Abstract
A feature shared by many inflammatory lung diseases is excessive neutrophilic infiltration. Neutrophil homing to airspaces involve multiple factors produced by several distinct cell types. Hepoxilin A3 is a neutrophil chemo-attractant produced by pathogen infected epithelial cells hypothesized to facilitate neutrophil breach of mucosal barriers. Using a Transwell model of lung epithelial barriers infected with P. aeruginosa, we explored the role of hepoxilin A3 in neutrophil trans-epithelial migration. Pharmacological inhibitors of enzymatic pathways necessary to generate hepoxilin A3, including phospholipase A2 and 12-lipoxygenase, potently interfere with P. aeruginosa-induced neutrophil trans-epithelial migration. Both transformed and primary human lung epithelial cells infected with P. aeruginosa generate hepoxilin A3 precursor arachidonic acid. All four known lipoxygenase enzymes capable of synthesizing hepoxilin A3 are expressed in lung epithelial cell lines, primary small airway epithelial cells, and human bronchial epithelial cells. Lung epithelial cells produce increased hepoxilin A3 and lipid derived neutrophil chemotactic activity in response to P. aeruginosa infection. Lipid derived chemotactic activity is soluble epoxide hydrolase sensitive, consistent with hepoxilin A3 serving a chemotactic role. Stable inhibitory structural analogues of hepoxilin A3 are capable of impeding P. aeruginosa-induced neutrophil trans-epithelial migration. Finally, intranasal infection of mice with P. aeruginosa promotes enhanced cellular infiltrate into the airspace as well as increased concentration of the 12-lipoxygenase metabolites hepoxilin A3 and 12-HETE. Data generated from multiple models herein provide further evidence that hepoxilin A3 is produced in response to lung pathogenic bacteria and functions to drive neutrophils across epithelial barriers.
Keywords: Neutrophils, Bacterial, Lipid Mediators, Chemotaxis, Inflammation, & Lung
Introduction
Infectious lung disease is a significant global health problem, and represents a particularly difficult challenge as antibiotic resistant pathogens emerge (1–4). During acute pneumonia, there is a rapid influx of innate immune cells to the lungs where they release caustic molecules that result in gross tissue damage (3, 5). Neutrophils are among the first responders in innate immunity, rapidly deployed to sites of infection to confront a variety of pathogens (5–7). Without appropriate resolution, however, neutrophils can exacerbate pathology during infectious and idiopathic inflammatory processes, such as the case in both acute and chronic lung diseases including pneumonia, cystic fibrosis, and severe bouts of asthma (3, 6–8).
To reach the airspace during infection, neutrophils must exit the circulatory system of the lung, navigate through the extracellular milieu, and ultimately cross the mucosal epithelial barrier. Evidence points to a tiered signaling hierarchy that mediates neutrophil adhesion, initial migration, deep tissue homing, and entry into organ spaces (9, 10). In this report, we examine the process by which neutrophils migrate across lung epithelial barriers. Our group previously identified the eicosanoid hepoxilin A3 (HXA3) as a potent chemoattractant that drives neutrophil migration across gut and lung epithelial barriers (11–15). HXA3 likely interacts directly with neutrophil receptors and induces chemotaxis without granule secretion or superoxide production (15–17).
A diverse array of biological function are exhibited by the group of lipid mediators known as eicosanoids, particularly during inflammatory processes (18). Eicosanoids such as HXA3 are generated through the liberation of arachidonic acid (ARA) from membrane phospholipids catalyzed by phospholipase A2 (PLA2) (16). Lipoxygenases represent an enzyme family that converts ARA into a discrete subset of bioactive lipid eicosanoids (19). The 12-lipoxygenase enzymatic activity (as defined by the site of oxygen addition to ARA) is a prerequisite for HXA3 synthesis, and is known to be exhibited by at least three lipoxygenase proteins identified in humans (16, 20, 21). The catalytic efficiency, however, varies between each enzyme and potentially between tissues where enzymes are expressed. Three human genes that encode enzymes with 12-lipoxygenase activity include alox12, alox15, and alox12B encoding 12-LO, 15-LO, and 12(R)-LO respectively (19, 22–25).
In the current study, we examined the prevalence of pathogen-induced, HXA3-mediated PMN trans-epithelial migration in a variety of lung models. We employed multiple human lung epithelial carcinoma cell lines, normal bronchial epithelial cells transformed by adenovirus, primary lung epithelial cells, and an in vivo mouse model of lung infection to explore the role of HXA3 in mediating PMN trans-epithelial migration. We analyzed lung epithelial cell models for expression of 12-lipoxygenases and evaluated the efficacy with which multiple distinct strategies to target HXA3 impact pathogen induced PMN trans-epithelial migration. We also explored whether mice produce 12-lipoxygenase products in the lung when infected with pathogen. Our studies herein provide further evidence for the hypothesis that HXA3 mediates PMN trans-epithelial migration in response to infection and likely contributes to PMN infiltration of the airspace.
Materials and Methods
Bacterial Strains
P. aeruginosa strain PAO1 and non-pathogenic E. coli K12 strain MC1000 were grown aerobically in LB-broth overnight at 37°C. For infection of epithelial cells, overnight cultures were washed once in HBSS and resuspended at a concentration of 6×107 bacteria/ml of HBSS.
Cell Culture
Source and description of lung epithelial cell lines maintained in various media with antibiotics are described in Table I. Polarized monolayers were grown on the underside of 0.33 cm2 collagen coated Transwell filters to study PMN migration in the physiological basolateral to apical direction as previously described (11–14). Epithelial barriers derived from each cell line were restrictive to the movement of small proteins as determined by measurement of horse radish peroxidase (HRP) flux across each epithelial barrier (12).
Table I.
List of Cell Lines Used in Study
| Cell Line | Description | Source | Media |
|---|---|---|---|
| H292 | Lung epithelial pulmonary mucoepidermoid carcinoma | ATCC* CRL-1848™ |
RPMI-1640* 10% HI-FBS |
| BEAS-2B | Normal bronchial epithelium that were transformed by an adenovirus | ATCC* CRL-9609™ |
DMEM*** 10% HI-FBS |
| A549 | Type II alveolar lung epithelial carcinoma | ATCC* CCL-185™ |
F-12K*** 10% HI-FBS |
| Calu-3 | Lung adenocarcinoma with epithelial morphology and phenotype | ATCC* HTB-55™ |
MEMα*** 10% HI-FBS |
| NHBE | Normal Human Bronchial Epithelium derived from healthy patient biopsy | Lonza** CC-2640 |
BEGM™** BulletKit® |
| SAEC | Small Airway Epithelial Cells derived from healthy patient biopsy | Lonza** CC-2647 |
SAGM™** BulletKit® |
American Type Tissue Culture Manassas, VA 20108 USA
Lonza Walkersville, Inc. Walkersvile, MD 21793-0127 USA
Invitrogen Corporation Carlsbad, CA 92008 USA
PMN Isolation
PMNs (polymorphonuclear leukocytes or neutrophils) were isolated from human blood treated with acid citrate/dextrose (MGH IRB protocol #: 1999-P-007782). The buffy coat was recovered by centrifugation. Plasma and mononuclear cells were removed by aspiration, and the majority of the red blood cells (RBCs) were removed using 2% gelatin sedimentation. Residual RBCs were removed by lysis in cold NH4Cl lysis buffer. After lysis, cells were washed, counted, and resuspended in HBSS(−) at a concentration of 5×107 cells/ml. (11–14).
PMN Trans-epithelial Migration Assay
Transwell inserts containing lung epithelial cell monolayers seeded on the underside were exposed to 25 μl of 6×107 bacteria/ml for 1 hr (11–14). After infection, PMNs (1×106) were added to the top (basolateral) chamber and incubated at 37°C for 2 hr. PMNs that fully migrated across the cell monolayer reaching the bottom (apical) chamber were quantified by the myeloperoxidase (MPO) assay. Uninfected lung epithelial monolayers or monolayers infected with non-pathogenic E. coli strain MC1000 serve as negative controls for PMN transmigration. Establishment of a concentration gradient of PMN chemo-attractant fMLP (100 nM added to apical chamber of uninfected monolayers at the same time that PMNs are added to the basolateral chamber) serves as a positive control for the ability of freshly isolated PMNs to migrate (26).
Cell Viability/Barrier Integrity Assays
The amount of lactic dehydrogenase (LDH) released into the supernatant with and without infection of PAO1 in the presence or absence of each of the inhibitors employed was quantified using the LDH based In Vitro Toxicology Assay Kit (Sigma, St. Louis MO). Barrier integrity of lung epithelial monolayers grown on Transwells was assayed by the horse radish peroxidase (HRP) flux assay as previously described (12).
Inhibitors
Lung epithelial cells were pre-treated 1 to 2 hours with each inhibitor followed by washing prior to infection and/or addition of PMNs for each assay. PLA2 inhibitor (ONO-RS-082), lipoxygenase inhibitor (CDC), DAG lipase inhibitor (RHC-80267) and cyclo-oxygenase inhibitor (NS-398) were purchased from Enzo Life Sciences, Inc. (Farmingdale, NY). For experiments involving stable HXA3 analogues (PN-II-218-36 & MV-I-237-20), analogues were added at various concentrations to the apical well of the infected Transwells at the same time that PMNs were added to the basolateral well and PMNs that migrated across the monolayers in the presence or absence of apically applied analogues were quantified by myeloperoxidase. Stable HXA3 ether analogues were synthesized in the Falck laboratory (manuscript in preparation). The inhibitors presented in this study did not affect cell viability either in the presence or absence of bacterial infection as assessed by both the LDH release assay and the barrier integrity assay (HRP flux). None of the compounds had any major effect on the amount of bacteria adhering to the lung epithelial monolayers.
