Abstract
Thiopurine S-methyltransferase (TPMT) catalyzes the S-methylation of aromatic and heterocyclic sulfhydryl compounds including thiopurine drugs such as 6-mercaptopurine, 6-thioguanine and azathioprine. TPMT activity exhibits genetic variation and shows tri-modal distribution with 89–94% of individuals possessing high activity, 6–11% intermediate activity and approximately 0.3% low activity. Patients with intermediate or deficient TPMT activity exposed to thiopurine drugs show severe hematopoietic toxicity. Three single nucleotide polymorphisms (SNPs) in TPMT (NM_000367.2:c.238G>C, NM_000367.2:c.460G>A and NM_000367.2:c.719A>G) define the most prevalent mutant alleles associated with loss of catalytic activity reported in several populations. The present study investigated, for the first time, the frequency distribution of these three SNPs of TPMT, their alleles and genotypes in a Southern Indian population. Peripheral blood was obtained from 326 individuals of a Southern Indian population, and genomic DNA was isolated from total peripheral white blood cells. The genotypes at the polymorphic loci were determined by allele-specific polymerase chain reaction, restriction fragment length polymorphism and confirmatory DNA sequencing. The estimated genotype frequency for homozygous TPMT*1/*1 was 97.24%, for heterozygous TPMT*1/*2 and TPMT*1/*3B, 0.61% each, and for heterozygous TPMT*1/*3C, 1.53%. The frequency of heterozygous mutants in the studied Indian population was 2.76%. This study demonstrated significant variations in TPMT gene polymorphisms in an Indian population in relation to other human populations and may help to predict both clinical efficacy and drug toxicity of thiopurine drugs.
Keywords: Thiopurine S-methyltransferase, polymorphisms, Indian population, pharmacogenetics, mercaptopurine, azathioprine, genetic testing
Introduction
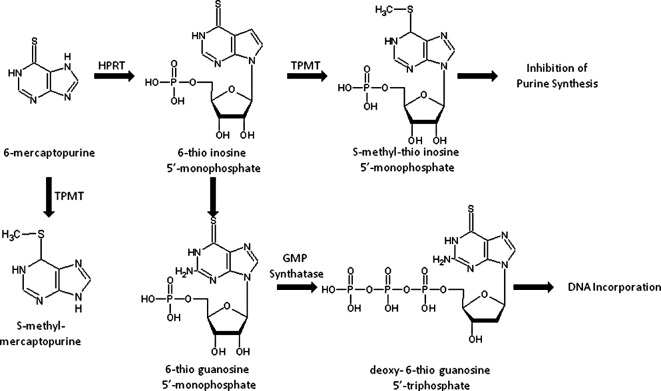
Thiopurine S-methyltransferase [TPMT: EC 2.1.1.67 (MIM 187680)] is a cytoplasmic enzyme present in both prokaryotes and eukaryotes. TPMT, first described by Remy (1), catalyzes the S-methylation of aromatic compounds including thiopurine drugs such as 6-mercaptopurine, 6-thioguanine and azathiopurine (2,3). Thiopurines are extensively used as chemotherapeutic agents in patients with neoplasia and auto-immune disorders. The prodrugs, thiopurines, are converted intracellularly to a thioguanine nucleotide form which exerts a cytotoxic effect. The first step in the intracellular activation of mercaptopurine involves being catalyzed by hypoxanthine-guanine phosphoribosyltransferase (HPRT), which yields thioinosine monophosphate (TIMP). Subsequently, TIMP is converted to thioguanine triphosphate nucleotides, which are incorporated into DNA. Excessive accumulation of thioguanine (TGN), a cytotoxic compound, induces severe cytotoxicity. Fig. 1 illustrates the drug metabolic pathway for thiopurine drugs by TPMT and the subsequent conversion into an inactive substance thereby decreasing the amount of prodrug available for active TGN (2).
Figure 1.
Pathway of thiopurine metabolism. TPMT, thiopurine S-methyltransferase; HPRT, hypoxanthine-guanine phosphoribosyltransferase.
The level of TPMT activity in human tissue is controlled by genetic polymorphisms responsible for individual differences in thiopurine toxicity and therapeutic efficacy (4–6). Several studies have reported that patients with low or undetectable TPMT activity are at high risk for severe toxicity when treated with conventional dosages of thiopurine drugs, while similar doses may be adequate to treat patients with high levels of enzyme activity (5,6). Due to these important clinical consequences, it is suggested that patients be screened for TPMT genotype to determine the enzyme activity before thiopurine therapy. The active gene for the enzyme is 34 kb in length, consisting of 10 exons, and is localized at 6q22.3 (4,7). The TPMT pseudogene has also been reported and mapped to human chromosome band 18q21.1 (8).
TPMT enzyme activity shows trimodal distribution with 89–94% of individuals possessing high activity, 6–11% intermediate activity and 0.3% low activity. The wild-type allele for high TPMT activity has been designated TPMT*1. Several variant alleles for TPMT (TPMT*2 to TPMT*20) have been reported in many ethnic groups. A list of known alleles of the TPMT gene is presented in Table I, with the type of variations and the loci. Among these, four mutant alleles namely TPMT*2, TPMT*3A, TPMT*3B and TPMT*3C have been identified as responsible for enzyme deficiency in several populations. Another sixteen allelic variants, TPMT*5 to TPMT*20 (Table I), have also been suggested to be associated with deficient TPMT activity (9–11).
