST-94J16 fusion protein
| Extract | HPL Activity
|
|
|---|---|---|
| HPOD | HPOT | |
| Δ A340 min−1 mg−1 protein | ||
| GST lysate | UDa | UD |
| GST-94J16 lysate | 1.1 ± 0.3 | 10.6 ± 1.6 |
| GST eluate | 0.2 ± 0.1 | 0.3 ± 0.2 |
| GST-94J16 eluate | 4.2 ± 0.8 | 43.8 ± 13.5 |
Substrate (either HPOD or HPOT) was added to E. coli lysate or affinity-purified eluate after induction of GST protein alone or a GST-94J16 fusion protein. HPL enzyme activity was measured as the rate of decrease in A340 (detection limit 0.004 unit min−1) in a coupled enzyme assay (Vick, 1991).
UD, Undetectable.
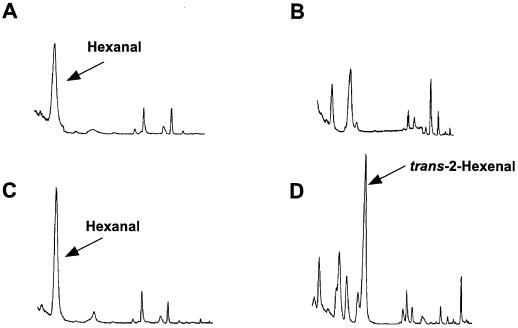
To establish C6-aldehyde production from the 94J16 gene product, we measured headspace volatiles produced from E. coli extracts containing either GST alone or the truncated 94J16 fusion protein in the presence of HPOT or HPOD substrate. Figure 5 demonstrates that in the presence of HPOD, the 94J16 bacterial extract produced only marginally more hexanal than the GST control. The presence of hexanal in the headspace of the GST control extracts indicated that there was some spontaneous cleavage of HPOD into the C6-aldehyde. The production of small quantities of hexanal was in keeping with the enzyme-activity data (Table I). In the presence of HPOT, a substantial peak was present at a retention time of 6.77 min in the total ion count chromatogram for the 94J16 bacterial extract that was not detectable in the corresponding chromatogram for the GST control extract (Fig. 5). This peak was identified as trans-2-hexenal by co-chromatography with commercial standards, as well as by MS. The immediate C6-aldehyde product of HPL activity with HPOT is thought to be cis-3-hexenal, which was subsequently isomerized in vivo to produce the trans-isomer. We have found, however, that with our methods of GC-MS analysis the cis-3-isomer is artificially isomerized to trans-2-hexenal during thermal desorption (J.M.C. Riley and J.E. Thompson, unpublished results). Levels of trans-2-hexenal produced by the 94J16 gene product from HPOT were approximately 12-fold higher than the levels of hexanal produced from HPOD (Table II), and the ratio of trans-2-hexenal:hexanal formation was in approximate agreement with the 10-fold-higher activity of the 94J16 gene product measured using the coupled-enzyme assay with HPOT relative to HPOD as the substrate (Table I).
Figure 5.
Total ion-count chromatogram of volatile compounds from the headspace of E. coli extracts containing GST protein alone (A and B) or 94J16 as a GST fusion protein (C and D). In A and C, HPOD was used as the substrate, and in B and D, HPOT was used as the substrate.
Table II.
C6-volatile production from E. coli extracts expressing either a GST protein or a GST-94J16 fusion protein
| Extract | Volatile Production
|
|
|---|---|---|
| Hexanal | trans-2-Hexenal | |
| μg min−1 mg−1 protein | ||
| GST eluate | 5.3 ± 1.0 | UDa |
| GST-94J16 eluate | 6.1 ± 0.6 | 9.8 ± 0.1 |
Headspace volatiles were measured by GC after the addition of HPOD or HPOT substrate to affinity-purified protein eluate. Quantity of C6 volatiles was measured relative to known quantities of standard compounds.
UD, Undetectable.