PGE2 E.I.A.: lung epithelial cells were grown to confluence in 24-well plates and used 5 to 7 days after seeding. Lung epithelial cells were pre-treated with inhibitors for 1 to 2 hrs, washed, and infected with 6×107 bacteria/ml for 1 hr at 37°C. Each well was washed three times in HBSS followed by incubation at 37°C for 2 hrs. Supernatents were collected and the amount of PGE2 in each well was quantified using the Prostaglandin E2 Express EIA Kit from Caymen Chemical (Ann Arbor, MI) (11).
Arachidonic Acid Release Assay
Lung epithelial cells were grown to confluence in 24-well plates and used 5 to 7 days after seeding (14). Cells were washed 3 times with PBS(−) and treated with media containing 0.2 μC/ml 3H-arachidonic acid (ARA), and incubated for 18–24 hrs. Cells were then washed 3 times to remove unincorporated 3H-ARA and treated with 0.5 ml of bacteria (6×107 bacteria/ml). Following infection, cells were incubated at 37°C for up to 6 hrs. Supernatants (100μl) were collected at 2, 4, and 6 hrs and radioactivity was measured by scintillation counting. After collection of supernatant, cells were lysed with 500 μl/well of 1% SDS, 1% Triton-X-100 and sampled (250μl) for measurement by scintillation counting. Data are displayed as the percentage of cpms measured in the supernatant at each time point of the total (% ARA release = [released/released + cell associated] × 100). Lung epithelial cell viability was maintained during the 6 hr infection as determined by the LDH release assay.
RT-PCR
Lung epithelial cells were grown in 6-well plates to confluence. Cells were lysed and RNA was purified using the Aurum total RNA mini kit (Biorad). The RNA concentration was standardized to 0.1 ug/ml. Before use, samples were treated with RQ1 DNAse (Promega) according to manufacture’s protocols. cDNA was generated from 2.5 μl clean RNA using the iScript kit (Invitrogen). cDNA (2 μl) was amplified using the iTaq PCR kit (Biorad) and primers specific to the genes gapdh, alox12, alox15, alox12b, and elox3 (Table II). Primers were synthesized at the MGH DNA Core facility. The amplified product was run on a 1.5% agarose gel containing 100ug/ml ethidium bromide (Biorad) and imaged under a UV light. The expected product sizes listed in Table II were confirmed using EZ Load 100 bp PCR Molecular Ruler (Biorad).
Table II.
List of Primers Used in Study
| Gene | Forward (5′ - 3′) | Reverse (5′ - 3′) | Size (bp) |
|---|---|---|---|
| GAPDH | CCCATCACCATCTTCCAGGA | GTTGTCATGGATGACCTTGG | 285 |
| alox12 | AAGCCCAAAGCTGTGCTAAA | TGCAGCAGGAGAGCTGAGTA | 381 |
| alox15 | GAACTCAAGGTGGAAGTACC | CTTCAGGCAGGCTCAGGACG | 190 |
| alox12B | ACCGTGCAGTGCCCTCAGGA | CCGGAGTGCCAGGGTCTCGT | 183 |
| eLox3 | GGCCCAGATCCCGGACACCT | GGGCTTCCTCTCTCCGCCCA | 267 |
Extraction of Lipids from Supernatants
Lung epithelial cells were seeded on 25 cm2 (SAEC) or 162 cm2 (H292 & BEAS-2B) flasks and grown to confluence. Confluent cell monolayers were treated with or without 6×107 bacteria/ml HBSS for 1 hr at 37°C, washed three times with HBSS, and incubated an additional 2 hrs at 37°C. Supernatants were collected acidified to pH 4.0 and, in experiments involving eicosanoid quantification, mixed with lipid extraction standards (LTB4-d4/15(S)-HETE-d8 obtained from CaymenChemical) (27, 28). Acidified supernatants were poured through a Supelco Discovery®; DSC-18 SPE Column and eluted with methanol. The lipid fraction suspended in methanol was dried under a stream of nitrogen to 100 μl methanol and stored at −80°C until processed further (11).
Measurement of lipid associated PMN chemo-attractant activity
Each extracted lipid sample for H292 lung epithelial cells with or without PAO1 infection (prepared in triplicate and stored at −80°C) was dried under a stream of nitrogen and re-suspended in 1.7 ml HBSS. One ml of re-suspended extracted lipid supernatants (referred to as straight) was added to the apical well of a Transwell containing H292 lung epithelial barrier. A volume of 0.6 ml re-suspended lipids was diluted in 1.2 ml HBSS (referred to as Diluted 1:3). One ml of diluted 1:3 resuspended lipids was added to the apical well of a Transwell containing H292 lung epithelial barrier. The remaining 0.8 ml diluted 1:3 re-suspended lipids were further diluted by adding 0.8 ml HBSS (referred to as Diluted 1:6). One ml of diluted 1:6 resuspended lipids was added to the apical well of a Transwell containing H292 lung epithelial barrier. The remaining 0.6 ml diluted 1:6 re-suspended lipids were further diluted by adding 0.6 ml HBSS (referred to as Diluted 1:12). One ml of diluted 1:12 resuspended lipids was added to the apical well of a Transwell containing H292 lung epithelial barrier. Lipids extracted from BEAS-2B and SAEC supernatants that have been treated either with or without PAO1 (prepared in triplicate and stored at −80°C) were re-suspended in 1 ml HBSS after drying remaining methanol with a stream of nitrogen. Re-suspended lipid extracts were then added to the apical well of a Transwell containing H292 lung epithelial barrier. PMNs (1×106) were added to the top (basolateral) chamber in a volume of 120 μl for all diluted re-suspended lipid samples that have been added to the bottom (apical) chamber. Transwells were then incubated at 37°C for 2 hr and PMNs that have fully migrated across the cell monolayer reaching the bottom (apical) chamber were quantified by the myeloperoxidase assay (11).
Assessment of soluble epoxide hydrolase sensitivity
Lipid extracts from the supernatant of lung epithelial cells in the presence or absence of PAO1 infection (prepared in triplicate and stored at −80°C) were re-suspended in 1 ml HBSS after removing methanol by drying under a stream of nitrogen. Each sample was split into two 0.5 ml fractions. To one of the two fractions, 3.5 μl of soluble epoxide hydrolase (sEH – 10 mg/ml) was added (29, 30). Fractions with and without sEH were mixed at 100 rpm for 2 hrs at 30°C. After incubation, 0.7 ml HBSS was added to each fraction, mixed and 1 ml mixed fraction was added to the apical well of a Transwell containing H292 lung epithelial barrier. PMNs (1×106) were added to the top (basolateral) chamber in a volume of 120 μl for all diluted re-suspended lipid samples with or without sEH treatment that have been added to the bottom (apical) chamber. Transwells were then incubated at 37°C for 2 hr and PMNs that have fully migrated across the cell monolayer reaching the bottom (apical) chamber were quantified by the myeloperoxidase assay.
Measurement and Quantification of Eicosanoids
HXA3 and 12-HETE were quantified after lipid extraction from either human lung epithelial cell supernatants or bronchoalveolar lavage (BAL) fluid of mice by LCMS/MS based lipidomics (11). In brief, extracted samples were analyzed by a triple quadruple linear ion trap LC/MS/MS system (MDS SCIEX 3200 QTRAP) equipped with a LUNA C18-2 mini-bore column using a mobile phase gradient of water/acetonitrile/acetic acid (72:28:0.01, v:v:v) and isopropanol/acetonitrile (60:40, v:v) with a 0.50 ml/flow rate (31). MS/MS analyses were carried out in negative ion mode and prominent fatty acid metabolites were quantified by multiple reaction monitoring (MRM mode) using established transitions for HXA3 (335→127 m/z), 12-HETE (319→179 m/z), LTB4-d4 (339→197 m/z) and 15-HETE-d8 (327→182 m/z). Calibration curves (1–1000 pg) and specific LC retention times for each compound were established with synthetic standards (Cayman Chemical, Ann Arbor, MI). Structures were confirmed for selected autacoids by MS/MS analyses using enhanced product ion mode with appropriate selection of the parent ion in quadrupole 1.
Mouse Model of Acute Pneumonia
C3H/HeN mice (6–8 weeks old) purchased from Harlan Laboratories were injected i.p. with anesthesia (Ketamine + Xylazine) prior to intra-nasal challenge with 4×108 CFU Pseudomonas aeruginosa (PAO1) or PBS control (10μl into each nostril) (32, 33). Five mice per treatment for each individual experiment were used. After 18 hrs, mice were euthanized by CO2 asphyxiation in accordance with an IACUC approved protocol.
Collection of BAL fluid
Broncholveolar lavage was performed at 18 hr post infection using 1 ml of 0.5 mM EDTA in PBS without calcium or magnesium and repeated three times. Fluid was centrifuged (220 × g) for 5 minutes and harvested cells were washed with ammonium chloride lysis buffer to remove RBCs. Total cell counts were determined by hemocytometer diluted in trypan blue containing buffer (32, 33). Myeloperoxidase activity of BAL fluid cells was assessed as previously described (12). For measurement of eicosanoids 12-HETE and HXA3, supernatants from centrifuged BAL fluid was subjected to the lipid extraction protocol with standards and quantified by LCMS/MS as described above.
