Abstract
FGFRL1 (FGFR5) is a member of the fibroblast growth factor receptor family, which lacks the intracellular tyrosine kinase domain required for signal transduction by trans-phosphorylation. Since it still contains a cytoplasmic domain of 100 amino acid residues with a peculiar histidine-rich element, it might be involved in an alternative pathway of downstream signaling. To get a clue about a possible mechanism, we compared the overall structure of FGFRL1 with all proteins from the UniProt databank. We found that the human genome encodes 42 structurally related proteins with a signal peptide, three Ig-like domains and a transmembrane domain. These proteins can be grouped in seven families, fibroblast growth factor receptors (FGFRs), Fc receptor-like proteins, IL-1 receptor-like proteins, killer cell Ig-like receptors (KIRs), nectin-like proteins, sialic acid binding lectins (SIGLECs) and signal regulatory proteins (SIRPs). The 7 families utilize four different strategies for signaling, namely a protein tyrosine kinase domain, a TIR (Toll/IL-1 receptor) domain, ITIM/ITAM motifs as well as carboxy-terminal peptides that interact with the PDZ domain of an adaptor protein. It remains to be determined whether FGFRL1 might also utilize one of these strategies for signaling.
Keywords: fibroblast growth factor receptor, FGFR-like 1, FGFR5, domain structure, immunoglobulin-like, nectin, review
Introduction
The fibroblast growth factor (FGF) signaling system controls a variety of cellular functions, including cell proliferation, differentiation, migration and apoptosis. In humans and mice, this system consists of 22 FGF ligands and 4 FGF receptors (FGFRs) (1–3). The FGFs are monomeric proteins that interact with heparan sulfate and bind, together with this glycosaminoglycan, to one or more of the FGFRs. Upon ligand binding, the FGFRs dimerize and trans-phosphorylate specific tyrosine residues in the cytoplasmic domain of the receptor. The signal is then passed on by various pathways involving Ras/MAP kinase, phospholipase Cγ, PI3-kinase and STAT.
All the FGFRs are expressed in the musculoskeletal system. FGFR1 and FGFR2 are particularly prevalent in bone. FGFR3 is found preferentially in cartilage and FGFR4 in muscle. It is therefore not surprising that germline mutations in the FGFR genes can cause a number of skeletal disorders, including craniosynostosis syndromes and chondrodysplasias (3–5). Somatic mutations in FGFRs can lead to unrestricted cellular growth and cancer. In fact, one third of all bladder carcinomas display nucleotide substitutions in FGFR3. Some hematological disorders such as chronic myeloproliferative diseases exhibit chromosomal translocations involving the FGFR1 gene at chromosome 8p11. Patients with multiple myelomas often exhibit translocations involving the FGFR3 locus at 4p16.
Ten years ago we described a fifth FGFR that we termed FGFRL1 (FGFR-like 1) (6). Independently, the same protein was discovered by two other research groups and termed FGFR5 (7,8). Similar to the classical FGFRs, the novel receptor contains an extracellular domain with three immunoglobulin(Ig)-like repeats and a single transmembrane domain. However, in contrast to the classical receptors, FGFRL1 lacks the protein tyrosine kinase domain but instead contains a short intracellular tail of 100 residues with a peculiar histidine-rich sequence. By Northern blotting and in situ hybridization experiments, FGFRL1 expression is detected at low levels in virtually all mesenchymal tissues and at higher levels in cartilage, bone and some muscles (9,10).
When produced in HEK293 cells or in Sf9 insect cells, the novel receptor binds FGF2 (7,9). Furthermore, recombinant FGFRL1 interacts strongly with heparin and heparan sulfate (9,11). Based on the interaction of the novel receptor with FGF ligands and heparin and on the absence of the tyrosine kinase domain, we speculated that FGFRL1 might function as a decoy receptor that modulates or inhibits FGF signaling (6,9). In fact, when over-expressed in MG63 osteosarcoma cells, it inhibits cell proliferation (9). In a luciferase system, it is able to reduce the activity of the FGF inducible responsive promoter element FIRE (12). Moreover, FGFRL1 expression is markedly increased during differentiation of myoblasts to myotubes, while it is barely expressed in undifferentiated cells (13). Taken together, these results suggest that FGFRL1 has a negative effect on cell proliferation and a positive effect on cell differentiation. Although the hypothesis of the decoy receptor is plausible and straightforward, it does not explain the existence of the relatively long intracellular domain with the peculiar histidine-rich sequence and several tyrosine motifs.
More information about the potential functions of FGFRL1 can be learned from experiments, in which its expression is specifically suppressed. Knock-down experiments with morpholino constructs in a Zebrafish model indicate that FGFRL1 is involved in gill cartilage development (14). Animals that have been injected with such morpholino constructs fail to properly form the pharyngeal arches. Our group recently demonstrated that mice with a targeted disruption of the FGFRL1 gene develop normally to term, but die immediately after birth due to severe respiratory distress (13). The respiratory problems are explained by the malformation of the diaphragm, which is not strong enough to inflate the lungs after birth. The knock-out animals also exhibit subtle bone alterations such as a dome-shaped head with a high front reminiscent of many human craniosynostosis syndromes (12). Another research group has generated similar FGFRL1 deficient mice and found alterations in the heart, especially in the ventricular valves, in addition to alterations in the diaphragm and the skull (15). The involvement of FGFRL1 in the formation of the skull was recently confirmed by the identification of the first human FGFRL1 mutation in a craniosynostosis patient (12). This patient displayed a 4 bp insertion in the last exon of the FGFRL1 gene that disrupted the reading frame of the intracellular domain. In contrast to wild-type protein, which was rapidly removed from the cell membrane and sorted to lysosomes, the mutant protein appeared to stay for a prolonged time at the plasma membrane where it interacted with FGF ligands (12).
