Summary
Inherited MC1R variants modulate MITF transcription factor signaling, which in turn affects tumor cell proliferation, apoptosis, and DNA repair. The aim of this BioGenoMEL collaborative study in 10 melanoma cohorts was to test the hypothesis that inherited variants thereby moderate survival expectation. A survival analysis in the largest cohort (Leeds) was carried out adjusting for factors known to impact on survival. The results were then compared with data from nine smaller cohorts. The absence of any consensus MC1R alleles was associated with a significantly lower risk of death in the Leeds set (HR, 0.64; 95% CI, 0.46–0.89) and overall in the 10 data sets (HR, 0.78; 95% CI, 0.65–0.94) with some support from the nine smaller data sets considered together (HR, 0.83; 95% CI, 0.67–1.04). The data are suggestive of a survival benefit for inherited MC1R variants in melanoma patients.
Keywords: MC1R, survival analysis, MITF, melanoma, forest plot
Significance
AJCC staging is of strong prognostic value for melanoma patients but only explains a proportion of the variance in survival. In some patients, there is evidence of a host immune response to the tumor both histologically and in the clinical manifestation of vitiligo, so that host/tumor interaction is postulated to be an additional factor which modifies prognosis. It is also likely that interaction between stromal tissues and cancer cells is important, genetically determined, and potentially modifiable. The new consortium BioGenoMEL seeks to bring together distinct patient cohorts to identify genes impacting on host/tumor interaction and therefore outcome, thereby improving our understanding of key biological pathways. This article is the first generated by BioGenoMEL as a consortium pooling data across multiple cohorts and provides evidence for a role for inherited MC1R variants in survival.
Introduction
Cutaneous melanoma is predominantly a cancer of white-skinned peoples, and those at increased risk include the very pale skinned (Gandini et al., 2005b), those with many melanocytic nevi (Gandini et al., 2005a), and those with a family history of melanoma (Gandini et al., 2005b). Although both fair hair and blue eyes are associated with increased susceptibility (Gudbjartsson et al., 2008), the pigmentary phenotypes most strongly associated are freckling, red hair, and a tendency to burn in the sun. These latter phenotypes are related to inherited variation in the gene coding for the melanocortin 1 receptor (MC1R) (Bishop et al., 2009) and the agouti locus (ASIP) (Brown et al., 2008; Gudbjartsson et al., 2008). The MC1R signals through a key pathway within melanocytes via the microphthalmia-associated transcription factor (MITF) to result in pigmentary changes: the default pigmentation is yellow/red (pheomelanin) (Beaumont et al., 2008), and signaling results in more black/brown pigment (eumelanin) synthesis. The agouti protein blocks this signaling, resulting in the default production of yellow/red pigment. It was recognized many years ago that some inherited variants in the MC1R gene are associated with red hair, and these have been classified by Duffy et al. (2004) into ‘R’ variants and ‘r’ variants, strongly and weakly associated with red hair, respectively.
Inherited MC1R variants are thought to increase melanoma risk as a result of consequent relative lack of eumelanin, but it is postulated that there are additional non-pigmentary effects (Robinson et al., 2010). The MITF transcription factor, expression of which is regulated by signaling through MC1R, has many target genes in addition to pigment biosynthesis enzymes, including genes that regulate DNA repair (APEX1) (Liu et al., 2009), the cell cycle (CDKN2A, CDK2) (Du et al., 2004; Loercher et al., 2005), apoptosis (BCL2) (McGill et al., 2002), and invasion (DIA1) (Carreira et al., 2006). The DNA repair gene apex nuclease 1, also known as apurinic endonuclease APEX1), is important in DNA repair responses to reactive oxygen species (ROS) and oxidative DNA base damage (Robinson et al., 2010). Kadekaro et al. (2010) showed that human melanocytes with two red hair color–associated MC1R alleles were resistant to α-melanocortin (α-MSH)-mediated DNA repair. The same group had earlier shown that MC1R activation mediated reduced oxidative DNA damage in melanocytes when exposed to UV radiation (Song et al., 2009).
The effect of MC1R variants on DNA repair and apoptosis may contribute to susceptibility but our hypothesis is that there may be additional effects on survival. There are published data to support this view. Overexpression of DNA repair pathways in melanoma has already been reported to be associated with metastasis and poor patient survival (Jewell et al., 2010; Winnepenninckx et al., 2006). This finding has led to the hypothesis that genetic stability conferred by high expression of DNA repair genes is required for metastasis formation (Sarasin and Kauffmann, 2008). Recent studies have provided support for the view that downstream effects of MITF (via APEX1) on apoptosis may be relevant to melanoma: Liu et al. (2009) showed that MITF-positive melanoma cell lines accumulated high levels of APEX1, and in another study, down-regulation of APEX1 using antisense resulted in apoptosis of melanoma cells in culture (Yang et al., 2005).
In summary, two of the well-established hallmarks of cancer are resistance to apoptosis/cell death and sustained proliferation (Hanahan and Weinberg, 2011); we hypothesized that melanoma cells carrying MC1R variants would have less of both, and additionally poorer DNA repair and therefore the patients would have better survival.
We tested this hypothesis by looking at Breslow thickness and overall survival (OS) in 10 melanoma cohorts in relation to MC1R genotype. These cohorts were collected by a new consortium called BioGenoMEL (http://www.biogenomel.eu). BioGenoMEL has been created to collaboratively identify small, inherited effects on survival, which potentially have profound biological significance.
Results
Description of the data sets
Table 1 gives the summary characteristics of the cohorts studied. Leeds (the test set) was the largest cohort at 751 eligible cases, the others ranging in size from 137 cases (Riga, Latvia) to 487 cases (Valencia, Spain). Figure 1 shows the Breslow thickness distribution (after exclusion of cases with tumors with thickness 0.75 mm or less): a wider range of thickness was seen particularly in some cohorts, particularly in the Latvian cohort. Figure 1 shows the median age at diagnosis, which was fairly well balanced between cohorts (range 50.0–61.5 yrs). Figure 1 also shows that the greatest difference among the cohorts apart from sample size was in the time period during which participants were recruited. In the combined data set, a strong association was seen between MC1R status and hair color (Table S2) as expected (Valverde et al., 1995).
1.
