Abstract
Experimental studies of translation have found that short genes tend to exhibit greater densities of ribosomes than long genes in eukaryotic species. It remains an open question whether the elevated ribosome density on short genes is due to faster initiation or slower elongation dynamics. Here, we address this question computationally using 5′-mRNA folding energy as a proxy for translation initiation rates and codon bias as a proxy for elongation rates. We report a significant trend toward reduced 5′-secondary structure in shorter coding sequences, suggesting that short genes initiate faster during translation. We also find a trend toward higher 5′-codon bias in short genes, suggesting that short genes elongate faster than long genes. Both of these trends hold across a diverse set of eukaryotic taxa. Thus, the elevated ribosome density on short eukaryotic genes is likely caused by differential rates of initiation, rather than differential rates of elongation.
Keywords: translation initiation, ribosome density, codon bias, gene length
Introduction
Synonymous sites in coding sequences have long been used as a neutral yardstick against which to compare amino acid changing substitutions, in the hope of detecting either purifying or positive selection on proteins (Kimura 1977; McDonald and Kreitman 1991; Goldman and Yang 1994; Muse and Gaut 1994). Nonetheless, synonymous mutations are known to experience selection in many cases (Andersson and Kurland 1990; Sawyer and Hartl 1992; Sharp et al. 1995; Duret 2002; Chamary et al. 2006; Hershberg and Petrov 2008; Sharp et al. 2010) for a variety of mechanisms, including the efficiency of gene translation, the stability of mRNAs (Shen et al. 1999; Duan et al. 2003; Capon et al. 2004; Chamary and Hurst 2005; Chamary et al. 2006; Shah and Gilchrist 2011) especially near the translation initiation site (Kudla et al. 2009; Gu et al. 2010; Keller et al. 2012), and the regulation of splicing, among others (Plotkin and Kudla 2011). The fact that synonymous mutations have phenotypic and fitness consequences complicates the interpretation of measures of selection, such as the ratio of substitution rates at synonymous and nonsynonymous sites, dN/dS (Kimura 1977; Goldman and Yang 1994; Muse and Gaut 1994; but see Hirsh et al. 2005).
Selection for translational efficiency remains the dominant explanation for systematic variation in codon usage among the genes in a genome, in diverse taxa (Plotkin and Kudla 2011). In accordance with this explanation, codon bias toward the most abundant iso-accepting tRNA species is generally strongest in those genes expressed at high levels, where efficiency would confer the greatest selective benefit to the cell. Nonetheless, the specific mechanisms by which codon bias confers relative fitness gains are actively debated (Shah and Gilchrist 2010; Plotkin and Kudla 2011).
Our understanding of the dynamics of gene translation, and the role of codon bias in translation, will benefit from new experimental techniques that parse the detailed kinetics of translation across the entire transcriptome. Especially promising are techniques that use high-throughput sequencing of ribosome-protected RNA to determine a “ribosomal footprint” on each mRNA (Ingolia et al. 2009, 2011; Guo et al. 2010; Oh et al. 2011; Bazzini et al. 2012; Brar et al. 2012; Li et al. 2012; Reid and Nicchitta 2012) with greater accuracy than earlier, polysome-based techniques (Arava et al. 2003). Among many other intriguing findings, these experiments have shown that the cell-wide average profile of ribosome densities in yeast exhibits a trend of decreasing ribosome density with codon position, from 5′ to 3′—an observation that has been explained, in part, by a trend toward less biased codon usage in the 5′-ends of genes, associated presumably with slower elongation and thus higher ribosome density (Tuller et al. 2010).
Aside from the 5′-ramp of elevated ribosome densities, sequencing (Ingolia et al. 2009) and polysome gradients in budding yeast (Arava et al. 2003) have also revealed another, possibly independent finding: shorter mRNAs tend to have a greater overall density of ribosomes than longer mRNAs. The same trend has been found in mouse, human, fruit fly, Arabidopsis, malaria, and fission yeast: shorter Open Reading Frames (ORFs) tend to exhibit more densely packed ribosomes (Cataldo et al. 1999; Branco-Price et al. 2005; Lackner et al. 2007; Qin et al. 2007; Hendrickson et al. 2009; Ingolia et al. 2009; Lacsina et al. 2011). There is debate about the cause of this trend. Some authors have attributed this relationship to a constant-length ramp of elevated 5′-density on all transcripts due to elongation dynamics (Ingolia et al. 2009) (so that shorter transcripts would be observed to have larger overall ribosome density); and others have attributed this trend to an increased rate of initiation in short yeast genes causing an increased density of ribosomes (Arava et al. 2003, 2005; Lackner et al. 2007). As a result, at present, it is unclear whether the greater overall density of ribosomes on short yeast genes is caused by a greater rate of initiation for such genes or a slower rate of early elongation in those genes.