The overall structure of FGFRL1 with its three Ig-like domains and the transmembrane segment is not unique. It is needless to say that Ig-like domains occur in all immunoglobulins but they are also found in a variety of other molecules, including cell adhesion proteins, cell surface receptors and muscle proteins (16,17). All Ig-like domains share a common core β-sandwich structure. According to sequence pattern and overall length, the Ig-like domains can be grouped into four sets: V (variable), C1 (constant-1), C2 (constant-2) and I (intermediate).
Here we have used a comprehensive bioinformatics approach to compare the domain structure of FGFRL1 with all proteins of the UniProt Databank. We found that the human genome encodes at least 42 molecules that exhibit a related domain structure with three extracellular Ig-like domains and a single transmembrane domain. Five of these molecules are involved in FGF signaling, 9 are involved in cell-cell contact at adherens junctions and 25 are involved in the control and modulation of the immune system. A detailed analysis of these proteins may yield valuable clues about a putative signaling mechanism utilized by FGFRL1.
Methods
Sequence searches were performed on the Vital-IT platform of the Swiss Institute of Bioinformatics (http://www.vital-it.ch) using the HitKeeper query language (18) and the MyHits interface (19). Pre-calculated hit lists on the human entries in the UniProtKB database (20) were queried for the presence of various Ig-domain descriptors originating from Pfam (21) (PF00047, PF07686, PF08205), SMART (22) (SM00409, SM00408), PROSITE (23) (PS50835) and Swiss-Prot features (20). Only proteins displaying the presence of exactly 3 Ig-like domains were kept for further analysis. The presence of transmembrane regions within these proteins was assessed using the transmembrane predictor software Phobius (24). The extracellular domains of the extracted proteins were compared pairwise with the extracellular domain of FGFRL1 using the program Gap and the scoring matrix Blosum62 (GCG, Accelrys Software Inc, Cambridge, UK). To enforce alignment over the entire length of the two sequences, the gap shift limits were set to 20.
Results and Discussion
Seven protein families with a related domain structure
To identify proteins with a domain structure similar to that of FGFRL1, we screened the UniProt databank for entries with three Ig-like domains and a single transmembrane domain. The major problem of this approach was the fact that no well defined domain descriptor for ‘Ig-like’ existed. In the first round of databank screening we therefore included all human proteins with domains conforming to at least one of the following descriptors, prf:IG_LIKE; iprsmart:SM00409.IG; iprpfam: PF00047.ig; iprpfam:PF07686.V-set; iprpfam:PF08205.C2-set_2; iprsmart:SM00408.IGc2; ft:DOMAIN_Ig-like_V; ft:DOMAIN_Ig-like_C2-type. Proteins with four or more Ig-like domains were then eliminated. According to this approach UniProt comprised a total of 3350 human proteins with 3 or less Ig-like domains. Of these, 52 had exactly 3 Ig-like domains and at least 1 transmembrane domain, 219 had exactly 3 Ig-like domains but no annotated transmembrane domain. The latter proteins were passed through a transmembrane predictor program (Phobius), which divided them into 140 proteins with a potential transmembrane domain and 79 proteins without. The remaining 192 proteins with 3 Ig-like domains and at least 1 transmembrane domain were manually inspected. Double hits, fragmented proteins as well as proteins with extracellular domains in addition to the 3 Ig-like domains were eliminated. This procedure yielded a list of 42 proteins that possessed exactly 3 Ig-like domains, a single transmembrane helix and an intracellular domain (Table I).
Table I.
Proteins encoded by the human genome that display a domain structure related to FGFRL1.