Hair color, median follow-up, and MC1R status of cases in each melanoma cohort
| MC1R [number (%)]b | Hair color [number (%)]b | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Center | Casesa | No. of deaths | Median follow-up (days) | −/− | r/− | R/− | r/r | R/r | R/R | ≥1 consensus alleles | No consensus alleles | Black/Brown | Blond | Red |
| Leeds | 751 | 157 | 2329 | 105 (14) | 134 (18) | 184 (25) | 54 (7) | 169 (22) | 105 (14) | 423 (56) | 328 (44) | 501 (67) | 144 (19) | 89 (12) |
| Valencia | 487 | 60 | 1458 | 171 (35) | 129 (26) | 88 (18) | 31 (6) | 51 (10) | 17 (3) | 388 (80) | 99 (20) | 377 (77) | 93 (19) | 17 (3) |
| Barcelona | 201 | 30 | 1058 | 82 (41) | 49 (24) | 42 (21) | 9 (4) | 15 (7) | 4 (2) | 173 (86) | 28 (14) | 147 (81) | 30 (17) | 4 (2) |
| Genoa | 140 | 18 | 1759.5 | 34 (24) | 40 (29) | 27 (19) | 8 (6) | 17 (12) | 14 (10) | 101 (72) | 39 (28) | 95 (69) | 25 (18) | 17 (12) |
| Vienna | 159 | 21 | 935 | 43 (27) | 52 (33) | 33 (21) | 8 (5) | 16 (10) | 7 (4) | 128 (81) | 31 (19) | 111 (70) | 38 (24) | 10 (6) |
| Paris | 407 | 88 | 1127 | 87 (21) | 121 (30) | 73 (18) | 41 (10) | 65 (16) | 20 (5) | 281 (69) | 126 (31) | 315 (78) | 67 (17) | 23 (6) |
| Essen | 218 | 91 | 1231.5 | 44 (20) | 51 (23) | 58 (27) | 12 (5) | 40 (18) | 13 (6) | 153 (70) | 65 (30) | – | – | – |
| Riga | 137 | 44 | 1018 | 46 (34) | 25 (18) | 32 (23) | 8 (6) | 18 (13) | 8 (6) | 103 (75) | 34 (25) | 128 (93) | 3 (2) | 6 (4) |
| Stockholm | 253 | 70 | 2689 | 45 (18) | 49 (19) | 77 (30) | 14 (6) | 37 (15) | 31 (12) | 171 (68) | 82 (32) | 43 (17) | 195 (77) | 15 (6) |
| Philadelphia | 307 | 36 | 2132 | 65 (21) | 62 (20) | 70 (23) | 22 (7) | 60 (20) | 28 (9) | 197 (64) | 110 (36) | 200 (65) | 71 (23) | 36 (12) |
| Total | 3060 | 615 | 1651 | 722 (24) | 712 (23) | 684 (22) | 207 (7) | 488 (16) | 247 (8) | 2118 (69) | 942 (31) | 1917 (68) | 666 (24) | 217 (8) |
Cases with MC1R, age, sex, and site information, Breslow thickness data >0.75 mm and a single primary melanoma recruited no more than 2 yrs after diagnosis.
Percentages may not total 100% because of rounding.
Figure 1.
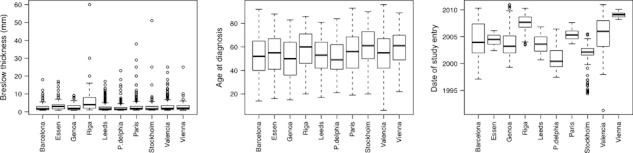
Box plots showing variation of Breslow thickness (thresholded > 0.75 mm), age of diagnosis, and date of study entry in the data taken for analysis from each of the 10 cohorts.
MC1R and tumor thickness
There was a small but significant inverse association between MC1R score and log Breslow thickness (estimate −0.02, P-value = 0.03) in cases whose tumor was thicker than 0.75 mm over all 10 cohorts, adjusted for center. This association was weaker, and not statistically significant, when the model was also adjusted for site of primary thickness (estimate −0.02, P-value = 0.1).
Analysis of the Leeds cohort survival data
The Kaplan–Meier curve looking at the relationship between hair color and survival in the Leeds cohort (in the 1397 cases in the cohort who had hair color and were eligible) is shown in Figure 2. The results of this analysis are consistent with the hypothesis: melanoma cases with black/brown hair had poorer outcome than those with blond hair or red hair (log rank test for a significant difference in outcome between the three groups, P = 0.02).
Figure 2.
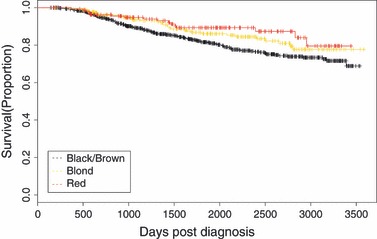
Kaplan–Meier curve showing differences in overall survival by hair color in the Leeds melanoma cohort (Black/brown = 965; Blond = 268; Red = 164; Log rank test, P = 0.02).
Results from the proportional hazards analysis of hair color, MC1R score, and agouti (ASIP) status in the Leeds data set are shown in Table 2. It can be seen that hair color in analyses adjusted for factors known to have an effect on outcome (age, sex and tumor thickness) was borderline significant as a determinant of OS (HR, 0.58; 95% CI, 0.35–0.97; P = 0.04, considering red hair compared with black/brown, adjusted for age, sex, Breslow thickness, and site of the primary). MC1R status was significantly associated with survival if considered as MC1R score (HR per point, 0.82; 95% CI, 0.72–0.94; P = 0.004), no consensus MC1R alleles versus one or more consensus alleles (HR, 0.64; 95% CI, 0.46–0.89; P = 0.008), and if MC1R score and ASIP were included together in a multivariable model (HR, 0.79; 95% CI, 0.69–0.91; P = 0.001).
Table 2.
Cox’s proportional hazards model for overall survival in the Leeds cohort
| Unadjusted HR (95% CI) | P-value | Adjusteda HR (95% CI) | P-value | |
|---|---|---|---|---|
| Blond versus black/brown | 0.70 (0.49–1.02) | 0.06 | 0.87 (0.60–1.26) | 0.5 |
| Red versus black/brown | 0.56 (0.34–0.92) | 0.02 | 0.58 (0.35–0.97) | 0.04 |
| MC1R score (per point) | 0.85 (0.74–0.96) | 0.009 | 0.82 (0.72–0.94) | 0.004 |
| MC1R no consensus alleles versus 1 or more consensus alleles | 0.65 (0.47–0.90) | 0.01 | 0.64 (0.46–0.89) | 0.008 |
| MC1R + ASIPb | ||||
| MC1R score (per point) | 0.82 (0.72–0.94) | 0.005 | 0.79 (0.69–0.91) | 0.001 |
| ASIP (per allele) | 0.63 (0.44–0.91) | 0.01 | 0.58 (0.40–0.85) | 0.005 |
Adjusted for age, sex, site of primary and Breslow thickness. Cases with tumors 0.75 mm or thinner and cases with multiple primary melanomas were excluded.