Against this backdrop of open questions, here we analyze the relationship between ORF length and measures of initiation and early elongation rates, across a diverse set of eukaryotic species. As a proxy for the initiation rate of a gene, we use the computationally predicted energy of its 5′-mRNA structure—a quantity that has been shown experimentally to correlate strongly with protein levels (Kudla et al. 2009) and which has been subject to natural selection in virtually all free-living (Gu et al. 2010; Tuller, Waldman, et al. 2010; Keller et al. 2012) and many viral species (Zhou and Wilke 2011). As a proxy for the early elongation rate of a gene, we use the codon adaptation index (CAI) (Sharp and Li 1987) of its early codons (Tuller et al. 2010). In general, by performing these analyses, we seek to understand whether the trend toward elevated ribosome densities in short genes (Cataldo et al. 1999; Arava et al. 2003, 2005; Branco-Price et al. 2005; Lackner et al. 2007; Qin et al. 2007; Hendrickson et al. 2009; Ingolia et al. 2009; Lacsina et al. 2011) is caused by faster initiation in those genes, slower early elongation in those genes, or both.
Results
Codon Bias, mRNA Structure, and ORF Length in Caenorhabditis elegans
We first investigated the relationship between ORF length and 5′-mRNA folding in the model species Caenorhabditis elegans, as well as the relationship between ORF length and 5′-codon bias. As described earlier, we use these two measures as proxies for the initiation rates and early elongation rates of genes. In particular, for each C. elegans transcript, we computed its predicted folding energy from nucleotide −4 to +37 (Kudla et al. 2009) relative to start, using RNAfold (Hofacker et al. 1994), and we computed the CAI of its first 50 codons. (We systematically explore alternative definitions of 5′-CAI later.)
We performed a Spearman rank correlation test between 5′-mRNA folding energy and ORF length, among the 29,857 transcripts in C. elegans (Assembly WS220). We similarly performed a rank correlation test between 5′-CAI values and ORF lengths. Our expectation was that compared with long genes, short genes should tend to have faster initiation rates and/or slower early elongation rates—to explain the tendency toward elevated ribosome densities on short genes (Cataldo et al. 1999; Arava et al. 2003, 2005; Branco-Price et al. 2005; Lackner et al. 2007; Qin et al. 2007; Hendrickson et al. 2009; Ingolia et al. 2009; Lacsina et al. 2011). Of these two alternative mechanisms, we might in principal expect the initiation-driven mechanism to be a stronger determinant of ribosome densities (Andersson and Kurland 1990; Bulmer 1991; Lackner et al. 2007).