Table I.
List of currently known mutant alleles of the thiopurine S-methyltransferase (TPMT) gene.
| Allele | Nucleotide change | Amino acid change | Exon position |
|---|---|---|---|
| TPMT*1 | Wild | Wild | |
| TPMT*1A | C178T | Silent | 1 |
| TPMT*1S | T474C | Silent | 6 |
| TPMT*2 | G238C | Ala80Pro | 5 |
| TPMT*3A | G460A | Ala154Thr | 7 |
| A719G | Try240Thr | 10 | |
| TPMT*3B | G460A | Ala154Thr | 7 |
| TPMT*3C | A719G | Try240Thr | 10 |
| TPMT*3D | G292T | Glu98-Stop | 5 |
| G460A | Ala154Thr | 7 | |
| A719G | Try240Thr | 10 | |
| TPMT*4 | G-A | Splice junction intron 9/exon 10 | |
| TPMT*5 | T146C | Leu49Ser | 4 |
| TPMT*6 | A539T | Tyr180phe | 8 |
| TPMT*7 | T681G | His227Glu | 10 |
| TPMT*8 | G644A | Arg215His | 10 |
| TPMT*9 | A356C | Lys119Thy | 5 |
| TPMT*10 | G430C | Gly144Arg | 7 |
| TPMT*11 | G395A | Cys132Thy | 6 |
| TPMT*12 | C374T | Ser125Leu | 6 |
| TPMT*13 | A83T | Glu28Val | 3 |
| TPMT*14 | A1G | Met1Val | 3 |
| TPMT*15 | G-A | Splice junction intron 7/exon 8 | |
| TPMT*16 | G488A | Arg163His | 7 |
| TPMT*17 | C124G | Gln42Glu | 3 |
| TPMT*18 | G211A | Gly71Arg | 4 |
| TPMT*19 | A356C | Lys122Thr | 5 |
| TPMT*20 | G106C | Gly36Ser | 3 |
The first identified variant allele, TPMT*2, contains a transversion c.238G>C, leading to substitution of p.Ala80Pro residue. It is a rare allele reported to be found in European Caucasians and African-Americans (12–14). The TPMT*3A mutant allele contains two nucleotide transitions, c.460G>A and c.719A>G, in the open reading frame, leading to the substitution of amino acids p.Ala154Thr and p.Tyr240Cys, respectively, and are found in African-American, European Caucasians and Southwest Asians. It is the most common allele among the European Caucasian population (13,15). TPMT*3B, that contains the transition c.460G>A, is a common allele in Caucasian populations. TPMT*3C contains transversion c.719A>G and is the most prevalent allele among the Chinese population. However, no similar data are available on TPMT polymorphisms in Indian populations. Identifying the most prevalent TPMT allele in Indian populations could facilitate the deployment of rapid DNA-based assays for patients before they are subjected to thiopurine drug therapy. Thus, the aim of the study was to determine the frequency of TPMT variant alleles in an Indian population in comparison to other populations. The present study focused on the detection of signature alleles for the TPMT gene, TPMT*2, *3A, *3B and *3C by using allele-specific (mutation-specific) oligonucleotide polymerase chain reaction (ASO-PCR), polymerase chain reaction, followed by restriction fragment length polymorphism (PCR-RFLP) analysis and by confirmatory DNA sequencing of the loci.
Materials and methods
Sample collection
Participants for the study were recruited randomly from among the population of Southern India. A total of 326 (176 males; 150 females) unrelated healthy Indian individuals were recruited, with a mean age of 31.4 years (range, 18–69 years). Venous blood (4 ml) was obtained from each participant in an EDTA vacutainer. The study was approved by the Institutional Ethics Committee of Manipal University as per the guidelines of the Indian Council of Medical Research, and written informed consent was obtained from all participants.
Isolation of genomic DNA and polymerase chain reaction
Genomic DNA was isolated from all the samples collected by the standard phenol-chloroform extraction method (16), and the three major TPMT polymorphisms were genotyped in each sample. The genotypes of the TPMT gene were analyzed for the three SNP loci, NM_000367.2:c.238G>C, NM_000367.2:c.460G>A and NM_000367.2:c.719A>G, to determine the frequencies of TPMT*2, *3A, *3B and *3C alleles and their genotypes. Genotyping of the NM_000367.2:c.238G>C locus was performed by ASO-PCR and those of NM_000367.2:c.460G>A and NM_000367.2:c.719A>G loci were performed by polymerase chain reaction, followed by PCR-RFLP. All PCR reactions were performed with 30–50 ng of DNA in 25 μl of the total reaction mixture containing 0.15 mM dNTPs, 20 pmol of each primer and 1 unit of Taq DNA polymerase.