MC1R and ASIP are included together as individual terms in the survival model.
Analysis of forest plots of the association of MC1R with overall survival
Figure 3 shows the forest plots for all the survival data. In the analyses adjusted for age and sex only, the nine other cohorts gave some support to the hypothesis that inheritance of MC1R variants was associated with improved outcome (HR, 0.82; 95% CI, 0.66–1.02; P = 0.08 for no consensus MC1R alleles versus one or more consensus alleles). This result attained statistical significance with the addition of the Leeds data (HR, 0.77; 95% CI, 0.64–0.93; P = 0.005).
Figure 3.
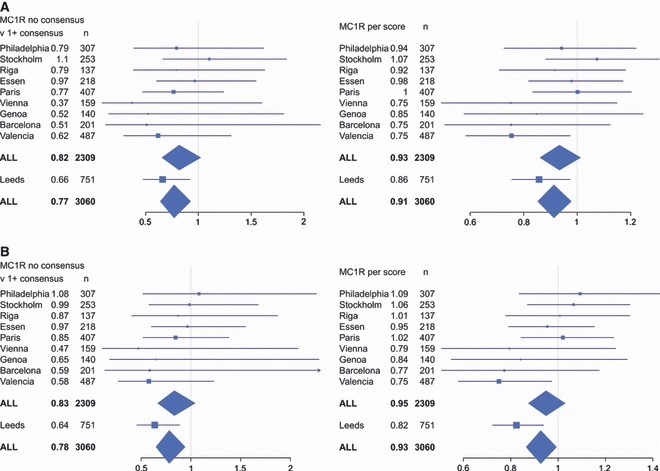
Forest plots of association of MC1R score with survival in each of the 10 melanoma cohorts and combined adjusted for (A) sex and age at diagnosis (B) site of primary, sex, age at diagnosis, and Breslow thickness.
In the analyses adjusted additionally for site of primary and Breslow thickness, we see similar patterns of association in the smaller nine cohorts (HR, 0.83; 95% CI, 0.67–1.04; P = 0.1 for no consensus MC1R alleles versus one or more consensus alleles) and in all 10 cohorts combined (HR, 0.78; 95% CI, 0.65–0.94; P = 0.009). This suggests that adjustment for Breslow thickness has only a small overall effect on the association of MC1R with outcome. To test this, we compared the results of the same analysis under a univariable model with one adjusted for Breslow thickness only. We found little difference in the estimated hazard ratio under both models (data not shown).
Individually, little change was observed in the magnitude and direction of hazard ratios for most of the cohorts with adjustment for site and thickness. However, in the Philadelphia cohort, the direction of effect was protective in the model adjusted for sex and age only (HR, 0.79; 95% CI, 0.39–1.62) but deleterious in the model adjusted for sex, age, site of primary and Breslow thickness (HR, 1.08; 95% CI, 0.52–2.26). There is a significant association between log Breslow thickness and MC1R score for these Philadelphia data even when adjusted for site of primary thickness (estimate −0.07, P = 0.02), suggesting that adjusting for Breslow thickness may be obscuring the true relationship between MC1R and outcome in the Philadelphia data set even though in the overall analysis it does not.
There is no evidence of significant study heterogeneity between the 10 cohorts in either the model using the MC1R score (Cochran’s Q test for heterogeneity P = 0.2) or the model comparing no consensus MC1R alleles versus one or more consensus alleles (Cochran’s Q test for heterogeneity P = 0.8). However, the clearest support for our hypothesis is seen in the second biggest study (Valencia), and there was also some suggestion of a stronger effect in the Mediterranean countries compared with others.
We tested the effect of ignoring the rare MC1R variants on the association of MC1R score with outcome and found that this had little effect on our overall conclusions. Details can be found in Supporting information.
Discussion
This study reports a survival analysis of a large cohort of melanoma patients from the UK, which identified an effect of hair color and inherited variants in the MC1R gene on OS. We sought to validate these findings in nine additional cohorts within the melanoma genetics consortium BioGenoMEL (http://www.biogenomel.eu).
The identification of hereditary variation moderating host/tumor interaction and therefore survival from cancer is predicted to require multiple large data sets to identify small but biologically important effects. Few or no studies of sufficient power have been performed to date. The strength of this collaborative study is the unique collection of multiple data sets from Europe and the USA, subject to a centralized analysis. A weakness is that although the fact that the majority of melanomas are diagnosed early with excellent survival is very good news in terms of patient outcome, this does reduce the power of the study, in that after exclusion of participants with tumors thinner than 0.75 mm, some of the truncated cohorts were individually small. An analysis of the prevalence of ‘R’MC1R variants in the 10 cohorts also shows significant differences among them. This was expected given the known variation in MC1R allele frequencies across Europe. The highest proportion with ‘R’ variants was seen in UK melanoma patients. High frequencies were also seen in the melanoma patients from Germany and Sweden. The proportion of ‘R’ variants in some studies was so low however, these had little individual power to test the hypothesis. Because we could only identify individuals with metastatic disease in a few of our cohorts, we could not comprehensively investigate the effect on the data of excluding individuals who presented with metastatic disease. We are not certain that omitting metastatic cases should be standard in survival analyses that use OS as an end point, given that it is known that there is considerable variation in the survival outcome of cases with stage III disease (Balch et al., 2001, 2010). There were limited data available on stage for the Valencia, Genoa, and Paris cohorts; in this limited data set, we saw that removing stage IV cases had little to no effect. In our analysis, we were unable to take into account effects of drug interventions such as dacarbazine. However no drug used at the time of cohort follow-up has been shown to have survival benefit, so we anticipate this information would not change our conclusions. Analysis was complicated by heterogeneity between each of the cohorts in geographic location, how cases were ascertained to the study and the time period in which cases were recruited. Our analysis assumes that the effect of MC1R is similar in each of the cohorts, which is why we stratified baseline risk for each cohort. It is known that average melanoma thickness has decreased over time in European cohorts (Crocetti and Carli, 2003; Garbe et al., 2000; Lipsker et al., 2007), and this could complicate comparisons of the 10 cohorts in our study. However, we did not see any evidence of an association in our own data (mostly collected from 1999 to 2010) between Breslow thickness and year of diagnosis. Cohorts differ in how and how often they obtain follow-up. As the number of deaths is one of the factors that determines power, cohorts that are followed up infrequently are not as informative as they potentially could be. However, because follow-up is continually updated, we anticipate that these cohorts will mature over time and we will have greater power to see associations with other germline variants in future studies. Another potential weakness of the study is that we did not perform centralized sequencing. Although Sanger sequencing is considered by many to be the ‘gold standard’ for mutation detection, the possibility of error arising from sequencing at different locations cannot be ruled out. However, we have seen excellent concordance in Sanger sequencing data from different centers, both in this study (sequencing in Leeds and Leiden of Leeds samples) and a previous study in which sequencing of CDKN2A across multiple centers within the GenoMEL genetics consortium was shown to be comparable (Harland et al., 2008).