Statistics
Data displayed for each figure are presented as a representative experiment with a mean (standard deviation) of at least three independent data points/condition. Each experiment was repeated multiple times yielding similar results. Statistical analysis was performed by a two-tailed unpaired Student’s t test for each internally controlled experiment and considered significant when p values are less than 0.05.
Results
A diversity of human lung epithelial cell models were examined to determine whether they were capable of forming barriers on Transwells and whether they facilitate pathogen induced PMN trans-epithelial migration. The lung epithelial models include both carcinoma cell lines as well as transformed normal bronchial epithelial cells (Table 1). All cell lines form functional barriers, confirmed by evidence for the restriction of HRP movement across each monolayer of A549, Calu-3, H292, and BEAS-2B cells grown on Transwells (data not shown) (12). To investigate whether P. aeruginosa (PAO1) is capable of instigating PMN trans-epithelial migration across each of these barriers, the apical surface of each monolayer was treated with HBSS or infected with either pathogenic PAO1 or non-pathogenic E. coli (MC1000). After 1 hr, non-adherent bacteria were washed away and PMNs were added to the basolateral side (12, 26). The number of PMNs that migrate from the basolateral to the apical well after incubation for 2 hrs was determined by quantification of MPO activity. We found a consistent trend in PMN trans-epithelial migration amongst all of the lung epithelial barriers tested (Fig. 1A). Few PMNs migrated across uninfected monolayers or monolayers infected with MC1000. Infection with PAO1, on the other hand, resulted in a robust and highly significant increase in the numbers of PMNs that migrated across lung epithelial barriers. We also observed that a significant number of PMNs migrated across uninfected monolayers derived from each of the cell lines in response to an imposed chemo-tactic gradient of fMLP. This served as a positive control for the ability of isolated PMNs to migrate towards a chemo-tactic gradient in each assay with each distinct cell line monolayer (26). These data are consistent with observations previously reported for A549 and Calu-3 cells, suggesting that PAO1 induced PMN trans-epithelial migration is a highly reproducible phenomena observed using a range of distinct lung epithelial cell barrier models (12, 14).
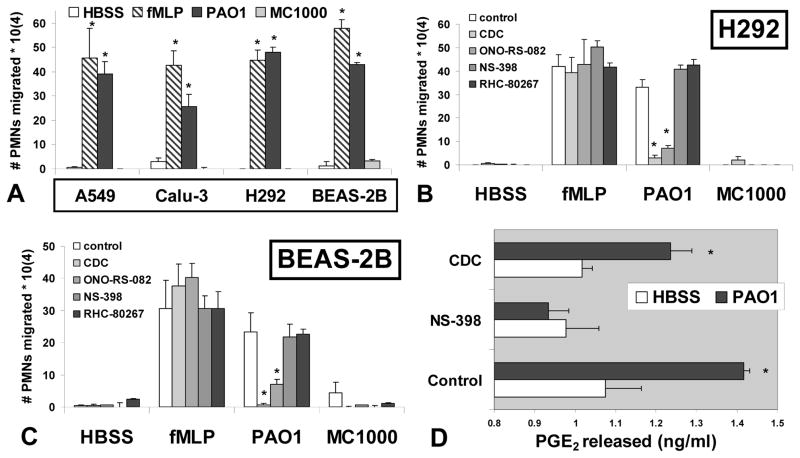
Figure 1. PMN migration across P. aeruginosa treated lung epithelial barriers is diminished following inhibition of the HXA3 synthetic pathway.
(A) Four distinct lung epithelial cell lines (Table I: A549, Calu-3, H292, and BEAS-2B) grown on Transwells to form epithelial barriers were treated with either P. aeruginosa strain PAO1 (black bars) or K12 E. coli strain MC1000 (gray bars). Monolayers incubated in HBSS alone serve as a negative control (white bars). A chemo-attractant gradient of fMLP (100 nM) was established to serve as a positive control for PMN movement (dashed bars). The symbol (*) represents a statistically significant difference compared to negative controls (HBSS and MC1000). (B & C) Transwells were pre-treated with pharmacological inhibitors CDC - 12-lipoxygenase inhibitor (50 μM), ONO-RS-082 - PLA2 inhibitor (5 μM), RHC-80267 - DAG lipase inhibitor (50 μM), NS-398 -COX inhibitor (50 μM) or vehicle control (1:1000 DMSO) prior to infection. The number of PMNs that migrate across lung epithelial barriers H292 (B) and BEAS-2B (C) was assessed in response to infection with PAO1, MC1000, or uninfected controls (HBSS/fMLP) with or without pre-treatment of each pharmacological inhibitor. The symbol (*) represents a statistically significant difference within a treatment group comparing inhibitor to vehicle control. (D) The amount of PGE2 released (ng/ml) by H292 cells in the presence or absence of PAO1 infection with or without pharmacological inhibitor pre-treatment was quantified by E.I.A.. The symbol (*) represents a statistically significant difference between uninfected (HBSS) and PAO1 infected within a treatment group. Statistical significance was determined using a two-tailed unpaired Student’s T-test (p < 0.05). Each data point represents the average of at least three separate wells. Each experiment was performed on at least three separate occasions yielding similar results.
Inhibitors of 12-lipoxygenases and phospholipase A2 (PLA2) were previously observed to potently interfere with PAO1 induced PMN migration across A549 barriers (11, 12, 14). To expand investigation of this phenomenon in alternative lung epithelial cell models, we established monolayers of a distinct carcinoma lung epithelial cell line (H292) and a transformed normal bronchial epithelial cell line (BEAS-2B) (Table 1). Prior to treatment with buffer or infection with bacteria, monolayers were treated for 1 to 2 hrs with pharmacological inhibitors of eicosanoid metabolic enzymes. ONO-RS-082 and CDC inhibit PLA2 and 12-lipoxygenases, respectively. Both enzymatic activities are crucial for the generation of the PMN chemo-attractant HXA3. The cyclo-oxygenase inhibitor NS-398 interferes with synthesis of a group of eicosanoids unrelated to the synthesis of HXA3 known as prostaglandins, which are bio-active, but do not serve as PMN chemo-attractants (34, 35). The diacyl glycerol (DAG) lipase inhibitor RHC-80267 was previously shown not to impact PAO1 induced epithelial migration across A549 monolayers (14). DAG lipase is an alternative means of generating the eicosanoid precursor ARA from DAG rather than from membrane phospholipids as is the case with PLA2 (36).
Pretreatment of H292 and BEAS-2B lung epithelial monolayers with either a 12-lipoxygnease inhibitor (CDC) (37) or a PLA2 inhibitor (ONO-RS-082) (38) potently blocked PAO1 induced PMN trans-epithelial migration, indicating a key role for these epithelial enzymatic activities in facilitating pathogen induced PMN trans-epithelial migration (fig 1B & 1C). Neither inhibitor interfered with migration across uninfected monolayers in response to an imposed fMLP gradient indicating that the general ability of PMNs to move across epithelial monolayers is not impacted by pre-treatment with either inhibitor. These studies are consistent with previous observations using A549 lung epithelial monolayers (12, 14). Also consistent with previous studies involving A549 monolayers, the DAG lipase inhibitor has no significant affect on PAO1 induced PMN migration across H292 and BEAS-2B monolayers (fig 1B & 1C) (14). The inhibitor of the cyclo-oxygenase pathway (NS-398), an eicosanoid metabolic pathway unrelated to HXA3 synthesis, exerted no significant impact on PAO1 induced PMN trans-epithelial migration (fig 1B & 1C). We have previously demonstrated that PAO1 infection of lung epithelial cells results in a significant increase in the release of prostaglandins, specifically prostaglandin E2 (PGE2) (11). Importantly, the concentration of NS-398 employed in the PMN migration experiments (fig. 1B & 1C) was sufficient to prevent PAO1 induced PGE2 release (fig 1D), confirming the effectiveness of the pharmacological inhibitor in our assay. Pre-treatment of H292 cells with the 12-lipoxygenase inhibitor CDC has no effect on PGE2 release as would be expected. CDC at an equivalent concentration is, however, capable of interfering with PAO1 induced HXA3 release as previously reported (12). Migration in response to HBSS alone or infection with MC1000 was minimal for all conditions tested (fig. 1B & 1C).
The rate limiting step in eicosanoid synthesis is often attributed to the generation of ARA (39, 40). We have previously demonstrated that infection of A549 lung epithelial cells with PAO1 result in a significant release of ARA (11, 14). We examined ARA release from a distinct transformed human lung epithelium cell line (H292) as well as from primary small airway epithelial cells (SAEC) after 2 hrs, 4 hrs, and 6 hrs stimulation with PAO1 or MC1000 (fig. 2A & 2B). The results indicated a significant elevation in ARA release by both H292 and SAEC epithelium treated with pathogenic PAO1 over time relative to untreated epithelial cells or epithelium treated with non-pathogenic MC1000 (Fig. 2A & 2B). These results are consistent with the hypothesis that PAO1 infection induces increased cellular generation of eicosanoids.