Molecular Characterization of Arabidopsis HPL
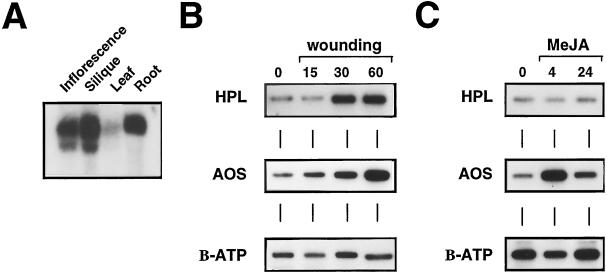
Northern-blot analysis was used to characterize the tissue-specific expression patterns for this gene in Arabidopsis. Figure 6A indicates that HPL had a defined pattern of gene expression. HPL was expressed at relatively high levels in mature inflorescence, at slightly lower levels in green silique and root tissue, and at substantially lower levels in green leaf tissue. This pattern of gene expression is somewhat surprising, given that HPL activity is associated with the production of “green-leaf” volatiles (Hatanaka, 1993). However, HPL cleavage products are present in significant quantities in most floral tissues (Loughrin et al., 1990; Knudsen et al., 1993), suggesting that they may be an important component of the collective floral scent. Similarly, Kamm and Buttery (1984) demonstrated that diseased roots of red clover contain significant quantities of hexanol and trans-2-hexenal, indicating that this tissue is also capable of producing C6 volatiles, which is in keeping with HPL gene expression in the root.
Figure 6.
Analysis of HPL gene-expression patterns by northern-blot and RT-PCR analysis. A, Northern-blot analysis of HPL mRNA accumulation in different tissues of Arabidopsis, including inflorescence, green silique, mature leaf, and root tissue. Total RNA was extracted from each tissue and 10 μg was separated on a formaldehyde gel before blotting onto nylon membrane and hybridization with a 94J16 (HPL) gene-specific probe. B, RT-PCR analysis of HPL, AOS, and β-ATPase expression after wounding of leaf tissue for 15, 30, or 60 min. Total RNA was extracted from intact or wounded tissue, treated with DNase, and subjected to a RT reaction with random hexanucleotide primers. Equal quantities of the RT products were PCR amplified for 20 cycles with gene-specific oligonucleotides corresponding to each gene. An aliquot of the PCR product was separated on an agarose gel, blotted, and hybridized with probes specific for each gene. C, RT-PCR analysis of HPL, AOS, and β-ATPase expression after exposure to MeJA for 4 or 24 h. RT-PCR conditions were as in B.
To characterize the expression of HPL in response to wounding and MeJA treatment we used RT-PCR, since we found message abundance to be too low in leaf tissue for definitive characterization by northern-blot analysis. Northern blots were also complicated by the presence of a second signal, slightly shorter than the predominant signal, that hybridized to the 94J16 probe, even under high-stringency conditions. Thus, to further ensure gene specificity, RT-PCR was used to provide a quantitative assessment of message levels. Preliminary optimization experiments found that by lowering the number of PCR cycles the reaction was quantitative (data not shown). A portion of the PCR reaction was separated by agarose-gel electrophoresis, blotted, and hybridized with gene-specific probes for 94J16, AT-AOS, or β-ATPase.
Release of C6 volatiles is associated with damaged plant tissue, so we quantified 94J16 mRNA levels after wounding to determine if the gene is wound induced. AT-AOS is wound inducible (Laudert et al., 1996), so mRNA levels for this gene were also included to serve as a positive control for wounding conditions. Figure 6B demonstrates that 94J16 mRNA quantity increases within 30 min after wounding, suggesting that transcription of this gene is stimulated in response to wounding. AOS mRNA also accumulated after wounding, as has been demonstrated previously (Laudert et al., 1996), but the constitutive control β-ATPase was unaffected by wound treatment. Wound induction of HPL is interesting in light of the fact that the C12 product of HPL gives rise to traumatin, a compound implicated in a signaling role during wounding (Zimmerman and Coudron, 1979). Recently, we demonstrated that C6 volatiles also signal a variety of defense-related genes in Arabidopsis (N.J. Bate and S.J. Rothstein, unpublished results), and C6 volatiles have also been shown to stimulate phytoalexin production in cotton (Zeringue, 1992). Thus, it appears that the wound-induced expression of HPL in tissue surrounding the wound site may result in the production of signal molecules to activate specific defense responses.