Finally, it is of concern that hair color might be graded differently in different populations, so what might be viewed as ‘blond’ in Latvia, for example, might be viewed differently in Sweden where most people are pale skinned. Indeed there was little evidence of an effect of hair color on survival in a meta-analysis of the individual nine smaller cohorts overall (data not shown). In collaborative studies of this type, there will always be genetic variation between the populations, which is difficult to correct for; this study was therefore of particular interest because some of that genetic variation was evident as a difference in phenotype. While the main purpose of the study was to investigate MC1R, this study also considers the general issues arising from the use of multiple data sets to look at outcome.
The analysis of the data from the largest cohort benefitted from its size, but also SNP typing for the agouti locus as well as MC1R. The survival analyses provided strong supportive evidence that increased numbers of MC1R‘R’ or ‘r’ variants have a protective effect on outcome, consistent with the hypothesis. Although the analysis based upon the number of variant ‘R’ or ‘r’ alleles is persuasive, there also appeared to be a deleterious effect of inheritance of one or more consensus MC1R alleles. Thus, these data suggest that in the presence of physiological signaling via the melanocortin receptor, biological processes downstream of MITF may result in poorer prognosis for melanoma patients. This is putatively through reduced apoptosis mediated by APEX1, greater double-strand break DNA repair, and/or increased cellular proliferation.
That there was some evidence in the 10 data sets of a relationship between MC1R variants, and thinner tumors is also supportive of the view that reduced proliferative effects may be seen in the presence of variant MC1R. We considered the possibility that differences in thickness may also reflect differences in ease of clinical diagnosis for patients with different skin type but published data suggest rather that diagnosis may be more readily missed in the very fair skinned (Cuellar et al., 2009) leading to the converse.
There was support for a protective effect of variant ‘R’ or ‘r’MC1R on death from melanoma from the other nine cohorts, although overall the result from those nine cohorts was not independently statistically significant. The forest plots in Figure 3 are presented within Europe ordered from the northern latitudes (where blond hair and fair skin are much more prevalent) to Mediterranean countries (where darker hair and skin types are more prevalent, as a result of the inheritance of different patterns of additional pigment genes) and where the proportion of the cohort with ‘R’ variants is much smaller. Although there was no statistically significant evidence of heterogeneity between cohorts, examination of the forest plot suggests that the protective effect of the consensus MC1R allele might be most obvious in the Mediterranean populations, although overall the Leeds cohort has significantly greater proportions of cases with ‘R’ variants than any other cohort. It is not clear therefore whether the limited variation between the cohorts is a result of chance, cohort size, or the co-inheritance of other pigment genes impacting on MC1R signaling.
Inherited MC1R variants have previously been suggested to increase the likelihood of somatic BRAF mutant tumors (Fargnoli et al., 2008; Landi et al., 2006), so we have considered the possibility that differences in survival associated with germline MC1R status might be related to somatic differences between the tumors. The data reported by Landi et al. however were not corroborated by others (Hacker et al., 2010; Thomas et al., 2010), and there are some (albeit small studies) which actually suggest that BRAF mutations are associated with an unfavorable prognosis (Long et al., 2011; Si et al., 2012). Although there is no clear evidence to support an effect of somatic tumor variation by MC1R status, it is difficult to exclude the possibility. This argues for the need to consider both germline and somatic genetic events in investigating host/tumor interaction, and this is the future intent of BioGenoMEL.
Agnostic genome-wide approaches remain the most likely to identify new biological pathways of relevance, although published results have been mixed. A recent genome-wide association study of 1145 patients with breast cancer, for example, failed to identify such genes (Azzato et al., 2010). A smaller genome-wide study, in 245 patients treated with chemotherapy for small cell lung cancer, however, appears to have identified inherited variation predictive of survival (Wu et al., 2010).
An alternative approach is to take a candidate gene approach but a recent review of 90 candidate gene studies performed in patients with lung cancer confirmed the folly of small-scale studies without validation. Nonetheless, the conclusion of the review was that a small set of potential biomarkers had been identified in this way (Horgan et al., 2011). The effect of inherited genetic variation on outcome from cancer is likely to be relatively small when compared with the effects of variation in somatic genetic changes. That these effects are predicted to be small does not diminish their potential for giving biological insights into host/tumor interaction and therefore approaches to adjuvant therapies.
In conclusion, both genome-wide and candidate gene studies may be needed to identify germline predictors of outcome, but it will clearly be necessary to work collaboratively within consortia such as BioGenoMEL. This will be particularly helpful to avoid a proliferation of small genetic studies producing contradictory results. This study illustrates the value of consortia, but reinforces the need for large studies even within consortia and the potential problems in looking at survival in genetically diverse populations.