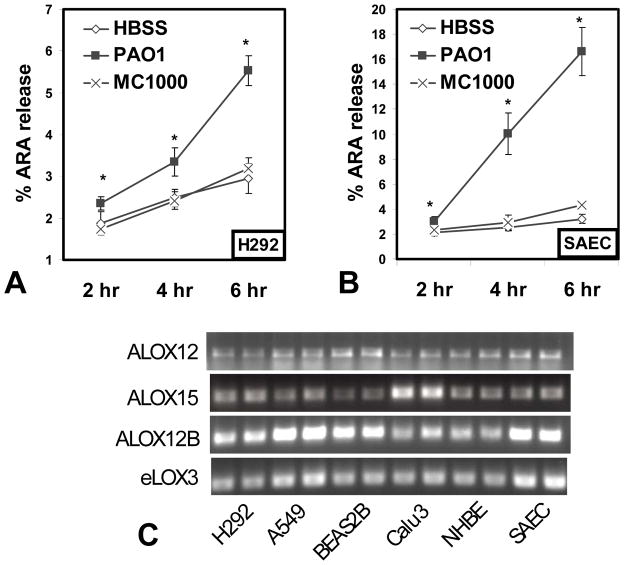
Figure 2. Arachidonic acid release following P. aeruginosa infection and lipoxygenase gene expression is lung epithelial cell models.
The lung epithelial cell line H292 (A) or primary lung epithelial cells SAEC (B) (Table I) were incubated overnight with 3H-ARA, followed by washing and treatment with HBSS alone (white diamonds), infection with P. aeruginosa strain PAO1 (black squares) or infection with K12 E. coli strain MC1000 (x symbol). Supernatants were sampled at 2 hr, 4 hr, and 6 hr post treatment/infection. The % ARA release represents the amount released into the supernatant as a percentage of the total (ARA released + cell associated ARA). The symbol (*) represents a statistically significant increase compared to HBSS control of the same time point using a two-tailed unpaired Student’s T-test (p < 0.05) and each data point represents an average of at least three separate wells. Each experiment was performed on at least three separate occasions yielding similar results. (C) RNA was extracted from a panel of human lung epithelial cells (Table I) to assess gene expression by RT-PCR, shown here in duplicate sample wells. The known human 12-lipoxygenases (alox12, alox15, and alox12B) and the hepoxilin synthase (elox3) were analyzed individually using gene specific primers (Table II).
Our studies suggest that 12-lipoxygenase activity is critical for PAO1 induced PMN trans-epithelial migration. There are three human lipoxygenases that exhibit 12-lipoxygenase activity including 12-LO (gene alox12), 12(R)-LO (gene alox12B), and 15-LO (gene alox15) (19, 21, 25). The 12-lipoxygenases convert ARA to HXA3 via the short-lived intermediate 12-HpETE (20). In the case of 12(R)-LO, an additional enzyme eLOX3 (gene elox3), serving as a hepoxilin synthase, is required to convert 12-HpETE to HXA3 (19, 22). A full panel of lung epithelial models described in Table I including multiple cell lines, primary normal human bronchial epithelial cells (NHBE), and primary small airway epithelial cells (SAECs) were evaluated for expression of all known lipoxygenase genes involved in direct synthesis of HXA3. Specific primers to amplify alox12, alox15, alox12B, and elox3 were designed and utilized to analyze gene expression by reverse transcription (RT)-PCR (Table II). We found that all four 12-lipoxygenase genes were expressed in each and every lung epithelial model investigated, suggesting that lung epithelial cells indeed express the genes necessary to synthesize the PMN chemo-attractant HXA3 (Fig. 2C).
Our hypothesis predicts that lung epithelial cells synthesize and release HXA3 in response to infection with PAO1 leading to directed PMN migration across infected epithelial barriers. Since HXA3 is an eicosanoid, its chemotactic potential would be expected to be enriched in the lipid extracted fraction of supernatants from lung epithelial cells infected with PAO1. To determine whether H292 lung epithelial cells produce increased lipid associated chemotactic activity in response to PAO1 infection, H292 cells were treated with PAO1 after which supernatants were collected, acidified, and passed through a C18 column followed by elution with methanol. Methanol fractions were dried under a stream of nitrogen and resuspended in HBSS for assessment of PMN chemotactic potential by establishing a gradient of the fraction at various dilutions across epithelial monolayers grown on Transwells (fig. 3A & B). PAO1 clearly induced significantly more lipid associated chemotactic activity when compared to either uninfected H292 cells (Fig. 3A) or H292 cells infected with the non-pathogenic E. coli strain MC1000 (Fig. 3B). The observation that PAO1 is capable of significantly increasing PMN chemotactic activity in the lipid fraction of lung epithelial cell supernatant was not restricted to the H292 cell line. The normal bronchial transformed cell line BEAS-2B (Fig. 3C) as well as the primary small airway epithelial cells (Fig. 3D) also released significantly greater lipid associated PMN chemotactic activity when cells were infected with PAO1 as assessed by quantifying the number of PMNs migrating across an H292 barrier in response to gradients of supernatant derived lipid enriched fractions from infected and uninfected BEAS-2B and SAEC cells.
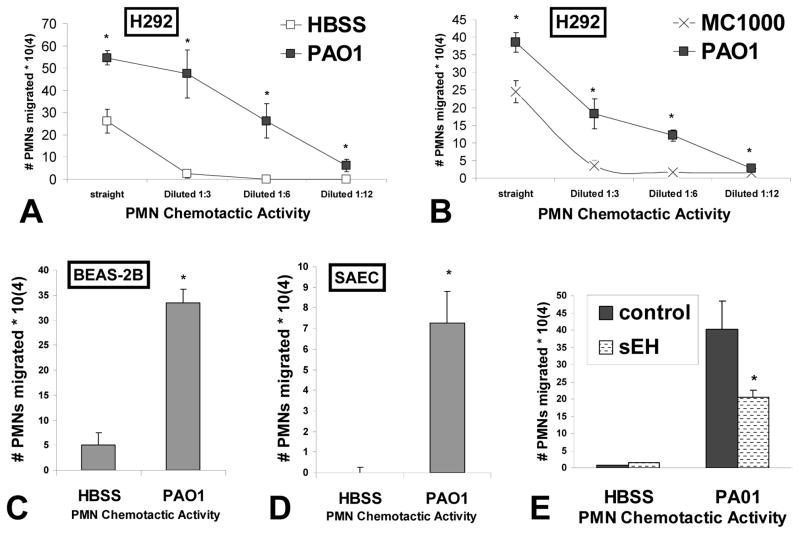
Figure 3. Lipid based chemotactic activity is increased in supernatants of various lung monolayers treated with P. aeruginosa and is sensitive to soluble epoxide hydrolase treatment.
The amount of PMN chemotactic activity was quantified in serial dilutions of lipids extracted from the supernatants of H292 cells infected with P. aeruginosa strain PAO1 (black squares) and compared with serial dilutions of lipid extracted supernatants from H292 cells in the absence of infection (HBSS, white squares) (A) or infected with K12 E. coli strain MC1000 (x symbol) (B). The amount of PMN chemotactic activity was quantified in lipids extracted from the supernatants of BEAS-2B (C) and primary lung epithelial cells SAEC (D) in the presence and absence of PAO1 infection. The symbol (*) represents a statistically significant difference compared to negative controls (HBSS and MC1000). (E) Lipids from the supernatants of uninfected (HBSS) and PAO1 infected H292 cells were extracted and split in two groups; a control group and a group subjected to treatment soluble epoxide hydrolase (sEH). Both groups were then evaluated for PMN chemotactic activity. The symbol (*) represent a statistically significant difference in PAO1 induced lipid extracted chemotactic activity when comparing sEH treated versus untreated. A two-tailed unpaired Student’s T-test (p < 0.05) was employed to determine statistical significance. Each data point represents the average of at least three separate wells. Experiments were performed on at least two separate occasions yielding similar results.
Our previous studies have demonstrated that the lung epithelial cell line A549 releases the PMN chemotactic eicosanoid HXA3 in response to infection with PAO1 (12). We observed herein that PAO1 infection of H292 cells results in a greater than 5-fold increase in the amount of HXA3 released as quantified by LCMS (2.6 pg +/− 1.9 uninfected versus 14.4 pg +/− 0.9 PAO1 infected). This observation is consistent with our finding of increased lipid associated chemotactic activity following PAO1 exposure (fig. 3A). To determine if HXA3 contributes to the increased lipid associated chemotactic activity described above, we employed the enzyme soluble epoxide hydrolase (sEH) (29). A unique structural feature of HXA3 amongst PMN lipid chemotactic factors is that HXA3 possesses an epoxide group (16). The enzyme sEH is capable of hydrolyzing the epoxide group and converting hepoxilins into trioxillins, which lack PMN chemotactic activity (41). Lipid associated fractions from conditioned supernatant of PAO1 treated and untreated H292 cells were subjected to treatment with sEH prior to analysis of chemotactic potential. As expected, lipid extracted conditioned supernatants from PAO1 treated H292 cells exhibited substantial chemotactic activity, whereas very little chemotactic activity was observed from lipid extracted conditioned supernatants from uninfected H292 cells (Fig. 3E). The amount of chemotactic activity derived from lipid extracted conditioned supernatants of PAO1 infected H292 cells was significantly reduced when pretreated with sEH, suggesting that chemotactic factor responsible for directing PMNs to move across epithelial monolayers was sensitive to sEH treatment (Fig 3E). Such results are consistent with the hypothesis that HXA3 is a lipid associated chemotactic factor responsible for PAO1 induced PMN trans-epithelial migration (12).