Allele-specific oligonucleotide polymerase chain reaction for NM_000367.2:c.238G>C
The NM_000367.2:c.238G>C polymorphism was analyzed by ASO-PCR to test for the presence of the TPMT*2 allele. DNA was amplified using a common reverse primer, 5′-TAAATAGGAACCATCGGAC AC-3′ and forward primer with the single allele discriminating base at their 3′ end used in a separate wild-type allele specific or mutant allele specific PCR reaction was either a) 5′-GTA TGATTTTATGCAGGTTTG-3′ or b) 5′-GTATGATTTTATG CAGGTTTC-3′, respectively, as previously reported. PCR thermal cycling conditions were: i) an initial 5-min denaturation at 94°C; ii) 34 cycles at 94°C (30 sec), 56°C (30 sec) and 72°C (60 sec); and iii) a final extension for 5 min at 72°C. Amplified 254-bp PCR product was visualized by electrophoresis using 2% of agarose gel containing ethidium bromide (0.5 μg/ml).
Genotyping of TPMT*3B and TPMT*3C by polymerase chain reaction-restriction fragment length polymorphism
The TPMT*3B (NM_000367.2:c.460G>A) polymorphism was genotyped by employing PCR-RFLP. The forward primer for PCR was 5′-AGGCAGCTAGGGAAAAAGAAAGGTG-3′ and reverse primer 5′-CAAGCCTTATAGCCTTACACCCAGG-3′. PCR thermal cycling conditions were: i) an initial 5-min denaturation at 94°C; ii) 34 cycles at 94°C (30 sec), 62°C (30 sec) and 72°C (60 sec); and iii) a final extension for 5 min at 72°C. The 694-bp PCR amplicon containing c.460G>A was digested with the restriction enzyme MwoI, and the DNA fragments were separated on a 2% agarose gel to discriminate the alleles. The wild-type allele contains the MwoI restriction site; however, the restriction site is absent in the mutant allele.
The analysis of the TPMT*3C polymorphism was performed by PCR-RFLP. DNA was amplified using the forward primer 5′-GAGACAGAGTTTCACCATCTTGG-3′ and reverse primer 5′-CAGGCTTTAGCATAATTTTCAAT TCCTC-3′. PCR thermal cycling conditions were: i) an initial 5-min denaturation at 94°C; ii) 34 cycles at 94°C (30 sec), 62°C (30 sec) and 72°C (60 sec); and iii) a final extension for 5 min at 72°C. The 373-bp PCR amplicon containing c.719A>G was digested with the restriction enzyme AccI, and the DNA fragments were separated on a 2% agarose gel to discriminate the alleles. The mutant type allele introduces an AccI restriction site which is absent in the wild-type allele.
DNA sequencing
The target TPMT PCR amplicons were sequenced in both forward and reverse directions for all the available genotypes for TPMT*2, *3B and *3C. Amplified PCR products were purified after eluting the fragments from the agarose gel using the standard isopropanol precipitation method, followed by direct sequencing using the Big Dye Terminator Cycle Sequencing Ready Reaction Mix Kit v3.1 (ABI, Foster City, CA, USA) according to the manufacturer's instructions. DNA sequencing was performed in an ABI, 3130 genetic analyzer, and the electropherograms of the DNA sequences in sense and antisense directions were analyzed for detection of mutations using the Bioinformatics Tools and Human Genome Nucleotide Reference Sequence NT_007592.14.
Statistical analysis
Subjects were genotyped, and allele frequencies were calculated by counting the alleles. The 95% confidence interval (CI) was calculated for all proportions of alleles and genotypes. The Chi-square test was performed to test for deviation from Hardy-Weinberg equilibrium for each locus.
Results
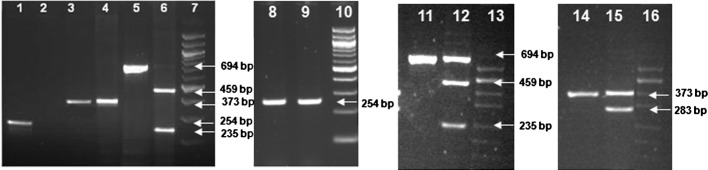
Our study determined the allele and genotype frequencies of the three TPMT gene polymorphisms, c.238G>C, c.460G>A and c.719A>G in a sample group of 326 individuals drawn from a Southern Indian population. Agarose gel electrophoresis images representative of genotyping experiment results by ASO-PCR and PCR-RFLP are depicted in Fig. 2. A summary of the results of allele and genotype frequencies at the three loci (c.238G>C, c.460G>A and c.719A>G) are presented with their Chi-square test P-values in Table II. The allele and genotype frequencies involving TPMT*1, *2, *3A, *3B and*3C alleles in the population sample were also calculated with 95% CI and are presented in Tables III and IV, respectively.
Figure 2.