MeJA is a compound thought to play a role in the activation of defense-related genes in response to wounding (Anderson, 1989; Creelman and Mullet, 1997). Treatment of Arabidopsis plants in sealed containers with 10 μm MeJA did not stimulate 94J16 mRNA levels over a 24-h period (Fig. 6C), whereas AOS mRNA was induced within 4 h. The mRNA levels were reduced somewhat in the β-ATPase control after 4 h, but by 24 h the levels had returned to untreated control levels. Avdiushko et al. (1995) reported that treatment with MeJA induced HPL enzyme activity and hexanal and trans-2-hexenal production, suggesting that if there are two HPL isozymes both are induced. However, much higher levels of MeJA were used by Avdiushko et al. (1995) than were used in this study (approximately 175-fold higher), and their findings indicated that Arabidopsis requires 5000-fold higher concentrations of MeJA treatment to achieve the same induction relative to cucumber (Avdiushko et al., 1995). It is possible that higher concentrations of MeJA may induce 94J16 or other HPL genes in Arabidopsis, but because the expression of a range of genes is induced by similar concentrations in a variety of species (Franceschi and Grimes, 1991; Farmer et al., 1992; Bolter, 1993; Benedetti et al., 1995), we conclude that, in a comparative sense, 94J16 is not responsive to MeJA.
Abbreviations:
- AOS
allene oxide synthase
- EST
expressed sequence tag
- GST
glutathione S-transferase
- HPL
hydroperoxide lyase
- HPOD
13S-hydroperoxy-9(Z),11(E)-octadecadienoic acid
- HPOT
13S-hydroperoxy-9(Z),11(E),15(Z)-octadecatrienoic acid
- MeJA
methyl jasmonate
- RACE
rapid amplification of cDNA ends
- RT
reverse transcriptase
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers W, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson JM (1989) Membrane-derived fatty acids as precursors to second messengers. In WF Smith, DJ Morr, eds, Second Messengers in Plant Growth and Development, Vol 6. Alan R Liss, New York, pp 181–212
- Archbold DD, Hamilton-Kemp TR, Barth MM, Langlois BE. Identifying natural volatile compounds that control gray mold (Botrytis cinera) during postharvest storage of strawberry, blackberry, and grape. J Agric Food Chem. 1997;45:4032–4037. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DM, Seidman JG, Smith JA, Struhl K (1987) Current Protocols in Molecular Biology. Wiley, New York
- Avdiushko S, Croft KPC, Brown GC, Jackson DM, Hamilton-Kemp TR, Hildebrand D. Effect of volatile methyl jasmonate on the oxylipin pathway in tobacco, cucumber, and Arabidopsis. Plant Physiol. 1995;109:1227–1230. doi: 10.1104/pp.109.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti CE, Xie D, Turner JG. COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 1995;109:567–572. doi: 10.1104/pp.109.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleé E, Joyard J. Envelope membranes from spinach chloroplasts are a site of metabolism of fatty acid hydroperoxides. Plant Physiol. 1996;110:445–454. doi: 10.1104/pp.110.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolter CJ. Methyl jasmonate induces papain inhibitor(s) in tomato leaves. Plant Physiol. 1993;103:1347–1353. doi: 10.1104/pp.103.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry M, Chua N-H. A nuclear gene encoding the beta subunit of the mitochondrial ATP synthase in Nicotiana plumbaginifolia. EMBO J. 1985;4:2159–2165. doi: 10.1002/j.1460-2075.1985.tb03910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Oligosaccharins, brassinolides, and jasmonates: nontraditional regulators of plant growth, development, and gene expression. Plant Cell. 1997;9:1211–1223. doi: 10.1105/tpc.9.7.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft KPC, Jüttner F, Slusarenko AJ. Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv phaseolicola. Plant Physiol. 1993;101:13–24. doi: 10.1104/pp.101.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Johnson RR, Ryan CA. Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 1992;98:995–1002. doi: 10.1104/pp.98.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi VR, Grimes HD. Induction of soybean vegetative storage proteins and anthocyanins by low-level atmospheric methyl jasmonate. Proc Natl Acad Sci USA. 1991;88:6745–6749. doi: 10.1073/pnas.88.15.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner HW, Dornbos DL, Jr, Desjardins A. Hexanal, trans-2-hexenal, and trans-2-nonenal inhibit soybean, Glycine max, seed germination. J Agric Food Chem. 1990;38:1316–1320. [Google Scholar]