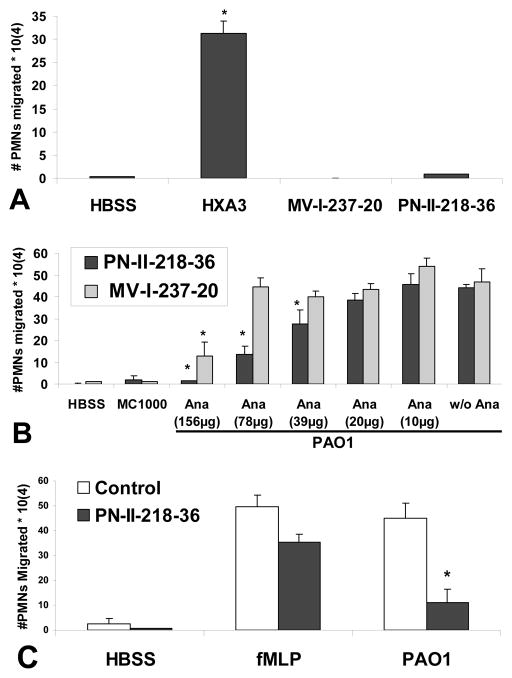
Our evidence thus far suggests that lung epithelial cells produce HXA3 in response to infection with P. aeruginosa and this molecule serves as a PMN chemo-attractant hypothesized to guide PMNs across epithelial monolayers. We next employed structural analogues of HXA3 whereby the epoxide was replaced with ether, designed to be stable yet non-functional. Elimination of the epoxide abolishes chemotactic activity based on observations that trioxilins, HXA3 metabolites lacking the epoxide, no longer possess PMN chemotactic activity (15). Further, it is believed that the epoxide group contributes to the relative instability of HXA3 and removal generates a more stable molecule (16). One of the analogues termed PN-II-218-36 was designed to competitively inhibit HXA3 chemotactic activity, as it is structurally identical to HXA3 with the exception of the epoxide replaced with ether. In light of the well known ω-hydroxylation of HXA3, a second compound MV-I-237-20 bearing an additional hydroxyl group on the C(20)-position was also prepared to address this contingency (42, 43). A gradient of HXA3 added exclusively to the apical well of lung epithelial monolayers drove greater than 30% PMNs added to the basolateral well across the monolayer (Fig. 4A), consistent with previous reports (12, 13). At the same concentration, neither HXA3 analogue exhibits any PMN chemotactic activity (Fig. 4A). When the compound PN-II-218-36 was added to the apical side of PAO1 infected lung epithelial monolayers, PMN trans-epithelial migration was impeded in a dose dependant manner with complete inhibition at the addition of 156 μg/ml PN-II-218-36 (Fig. 4B, black bars). The structurally less similar analogue MV-I-237-20 was not as effective at interfering with PMN trans-epithelial migration (Fig. 4B, gray bars). The effect of PN-II-218-36 appears to be selective to PAO1 induced migration as PMNs migrating towards an fMLP gradient were significantly less impacted by the presence of analogue PN-II-218-36 (Fig. 4C).
Figure 4. Stable analogues of HXA3 lacking epoxide exhibit an inhibitory effect on P. aeruginosa induced PMN trans-epithelial migration.
The ability of an established gradient of HXA3 (2.5 μg/ml) and HXA3 structural analogues (MV-I-237-20 & PN-II-218-36 (500 μg/ml)) across H292 epithelial barriers to promote PMN trans-epithelial migration is evaluated (A). For (A), the symbol (*) represent a statistically significant difference when compared to no gradient (HBSS) using A two-tailed unpaired Student’s T-test (p < 0.05). Structural analogues MV-I-237-20 and PN-II-218-36, at a range of doses, were added to the apical side at the same time PMNs were added to the basolateral side to determine if either analogue impacts P. aeruginosa strain PAO1 induced PMN trans-epithelial migration (B). The ability of PN-II-218-36 (110 μg) to influence trans-epithelial PMN migration in response to either an established gradient of fMLP (100 nM) or infection with PAO1 is evaluated (C). The symbol (*) represent a statistically significant difference when compared to PAO1 induced trans-epithelial migration in the absence of analogue using a two-tailed unpaired Student’s T-test (p < 0.05). Each data point represents the average of at least three separate wells. Experiments were performed on at least two separate occasions yielding similar results.
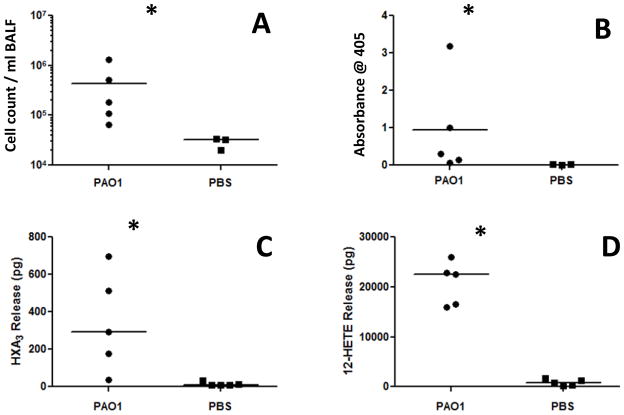
P. aeruginosa induced acute pneumonia models have been widely employed to investigate various aspects of PMN recruitment into the airspace (44–53). Our in vitro system models migration of PMNs across the airway epithelial barrier, an event representing the final step in PMN emigration from circulation to the airspace during acute infectious pneumonia. Since we hypothesize that HXA3 is the key chemo-attractant for directing PMNs across the lung epithelial barrier during infection with PAO1, we next investigated whether HXA3 is produced in the airspace during in vivo infection. C3H/HeN mice were challenged with 4×108 CFU PAO1 or mock infection (PBS) (10μl into each nostril) and sacrificed 18 hrs post infection. Bronchoalveolar lavage (BAL) fluid was collected to determine the number of cells and the amount of MPO activity present. In addition, the quantity of 12-LO synthesized eicosanoids including 12-HETE and HXA3 was measured in the BAL fluid. There was a significant increase in the number of cells and MPO activity in PAO1 infected mice suggesting substantial PMN recruitment to the airspace consistent with previous studies (figure 5A & B) (32, 33). This observation correlated with a highly significant increase in 12-LO eicosanoids in the BAL fluid of infected mice, both PMN chemo-attractant HXA3, and the more stable 12-LO metabolite 12-HETE (figure 5C & D).
Figure 5. The number of PMNs and the concentration of 12-lipoxygenase products are increased in the BAL fluid in response to Pseudomonas aeruginosa intranasal infection.
Mice were challenged via intranasal inoculation with P. aeruginosa strain PAO1 (5 mice) or PBS (5 mice) and sacrificed after 18 hours. BAL fluid was collected and analyzed for cellular infiltrate, myeloperoxidase activity, and 12-lipoxygenase eicosanoids. The quantity of cellular infiltration (A) MPO activity (B) HXA3 (C) and 12-HETE (D) present in the airspace after infection with PA01 was compared to PBS mock infection control and differences from control reaching statistical significance using a two-tailed unpaired Student’s T-test (p < 0.05) are represent by (*). In vivo experiments were performed on two separate occasions yielding similar results.
Discussion
PMN infiltration of the airway mucosa and accumulation in the airspace, if excessive and prolonged, can be severely detrimental to health during airway inflammatory disease states (5–7). Although recruitment of PMNs to mucosal surfaces is critical during infection to provide pathogen clearance, pathology from such infections often reflects indiscriminant and potent actions of PMNs on host tissue (52). Moreover, in the case of airway inflammation not directly associated with microbial threats, any potential benefits derived from PMN presence in the airspace would appear to be outweighed by their propensity to cause host tissue damage. Therefore, a thorough understanding of the mechanism by which PMNs not only exit the microvasculature, but also specifically how they cross mucosal epithelial barriers during disease may lead to the development of therapies geared towards reducing the numbers of PMNs that breach the mucosal barrier and collect in the airspace (6, 10).
A plethora of host derived chemo-attractants are described that engage PMNs in an effort to relay them to the airspace (54). These include protein CXC chemokines (CXCL1-3, 5, & 8), CXC-like extracellular matrix breakdown product N-acetyl Pro-Gly-Pro (PGP), protein fragment complement component C5a, and lipid mediators leukotriene B4 (LTB4) & platelet activating factor (PAF) (55–58). Despite the wealth of knowledge regarding these chemo-attractants and their receptors, a clear understanding of their respective roles in PMN recruitment following infection in vivo is unclear. CXC chemokines are released basolaterally from infected mucosal epithelial cells where they imprint underlying extracellular matrix enabling directed PMN movement through sub-epithelial space (10, 26, 59, 60). Evidence from in vivo modeling notes a requirement for CXC chemokines such as CXCL5 and CXC-like PGP in recruitment of PMNs to the airspace, likely by directing PMNs across the endothelial barrier and through the tissue space leading up to the epithelial barrier (55–58, 60, 61). Chemo-attractants LTB4, PAF, and C5a are found in the infected lung, but a discrete role in the orchestration of PMN movement from blood to airspace has not been elucidated (62, 63). The bacterial tri-peptide fMLP triggers anti-bacterial responses once primed PMNs arrive at the site of infection, but is unlikely involved in recruitment (26, 57).