Images of PCR amplicons containing SNP loci of TPMT, NM_000367.2:c.238G>C, NM_000367.2:c.460G>A and NM_000367.2:c.719A>G, and their restriction fragments after agarose gel electrophoresis. All the lanes in the consecutive gel images are numbered sequentially from 1 to 16. Sizes of the DNA fragments are indicated by arrows shown in the image of 1.5% agarose gel electrophoresis. Lanes 7, 10, 13 and 16 show DNA size markers; 7 and 11 are 100-bp DNA ladder, and lanes 13 and 16 are pUC/Hind III DNA ladder. Lanes 1–6 show the wild-type alleles of the three TPMT polymorphisms (NM_000367.2:c.238G>C, NM_000367.2:c.719A>G and NM_000367.2:c.460G>A) and their restriction enzyme digestion patterns; lane 1 shows the allele-specific amplicon representing the wild-type c.238G allele; lane 2, which is blank, corresponds to the mutant allele-specific reaction; lanes 3 and 4 represent PCR amplicon with c.719A allele and the AccI restriction enzyme-treated and undigested PCR amplicons, respectively; lanes 5 and 6 show the wild-type c.450G PCR amplicon and its MwoI restriction endonuclease digested fragments of sizes 459 and 235 bp, respectively. Lanes 8–16 show the gel images of the PCR amplicons with mutant alleles at the three TPMT SNP loci, NM_000367.2:c.238G>C, NM_000367.2:c.460G>A and NM_000367.2:c.719A>G, respectively. Lanes 8 and 9 show allele-specific PCR amplicons of 254 bp from a heterozygote for the c.238G>C polymorphism. Lane 11 shows PCR amplicon of the heterozygous c.450G>A and the MwoI digested fragments of the amplicon giving DNA fragments of 559 and 235 bp, respectively, along with the wild-type undigested fragment of size 694 bp (lane 12). Lane 14 depicts the PCR amplicon of a heterozygote of c.719A>G SNP and the AccI restriction enzyme-treated undigested mutant allele, and the PCR fragments with wild-type allele digested to fragments of 283 bp and a smaller one of 90 bp that is not shown in the gel (lane 15).
Table II.
Distribution of genotypes and alleles of thiopurine S-methyltransferase, 238G>C, 460G>A and 719A>G single nucleotide polymorphismsa, in a Southern Indian population, tested for Hardy-Weinberg equilibrium.
| Locus | Genotype counts (Frequency %) | Allele counts (Frequency %) | P-valueb | |||
|---|---|---|---|---|---|---|
| c.238G>C (rs1800462) | GG | GC | CC | G | C | |
|
|
|
|||||
| 324 (99.39) | 2 (0.61) | 0 | 650 (99.69) | 2 (0.31) | 0.96 | |
| c.460G>A (rs1800460) | GG | GA | AA | G | A | |
|
|
|
|||||
| 324 (99.39) | 2 (0.61) | 0 | 650 (99.69) | 2 (0.31) | 0.96 | |
| c.719A>G (rs1142345) | AA | AG | GG | A | G | |
|
|
|
|||||
| 321 (98.47) | 5 (1.53) | 0 | 647 (99.23) | 5 (0.77) | 0.89 | |
Ref. seq: NM_000367.2, based on cDNA.
P<0.05 is considered significant (Chi-square test for Hardy-Weinberg equilibrium).
Table III.
Functional variants of thiopurine S-methyltransferase (TPMT) gene alleles and their distribution in a Southern Indian population with their 95% confidence interval (CI).
| Type of allele | No. of alleles | Percentage of allele | 95% CI |
|---|---|---|---|
| TPMT*1 | 643 | 98.62 | 97.7–99.5 |
| TPMT*2 | 2 | 0.31 | 0.0–0.7 |
| TPMT*3A | 0 | 0.00 | - |
| TPMT*3B | 2 | 0.31 | 0.0–0.7 |
| TPMT*3C | 5 | 0.76 | 0.1–1.4 |
| Total | 652 | 100.00 |
Table IV.
Distribution of genotype frequencies of thiopurine S-methyltransferase (TPMT) gene functional variants (involving TPMT*1, *2, *3A, *3B and *3C alleles) in a Southern Indian population and their Chi-square test result for Hardy-Weinberg equilibrium.
| Genotype | No. of samples | Frequency (%) | 95% CI |
|---|---|---|---|
| Distribution of genotypes for individual loci | |||
| TPMT*1/TPMT*1 | 317 | 97.24 | 95.5–99.0 |
| TPMT*1/TPMT*2 | 2 | 0.61 | 0.0–1.5 |
| TPMT*1/TPMT*3A | 0 | 0.00 | 0 |
| TPMT*1/TPMT*3B | 2 | 0.61 | 0.0–1.5 |
| TPMT*1/TPMT*3C | 5 | 1.53 | 0.2–2.9 |
| Total | 326 | 100.00 | |
| Distribution of genotypes for all loci | |||
| Homozygous wild (Total) | 317 | 97.24 | 95.5–99.0 |
| Heterozygous mutant (Total) | 9 | 2.76 | 0.98–4.5 |
| Homozygous mutant | 0 | 0.00 | 0 |
| Total | 326 | 100.00 |
Result of the Chi-square test for Hardy-Weinberg equilibrium for wild-type vs. all mutantsa, χ2 =0.06; P=0.8.
P<0.05 is considered significant, or P<0.05 is not consistent with Hardy-Weinberg equilibrium. CI, confidence interval.