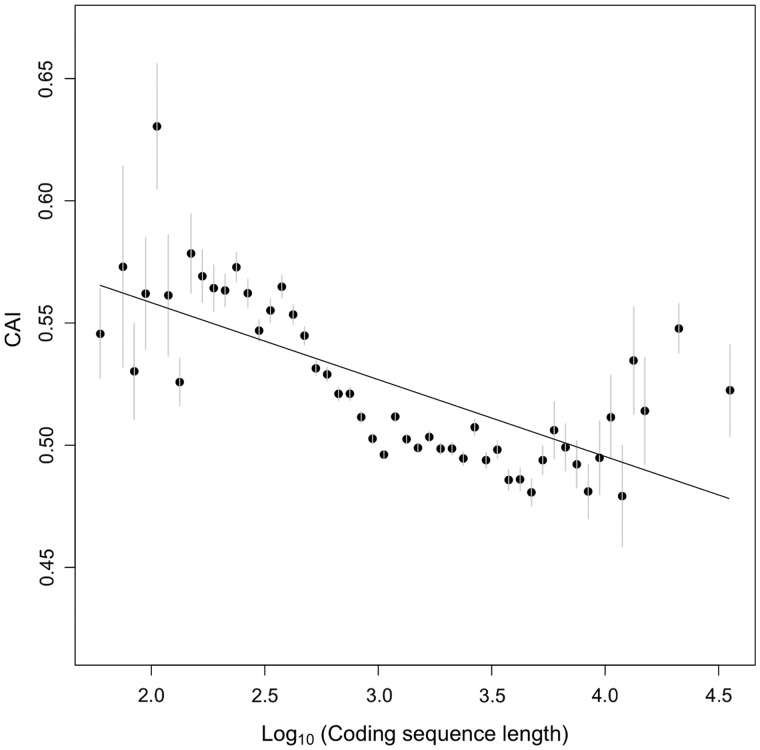
In accordance with these expectations, we found a significant negative rank correlation (Spearman rho = −0.12, P < 7 × 10−90) between 5′-mRNA folding energy and ORF length, indicating a tendency toward weaker mRNA structure and presumably faster initiation in short C. elegans genes (fig. 1). On the other hand, we also found a significant negative rank correlation (Spearman rho = −0.16, P < 5 × 10−179) between 5′-CAI and length, suggesting shorter genes tend to have faster early elongate rates (fig. 2). Given that shorter genes have higher CAI and hence faster elongation rates, we would expect a lower ribosomal density for shorter genes contrary to the observed patterns. As a result, we conclude that higher ribosomal densities of shorter genes are most likely explained by faster initiation rates as shown by weaker 5′-mRNA secondary structures.
Fig. 1.—
Short C. elegans genes have higher 5′-mRNA folding energies than long C. elegans genes, suggesting faster initiation in short genes. Genes have been binned according to their log (ORF length), with dots showing the mean computed 5′-mRNA folding energy in each bin and lines showing ±1 standard deviation. The solid line shows best-fit regression (Spearman rho = −0.12, P < 7 × 10−90).
Fig. 2.—
Short C. elegans genes have higher 5′-CAIs than long C. elegans genes, suggesting faster elongation in short genes. Genes have been binned according to their log (ORF length), with dots showing the mean computed 5′-CAI in each bin and lines showing ±1 standard deviation. The solid line shows best-fit regression (Spearman rho = −0.16, P < 5 × 10−179).
Codon Bias, mRNA Structure, and ORF Length in 120 Eukaryotic Species
Given our results in C. elegans, we then asked how broadly these trends in gene length and 5′-mRNA structure hold across eukaryotes. We repeated the 5′-mRNA folding energy calculations in 120 eukaryote species and the 5′-CAI calculations in 89 of those species for which a reliable reference set of genes was available for computing CAI. (The sets of species used in 5′-mRNA folding energy and 5′-CAI calculations are listed in supplementary table S1, Supplementary Material online). The results of these calculations and their correlations with ORF length are summarized in table 1.
Table 1.
Most Eukaryotic Species Show a Tendency Toward Weak 5′-mRNA Structure and High 5′-Codon Bias in Shorter Genes
| Correlations with ORF Length | 5′ Free Energy (120 Species) | 5′-CAI (89 Species) |
|---|---|---|
| % Species with negative correlation | 82 | 83 |
| % Species with significant negative correlation | 73 | 67 |
| % Species with positive correlation | 18 | 17 |
| % Species with significant positive correlation | 11 | 15 |
| Two-sided binomial P value | 1.2 × 10−12 | 1.5 × 10−10 |
Note.—In particular, there is a negative rank correlation between 5′-mRNA folding energy and ORF length in 82% of the 120 eukaryotic species tested, and similarly, a negative rank correlation between 5′-CAI and -ORF length in 83% of the 89 species tested. The overall tendency toward negative correlations is highly significant, in both cases.