Using a multifaceted in vitro model, we previously identified the eicosanoid HXA3 as the key chemotactic signal that mediates directed migration of PMNs across epithelial barriers (12, 15, 60). Identification of HXA3 as mediator of PMN trans-epithelial migration thus reveals an additional, but uniquely important chemo-attractant hypothesized to facilitate PMN recruitment to the airspace during infection. Studies reported herein provide further support both in vitro and in vivo for the involvement of HXA3 in facilitating trans-epithelial migration of PMNs in the lung.
As described above, we demonstrate that the phenomena of P. aeruginosa induced PMN migration across lung epithelial barriers is shared amongst a range of distinct lung epithelial cell barrier models. P. aeruginosa induced PMN trans-epithelial migration represents both a phospholipase A2 and 12-lipoxygenase dependant event as evidenced by failure of PMNs to migrate in response to epithelial infection following pretreatment with specific inhibitors of each of these enzymatic pathways. PLA2 and 12-lipoxygenase enzymatic pathways are responsible for mediating the synthesis of HXA3 (16, 60). At present, it is unclear which specific PLA2 or 12-lipoxygenase isoform is critical for lung epithelial cells to synthesize HXA3 in response to P. aeruginosa infection. Our previous studies suggest that the calcium dependant PLA2 isoform cytosolic PLA2-α (c PLA2-α), generally thought to be the major PLA2 isoform responsible for eicosanoid generation, is not involved in HXA3 synthesis despite being activated in lung epithelial cells upon infection with P. aeruginosa and serving a key role in the generation of the eicosanoid prostaglandin E2 (11).
Since 12-lipoxygenase activity has been described to be present in more than a one enzyme, multiple enzymatic sources for HXA3 generation can be considered (19, 64). In the context of human lipoxygenases, there are three known enzymes with 12-lipoxygenase activity: 12-LO, 12(R)-LO, and 15-LO encoded by the genes alox12, alox12B, and alox15, respectively (19, 21, 25). 12(R)-LO additionally requires the hepoxilin synthetic enzyme eLOX3 encoded by the gene elox3 to generate HXA3 (22). We observed herein that all lung epithelial models, including primary small airway epithelial cells and primary bronchial epithelial cells, express all four known human lipoxygenase genes that encode enzymes with the capacity for HXA3 synthesis. While these different lipoxygenase enzymes catalyze HXA3 formation at different rates, each of these expressed genes has the potential to encode enzymes involved in the production of HXA3 by lung epithelial cells in response to treatment with P. aeruginosa. Dissecting the lipoxygenase family in an inflammatory context to determine which lipoxygenase enzyme(s) are critical contributors to the generation of HXA3 represents an important future pursuit (19, 21, 64).
We have previously demonstrated that A549 lung epithelial cells produce HXA3 in response to P. aeruginosa and gradients of HXA3 established across A549 barriers facilitate PMN trans-epithelial migration (12, 13). We found in the current investigation that infection of the H292 cell line also resulted in the release of HXA3 and gradients of HXA3 across H292 monolayers drive PMN trans-epithelial migration. In addition to multiple lung epithelial cell lines, we observed that primary lung epithelial cells release the universal eicosanoid precursor ARA in response to infection with P. aeruginosa in a time dependent manner. Increased release of ARA is supportive of the hypothesis that enhanced production of eicosanoids such as HXA3 occurs in response to pathogen infection. Since our hypothesis assumes that HXA3 is released in response to infection and this PMN chemo-attractant drives trans-epithelial migration, we would predict that the lipid enriched fraction of the supernatant from P. aeruginosa infected lung epithelial cells would possess abundant chemotactic activity. This was indeed the case as multiple lung epithelial cell lines, including primary small airway epithelial cells, exhibited a significant increase in lipid fraction associated PMN chemotactic activity in response to infection with P. aeruginosa. Furthermore, this lipid associated PMN chemotactic activity was diminished when exposed to soluble epoxide hydrolase (sEH). HXA3 is distinguishable from other lipid chemo-attractants such as LTB4 and PAF in that HXA3 possesses an epoxide within its structure that is critical for activity and vulnerable to the actions of sEH (16, 29, 41). Clearly, P. aeruginosa infection of lung epithelial cells results in the increased production of lipid associated PMN chemotactic activity that is sensitive to sEH and this observation is consistent with an increased presence of functional HXA3 in the supernatants of infected lung epithelial cells.
To further probe the hypothesis that HXA3 mediates P. aeruginosa induced PMN trans-epithelial migration, we employed structural analogues of HXA3 that lack the critical epoxide. Both HXA3 structural analogues, whereby the epoxide is replaced with an ether, exhibit no PMN chemotactic activity, which is in direct contrast to native HXA3 that exhibits abundant PMN chemotactic activity. Each analogue does however display antagonistic activity. Addition of the epoxide deficient HXA3 structural analogues significantly impedes P. aeruginosa induced PMN trans-epithelial migration. It is unclear how exactly the analogues interfere with P. aeruginosa induced PMN trans-epithelial migration. We speculate that such analogues may serve as competitive inhibitors of HXA3. In the P. aeruginosa induced PMN trans-epithelial migration assays, we hypothesize that HXA3 is produced by the lung epithelial cells in response to infection and this drives PMNs across the epithelial monolayer. Addition of inhibitory analogues might interfere with the ability of native HXA3 to interact with PMNs and promote chemotaxis. Consistent with the notion is the observation that HXA3 structural analogues were more effective at interfering with P. aeruginosa induced PMN trans-epithelial migration than with PMN migration in response to a gradient of the peptide chemo-attractant fMLP. Little is currently known regarding the receptor that HXA3 interacts with on PMNs to evoke a chemotactic response. Future studies using native HXA3 and the inhibitory analogues described above will greatly assist in addressing this important issue.
Investigators have explored many aspects of PMN airspace recruitment using P. aeruginosa infected mice. Such studies have established roles for key adhesion molecules, cytokines, chemo-attractants, toll-like receptors, signaling components, and transcription factors in the multifaceted process of emigration of PMNs from bloodstream to airway (44–53). Multiple studies have now convincingly established HXA3 as a PMN chemo-attractant (12, 13, 15, 17, 60). We have shown that airway epithelial cells secrete HXA3 in response to infection and hypothesize that HXA3 is the key chemo-attractant required to facilitate PMN breach of the mucosal barrier. To extend our in vitro observations into the context of an in vivo model, we examined whether an increase in the concentration of HXA3 occurs in the airspace of mice in association with PMN accumulation subsequent to infection. Using a well established model of acute bacterial pneumonia in mice, we observed that PMNs accumulate in the airspace following intra-nasal exposure to P. aeruginosa, consistent with previous observations. Further, we observed a significant increase in the presence of eicosanoids derived from the 12-lipoxygenase enzymatic pathway including 12-HETE and HXA3. Therefore, our results suggest that in addition to an increase in the number of PMNs in the airspace in response to P. aeruginosa infection, there is also an increase in the concentration of HXA3 measured in the BAL fluid. Although increased presence of HXA3 located within the airspace and associated with greater numbers of transmigrated PMNs suggests the potential for HXA3 in serving a role in driving PMNs across the mucosa into the airspace, such an observation is insufficient to demonstrate a cause and effect relationship. However the novel finding that in vivo infection of the lung with P. aeruginosa results in release of HXA3 in the airspace in combination with the substantial evidence from in vitro modeling demonstrating the role of HXA3 in driving PMN trans-epithelial migration encourages further investigation of this potentially important inflammatory mechanism (11–15, 17, 60). Future studies will examine if reducing HXA3 synthesis and/or activity in vivo will have an impact on the numbers of PMNs that accumulate in the airspace following infection.
In conclusion, our study herein provides further evidence for the role of HXA3 in PMN recruitment to the airspace, particularly with respect to its chemotactic activity in guiding PMNs across the mucosal barrier. Based on our studies to date, interfering with the synthesis and/or function of the eicosanoid chemo-attractant HXA3 represents a potentially compelling mechanistic target for developing and exploring a novel class of pharmaceutical compounds with significant untapped therapeutic potential towards alleviating PMN mediated mucosal surface injury during lung disease. Eicosanoid subsets including leukotrienes and prostaglandins have been exploited as therapeutic targets to alleviate overzealous inflammation (65). Multiple leukotriene inhibitors are available for treatment of allergies and asthma including 5-LO inhibitor Zileuton (66). Ibuprofen, a cyclo-oxygenase inhibitor, is an anti-inflammatory therapy used for many ailments including cystic fibrosis (67). There are few, if any, studies exploring the therapeutic benefits of 12-LO inhibition or HXA3 neutralization in lung disease. There are many circumstances where controlling PMN infiltration into the airspace could alleviate inflammatory damage and improve lung function. For disease processes as diverse as pneumonia, cystic fibrosis and asthma, such a targeted therapeutic strategy could prove effective. Further exploration of the role of HXA3 in PMN recruitment is warranted in order to realize this novel potential therapeutic benefit.
Acknowledgments
We thank Vindhra Mani and Kyle Seamon for technical assistance with LC/MS/MS analyses. We also thank Dr. Ronald E. Kleinman MD, Physician in Chief of Massachusetts General Hospital for Children and Dr. W. Allan Walker, MD, Director of the Mucosal Immunology Laboratory at Massachusetts General Hospital for continued support.