Of the 326 healthy Indian participants investigated in this study, 317 were found to be homozygous for TPMT*1/*1 (97.24%; 95% CI, 95.5–99.0). Two participants were heterozygous for the alleles TPMT*1/*2 (0.61%; 95% CI, 0–1.5), 2 were heterozygous for alleles TPMT*1/*3B (0.61%; 95% CI, 0–1.5) and 5 were heterozygous for alleles TPMT*1/*3C (1.53%; 95% CI, 0.2–2.9) among all the individuals of the group. The TPMT*3A allele was not found in individuals from the investigated population in this study, and no individual was found homozygous for mutant alleles (Table IV). The allele and genotype frequencies did not show any deviation from Hardy-Weinberg equilibrium as shown by the Chi-square test results (χ2=0.06; P=0.8).
Discussion
TPMT catalyzes the S-methylation of thiopurine drugs such as 6-mercaptopurine and 6-thioguanine to form inactive metabolites, a pathway that competes with xanthine oxidase-mediated conversion of 6-MP to the inactive 6-thiouric acid and HPRT-mediated conversion of 6-MP into the active 6-thioguanine nucleotide. Patients with inherited low levels of TPMT enzyme activity are at increased risk for thiopurine-induced toxicity when treated with thiopurine drugs such as 6-mercaptopurine, 6-thioguanine and azathioprine; and those with inherited higher levels of enzyme activity may be treated inadequately. Since these drugs have a narrow therapeutic window, toxicities such as life-threatening myelosuppression assume great importance. The TPMT genetic polymorphism represents a good example of the well established clinical importance of pharmacogenetic variation of a drug-metabolizing enzyme. The prevalence of various TPMT alleles in the Caucasian population is reported to be 0.3% which occur as homozygous mutants, 11% which occur as heterozygous and 89% as homozygous wild-types (17,18). Homozygous-mutated individuals are at increased risk for potentially life-threatening toxicity when exposed to standard doses of thiopurine drugs, while heterozygous carriers are advised to begin therapy with 50–60% of the recommended dose (19). The molecular basis of TPMT deficiency is well understood, where TPMT*2, *3A, *3B and *3C are the most common variants detected in more than 80% of individuals with low or intermediate TPMT activity (20,21). The frequencies of these alleles in various populations of the world that are available in published literature have been summarized in Table V.
Table V.
Frequencies (%) of thiopurine S-methyltransferase (TPMT)*2, *3A, *3B and *3C alleles reported in different populations of the world.
| Population | No. of participants (n) | TPMT*2 | TPMT*3A | TPMT*3B | TPMT*3C | Reference |
|---|---|---|---|---|---|---|
| Indians | 326 | 0.61 | 0.0 | 0.61 | 1.53 | Present study |
| African American | 248 | 0.40 | 0.8 | N/D | 2.40 | (9) |
| American Caucasian | 282 | 0.20 | 3.2 | N/D | 0.20 | (9) |
| Brazilian | 306 | 0.80 | 2.0 | N/D | 2.50 | (26) |
| Kenya | 101 | 0.00 | 0.0 | 0.00 | 5.40 | (27) |
| British Caucasian | 199 | 0.50 | 4.5 | 0.00 | 0.30 | (28) |
| South West Asian | 99 | 0.00 | 1.0 | 0.00 | 0.00 | (28) |
| Egyptian | 200 | 0.00 | 0.3 | 0.00 | 1.30 | (30) |
| French Caucasian | 469 | 3.00 | 0.7 | N/D | 0.40 | (31) |
| Italian | 103 | 0.50 | 3.9 | N/D | 1.00 | (32) |
| Norwegian Caucasian | 66 | 0.00 | 3.4 | N/D | 0.30 | (35) |
| Saami | 194 | 0.00 | 0.0 | N/D | 3.30 | (35) |
| Polish | 358 | 0.40 | 2.7 | 0.00 | 0.10 | (37) |
| South East Asian | 300 | 0.00 | 0.0 | 0.00 | 1.00 | (38) |
| Swedish | 800 | 0.10 | 3.8 | 0.10 | 0.40 | (39) |
| Thai | 200 | 0.00 | 0.0 | 0.00 | 5.00 | (40) |
N/D, not determined.