Methods
Data collection
The Leeds melanoma cohort
The UK Multicenter Research Ethics Committee (MREC) and the Patient Information Advisory Group (the predecessor of the National Information Governance Board) approved the study. Population-ascertained incident melanoma cases were recruited to a case–control study in a geographically defined area of the UK (Yorkshire and the Northern region south of the River Tyne) (67% participation rate); 960 cases (aged 18–76 yrs) were recruited in the period from September 2000 to December 2005, as described previously (Falchi et al., 2009; Randerson-Moor et al., 2009). Recruitment (and therefore blood sampling) took place wherever possible 3–6 months after diagnosis. Age, sex, natural hair color at age 18 yrs, propensity to burn, ability to tan, skin color of inside upper arm, and freckling as a child using Gallagher’s freckle chart (Lee et al., 2005) were self-reported. Recruitment to the cohort has continued beyond 2005, but MC1R genotyping is not yet available for the more recent samples. The Leeds cohort contains 1954 cases in total, and MC1R genotyping was available for 966 of these cases. After applying the selection criteria applied to all cohorts and listed below in the section on Statistical analysis, 751 cases were eligible for the survival analysis. Information on hair color was available for 1659 cases, and after applying the same selection criteria, 1397 cases were eligible.
BioGenoMEL cohorts
Table 1 shows comparative data on the nine additional cohorts contributed by members of the BioGenoMEL consortium. These cohorts have been collected in the period 1991 to the present day by research groups in Southern Europe [centers in Barcelona, Valencia and Genoa (Goldstein et al., 2007; Pastorino et al., 2008; Scherer et al., 2009)], Central Europe (Vienna), Northern Europe [Paris and Essen (Guedj et al., 2008; Scherer et al., 2009)], far Northern Europe [Riga and Stockholm (Hoiom et al., 2009)], and the USA [Philadelphia (Kanetsky et al., 2010)]. In the Leeds, cohort disease/vital status is established through annual follow-up, inquiry from the GPs, cancer registry data, and by extraction of clinical notes. Attainment of follow-up data was from equivalent sources in other centers. Details can be found in Table S3.
MC1R sequencing
In nine centers, the whole MC1R gene was sequenced, and in one center (Sweden), the most common variants were genotyped (see below).
For the Leeds samples, standard PCR techniques were used to amplify the entire 954-nucleotide coding region of the single-exon MC1R gene, plus the surrounding untranslated regions, either as a single large amplicon or in smaller overlapping amplicons. Purified PCR products were sequenced using sequencing primers spanning the MC1R gene. Sequencing reactions were performed using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Warrington, UK) using standard sequencing conditions. The sequencing reaction products were run on an ABI prism 3130XL Genetic Analyzer (Applied Biosystems).
Sequence data were analyzed using SeqScape (Applied Biosystems) or CodonCode Aligner software (CodonCode Corp., Dedham, MA, USA). Compiled sequence data were double scored and checked by a second analyst. MC1R polymorphisms were identified by comparison with the MC1R consensus sequence (NCBI accession no. NM_002386). Primer sequences for PCR and sequencing are available on request. There was no centralized sequencing between groups, most groups had previously sequenced MC1R for other analyses addressed to understanding genetic susceptibility to melanoma (Demenais et al., 2010; Ghiorzo et al., 2009; Kanetsky et al., 2004; Matichard et al., 2004; Scherer et al., 2009). Details can be found in Table S3. Most centers used similar standard sequencing techniques to screen for MC1R variants, as described above.
In the Leeds samples, one of 31 plates was genotyped in both Leeds and Leiden to test for errors in the genotyping process. There was 100% concordance between calls for these samples.
Swedish samples were analyzed using a Protease-mediated Allele-Specific Extension (PrASE) method, specific to 21 of the most common MC1R variants in European populations (Kaller et al., 2005).
MC1R analysis
As MC1R variants are numerous and are thought to have a variable impact on signaling through MITF, the analytic approach was considered carefully. We based the analysis upon a published and widely adopted classification system using the ‘r’, ‘R’ nomenclature first described by Duffy et al. (2004). Figure 4 shows a flowchart that explains the classification system we implemented, and a detailed explanation is provided in Supporting information.
Figure 4.
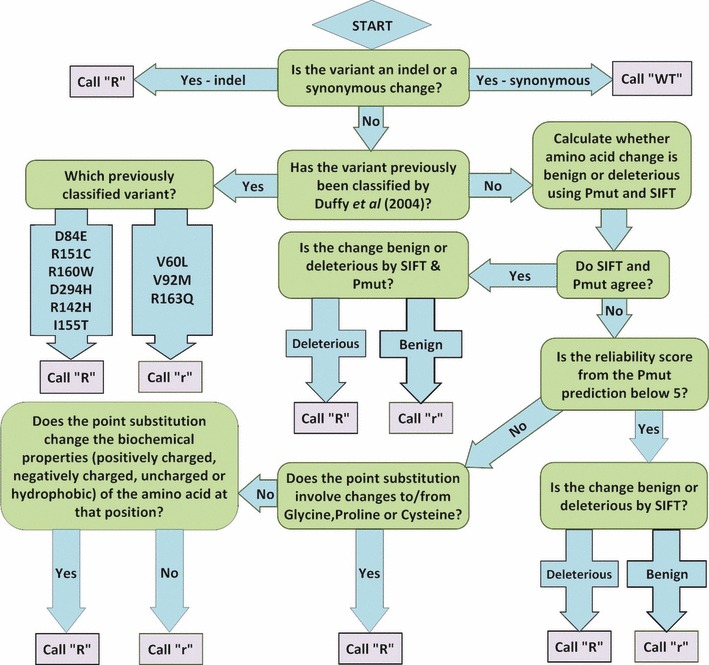
Flowchart showing how MC1R variants are called ‘r’ or ‘R’ using SIFT and PMut to call rare variants (see Methods).
The classification was turned into a numerical score in the range 0–4 by summing across the two alleles, giving a value of 1 to ‘r’ and 2 to ‘R’ variants. Thus, individuals with two copies of the consensus sequence (−/−) scored 0, and individuals with two ‘R’ variants (R/R) scored 4. It has been suggested previously that red hair color alleles may act in a recessive manner and that one fully functional copy of MC1R may be sufficient to provide normal function (Beaumont et al., 2008). Therefore, we also looked at a second classification system contrasting individuals who have no consensus alleles (r/r, R/r, R/R) with those who have one or more consensus alleles (−/−, r/−, R/−).
Statistical analysis
To test the hypothesis that individuals with MC1R variants have thinner tumors, a linear regression analysis was conducted, regressing the natural logarithm of Breslow thickness on the MC1R score, adjusting for center, using the ‘lm’ routine in r 2.10.1 (R Development Core Team, 2010).