Abbreviations used in this paper
- PMNs
polymorphonuclear cells (i.e. neutrophils)
- HXA3
hepoxilin A3
- LO
lipoxygenase
- CDC
cinnamyl 3,4-dihydroxy-α-cyanocinnamate
- PGE2
prostaglandin E2
- ARA
Arachidonic acid
- sEH
soluble epoxide hydrolase
- Ana
analogue
- BAL fluid
bronchial alveolar lavage fluid
- MPO
myloperoxidase
- 12-HETE
12-Hydroxyeicosa-5Z,8Z,10E,14Z-tetraenoic acid
Footnotes
This work was supported financially by NIH (1 R01 AI095338-01A1) & (5K22AI065425-02) as well as the Cystic Fibrosis Foundation (HURLEY08G0). KG was supported by NIH EY016136. CM & BDH were supported by NIEHS R01 ES002710, NIAID R01 AI091699, and BDH is a George and Judy Marcus Senior Fellow of the American Asthma Foundation. JRF derived financial support from NIH RO1 GM31278 and the Robert A. Welch Foundation (GL625910). BAM is supported by NIH DK 56754 and DK 33506
References
- 1.DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119:2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizgerd JP. Lung infection--a public health priority. PLoS Med. 2006;3:e76. doi: 10.1371/journal.pmed.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntosh K. Community-acquired pneumonia in children. N Engl J Med. 2002;346:429–437. doi: 10.1056/NEJMra011994. [DOI] [PubMed] [Google Scholar]
- 4.Restrepo MI, Anzueto A. The role of gram-negative bacteria in healthcare-associated pneumonia. Semin Respir Crit Care Med. 2009;30:61–66. doi: 10.1055/s-0028-1119810. [DOI] [PubMed] [Google Scholar]
- 5.Weiss SJ. Tissue destruction by neutrophils. N Engl J Med. 1989;320:365–376. doi: 10.1056/NEJM198902093200606. [DOI] [PubMed] [Google Scholar]
- 6.Burns AR, Smith CW, Walker DC. Unique structural features that influence neutrophil emigration into the lung. Physiol Rev. 2003;83:309–336. doi: 10.1152/physrev.00023.2002. [DOI] [PubMed] [Google Scholar]
- 7.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macdowell AL, Peters SP. Neutrophils in asthma. Curr Allergy Asthma Rep. 2007;7:464–468. doi: 10.1007/s11882-007-0071-6. [DOI] [PubMed] [Google Scholar]
- 9.McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CC, Beck PL, Muruve DA, Kubes P. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- 10.Sanz MJ, Kubes P. Neutrophil-active chemokines in in vivo imaging of neutrophil trafficking. Eur J Immunol. 42:278–283. doi: 10.1002/eji.201142231. [DOI] [PubMed] [Google Scholar]
- 11.Hurley BP, Pirzai W, Mumy KL, Gronert K, McCormick BA. Selective eicosanoid-generating capacity of cytoplasmic phospholipase A2 in Pseudomonas aeruginosa-infected epithelial cells. Am J Physiol Lung Cell Mol Physiol. 300:L286–294. doi: 10.1152/ajplung.00147.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley BP, Siccardi D, Mrsny RJ, McCormick BA. Polymorphonuclear cell transmigration induced by Pseudomonas aeruginosa requires the eicosanoid hepoxilin A3. J Immunol. 2004;173:5712–5720. doi: 10.4049/jimmunol.173.9.5712. [DOI] [PubMed] [Google Scholar]
- 13.Hurley BP, Sin A, McCormick BA. Adhesion molecules involved in hepoxilin A3-mediated neutrophil transepithelial migration. Clin Exp Immunol. 2008;151:297–305. doi: 10.1111/j.1365-2249.2007.03551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley BP, Williams NL, McCormick BA. Involvement of phospholipase A2 in Pseudomonas aeruginosa-mediated PMN transepithelial migration. Am J Physiol Lung Cell Mol Physiol. 2006;290:L703–L709. doi: 10.1152/ajplung.00390.2005. [DOI] [PubMed] [Google Scholar]
- 15.Mrsny RJ, Gewirtz AT, Siccardi D, Savidge T, Hurley BP, Madara JL, McCormick BA. Identification of hepoxilin A3 in inflammatory events: a required role in neutrophil migration across intestinal epithelia. Proc Natl Acad Sci U S A. 2004;101:7421–7426. doi: 10.1073/pnas.0400832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pace-Asciak CR, Reynaud D, Demin P, Nigam S. The hepoxilins. A review. Adv Exp Med Biol. 1999;447:123–132. [PubMed] [Google Scholar]
- 17.Sutherland M, Schewe T, Nigam S. Biological actions of the free acid of hepoxilin A3 on human neutrophils. Biochem Pharmacol. 2000;59:435–440. doi: 10.1016/s0006-2952(99)00345-7. [DOI] [PubMed] [Google Scholar]
- 18.Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobrian AD, Lieb DC, Cole BK, Taylor-Fishwick DA, Chakrabarti SK, Nadler JL. Functional and pathological roles of the 12- and 15-lipoxygenases. Prog Lipid Res. 50:115–131. doi: 10.1016/j.plipres.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nigam S, Patabhiraman S, Ciccoli R, Ishdorj G, Schwarz K, Petrucev B, Kuhn H, Haeggstrom JZ. The rat leukocyte-type 12-lipoxygenase exhibits an intrinsic hepoxilin A3 synthase activity. J Biol Chem. 2004;279:29023–29030. doi: 10.1074/jbc.M307576200. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimoto T, Takahashi Y. Arachidonate 12-lipoxygenases. Prostaglandins Other Lipid Mediat. 2002;68–69:245–262. doi: 10.1016/s0090-6980(02)00034-5. [DOI] [PubMed] [Google Scholar]
- 22.Yu Z, Schneider C, Boeglin WE, Marnett LJ, Brash AR. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc Natl Acad Sci U S A. 2003;100:9162–9167. doi: 10.1073/pnas.1633612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z, Schneider C, Boeglin WE, Brash AR. Mutations associated with a congenital form of ichthyosis (NCIE) inactivate the epidermal lipoxygenases 12R-LOX and eLOX3. Biochim Biophys Acta 1686. 2005;3:238–247. doi: 10.1016/j.bbalip.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn H, Walther M, Kuban RJ. Mammalian arachidonate 15-lipoxygenases structure, function, and biological implications. Prostaglandins Other Lipid Mediat. 2002;68–69:263–290. doi: 10.1016/s0090-6980(02)00035-7. [DOI] [PubMed] [Google Scholar]
- 25.McDonnell M, Li H, Funk CD. Characterization of epidermal 12(S) and 12(R) lipoxygenases. Adv Exp Med Biol. 2002;507:147–153. doi: 10.1007/978-1-4615-0193-0_23. [DOI] [PubMed] [Google Scholar]
- 26.McCormick BA, Hofman PM, Kim J, Carnes DK, Miller SI, Madara JL. Surface attachment of Salmonella typhimurium to intestinal epithelia imprints the subepithelial matrix with gradients chemotactic for neutrophils. J Cell Biol. 1995;131:1599–1608. doi: 10.1083/jcb.131.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leedom AJ, Sullivan AB, Dong B, Lau D, Gronert K. Endogenous LXA4 circuits are determinants of pathological angiogenesis in response to chronic injury. Am J Pathol. 176:74–84. doi: 10.2353/ajpath.2010.090678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shahabi S, Hassan ZM, Jazani NH. Post heat shock tolerance: a neuroimmunological anti-inflammatory phenomenon. J Inflamm (Lond) 2009;6:7. doi: 10.1186/1476-9255-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morisseau C, Hammock BD. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol. 2005;45:311–333. doi: 10.1146/annurev.pharmtox.45.120403.095920. [DOI] [PubMed] [Google Scholar]
- 30.Newman JW, Morisseau C, Harris TR, Hammock BD. The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity. Proc Natl Acad Sci U S A. 2003;100:1558–1563. doi: 10.1073/pnas.0437724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang SB, Hu KM, Seamon KJ, Mani V, Chen Y, Gronert K. Estrogen negatively regulates epithelial wound healing and protective lipid mediator circuits in the cornea. FASEB J. 26:1506–1516. doi: 10.1096/fj.11-198036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Priebe GP, Meluleni GJ, Coleman FT, Goldberg JB, Pier GB. Protection against fatal Pseudomonas aeruginosa pneumonia in mice after nasal immunization with a live, attenuated aroA deletion mutant. Infect Immun. 2003;71:1453–1461. doi: 10.1128/IAI.71.3.1453-1461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priebe GP, Walsh RL, Cederroth TA, Kamei A, Coutinho-Sledge YS, Goldberg JB, Pier GB. IL-17 is a critical component of vaccine-induced protection against lung infection by lipopolysaccharide-heterologous strains of Pseudomonas aeruginosa. J Immunol. 2008;181:4965–4975. doi: 10.4049/jimmunol.181.7.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 35.Vancheri C, Mastruzzo C, Sortino MA, Crimi N. The lung as a privileged site for the beneficial actions of PGE2. Trends Immunol. 2004;25:40–46. doi: 10.1016/j.it.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Burgoyne RD, Morgan A. The control of free arachidonic acid levels. Trends Biochem Sci. 1990;15:365–366. doi: 10.1016/0968-0004(90)90227-3. [DOI] [PubMed] [Google Scholar]