- Gardner HW, Weisleder D, Plattner RD. Hydroperoxide lyase and other hydroperoxide-metabolizing activity in tissues of soybean, Glycine max. Plant Physiol. 1991;97:1059–1072. doi: 10.1104/pp.97.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring DR, Banks P, Beversdorf WD, Rothstein SJ. Use of the polymerase chain reaction to isolate an S-locus glycoprotein cDNA introgressed from Brassica campestris into B. napus ssp. oleifera. Mol Gen Genet. 1992;234:185–192. doi: 10.1007/BF00283838. [DOI] [PubMed] [Google Scholar]
- Hatanaka A. The biogeneration of green odour by green leaves. Phytochemistry. 1993;34:1201–1218. [Google Scholar]
- Kamm JA, Buttery RG. Root volatile components of red clover: identification and bioassay with the clover root borer (Coleoptera:Scolytidae) Environ Entomol. 1984;13:1427–1430. [Google Scholar]
- Knudsen JT, Tollsten L, Bergstrom LG. Floral scents: a checklist of volatile compounds isolated by head-space techniques. Phytochemistry. 1993;33:253–280. [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Holländer-Czytko H, Weiler EW. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol. 1996;31:323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- Loughrin JH, Hamilton-Kemp TR, Andersen RA, Hildebrand DF. Volatiles from flowers of Nicotiana sylvestris, N. otophora and Malus × domestica: headspace components and day/night changes in their relative concentrations. Phytochemistry. 1990;29:2473–2477. [Google Scholar]
- Matsui K, Shibutani M, Hase T, Kajiwara T. Bell pepper fruit fatty acid hydroperoxide lyase is a cytochrome P450 (CYP74B) FEBS Lett. 1996;394:21–24. doi: 10.1016/0014-5793(96)00924-6. [DOI] [PubMed] [Google Scholar]
- Matsui K, Toyota H, Kajiwara T, Kakuno T, Hatanaka A. Fatty acid hydroperoxide cleaving enzyme, hydroperoxide lyase, from tea leaves. Phytochemistry. 1991;30:2109–2113. [Google Scholar]
- Pan Z, Durst F, Werck-Reichhart D, Gardner HW, Camara B, Cornish K, Backhaus RA. The major protein of guayule rubber particles is a cytochrome P450. J Biol Chem. 1995;270:8487–8494. doi: 10.1074/jbc.270.15.8487. [DOI] [PubMed] [Google Scholar]
- Riley JCM, Willemot C, Thompson JE. Lipoxygenase and hydroperoxide lyase activities in ripening tomato fruit. Postharvest Biol Technol. 1996;7:97–107. [Google Scholar]
- Song W-C, Funk CD, Brash AR. Molecular cloning of an allene oxide synthase: a cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc Natl Acad Sci USA. 1993;90:8519–8523. doi: 10.1073/pnas.90.18.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA. A spectrophotometric assay for hydroperoxide lyase. Lipids. 1991;26:315–320. [Google Scholar]
- Vick BA, Zimmerman DC. Lipoxygenase and hydroperoxide lyase in germinating watermelon seedlings. Plant Physiol. 1976;57:780–788. doi: 10.1104/pp.57.5.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Heijne G, Steppuhn J, Herrman RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–546. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Zeringue HJ., Jr Effects of C6–C10 alkenals and alkanals on eliciting a defence response in the developing cotton boll. Phytochemistry. 1992;31:2305–2308. [Google Scholar]
- Zimmerman DC, Coudron CA. Identification of traumatin, a wound hormone, as 12-oxo-trans-10-dodecanoid acid. Plant Physiol. 1979;63:536–541. doi: 10.1104/pp.63.3.536. [DOI] [PMC free article] [PubMed] [Google Scholar]