Table 1 summarizes the proportion of species tested that exhibit a negative rank correlation between 5′-mRNA folding energy and ORF length or between 5′-CAI and ORF length. In addition, we report the proportion of species that feature a significant negative correlation, at the 5% significance level. As summarized in table 1, the results found in C. elegans hold very broadly across eukaryotes: approximately 80% of tested eukaryotes exhibit negative correlations between mRNA folding and length and between 5′-CAI and length. The preponderance of significant negative correlations with ORF length among eukaryotes is itself highly significant, for both 5′-mRNA folding energy (binomial P < 10−11) and 5′-CAI (binomial P < 10−9)—suggesting a systematic eukaryotic trend toward faster translation initiation and faster early elongation in short versus long genes. Thus, our results suggest that the higher ribosome density observed in shorter eukaryotes genes is likely due to faster initiation rates in shorter genes.
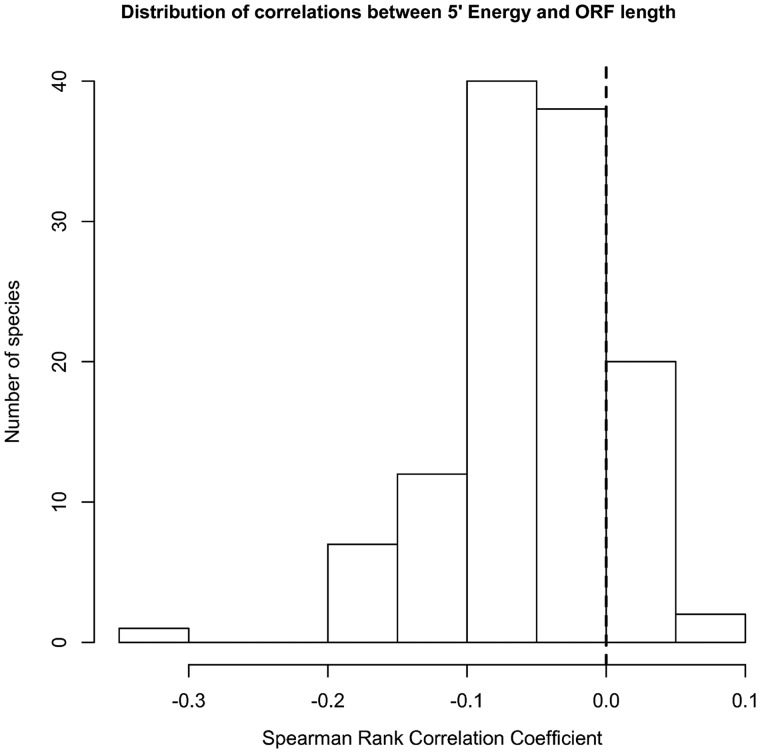
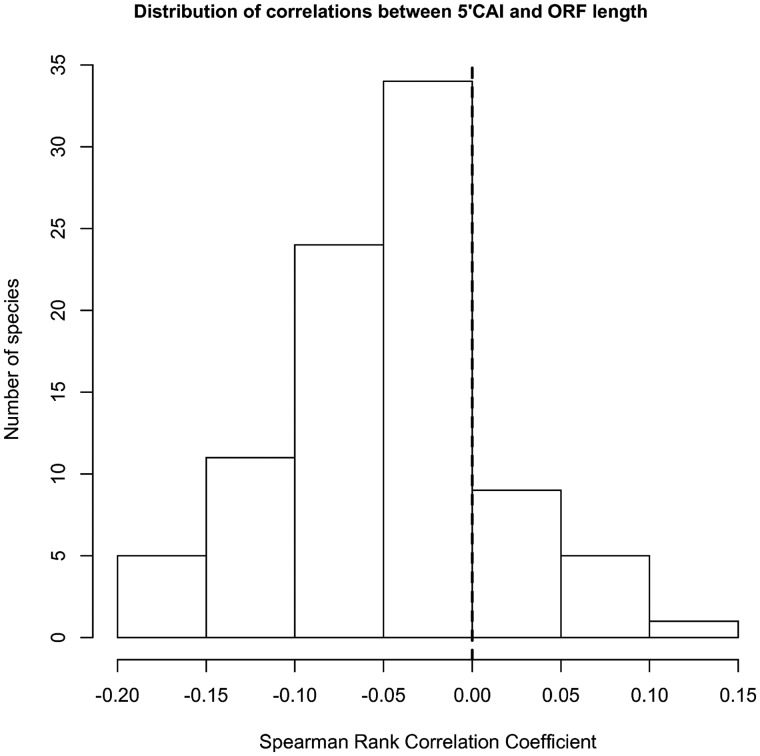
The distribution of correlations for energy and CAI are presented in figures 3 and 4, and the complete results for each species used in the energy and CAI calculations are presented in supplementary tables S2 and S3, Supplementary Material online, respectively.