| Gene symbol | Common name | Accession number | Order of domains | Extracellular residues (Total amino acids) | Sequence identity |
|---|---|---|---|---|---|
| FGFRL1 | FGF receptor -like 1 (FGFR5) | Q8N441 | C2, C2, C2, TM, His | 25–378 (504) | 100 |
| FGFR1 | FGF receptor 1 | P11362 | C2, C2, C2, TM, kinase | 22–376 (822) | 32 |
| FGFR2 | FGF receptor 2 | P21802 | C2, C2, C2, TM, kinase | 22–377 (821) | 33 |
| FGFR3 | FGF receptor 3 | P22607 | C2, C2, C2, TM, kinase | 23–375 (806) | 35 |
| FGFR4 | FGF receptor 4 | P22455 | C2, C2, C2, TM, kinase | 22–369 (802) | 34 |
| FCGR1A | Fcγ receptor 1 | P12314 | C2, C2, C2, TM | 16–292 (374) | 18 |
| FCRL1 | Fc receptor -like 1 | Q96LA6 | C2, C2, C2, TM, ITAM | 17–307 (429) | 24 |
| FCRL6 | Fc receptor -like 6 | Q6DN72 | C2, C2, C2, TM, ITIM | 20–307 (434) | 22 |
| IL-1R1 | IL-1 receptor 1 | P14778 | C2, C2, C2, TM, TIR | 18–336 (569) | 22 |
| IL-1R2 | IL-1 receptor 2 | P27930 | C2, C2, C2, TM | 14–343 (398) | 20 |
| IL-18R1 | IL-18 receptor | Q13478 | C2, C2, C2, TM, TIR | 19–329 (541) | 18 |
| IL-1RL1 | IL-1 receptor -like 1 | Q01638 | C2, C2, C2, TM, TIR | 19–328 (556) | 19 |
| IL-1RL2 | IL-1 receptor -like 2 | Q9HB29 | C2, C2, C2, TM, TIR | 20–335 (575) | 19 |
| IL-1RAP | IL-1 receptor accessory protein | Q9NPH3 | C2, C2, C2, TM, TIR | 21–367 (570) | 23 |
| IL-18RAP | IL-18 receptor accessory protein | O95256 | C2, C2, C2, TM, TIR | 20–356 (599) | 21 |
| IL-1RAPL1 | IL-1 receptor accessory protein-like 1 | Q9NZN1 | C2, C2, C2, TM, TIR | 19–357 (696) | 21 |
| IL-1RAPL2 | IL-1 receptor accessory protein-like 2 | Q9NP60 | C2, C2, C2, TM, TIR | 17–354 (686) | 20 |
| KIR3DL1 | Killer cell Ig-like receptor 3DL1 | P43629 | C2, C2, C2, TM, ITIM | 22–340 (444) | 20 |
| KIR3DL2 | Killer cell Ig-like receptor 3DL2 | P43630 | C2, C2, C2, TM, ITIM | 22–340 (455) | 21 |
| KIR3DL3 | Killer cell Ig-like receptor 3DL3 | Q8N743 | C2, C2, C2, TM, ITIM | 26–322 (410) | 22 |
| KIR3DS1 | Killer cell Ig-like receptor 3DS1 | Q14943 | C2, C2, C2, TM | 22–340 (387) | 21 |
| PVR | Poliovirus receptor (nectin-like 5) | P15151 | V, C2, C2, TM | 21–343 (417) | 25 |
| PVRL1 | Poliovirus receptor-related P1 (nectin 1) | Q15223 | V, C2, C2, TM | 31–355 (517) | 19 |
| PVRL2 | Poliovirus receptor-related P2 (nectin 2) | Q92692 | V, C2, C2, TM | 32–360 (538) | 20 |
| PVRL3 | Poliovirus receptor-related P3 (nectin 3) | Q9NQS3 | V, C2, C2, TM | 58–404 (549) | 26 |
| PVRL4 | Poliovirus receptor-related P4 (nectin 4) | Q96NY8 | V, C2, C2, TM | 32–349 (510) | 22 |
| CADM1 | Cell adhesion molecule 1 (nectin-like 2) | Q9BY67 | V, C2, C2, TM | 45–374 (442) | 21 |
| CADM2 | Cell adhesion molecule 2 (nectin-like 3) | Q8N3J6 | V, C2, C2, TM | 25–367 (435) | 21 |
| CADM3 | Cell adhesion molecule 3 (nectin-like 1) | Q8N126 | V, C2, C2, TM | 25–330 (398) | 24 |
| CADM4 | Cell adhesion molecule 4 (nectin-like 4) | Q8NFZ8 | V, C2, C2, TM | 21–324 (388) | 21 |
| SIGLEC6 | Sialic acid-binding Ig-like lectin 6 | O43699 | V, C2, C2, TM, ITIM | 27–347 (453) | 20 |
| SIGLEC7 | Sialic acid-binding Ig-like lectin 7 | Q9Y286 | V, C2, C2, TM, ITIM | 19–353 (467) | 23 |
| SIGLEC8 | Sialic acid-binding Ig-like lectin 8 | Q9NYZ4 | V, C2, C2, TM, ITIM | 17–363 (499) | 24 |
| SIGLEC9 | Sialic acid-binding Ig-like lectin 9 | Q9Y336 | V, C2, C2, TM, ITIM | 18–348 (463) | 22 |
| SIGLEC14 | Sialic acid-binding Ig-like lectin 14 | Q08ET2 | V, C2, C2, TM | 17–358 (396) | 20 |
| SIRPA | Tyrosine phosphatase substrate 1 | P78324 | V, C1, C1, TM, ITIM | 27–372 (503) | 20 |
| SIRPB1 | Signal-regulatory protein β1 | O00241 | V, C1, C1, TM | 27–371 (398) | 20 |
| SIRPG | Signal-regulatory protein γ | Q9P1W8 | V, C1, C1, TM | 29–360 (387) | 22 |
| AGER | Advanced glycosylation end product receptor | Q15109 | V, C2, C2, TM | 23–342 (404) | 24 |
| BSG | Basigin | P35613 | (C2), C2, V, TM | 22–323 (385) | 21 |
| CD96 | T-cell surface protein tactile | P40200 | V, V, C2, TM | 22–519 (585) | 21 |
| HHLA2 | HERV-H LTR-associating protein 2 | Q9UM44 | V, C1, V, TM | 23–344 (414) | 24 |
His, histidine-rich; V, variable; C1, constant-1; C2, constant-2; TIR, Toll/IL-1 receptor; ITIM, immunoreceptor tyrosine-based inhibition motif; ITAM, immunoreceptor tyrosine-based activation and TM, transmembrane domain.