We defined survival time as the period between the date of surgical excision of the primary and date of death or last date of follow-up (at which point records were censored). Kaplan–Meier curves were drawn to investigate differences in OS with respect to hair color (classified as black/brown, blond, or red) and MC1R status in the Leeds cohort, using the ‘survfit’ routine in the ‘survival’ package in r. To test for a significant different in outcome between the three hair color groups in the Leeds cohort, a log rank test was performed using the ‘survdiff’ routine in the ‘survival’ package in r.
Multivariate survival analyses were performed using Cox’s proportional hazards model. Models were fitted using the ‘coxph’ routine in the ‘survival’ package in r. Hazard ratio estimates were calculated for the effect of hair color and MC1R on OS adjusted for Breslow thickness, sex, site of melanoma (head/neck, trunk, limbs or other), and age of diagnosis. We also report models adjusted only for age of diagnosis and sex for the MC1R analyses. We did not adjust for tumor ulceration because of incomplete data for this variable across all the cohorts. We could not adjust for AJCC staging for the same reason. We tested the effect of adding stage in three cohorts for which we had data and we found that including stage did not change the association of MC1R with outcome. To test for study heterogeneity, we performed Cochran’s Q test. To do this, we first took relevant point estimates and standard errors for each study from the fitted Cox’s proportional hazards models. These data were then used to construct a meta-analysis assuming fixed effects using the ‘metaan’ function in Stata version 10, which reports the result of Cochran’s Q test.
We excluded cases with thin tumors (0.75 mm or less) from all analyses on the basis that these cases have an excellent prognosis and add little information to the estimation of the effect of predictors of survival. We did not test for CDKN2A mutation carrier status in the 10 melanoma cohorts, but we expect that because the cohorts were not ascertained on the basis of family history that these would be rare and account for 2% of the cases (based on our own unpublished data). A modest number of cases with nodal or metastatic disease were included in the analysis; nodal primaries were assigned a Breslow thickness of 4 mm. We also excluded individuals with multiple primary melanomas and prevalent cases that were recruited in a proportion of the studies. It has been shown that introducing cases into a study a long time after diagnosis (left truncated data) can introduce bias into survival analysis (Cnaan and Ryan, 1989). Each center was therefore asked to provide information of the date of study entry for each case to determine whether the case was recruited within 2 yrs of diagnosis, which we defined as ‘incident’ as opposed to ‘prevalent’ cases. We investigated whether it was possible to incorporate prevalent cases into our study by measuring survival from the date of study entry [as described by Azzato et al. (2009)] but we found the proportional hazards assumption was violated in this model (Schoenfeld global test for proportional hazards in the model containing MC1R score, P = 0.002). It is important that the proportional hazards assumption holds when including prevalent cases in this way (Azzato et al., 2009) so we discounted them from further analyses and focused efforts on cases recruited within 2 yrs of diagnosis. A breakdown of how cases were excluded can be found in Table S4.
The hypothesis that MC1R variants might be associated with survival was first tested by evaluating hair color and survival. Hair color is determined by a number of pigment genes, but red hair is predominantly (but not exclusively) determined by MC1R. Support for the hypothesis was seen, in that those with red hair had better survival (see Figure 2, and Table 2), and therefore the association between MC1R status and OS was also investigated in each of the 10 data sets. We created a combined estimate for the nine smaller data sets by including study as a stratification variable in the model.
The agouti signaling protein (coded by ASIP, a melanoma susceptibility locus) is a competitive agonist of melanocyte-stimulating hormone (MSH) which binds to the melanocortin receptor, so that inheritance of the risk allele at ASIP results in reversion to the null (pheomelanin) phenotype. Hence, it is postulated that inheritance of risk alleles at ASIP has a similar effect to variant MC1R on MITF signaling. Therefore, in the Leeds data set, where we had both MC1R sequence data and ASIP SNP genotyping (for the single nucleotide polymorphism rs4911442), we included the ASIP and MC1R data together in a separate model to investigate the combined effect of the two on survival.
Forest plots were used to compare the hazard ratio (HR) estimates across studies; HR estimates for MC1R score and for no consensus MC1R alleles versus one or more consensus MC1R alleles were plotted for each center, alongside a pooled estimate.
The relationship between the age variable and the log hazard for survival was suspected to be nonlinear. Details of how we tested this are provided in Supporting information.
Acknowledgments
The collection of samples in the Melanoma Cohort Study was funded by Cancer Research UK (project grant C8216/A6129, programme awards C588/A4994 and C588/A10589, and Centre Award C37059/A11941) and by the NIH (R01 CA83115). The collection of samples from Valencia were obtained from the Biobank of the Fundación Instituto Valenciano de Oncología. The Barcelona team research has been supported by Fondo de Investigaciones Sanitarias (grants 05/0302, 06/0265 and 09/1393). Zighereda Ogbah received financial support provided by Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR). The project also received support from the European Commission under the 6th Framework Programme, Contract nr: LSHC-CT-2006-018702 (GenoMEL) and by the National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115).
Stockholm team additional funding: The Swedish Cancer Society, The Radiumhemmet Research Funds, The Karolinska Institutet Research Funds. Riga team additional funding: ESF Project No.1DP/1.1.1.2.0/09/APIA/VIAA/150, Latvian Council of Science project No.10.0010.8. The Paris team (Melan-cohort) was sponsored by the ‘Assistance Publique Hôpitaux de Paris’ (AP-HP) (Departement of Clinical Research and Development) and was funded by a grant of French Ministry of Health (Programme Hospitalier de Recherche Clinique, AOR02089 AOM06200). The authors thank Amel Ouslimani (Département de la Recherche Clinique et du Développement), Philippe Aegerter, Claire Ribet and véronique Clérisse (Paris-Ouest Clinical Research Unit) for monitoring and logistic aspects. Genoa team additional funding: Fondazione CARIGE 2010, IMI and ACM 2011, Italian Ministry of Health DGRST.4/4235-P1.9.A.B. Dirk Schadendorf and Rajiv Kumar: Funding by DFG (Scha 422/11-1).
The Penn study was supported by the National Cancer Institute (CA80700 and CA092428) and CA016520 to the Abramson Cancer Center of the University of Pennsylvania in support of the Melanoma Core. Financial support for the Vienna team: Jubiläumsfonds by the Österreichische Nationalbank (project number 12161 and 13036) and the Hans und Blanca Moser Stiftung. The BioGenoMEL consortium is supported a grant from the Skin Cancer Research Fund (SCaRF) http://www.skin-cancer-research-fund.org.uk.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Data S1. Methods.