To date, no studies have been carried out in the Southern Indian population to determine the frequencies of the TPMT alleles that are important in thiopurine drugs pharmacogenomics. In this study, we did not include an analysis for other alleles including rare variants such as TPMT*3D, *4 and *20. The promoter polymorphisms were also ignored due to their minor effect on the modulation of TPMT activity compared with variation in the open reading frame. In this study, the TPMT*2 and *3B variants were found at a low allele frequency of 0.31% for each. No TPMT*3A allele was observed in the subjects studied that appeared to be significant. However, the absence of the TPMT*3A allele in this population needs to be confirmed through study of an appropriately large sample of the population. The most predominant mutant allele found in the Southern Indian population in this study was the TPMT*3C allele with a frequency of 0.76%, representing 55.6% of the mutant alleles investigated. Therefore, a test for this mutation alone in this population may prospectively identify approximately half the proportion of individuals at risk of hematopoietic toxicity in the event of administration of a regular dose of thiopurine drugs. From an anthropological point of view, the TPMT*3C allele was found in a Chinese (22) and in a Ghanaian population (23). It was the first TPMT mutation to arise in humans, and this mutation is also found in Caucasians, either alone in the TPMT*3C allele or in combination with the TPMT*3B allele in the form of TPMT*3A (24). It has been postulated that the ancestral TPMT*3C allele founded prior to the divergence of African and non-African populations was likely to have evolved to contain a second mutation giving rise to the TPMT*3A allele which predominated in American and European Caucasians. The presence of the TPMT*3A allele in African-Americans may be explained on the basis of genome integration due to interethnic admixture (9,25,26). Inhibitors of the activity of the enzyme may also have adverse consequences for carriers of these mutant alleles, whether homozygous or heterozygous. Furthermore, it should be noted that the true significance of this human genetic variation may be better explained in relation to the S-methylation of some endogenous substrates yet to be identified, as opposed to the metabolism of any group of drugs (41).
Our study was the first major study designed to analyze the genetic polymorphisms of TPMT in 326 randomly selected unrelated South Indian subjects from different locations in this region of the country representing the heterogeneous ethnic background of the population. Our study established the frequencies of the TPMT alleles. Overall, 2.76% of the Indian subjects were found to be heterozygous variants for TPMT alleles. Conformity for Hardy-Weinberg equilibrium for the loci suggest that no significant genotyping error was involved (Table II), that allele-based genetic effects may not have been biased and that the mutations are not relatively recent. Comparison of the mutation frequencies obtained in this study with those reported in different ethnic groups shows considerable difference between the Indian and other populations (Table V). The present study delineates more data on variations in the TPMT gene in Indian populations. Identification of such variant allele frequencies may help to predict both clinical efficacy and drug toxicity of thiopurine drugs in Indian populations and to introduce appropriate genetic screening protocols.
Acknowledgments
This study was supported by the Technology Information Forecasting and Assessment Council-Centre of Relevance and Excellence (TIFAC-CORE) in Pharmacogenomics, at the Manipal Life Sciences Centre, Manipal University, India.
References
- 1.Remy CN. Metabolism of thiopyrimidines and thiopurines. S-Methylation with S-adenosylmethionine transmethylase and catabolism in mammalian tissues. J Biol Chem. 1963;238:1078–1084. [PubMed] [Google Scholar]
- 2.Tidd DM, Paterson AR. A biochemical mechanism for the delayed cytotoxic reaction of 6-mercaptopurine. Cancer Res. 1974;34:738–746. [PubMed] [Google Scholar]
- 3.Lennard L, van Loon JA, Lilleyman JS, Weinshilboum RM. Thiopurine pharmacogenetics in leukemia: correlation of erythrocyte thiopurine methyltransferase activity and 6-thioguanine nucleotide concentrations. Clin Pharmacol Ther. 1987;41:18–25. doi: 10.1038/clpt.1987.4. [DOI] [PubMed] [Google Scholar]
- 4.Krynetski EY, Evans WE. Drug methylation in cancer therapy: lessons from the TPMT polymorphism. Oncogene. 2003;22:7403–7413. doi: 10.1038/sj.onc.1206944. [DOI] [PubMed] [Google Scholar]
- 5.Woodson LC, Dunnette JH, Weinshilboum RM. Pharmacogenetics of human thiopurine methyltransferase: kidney-erythrocyte correlation and immunotitration studies. J Pharmacol Exp Ther. 1982;222:174–181. [PubMed] [Google Scholar]