It is obvious that this list might still not be complete. There are several entries in UniProt that are ill-defined, mainly entries that were automatically derived from TREMBL. Moreover, there are proteins with four or more Ig-like domains that may also exist in an alternatively spliced form with three Ig-like domains. Furthermore, some generally accepted Ig-like domains might not be picked up correctly by the Ig-domain descriptors mentioned above.
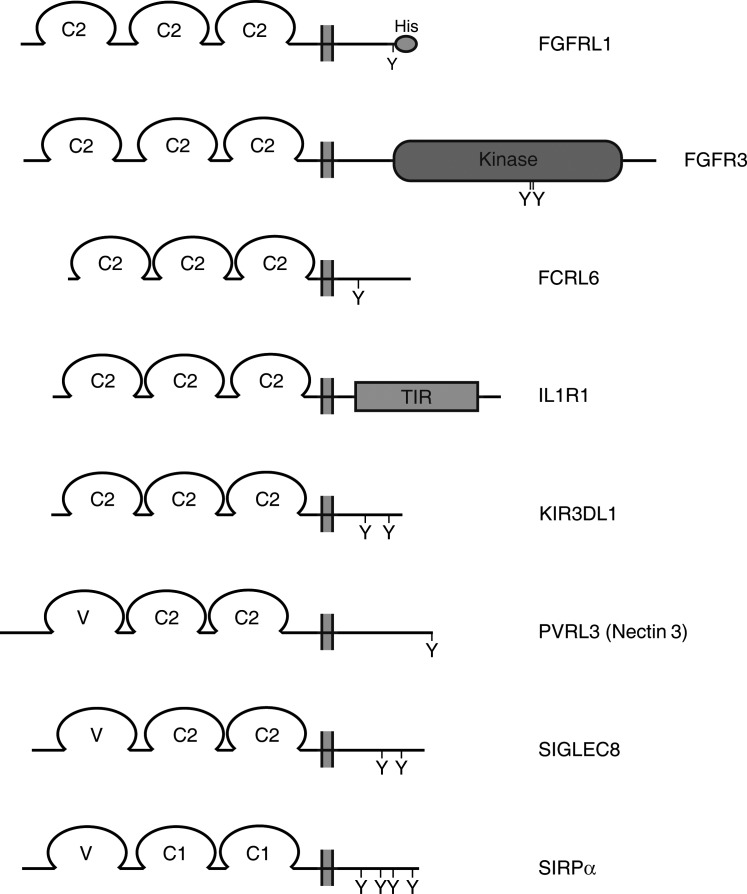
The 42 proteins from Table I fall into 7 protein families: FGF receptors, Fc receptor-like proteins, IL-1 receptor-like proteins, natural killer cell Ig-like receptors, nectin-like proteins, sialic acid binding Ig-like lectins and signal-regulatory proteins. By definition, all these proteins contain a cleavable signal peptide, followed by three Ig-like domains and a transmembrane domain (Fig. 1). Consequently, the size of the extracellular domain of these proteins varies between 300–350 amino acid residues. When compared with the extracellular domain of FGFRL1, this region shares 18–26% sequence identity with the exception of the extracellular domains from FGFRs, which display 32–35% sequence identity (Table I). In the latter case, the similarity increases to 39–42% if conservative amino acid replacements are included, underlining the previous conclusion that FGFRL1 is most closely related to the FGFRs. The size of the intracellular domain varies between 4 (SIRPγ) and 425 (FGFR1) amino acid residues. For down-stream signaling, a protein must utilize this domain in order to trigger a certain pathway. This domain may therefore provide valuable clues about a putative signaling mechanism utilized by FGFRL1.
Figure 1.
Domain structure of transmembrane proteins that display an overall structure similar to FGFRL1. Three Ig-like domains are indicated by their affiliation to a particular set (C1, C2 and V). The intracellular domains are labeled (His, histidine-rich; kinase, protein tyrosine kinase and TIR, Toll/IL-1 receptor domain). The position of tyrosine residues involved in downstream signaling is marked by Y. One representative example from each protein family is listed in alphabetical order, with FGFRL1 and FGFR3 being shown at the top.
In the following, we will briefly summarize the major properties and signaling mechanisms of the different protein families. The FGFR family has already been described in the introduction in some detail. For a comprehensive survey of an individual protein, the reader is referred to more detailed reviews of a particular family.