Table S1. MC1R calls including andexcluding rare variants determined using the bioinformaticanalysis. Variant calls based only on the nine variantsdescribed in (Duffy et al. 2004) are in italics, variantcalls based upon inclusion of information taken from rare variantsare in bold.
Table S2. MC1R status and hair colour inthe combined dataset. MC1R is strongly associated withthe hair colour phenotype (Pearson’s Chi-squared test<0.0001).
Table S3. MC1R Genotyping method, patient accrual and follow up in each group.
Table S4. Breakdown of the exclusion of cases ineligible for analysis by cohort.
References
- Azzato EM, Greenberg D, Shah M, Blows F, Driver KE, Caporaso NE, Pharoah PD. Prevalent cases in observational studies of cancer survival: do they bias hazard ratio estimates? Br. J. Cancer. 2009;100:1806–1811. doi: 10.1038/sj.bjc.6605062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzato EM, Pharoah PD, Harrington P, et al. A genome-wide association study of prognosis in breast cancer. Cancer Epidemiol. Biomarkers Prev. 2010;19:1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch CM, Soong SJ, Gershenwald JE, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J. Clin. Oncol. 2001;19:3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, et al. Multivariate analysis of prognostic factors among 2,313 patients with stage III melanoma: comparison of nodal micrometastases versus macrometastases. J. Clin. Oncol. 2010;28:2452–2459. doi: 10.1200/JCO.2009.27.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont KA, Shekar SN, Cook AL, Duffy DL, Sturm RA. Red hair is the null phenotype of MC1R. Hum. Mutat. 2008;29:E88–E94. doi: 10.1002/humu.20788. [DOI] [PubMed] [Google Scholar]
- Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat. Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnaan A, Ryan L. Survival analysis in natural history studies of disease. Stat. Med. 1989;8:1255–1268. doi: 10.1002/sim.4780081009. [DOI] [PubMed] [Google Scholar]
- Crocetti E, Carli P. Changes from mid-1980s to late 1990s among clinical and demographic correlates of melanoma thickness. Eur. J. Dermatol. 2003;13:72–75. [PubMed] [Google Scholar]
- Cuellar F, Puig S, Kolm I, Puig-Butille J, Zaballos P, Marti-Laborda R, Badenas C, Malvehy J. Dermoscopic features of melanomas associated with MC1R variants in Spanish CDKN2A mutation carriers. Br. J. Dermatol. 2009;160:48–53. doi: 10.1111/j.1365-2133.2008.08826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demenais F, Mohamdi H, Chaudru V, et al. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: a GenoMEL study. J. Natl Cancer Inst. 2010;102:1568–1583. doi: 10.1093/jnci/djq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Duffy DL, Box NF, Chen W, Palmer JS, Montgomery GW, James MR, Hayward NK, Martin NG, Sturm RA. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum. Mol. Genet. 2004;13:447–461. doi: 10.1093/hmg/ddh043. [DOI] [PubMed] [Google Scholar]
- Falchi M, Bataille V, Hayward NK, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargnoli MC, Pike K, Pfeiffer RM, et al. MC1R variants increase risk of melanomas harboring BRAF mutations. J. Invest. Dermatol. 2008;128:2485–2490. doi: 10.1038/jid.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandini S, Sera F, Cattaruzza MS, Pasquini P, Abeni D, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur. J. Cancer. 2005a;41:28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Gandini S, Sera F, Cattaruzza MS, Pasquini P, Zanetti R, Masini C, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur. J. Cancer. 2005b;41:2040–2059. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Garbe C, McLeod GR, Buettner PG. Time trends of cutaneous melanoma in Queensland, Australia and Central Europe. Cancer. 2000;89:1269–1278. doi: 10.1002/1097-0142(20000915)89:6<1269::aid-cncr11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Ghiorzo P, Pastorino L, Pizzichetta MA, et al. CDKN2A and MC1R analysis in amelanotic and pigmented melanoma. Melanoma Res. 2009;19:142–145. doi: 10.1097/CMR.0b013e32832a1e18. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Chaudru V, Ghiorzo P, et al. Cutaneous phenotype and MC1R variants as modifying factors for the development of melanoma in CDKN2A G101W mutation carriers from 4 countries. Int. J. Cancer. 2007;121:825–831. doi: 10.1002/ijc.22712. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- Guedj M, Bourillon A, Combadieres C, et al. Variants of the MATP/SLC45A2 gene are protective for melanoma in the French population. Hum. Mutat. 2008;29:1154–1160. doi: 10.1002/humu.20823. [DOI] [PubMed] [Google Scholar]
- Hacker E, Hayward NK, Dumenil T, James MR, Whiteman DC. The association between MC1R genotype and BRAF mutation status in cutaneous melanoma: findings from an Australian population. J. Invest. Dermatol. 2010;130:241–248. doi: 10.1038/jid.2009.182. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harland M, Goldstein AM, Kukalizch K, et al. A comparison of CDKN2A mutation detection within the Melanoma Genetics Consortium (GenoMEL) Eur. J. Cancer. 2008;44:1269–1274. doi: 10.1016/j.ejca.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiom V, Tuominen R, Kaller M, Linden D, Ahmadian A, Mansson-Brahme E, Egyhazi S, Sjoberg K, Lundeberg J, Hansson J. MC1R variation and melanoma risk in the Swedish population in relation to clinical and pathological parameters. Pigment Cell Melanoma Res. 2009;22:196–204. doi: 10.1111/j.1755-148X.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- Horgan AM, Yang B, Azad AK, Amir E, John T, Cescon DW, Wheatley-Price P, Hung RJ, Shepherd FA, Liu G. Pharmacogenetic and germline prognostic markers of lung cancer. J. Thorac. Oncol. 2011;6:296–304. doi: 10.1097/JTO.0b013e3181ffe909. [DOI] [PubMed] [Google Scholar]