Fig. 3.—
The distribution of Spearman rank correlation coefficients between 5′-energy and -ORF length in 120 eukaryotic species.
Fig. 4.—
The distribution of Spearman rank correlation coefficients between 5′-CAI and ORF length in 89 eukaryotic species.
Weak 5′-mRNA Folding in Short Genes, Controlling for 5′-CAI
In the previous sections, we have established a systematic trend toward weaker 5′-mRNA structure in short genes, as opposed to long genes; and we argued that the resulting increase in initiation rates is responsible for the greater density of ribosomes typically found in short eukaryotic genes. Nonetheless, we have also found a trend toward increased CAI in the same region, in short genes—and so the possibility remains that some subtle patterns of 5′-CAI might be responsible for the trend observed in mRNA structure. To resolve this issue, we have performed a randomization procedure that isolates the effects of synonymous codons on 5′-mRNA structure, controlling for 5′-CAI.
For each species, we randomly shuffled the first 50 codons of each coding sequence, and we repeated this process 100 times for each gene. In each such permutation, the 5′-CAI of the gene is preserved, whereas the mRNA structure is possibly perturbed. We then computed the quantile of the 5′-mRNA folding energy for the true gene sequence with respect to this null distribution of permuted sequences. Because our hypothesis is that shorter genes are under selection for weaker 5′-mRNA folding (i.e., higher energy) regardless of 5′-CAI, we expect a higher quantile for shorter genes. We tested this expectation by computing the Spearman rank correlation between the length of each ORF in the genome and the quantile of its true mRNA folding energy compared with the null distribution.
As listed in table 2, we observed a negative rank correlation between the energy quantile and the ORF length in the great majority species (binomial P value < 6 × 10−15)—indicating that the trend toward weak mRNA structure in short genes holds even after controlling for 5′-CAI. These analyses substantiate our hypothesis that shorter eukaryotic genes are under selection to have faster translation initiation rates, achieved through weaker 5′-mRNA folding.
Table 2.
Most Species Exhibit a Tendency Toward Weak 5′ Free Energy in Short Genes, Even After Controlling for 5′-CAI
| Correlation between ORF Length and Quantile of Observed 5′ Free Energy | % Species (of 120 Tested) |
|---|---|
| Negative correlation | 84 |
| Significant negative correlation | 65 |
| Positive correlation | 16 |
| Significant positive correlation | 2.5 |
| One-sided binomial P value | 5.38 × 10−15 |
Note.—In the majority of species tested, we find a negative rank correlation between ORF length and the quantile of the observed 5′-mRNA free energy among the free energies of permuted sequences that retain the same 5′-CAI value. The tendency toward negative correlations across species is highly significant.
Robustness of Results
In the preceding analyses, we calculated 5′-CAI using the first 50 codons of each ORF. We chose this region to coincide as much as possible with the ramp of slow codons reported by Tuller et al. (2010). We repeated the 5′-CAI calculations using the first 13, 15, 20, 30, 40, and 60 codons and obtained similar qualitative results in each case (supplementary table S4, Supplementary Material online). The ribosomal density on a gene might be affected by codons beyond the 5′ region of gene as well. For instance, slow codons in the middle or end of a gene might cause a bottleneck for ribosomes, leading to higher ribosomal densities irrespective of the codon composition in the 5′ region. As a result, we also verified the robustness of our results by considering the CAI of entire ORF, producing the same qualitative, but slightly weaker, result (36% positive correlations, 64% negative correlations, two-sided Binomial P value < 0.011. For the complete tabulation of these results see supplementary table S8, Supplementary Material online.
Another potential concern that may arise from our 5′-CAI calculation is that we excluded sequences shorter than 51 codons. Is it possible that the sequences shorter than 51 codons could have a different CAI pattern and somehow diluted the observed CAI pattern? To answer this question, we modified the definition of 5′-CAI to include coding sequences shorter than 51 codons long, by computing the geometric mean of the relative adaptiveness of all the nonstop codons in the sequence. Again, this did not change our qualitative results (supplementary table S5, Supplementary Material online).