Fc Receptor-like molecules
Fc receptors are molecules found on the surface of various cells from the immune system, including macrophages, neutrophils, mast cells and natural killer cells (25). They bind to the Fc region of antibodies that are attached to invading pathogens or infected cells. There are Fc receptors for all types of antibodies but only the Fcγ receptor I resembles FGFRL1 with its 3 Ig-like domains. Besides the classical Fc receptors, there are six Fc receptor-like molecules that occur preferentially on the surface of B cells. Two of these proteins possess 3 Ig-like domains (FCRL1 and FCRL6) and are therefore included in Table I. So far there is no direct evidence to support a role for the Fc receptor-like molecules as Ig-binding receptors. However, these molecules contain cytoplasmic tails that harbor immunoreceptor tyrosine-based activation (ITAM) and inhibition (ITIM) motifs. Through these motifs they deliver activating or inhibiting signals to the interior of the cells (see below), suggesting that these receptors play a role in regulating activation and differentiation of B cells and other cells. Fcγ receptor I does not have an ITAM but it still transmits activating signals to the interior of the cells by interacting with another protein that does possess an ITAM.
The interleukin-1 receptor family
Inflammatory cytokines such as IL-1 and IL-18 mediate their effects through specific transmembrane receptors present on the surface of target cells (26,27). These receptors possess three Ig-like domains in their extracellular part and a signaling domain termed TIR (Toll/IL-1 receptor) in their intracellular part. Altogether, there are nine related IL-1 receptor-like molecules. The receptor for IL-1 is a dimer consisting of IL-1R1 and IL-1RAP (IL-1 receptor accessory protein), the receptor for IL-18 a dimer of IL-18R and IL-18RAP. Other cytokines such as IL-33 and IL-1F6 bind to dimeric receptors consisting of various combinations of the other chains listed in Table I. Upon ligand binding cytosolic adaptor proteins such as MyD88 are recruited to the intracellular TIR domain of the receptors and these adaptor proteins trigger down-stream signaling by phosphorylation. IL-1 receptor II (IL1R2) is a negatively acting receptor that lacks the intracellular TIR domain. It still forms dimers with IL1R1 but does not take part in signaling. Since it can bind IL-1 it appears to act as a decoy receptor.
Killer cell Ig-like receptors (KIRs)
KIRs are transmembrane proteins expressed by natural killer cells and some T cells (28,29). The genes for the KIRs are found in a cluster on human chromosome 19q13.4 within the leukocyte receptor complex. The KIR proteins are classified by the number of Ig-like domains (2D or 3D) and by the presence of a long (L) or a short (S) cytoplasmic domain. KIR proteins with the longer cytoplasmic domain elicit inhibitory signals upon ligand binding via the ITIM motif introduced above. KIR proteins with the shorter cytoplasmic domain lack the ITIM motif. Instead they associate with the tyrosine kinase binding protein TYRO to transduce activating signals to the cells. The ligands for KIR proteins are members of the HLA class I antigens. KIR proteins are therefore thought to play an important role in the regulation of the immune response. Besides the genes for three KIR3DL proteins and one KIR3DS protein, there are two KIR3D genes (KIR3DP1, KIR3DX1) not listed here that may represent pseudogenes.
Nectins and nectin-like proteins
Nectins (nectin 1–4) and nectin-like proteins (nectin-like 1–5) constitute a family of 9 cell surface proteins that are involved in cell adhesion, migration and proliferation (30,31). The nectins form homodimers on the plasma membrane of various cell types and interact in trans with nectins and nectin-like proteins expressed on neighboring cells. These interactions result in calcium-independent cell adhesion at cell-cell adherens junctions and lead to the reorganization of the actin cytoskeleton. For the interaction with the cytoskeleton, the nectins bind with their intracellular, C-terminal end to afadin. Afadin in turn interacts with filamentous actin. Nectin-like protein 5 has a unique additional function at the leading edge of migrating cells where it interacts with integrin αvβ3 to facilitate cell movement. Some nectins are important ways of entry for viruses into cells. Nectin-1 is the receptor for herpes virus, nectin-like 5 is the receptor for poliovirus.
Sialic acid binding Ig-like lectins (SIGLECs)
SIGLECs are cell surface receptors that bind carbohydrates, especially sialic acid from glycoproteins of other cells, but also from viruses and bacteria (32,33). The SIGLECs are primarily, but not exclusively, expressed by hematopoietic cells. They contain one amino-terminal V-set Ig-like domain that interacts with the carbohydrates and a variable number of C2-set Ig-like domains. The human genome contains 14 different SIGLEC genes (SIGLEC1-15, SIGLEC13 is found in chimpanzee and baboon, but not in humans) and a similar number of SIGLEC pseudogenes. Only 5 SIGLECs are listed in Table I because only these proteins contain exactly 3 Ig-like domains. Most SIGLECs possess one or more copies of the ITIM element in their cytoplasmic domain. This motif is phosphorylated by cytoplasmic tyrosine kinases and plays a key role in cell signaling (see below). Consequently, the SIGLECs are believed to have a function in the activation of cells from the immune system.
Signal-regulatory proteins (SIRPs)
The SIRPs form a family of related proteins that are mainly expressed on the surface of myeloid cells (34). There are three family members, SIRPα, SIRPβ and SIRPγ. In the human population, the genes for the SIRPs appear to be highly polymorph. The cytoplasmic domain of SIRPα contains four tyrosine residues that conform loosely to the ITIM motif and can become phosphorylated. The phosphorylated motifs associate with tyrosine phosphatases and induce inhibitory functions in cell signaling. SIRPβ has a very short cytoplasmic region of 6 amino acids and lacks the ITIM motif. Instead it contains a lysine residue that interacts with a protein called DAP12 which in turn transmits activating signals. Because the two proteins elicit opposing functions, they are also referred to as paired receptors. SIRPγ has only 4 amino acid residues in its cytoplasmic domain and does not appear to transmit any signals.