- 6.Krynetski EY, Tai HL, Yates CR, Fessing MY, Loennechen T, Schuetz JD, Relling MV, Evans WE. Genetic polymorphism of thiopurine S-methyltransferase: clinical importance and molecular mechanisms. Pharmacogenetics. 1996;6:279–290. doi: 10.1097/00008571-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Snow JL, Gibson LE. The role of genetic variation in thiopurine methyltransferase activity and the efficacy and/or side effects of azathioprine therapy in dermatologic patients. Arch Dermatol. 1995;131:193–197. [PubMed] [Google Scholar]
- 8.Lee D, Szumlanski C, Houtman J, Honchel R, Rojas K, Overhauser J, Wieben ED, Weinshilboum RM. Thiopurine methyltransferase pharmacogenetics. Cloning of human liver cDNA and a processed pseudogene on human chromosome 18q21.1. Drug Metab Dispos. 1995;23:398–405. [PubMed] [Google Scholar]
- 9.Hon YY, Fessing MY, Pui C-H, Relling MV, Krynetski EY, Evans WE. Polymorphism of the thiopurine S-methyltransferase gene in African-Americans. Hum Mol Genet. 1999;8:371–376. doi: 10.1093/hmg/8.2.371. [DOI] [PubMed] [Google Scholar]
- 10.Evans WE, Hon YY, Bomgaars L, Coutre S, Holdsworth M, Janco R, Kalwinsky D, Keller F, Khatib Z, Margolin J, Murray J, Quinn J, Ravindranath Y, Ritchey K, Roberts W, Rogers ZR, Schiff D, Steuber C, Tucci F, Kornegay N, Krynetski EY, Relling MV. Preponderance of thiopurine S-methyltransferase deficiency and heterozygosity among patients intolerant to mercaptopurine or azathioprine. J Clin Oncol. 2001;19:2293–2301. doi: 10.1200/JCO.2001.19.8.2293. [DOI] [PubMed] [Google Scholar]
- 11.Lindqvist M, Haglund S, Almer S, Peterson C, Taipalensu J, Hertervig E, Lyrenäs E, Söderkvist P. Identification of two novel sequence variants affecting thiopurine methyltransferase enzyme activity. Pharmacogenetics. 2004;14:261–265. doi: 10.1097/00008571-200404000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Krynetski EY, Schuetz JD, Galpin AJ, Pui C-H, Relling MV, Evans WE. A single point mutation leading to loss of catalytic activity in human thiopurine S-methyltransferase. Proc Natl Acad Sci USA. 1995;92:949–953. doi: 10.1073/pnas.92.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tai HL, Krynetski EY, Yates CR, Loennechen T, Fessing MY, Krynetskaia NF, Evans WE. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- 14.Otterness D, Szumlanski C, Lennard L, Klemetsdal B, Aarbakke J, Park-Hah JO, Iven H, Schmiegelow K, Branum E, O'Brien J, Weinshilboum R. Human thiopurine methyltransferase pharmacogenetics: gene sequence polymorphisms. Clin Pharmacol Ther. 1997;62:60–73. doi: 10.1016/S0009-9236(97)90152-1. [DOI] [PubMed] [Google Scholar]
- 15.Otterness DM, Szumlanski CL, Wood TC, Weinshilboum RM. Human thiopurine methyltransferase pharmacogenetics. Kindred with a terminal exon splice junction mutation that results in loss of activity. J Clin Invest. 1998;101:1036–1044. doi: 10.1172/JCI1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. 2nd edition. Cold Spring Harbor; New York: 1989. Analysis and cloning of eukaryotic genomic DNA; pp. 9.16–9.23. A Laboratory Manual. [Google Scholar]
- 17.Lennard L, Lilleyman JS, van Loon J, Weinshilboum RM. Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet. 1990;336:225–229. doi: 10.1016/0140-6736(90)91745-v. [DOI] [PubMed] [Google Scholar]
- 18.Weinshilboum R. Thiopurine pharmacogenetics: clinical and molecular studies of thiopurine methyltransferase. Drug Metab Dispos. 2001;29:601–605. [PubMed] [Google Scholar]
- 19.Coulthard SA, Hogarth LA, Little M, Matheson EC, Redfern CPF, Minto L, Hall AG. The effect of thiopurine methyltransferase expression on sensitivity to thiopurine drugs. Mol Pharmacol. 2002;62:102–109. doi: 10.1124/mol.62.1.102. [DOI] [PubMed] [Google Scholar]
- 20.Yates CR, Krynetski EY, Loennechen T, Fessing MY, Tai HL, Pui CH, Relling MV, Evans WE. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997;126:608–614. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 21.Black AJ, McLeod HL, Capell HA, Powrie RH, Matowe LK, Pritchard SC, Collie-Duguid ESR, Reid RM. Thiopurine methyltransferase genotype predicts therapy-limiting severe toxicity from azathioprine. Ann Intern Med. 1998;129:716–718. doi: 10.7326/0003-4819-129-9-199811010-00007. [DOI] [PubMed] [Google Scholar]
- 22.Kham SK, Tan PL, Tay AH, Heng CK, Yeoh AE, Quah TC. Thiopurine methyltransferase polymorphisms in a multiracial Asian population and children with acute lymphoblastic leukemia. J Pediatr Hematol Oncol. 2002;24:353–359. doi: 10.1097/00043426-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Ameyaw MM, Collie-Duguid ES, Powrie RH, Ofori-Adjei D, McLeod HL. Thiopurine methyltransferase alleles in British and Ghanaian populations. Hum Mol Genet. 1999;8:367–370. doi: 10.1093/hmg/8.2.367. [DOI] [PubMed] [Google Scholar]