- 37.Cho H, Ueda M, Tamaoka M, Hamaguchi M, Aisaka K, Kiso Y, Inoue T, Ogino R, Tatsuoka T, Ishihara T, et al. Novel caffeic acid derivatives: extremely potent inhibitors of 12-lipoxygenase. J Med Chem. 1991;34:1503–1505. doi: 10.1021/jm00108a039. [DOI] [PubMed] [Google Scholar]
- 38.Kurosawa M, Hisada T, Ishizuka T. Effect of phospholipase A2 inhibitor ONO-RS-082 on substance P-induced histamine release from rat peritoneal mast cells. Int Arch Allergy Immunol. 1992;97:226–228. doi: 10.1159/000236123. [DOI] [PubMed] [Google Scholar]
- 39.Hurley BP, McCormick BA. Multiple roles of phospholipase A2 during lung infection and inflammation. Infect Immun. 2008;76:2259–2272. doi: 10.1128/IAI.00059-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami M, Kudo I. Phospholipase A2. J Biochem. 2002;131:285–292. doi: 10.1093/oxfordjournals.jbchem.a003101. [DOI] [PubMed] [Google Scholar]
- 41.Cronin A, Decker M, Arand M. Mammalian soluble epoxide hydrolase is identical to liver hepoxilin hydrolase. J Lipid Res. doi: 10.1194/jlr.M009639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pace-Asciak CR, Reynaud D, Rounova O, Demin P, Pivnitsky KK. Hepoxilin A3 is metabolized into its omega-hydroxy metabolite by human neutrophils. Adv Exp Med Biol. 1999;469:535–538. doi: 10.1007/978-1-4615-4793-8_78. [DOI] [PubMed] [Google Scholar]
- 43.Reynaud D, Pace-Asciak CR. Docosahexaenoic acid causes accumulation of free arachidonic acid in rat pineal gland and hippocampus to form hepoxilins from both substrates. Biochim Biophys Acta. 1997;1346:305–316. doi: 10.1016/s0005-2760(97)00041-6. [DOI] [PubMed] [Google Scholar]
- 44.Power MR, Peng Y, Maydanski E, Marshall JS, Lin TJ. The development of early host response to Pseudomonas aeruginosa lung infection is critically dependent on myeloid differentiation factor 88 in mice. J Biol Chem. 2004;279:49315–49322. doi: 10.1074/jbc.M402111200. [DOI] [PubMed] [Google Scholar]
- 45.Prince AS, Mizgerd JP, Wiener-Kronish J, Bhattacharya J. Cell signaling underlying the pathophysiology of pneumonia. Am J Physiol Lung Cell Mol Physiol. 2006;291:L297–300. doi: 10.1152/ajplung.00138.2006. [DOI] [PubMed] [Google Scholar]
- 46.Qin L, Quinlan WM, Doyle NA, Graham L, Sligh JE, Takei F, Beaudet AL, Doerschuk CM. The roles of CD11/CD18 and ICAM-1 in acute Pseudomonas aeruginosa-induced pneumonia in mice. J Immunol. 1996;157:5016–5021. [PubMed] [Google Scholar]
- 47.Sadikot RT, Zeng H, Joo M, Everhart MB, Sherrill TP, Li B, Cheng DS, Yull FE, Christman JW, Blackwell TS. Targeted immunomodulation of the NF-kappaB pathway in airway epithelium impacts host defense against Pseudomonas aeruginosa. J Immunol. 2006;176:4923–4930. doi: 10.4049/jimmunol.176.8.4923. [DOI] [PubMed] [Google Scholar]
- 48.Schultz MJ, Rijneveld AW, Florquin S, Edwards CK, Dinarello CA, van der Poll T. Role of interleukin-1 in the pulmonary immune response during Pseudomonas aeruginosa pneumonia. Am J Physiol Lung Cell Mol Physiol. 2002;282:L285–290. doi: 10.1152/ajplung.00461.2000. [DOI] [PubMed] [Google Scholar]
- 49.Skerrett SJ, Liggitt HD, Hajjar AM, Wilson CB. Cutting edge: myeloid differentiation factor 88 is essential for pulmonary host defense against Pseudomonas aeruginosa but not Staphylococcus aureus. J Immunol. 2004;172:3377–3381. doi: 10.4049/jimmunol.172.6.3377. [DOI] [PubMed] [Google Scholar]
- 50.Skerrett SJ, Wilson CB, Liggitt HD, Hajjar AM. Redundant Toll-like receptor signaling in the pulmonary host response to Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol. 2007;292:L312–322. doi: 10.1152/ajplung.00250.2006. [DOI] [PubMed] [Google Scholar]
- 51.Sun L, Guo RF, Newstead MW, Standiford TJ, Macariola DR, Shanley TP. Effect of IL-10 on Neutrophil Recruitment and Survival after Pseudomonas aeruginosa Challenge. Am J Respir Cell Mol Biol. 2008 doi: 10.1165/rcmb.2008-0202OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koh AY, Priebe GP, Ray C, Van Rooijen N, Pier GB. Inescapable need for neutrophils as mediators of cellular innate immunity to acute Pseudomonas aeruginosa pneumonia. Infect Immun. 2009;77:5300–5310. doi: 10.1128/IAI.00501-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen SM, Cheng DS, Williams BJ, Sherrill TP, Han W, Chont M, Saint-Jean L, Christman JW, Sadikot RT, Yull FE, Blackwell TS. The nuclear factor kappa-B pathway in airway epithelium regulates neutrophil recruitment and host defence following Pseudomonas aeruginosa infection. Clin Exp Immunol. 2008;153:420–428. doi: 10.1111/j.1365-2249.2008.03707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rollins BJ. Where the confusion began: cloning the first chemokine receptors. J Immunol. 2009;183:2893–2894. doi: 10.4049/jimmunol.0990065. [DOI] [PubMed] [Google Scholar]
- 55.Chun J, Prince A. Ca2+ signaling in airway epithelial cells facilitates leukocyte recruitment and transepithelial migration. J Leukoc Biol. 2009;86:1135–1144. doi: 10.1189/jlb.0209072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Craig A, Mai J, Cai S, Jeyaseelan S. Neutrophil recruitment to the lungs during bacterial pneumonia. Infect Immun. 2009;77:568–575. doi: 10.1128/IAI.00832-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stillie R, Farooq SM, Gordon JR, Stadnyk AW. The functional significance behind expressing two IL-8 receptor types on PMN. J Leukoc Biol. 2009;86:529–543. doi: 10.1189/jlb.0208125. [DOI] [PubMed] [Google Scholar]
- 58.Weathington NM, van Houwelingen AH, Noerager BD, Jackson PL, Kraneveld AD, Galin FS, Folkerts G, Nijkamp FP, Blalock JE. A novel peptide CXCR ligand derived from extracellular matrix degradation during airway inflammation. Nat Med. 2006;12:317–323. doi: 10.1038/nm1361. [DOI] [PubMed] [Google Scholar]
- 59.Farberman MM, Ibricevic A, Joseph TD, Akers KT, Garcia-Medina R, Crosby S, Clarke LL, Brody SL, Ferkol TW. The Effect of Polarized Release of CXC-chemokines from Wild-type and Cystic Fibrosis Murine Airway Epithelial Cells. Am J Respir Cell Mol Biol. doi: 10.1165/rcmb.2009-0249OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCormick BA. Bacterial-induced hepoxilin A3 secretion as a pro-inflammatory mediator. FEBS J. 2007;274:3513–3518. doi: 10.1111/j.1742-4658.2007.05911.x. [DOI] [PubMed] [Google Scholar]
- 61.Mizgerd JP, Skerrett SJ. Animal models of human pneumonia. Am J Physiol Lung Cell Mol Physiol. 2008;294:L387–398. doi: 10.1152/ajplung.00330.2007. [DOI] [PubMed] [Google Scholar]
- 62.Bodini A, D’Orazio C, Peroni D, Corradi M, Folesani G, Baraldi E, Assael BM, Boner A, Piacentini GL. Biomarkers of neutrophilic inflammation in exhaled air of cystic fibrosis children with bacterial airway infections. Pediatr Pulmonol. 2005;40:494–499. doi: 10.1002/ppul.20336. [DOI] [PubMed] [Google Scholar]
- 63.Mackerness KJ, Jenkins GR, Bush A, Jose PJ. Characterisation of the range of neutrophil stimulating mediators in cystic fibrosis sputum. Thorax. 2008;63:614–620. doi: 10.1136/thx.2007.089359. [DOI] [PubMed] [Google Scholar]
- 64.Funk CD, Chen XS, Johnson EN, Zhao L. Lipoxygenase genes and their targeted disruption. Prostaglandins Other Lipid Mediat. 2002;68–69:303–312. doi: 10.1016/s0090-6980(02)00036-9. [DOI] [PubMed] [Google Scholar]
- 65.Yoshikai Y. Roles of prostaglandins and leukotrienes in acute inflammation caused by bacterial infection. Curr Opin Infect Dis. 2001;14:257–263. doi: 10.1097/00001432-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 66.Berger W, De Chandt MT, Cairns CB. Zileuton: clinical implications of 5-Lipoxygenase inhibition in severe airway disease. Int J Clin Pract. 2007;61:663–676. doi: 10.1111/j.1742-1241.2007.01320.x. [DOI] [PubMed] [Google Scholar]
- 67.Konstan MW. Ibuprofen therapy for cystic fibrosis lung disease: revisited. Curr Opin Pulm Med. 2008;14:567–573. doi: 10.1097/MCP.0b013e32831311e8. [DOI] [PubMed] [Google Scholar]