Other proteins
The remaining four proteins do not appear to belong to any of the above mentioned protein families. AGER is a receptor for advanced glycosylation end products, which may occur by non-enzymatic glucosidation of α- and ε-amino groups with blood sugars (35). Owing to its extracellular Ig-like domains (V, C2, C2) and its sugar binding properties, it resembles the SIGLECs. Basigin (CD147) is thought to have a function in intercellular recognition (36). It interacts with many different ligands, including certain cyclophilins and integrins. CD96 (tactile) plays a role in the adhesive interactions of T-cells and natural killer cells during the late phase of the immune response (37). It may also be involved in antigen presentation. Finally, HHLA2 has been identified by virtue of several EST and cDNA clones (38). Its gene is associated with the human endogenous retrovirus H family long terminal repeat (HERV-H LTR).
ITIM/ITAM containing receptors
When the functions and signaling mechanisms of the seven protein families are compared it becomes evident that at least some members of each family are equipped with intracellular domains or motifs that are directly or indirectly involved in down-stream signaling (Fig. 1). The classical FGFRs harbor a well-defined tyrosine kinase domain of approximately 300 amino acid residues that phosphorylates strategic tyrosine residues in the C-terminal domain of an adjacent receptor. FGFRL1 is clearly lacking such a tyrosine kinase domain. On the other hand, the IL-1 receptor I contains an intracellular TIR domain of approximately 200 amino acid residues that interacts with a multifunctional adaptor protein (MyD88), which in turn recruits signaling molecules including tyrosine kinases to the plasma membrane. The intracellular domain of FGFRL1 is shorter than that of IL-1 receptor I and does not display any conserved, well-defined domain like TIR. One will therefore have to focus on conserved, short sequence motifs rather than extended domains. In fact, such motifs are found in many of the seven protein families, namely in the Fc receptor-like proteins, the KIRs, the SIGLECs and the SIRPs. Members of these families harbor specific sequence motifs termed ITAM (immunoreceptor tyrosine-based activation motifs) and ITIM (immunoreceptor tyrosine-based inhibition motifs) that are involved in down-stream signaling.
The ITAM is a tandem motif that conforms to the amino acid sequence YxxL/I(x)6–12YxxL/I, whereas the ITIM is a single motif that conforms to the sequence S/I/V/LxYxxI/V/L (39–41). After ligand binding, ITAM-bearing receptors become phosphorylated by Src family kinases. The phosphorylated tyrosines serve subsequently as docking sites for the recruitment of other tyrosine kinases such as Syk or ZAP-70, resulting in the activation of several down-stream signaling pathways. ITIM-bearing receptors can also become phosphorylated after ligand binding by Src family kinases but they elicit opposing effects. The phosphorylated ITIMs bind SH2-containg phosphatases such as SHP-1 (phosphotyrosine phosphatase-1), SHP-2 and SHIP (inositol phosphatase). These enzymes can dephosphorylate the cytoplasmic domains of adjacent receptors, thereby inhibiting activation of the cells.
The ITAM/ITIM signaling motifs reveal some similarity to a tandem tyrosin-based motif occurring in the FGFRL1 receptor, PKLYPKLYTDI. We have demonstrated that this tyrosine based motif is responsible for intracellular sorting of FGFRL1 (12). When the tyrosines were changed to alanine by in vitro mutagenesis, the mutated protein stayed for a prolonged time at the cell membrane, whereas the wild-type protein was rapidly removed from the cell membrane and sorted to lysosomes. It is intriguing to speculate that the same tandem tyrosine-based motif could also become phosphorylated by an intracellular kinase. The phosphorylated motif would then bind SH2 containing phosphatases, which in turn could down-regulate the activity of adjacent FGFRs.
We have tried to check this possibility. Phosphorylation of FGFRL1 was investigated in cells, before and after stimulation by FGF2 or EGF, with pan-phosphotyrosine specific antibodies (Steinberg and Trueb, unpublished observation). However, we have not been able to detect any phosphorylation of FGFRL1. It is therefore unlikely that FGFRL1 is involved in a signaling pathway with SH2 domain containing adaptor proteins that would bind to its C-terminal end. Nevertheless, this possibility should be kept in mind when the signaling mechanism of the novel receptor is investigated.
Reorganization of the cytoskeleton
If FGFRL1 has no tyrosine kinase domain similar to the FGFRs, no TIR domain similar to the IL-1 receptor and no ITAM/ITIM signaling motif similar to many other receptors of the immune system, how might it participate in down-stream signaling? One group of proteins that still remains for comparison are the nectin and nectin-like proteins. In fact, with 26% sequence identity (33% sequence similarity if conservative amino acid substitutions are included), nectin-3 displays, after the classical FGFRs, the best similarity to FGFRL1 of the 42 proteins listed in Table I. Like the nectins, FGFRL1 forms homodimers on the cell membrane as demonstrated by FRET measurements and by immunoprecipitation experiments (11). Furthermore, FGFRL1 occurs preferentially at cell-cell contact sites similar to the nectins.