- 24.Leipold G, Schütz E, Haas JP, Oellerich M. Azathioprine-induced severe pancytopenia due to a homozygous two-point mutation of the thiopurine methyltransferase gene in a patient with juvenile HLA-B27-associated spondylarthritis. Arthritis Rheum. 1997;40:1896–1898. doi: 10.1002/art.1780401026. [DOI] [PubMed] [Google Scholar]
- 25.Laróvere LE, de Kremer RD, Lambooy LH, de Abreu RA. Genetic polymorphism of thiopurine S-methyltransferase in Argentina. Ann Clin Biochem. 2003;40:388–393. doi: 10.1258/000456303766477039. [DOI] [PubMed] [Google Scholar]
- 26.Reis M, Santoro A, Suarez-Kurtz G. Thiopurine methyltransferase phenotypes and genotypes in Brazilians. Pharmacogenetics. 2003;13:371–373. doi: 10.1097/00008571-200306000-00009. [DOI] [PubMed] [Google Scholar]
- 27.McLeod HL, Pritchard SC, Githang'a J, Indalo A, Ameyaw MM, Powrie RH, Booth L, Collie-Duguid ES. Ethnic differences in thiopurine methyltransferase pharmacogenetics: evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics. 1999;9:773–776. doi: 10.1097/00008571-199912000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Collie-Duguid ES, Pritchard SC, Powrie RH, Sludden J, Collier DA, Li T, McLeod HL. The frequency and distribution of thiopurine methyltransferase alleles in Caucasian and Asian populations. Pharmacogenetics. 1999;9:37–42. doi: 10.1097/00008571-199902000-00006. [DOI] [PubMed] [Google Scholar]
- 29.Isaza C, Henao J, López AM, Cacabelos R. Allelic variants of the thiopurine methyltransferase (TPMT) gene in the Colombian population. Methods Find Exp Clin Pharmacol. 2003;25:423–429. doi: 10.1358/mf.2003.25.6.769646. [DOI] [PubMed] [Google Scholar]
- 30.Hamdy SI, Hiratsuka M, Narahara K, Endo N, El-Enany M, Moursi N, Ahmed MS-E, Mizugaki M. Genotype and allele frequencies of TPMT, NAT2, GST, SULT1A1 and MDR-1 in the Egyptian population. Br J Clin Pharmacol. 2003;55:560–569. doi: 10.1046/j.1365-2125.2003.01786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganiere-Monteil C, Medard Y, Lejus C, Bruneau B, Pineau A, Fenneteau O, Bourin M, Jacqz-Aigrain E. Phenotype and genotype for thiopurine methyltransferase activity in the French Caucasian population: impact of age. Eur J Clin Pharmacol. 2004;60:89–96. doi: 10.1007/s00228-004-0732-5. [DOI] [PubMed] [Google Scholar]
- 32.Rossi AM, Bianchi M, Guarnieri C, Barale R, Pacifici GM. Genotype-phenotype correlation for thiopurine S-methyltransferase in healthy Italian subjects. Eur J Clin Pharmacol. 2001;57:51–54. doi: 10.1007/s002280000246. [DOI] [PubMed] [Google Scholar]
- 33.Hiratsuka M, Inoue T, Omori F, Agatsuma Y, Kishikawa Y, Mizugaki M. Detection assay of rare variants of the thiopurine methyltransferase gene by PCR-RFLP using a mismatch primer in a Japanese population. Biol Pharm Bull. 2000;23:1090–1093. doi: 10.1248/bpb.23.1090. [DOI] [PubMed] [Google Scholar]
- 34.Wei H, Zhou S, Li C, Zhang J, Wu J, Huang M. Phenotyping and genotyping studies of thiopurine S-methyltransferase in Kazaks. Pharm Res. 2005;22:1762–1766. doi: 10.1007/s11095-005-7095-1. [DOI] [PubMed] [Google Scholar]
- 35.Loennechen T, Utsi E, Hartz I, Lysaa R, Kildalsen H, Aarbakke J. Detection of one single mutation predicts thiopurine S-methyltransferase activity in a population of Saami in northern Norway. Clin Pharmacol Ther. 2001;70:183–188. doi: 10.1067/mcp.2001.117445. [DOI] [PubMed] [Google Scholar]
- 36.Alves S, Prata MJ, Ferreira F, Amorim A. Thiopurine methyltransferase pharmacogenetics: alternative molecular diagnosis and preliminary data from Northern Portugal. Pharmacogenetics. 1999;9:257–261. [PubMed] [Google Scholar]
- 37.Kurzawski M, Gawronska-Szklarz B, Drozdzik M. Frequency distribution of thiopurine S-methyltransferase alleles in a polish population. Ther Drug Monit. 2004;26:541–545. doi: 10.1097/00007691-200410000-00013. [DOI] [PubMed] [Google Scholar]
- 38.Chang JG, Lee LS, Chen CM, Shih MC, Wu MC, Tsai FJ, Liang DC. Molecular analysis of thiopurine S-methyltransferase alleles in South-east Asian populations. Pharmacogenetics. 2002;12:191–195. doi: 10.1097/00008571-200204000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Haglund S, Lindqvist M, Almer S, Peterson C, Taipalensuu J. Pyrosequencing of TPMT alleles in a general Swedish population and in patients with inflammatory bowel disease. Clin Chem. 2004;50:288–295. doi: 10.1373/clinchem.2003.023846. [DOI] [PubMed] [Google Scholar]
- 40.Srimartpirom S, Tassaneeyakul W, Kukongviriyapan V, Tassaneeyakul W. Thiopurine S-methyltransferase genetic polymorphism in the Thai population. Br J Clin Pharmacol. 2004;58:66–70. doi: 10.1111/j.1365-2125.2004.02112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vesell ES. Therapeutic lessons from pharmacogenetics. Ann Intern Med. 1997;126:653–655. doi: 10.7326/0003-4819-126-8-199704150-00012. [DOI] [PubMed] [Google Scholar]