- Jewell R, Conway C, Mitra A, et al. Patterns of expression of DNA repair genes and relapse from melanoma. Clin. Cancer Res. 2010;16:5211–5221. doi: 10.1158/1078-0432.CCR-10-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadekaro AL, Leachman S, Kavanagh RJ, et al. Melanocortin 1 receptor genotype: an important determinant of the damage response of melanocytes to ultraviolet radiation. FASEB J. 2010;24:3850–3860. doi: 10.1096/fj.10-158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaller M, Tuominen R, Ahmadian A, Magnusson V, Egyhazi S, Hansson J, Lundeberg J. Detection of MC1R polymorphisms with protease-mediated allele-specific extension as an alternative to direct sequencing. Clin. Chem. 2005;51:2388–2391. doi: 10.1373/clinchem.2005.056820. [DOI] [PubMed] [Google Scholar]
- Kanetsky PA, Ge F, Najarian D, Swoyer J, Panossian S, Schuchter L, Holmes R, Guerry D, Rebbeck TR. Assessment of polymorphic variants in the melanocortin-1 receptor gene with cutaneous pigmentation using an evolutionary approach. Cancer Epidemiol. Biomarkers Prev. 2004;13:808–819. [PubMed] [Google Scholar]
- Kanetsky PA, Panossian S, Elder DE, Guerry D, Ming ME, Schuchter L, Rebbeck TR. Does MC1R genotype convey information about melanoma risk beyond risk phenotypes? Cancer. 2010;116:2416–2428. doi: 10.1002/cncr.24994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Bauer J, Pfeiffer RM, Elder DE, Hulley B, Minghetti P, Calista D, Kanetsky PA, Pinkel D, Bastian BC. MC1R germline variants confer risk for BRAF-mutant melanoma. Science. 2006;313:521–522. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- Lee TK, Rivers JK, Gallagher RP. Site-specific protective effect of broad-spectrum sunscreen on nevus development among white schoolchildren in a randomized trial. J. Am. Acad. Dermatol. 2005;52:786–792. doi: 10.1016/j.jaad.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Lipsker D, Engel F, Cribier B, Velten M, Hedelin G. Trends in melanoma epidemiology suggest three different types of melanoma. Br. J. Dermatol. 2007;157:338–343. doi: 10.1111/j.1365-2133.2007.08029.x. [DOI] [PubMed] [Google Scholar]
- Liu F, Fu Y, Meyskens FL., Jr MiTF regulates cellular response to reactive oxygen species through transcriptional regulation of APE-1/Ref-1. J. Invest. Dermatol. 2009;129:422–431. doi: 10.1038/jid.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J. Cell Biol. 2005;168:35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, Hughes TM, Thompson JF, Scolyer RA, Kefford RF. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J. Clin. Oncol. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- Matichard E, Verpillat P, Meziani R, et al. Melanocortin 1 receptor (MC1R) gene variants may increase the risk of melanoma in France independently of clinical risk factors and UV exposure. J. Med. Genet. 2004;41:e13. doi: 10.1136/jmg.2003.011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Pastorino L, Bonelli L, Ghiorzo P, et al. CDKN2A mutations and MC1R variants in Italian patients with single or multiple primary melanoma. Pigment Cell Melanoma Res. 2008;21:700–709. doi: 10.1111/j.1755-148X.2008.00512.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2009. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org.
- Randerson-Moor JA, Taylor JC, Elliott F, et al. Vitamin D receptor gene polymorphisms, serum 25-hydroxyvitamin D levels, and melanoma: UK case–control comparisons and a meta-analysis of published VDR data. Eur. J. Cancer. 2009;45:3271–3281. doi: 10.1016/j.ejca.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S, Dixon S, August S, Diffey B, Wakamatsu K, Ito S, Friedmann PS, Healy E. Protection against UVR involves MC1R-mediated non-pigmentary and pigmentary mechanisms in vivo. J. Invest. Dermatol. 2010;130:1904–1913. doi: 10.1038/jid.2010.48. [DOI] [PubMed] [Google Scholar]
- Sarasin A, Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat. Res. 2008;659:49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Scherer D, Nagore E, Bermejo JL, Figl A, Botella-Estrada R, Thirumaran RK, Angelini S, Hemminki K, Schadendorf D, Kumar R. Melanocortin receptor 1 variants and melanoma risk: a study of 2 European populations. Int. J. Cancer. 2009;125:1868–1875. doi: 10.1002/ijc.24548. [DOI] [PubMed] [Google Scholar]
- Si L, Kong Y, Xu X, Flaherty KT, Sheng X, Cui C, Chi Z, Li S, Mao L, Guo J. Prevalence of BRAF V600E mutation in Chinese melanoma patients: large scale analysis of BRAF and NRAS mutations in a 432-case cohort. Eur. J. Cancer. 2012;48:94–100. doi: 10.1016/j.ejca.2011.06.056. [DOI] [PubMed] [Google Scholar]
- Song X, Mosby N, Yang J, Xu A, Abdel-Malek Z, Kadekaro AL. alpha-MSH activates immediate defense responses to UV-induced oxidative stress in human melanocytes. Pigment Cell Melanoma Res. 2009;22:809–818. doi: 10.1111/j.1755-148X.2009.00615.x. [DOI] [PubMed] [Google Scholar]
- Thomas NE, Kanetsky PA, Edmiston SN, et al. Relationship between germline MC1R variants and BRAF-mutant melanoma in a North Carolina population-based study. J. Invest. Dermatol. 2010;130:1463–1465. doi: 10.1038/jid.2009.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat. Genet. 1995;11:328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J. Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- Wu C, Xu B, Yuan P, et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res. 2010;70:9721–9729. doi: 10.1158/0008-5472.CAN-10-1493. [DOI] [PubMed] [Google Scholar]
- Yang S, Irani K, Heffron SE, Jurnak F, Meyskens FL., Jr lterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE/Ref-1) in human melanoma and identification of the therapeutic potential of resveratrol as an APE/Ref-1 inhibitor. Mol. Cancer Ther. 2005;4:1923–1935. doi: 10.1158/1535-7163.MCT-05-0229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article:
Data S1. Methods.
Table S1. MC1R calls including andexcluding rare variants determined using the bioinformaticanalysis. Variant calls based only on the nine variantsdescribed in (Duffy et al. 2004) are in italics, variantcalls based upon inclusion of information taken from rare variantsare in bold.
Table S2. MC1R status and hair colour inthe combined dataset. MC1R is strongly associated withthe hair colour phenotype (Pearson’s Chi-squared test<0.0001).
Table S3. MC1R Genotyping method, patient accrual and follow up in each group.
Table S4. Breakdown of the exclusion of cases ineligible for analysis by cohort.