Nectins are involved in the initial steps of the formation of intercellular adherens junctions (30,31). For this purpose, they interact with a variety of peripheral membrane proteins including the actin filament-binding protein afadin and the cell polarity protein Par-3. These interactions are achieved via binding of the carboxy-terminal end of the nectins (amino acid residues E/A-X-Y-V) with the PDZ domains of afadin and Par-3. Afadin in turn can bind to F-actin and link the nectins in this way to the cytoskeleton. In addition to its PDZ and actin-binding domains, afadin has other interaction domains, such as two Ras association domains, a forkhead-associated domain, a DIL domain and three proline-rich domains. Likewise, Par-3 possesses three PDZ domains and two CR domains. By virtue of interaction with such versatile adaptor proteins, the nectins can indirectly induce the activation of several intracellular signaling molecules, including Rap1, Cdc42 and Rac, and trigger the reorganization of the cytoskeleton.
It is of interest to note that FGFRL1 exhibits a C-terminal sequence with a tyrosine residue (H-Y-Q-C) that is somewhat related to that of the nectins. Together with the histidine-rich sequence, this motif is the only element of the intracellular domain that is conserved among different species, from mammals to fish and lancelet. We do not know whether this C-terminal sequence can indeed be recognized by a specific PDZ domain. On the other hand, PDZ recognition motifs that interact with similar C-terminal sequences have been described in the literature, such as the PDZ domain of the adaptor protein Mint1 that recognizes the C-terminal sequence D-H-W-C of a calcium channel (42,43). It remains to be determined whether cells express an adaptor protein with a PDZ domain that specifically interacts with the C-terminus of FGFRL1. Such a protein might anchor FGFRL1 to the cytoskeleton or to another intracellular protein complex.
Conclusions
The domain structure of FGFRL1 with its 3 Ig-like domains and the transmembrane segment is not unique but occurs in more than 40 human proteins, which can be grouped in 7 families. Most of these proteins play a role in the immune system, but there are also some that serve an important function in cell-cell adhesion at adherens junctions. A great deal of these proteins forms homodimers on cell surfaces and acts as some kind of receptors. The ligands for these receptors are either cytokines, antibodies, carbohydrate chains or cell surface molecules. A structure with three Ig-like domains appears to be particularly well suited for recognition and interaction. In the adaptive immune system, this structure is employed to recognize a nearly unlimited number of antigens.
FGFRL1 shares many similarities with the prototype of such a 3 Ig-domain receptor. It also forms constitutive dimers on the cell surface and it has the ability to bind heparin chains and FGF ligands. During evolution, the other Ig-domain proteins have adapted various possibilities to signal their ligand binding state to the interior of the cells. For this purpose, some make use of strategic tyrosine motifs that become phosphorylated and provide binding sites for SH2 domain containing adaptor proteins, or alternatively some interact through their C-terminal end with a PDZ domain protein, which in turn interacts with the cytoskeleton. FGFRL1 displays some parallels in this respect. It possesses several tyrosine motifs in its C-terminal domain that could theoretically become phosphorylated upon ligand binding or that might interact with a PDZ domain protein which links it to the cytoskeleton. To date there is no evidence for the first possibility. We therefore favor the second possibility of an interaction with a PDZ domain protein. In this way, FGFRL1 might be able to signal information from the outside of the cell to the interior although it is lacking the tyrosine kinase domain typically associated with the classical FGFRs. Thus, the genome-wide comparison of FGFRL1 with structurally related proteins from the UniProt databank has opened new avenues for research that can now be tackled experimentally.
Acknowledgments
This study was supported by grants from the Swiss National Science Foundation (3100A0-113806), the Swiss Foundation for Research on Muscular Diseases and the Olga Mayenfisch Foundation.
References
- 1.Itoh N. The Fgf families in humans, mice, and zebrafish: their evolutional processes and roles in development, metabolism, and disease. Biol Pharm Bull. 2007;30:1819–1825. doi: 10.1248/bpb.30.1819. [DOI] [PubMed] [Google Scholar]
- 2.Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Wilkie AO. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 2005;16:187–203. doi: 10.1016/j.cytogfr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Coumoul X, Deng CX. Roles of FGF receptors in mammalian development and congenital diseases. Birth Defects Res C Embryo Today. 2003;69:286–304. doi: 10.1002/bdrc.10025. [DOI] [PubMed] [Google Scholar]
- 6.Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- 7.Sleeman M, Fraser J, McDonald M, Yuan S, White D, Grandison P, Kumble K, Watson JD, Murison JG. Identification of a new fibroblast growth factor receptor, FGFR5. Gene. 2001;271:171–182. doi: 10.1016/s0378-1119(01)00518-2. [DOI] [PubMed] [Google Scholar]
- 8.Kim I, Moon S-O, Yu K-H, Kim U-H, Koh GY. A novel fibroblast growth factor receptor-5 preferentially expressed in the pancreas. Biochim Biophys Acta. 2001;1518:152–156. doi: 10.1016/s0167-4781(00)00282-7. [DOI] [PubMed] [Google Scholar]
- 9.Trueb B, Zhuang L, Taeschler S, Wiedemann M. Characterization of FGFRL1, a novel FGF receptor preferentially expressed in cartilage. J Biol Chem. 2003;278:33857–33865. doi: 10.1074/jbc.M300281200. [DOI] [PubMed] [Google Scholar]
- 10.Trueb B, Taeschler S. Expression of FGFRL1, a novel fibroblast growth factor receptor, during embryonic development. Int J Mol Med. 2006;17:617–620. [PubMed] [Google Scholar]
- 11.Rieckmann T, Kotevic I, Trueb B. The cell surface receptor FGFRL1 forms constitutive dimers that promote cell adhesion. Exp Cell Res. 2008;314:1071–1081. doi: 10.1016/j.yexcr.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 12.Rieckmann T, Zhuang L, Flück CE, Trueb B. Characterization of the first FGFRL1 mutation identified in a craniosynostosis patient. Biochim Biophys Acta. 2009;1792:112–121. doi: 10.1016/j.bbadis.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Baertschi S, Zhuang L, Trueb B. Mice with a targeted disruption of the Fgfrl1 gene die at birth due to alterations in the diaphragm. FEBS J. 2007;274:6241–6253. doi: 10.1111/j.1742-4658.2007.06143.x. [DOI] [PubMed] [Google Scholar]
- 14.Hall C, Flores MV, Murison G, Crosier K, Crosier P. An essential role for zebrafish Fgfrl1 during gill cartilage development. Mech Dev. 2006;123:925–940. doi: 10.1016/j.mod.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, Te Welscher P. Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis Model Mech. 2009;2:283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barclay AN. Ig-like domains: evolution from simple interaction molecules to sophisticated antigen recognition. Proc Natl Acad Sci USA. 1999;96:14672–14674. doi: 10.1073/pnas.96.26.14672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith DK, Xue H. Sequence profiles of immunoglobulin and immunoglobulin-like domains. J Mol Biol. 1997;12:530–545. doi: 10.1006/jmbi.1997.1432. [DOI] [PubMed] [Google Scholar]
- 18.Hau J, Muller M, Pagni M. HitKeeper, a generic software package for hit list management. Source Code Biol Med. 2007;28:2:2. doi: 10.1186/1751-0473-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:W433–W437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJA. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Davis RS. Fc receptor-like molecules. Annu Rev Immunol. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- 26.O’Neill LA. The interleukin-1 receptor/Toll-like receptor super-family: 10 years of progress. Immunol Rev. 2008;226:10–18. doi: 10.1111/j.1600-065X.2008.00701.x. [DOI] [PubMed] [Google Scholar]
- 27.Boraschi D, Tagliabue A. The interleukin-1 receptor family. Vitam Horm. 2006;74:229–254. doi: 10.1016/S0083-6729(06)74009-2. [DOI] [PubMed] [Google Scholar]
- 28.Gardiner CM. Killer cell immunoglobulin-like receptors on NK cells: the how, where and why. Int J Immunogenet. 2008;35:1–8. doi: 10.1111/j.1744-313X.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- 29.Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyoshi J, Takai Y. Nectin and nectin-like molecules: biology and pathology. Am J Nephrol. 2007;27:590–604. doi: 10.1159/000108103. [DOI] [PubMed] [Google Scholar]
- 31.Rikitake Y, Takai Y. Interactions of the cell adhesion molecule nectin with transmembrane and peripheral membrane proteins for pleiotropic functions. Cell Mol Life Sci. 2008;65:253–263. doi: 10.1007/s00018-007-7290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crocker PR, Redelinghuys P. Siglecs as positive and negative regulators of the immune system. Biochem Soc Trans. 2008;36:1467–1471. doi: 10.1042/BST0361467. [DOI] [PubMed] [Google Scholar]
- 33.Von Gunten S, Bochner BS. Basic and clinical immunology of Siglecs. Ann N Y Acad Sci. 2008;1143:61–82. doi: 10.1196/annals.1443.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. 2006;6:457–464. doi: 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- 35.Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- 36.Yurchenko V, Constant S, Bukrinsky M. Dealing with the family: CD147 interactions with cyclophilins. Immunology. 2006;117:301–309. doi: 10.1111/j.1365-2567.2005.02316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155) J Immunol. 2004;172:3994–3998. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- 38.Mager DL, Hunter DG, Schertzer M, Freeman JD. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3) Genomics. 1999;59:255–263. doi: 10.1006/geno.1999.5877. [DOI] [PubMed] [Google Scholar]
- 39.Barrow AD, Trowsdale J. You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur J Immunol. 2006;36:1646–1653. doi: 10.1002/eji.200636195. [DOI] [PubMed] [Google Scholar]
- 40.Daeron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- 41.Pinheiro da Silva F, Aloulou M, Benhamou M, Monteiro RC. Inhibitory ITAMs: a matter of life and death. Trends Immunol. 2008;29:366–373. doi: 10.1016/j.it.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Bezprozvanny I, Maximov A. Classification of PDZ domains. FEBS Lett. 2001;509:457–462. doi: 10.1016/s0014-5793(01)03214-8. [DOI] [PubMed] [Google Scholar]
- 43.Maximov A, Sudhof TC, Bezprozvanny I. Association of neuronal calcium channels with modular adaptor proteins. J Biol Chem. 1999;274:24453–24456. doi: 10.1074/jbc.274.35.24453. [DOI] [PubMed] [Google Scholar]