Discussion
We have reported a strong trend toward weaker 5′-mRNA structure in short genes, when compared with long genes, among eukaryotic species. Moreover, we also observed a trend toward higher 5′-codon bias in short versus long genes—indicating that elongation dynamics driven by codon bias is unlikely to be the cause of higher ribosomal densities on short genes. For each individual species, the correlation between ORF length and 5′-mRNA folding energy/5′-CAI is usually statistically significant but not strong. Nonetheless, the trend of reduced 5′-secondary structure in short coding sequences was observed in the majority of eukaryotic species (82%) tested. The statistical significance of this trend is extraordinarily strong and so too is the biological significance: more than three-quarters of eukaryotic species exhibit reduced 5′-mRNA structure in short genes.
To the extent that 5′-mRNA structure modulates initiation (Bettany et al. 1989; de Smit and van Duin 1990; Eyre-Walker and Bulmer 1993; Kudla et al. 2009; Gu et al. 2010; Keller et al. 2012), our results suggest that faster initiation is responsible for the empirical observation in diverse eukaryotes (Cataldo et al. 1999; Arava et al. 2003; Branco-Price et al. 2005; Lackner et al. 2007; Qin et al. 2007; Hendrickson et al. 2009; Lacsina et al. 2011) that short mRNAs are more densely packed with ribosomes than long mRNAs.
Our analyses across a diverse set of eukaryotic species substantiates several authors’ interpretation of patterns of ribosomal densities and ORF length, which have been attributed to initiation-driven mechanisms as opposed to elongation effects (Arava et al. 2003, 2005; Lackner et al. 2007). Our results confirm that the effects of initiation, modulated by ribosomal binding to the 5′-end of mRNA and scanning to start codon, strongly outweigh those of elongation dynamics, modulated by codon bias. This view is in contrast with other studies that propose a dominant role of codon usage in shaping ribosomal occupancies (Tuller et al. 2010). Nonetheless, our results do not directly contradict those of Tuller et al. (2010), however, because those authors considered relative codon usage within each ORF, whereas we have studied absolute codon usage across different ORFs.
Other factors such as protein folding (Kimchi-Sarfaty et al. 2007) and sequence similarity to ribosome binding sites (Li et al. 2012) may also influence ribosome density. However, such effects are generally not considered as major determinants in shaping overall ribosome density (Plotkin and Kudla 2011; Li et al. 2012). These factors, which are difficult to quantify systematically, are probably less likely to show systematic trends with respect to ORF length, such as those we have observed for 5′-CAI and 5′-mRNA secondary structure.
It is interesting to ask whether there are any commonalities among the 22 “counterexample” species in which we observed a positive rank correlation between 5′-energy and ORF length. What differentiates these organisms from the other eukaryotes we have studied? To answer this question, we examined the phylogenetic relationship of all the studied species and the distribution along this phylogeny of those 22 species exhibiting a positive rank correlation between ORF length and 5′ free energy (supplementary fig. S1, Supplementary Material online). Although a few of these counter examples are clearly closely related sister species, overall these 22 species are distributed relatively uniformly among eukaryotes, as opposed to being mostly monophyletic. And so we do not find any obvious commonality among these species with respect to their evolutionary history and, likely, ecological contexts.
Our results on systematically weaker 5′-mRNA structure in short genes beg the question: why should short genes experience selection for fast translation initiation? It has been suggested that highly expressed genes are shorter in many eukaryotes (Eyre-Walker 1996; Duret and Mouchiroud 1999; Eisenberg and Levanon 2003; Rao et al. 2010), also short genes are enriched for constitutively expressed housekeeping and ribosomal genes (Hurowitz and Brown 2003), which must produce protein as rapidly as possible. This alone might explain why short genes experience selection for faster initiation (Reuveni et al. 2011). In addition, housekeeping genes tend to have shorter 5′-untranslated regions (UTRs) and are under weaker post-transcriptional regulation (Hurowitz and Brown 2003; David et al. 2006; Lin and Li 2012). The probability of successful ribosomal binding and scanning on an mRNA may depend on the length of its 5′-UTRs. As a result, genes that require post-transcriptional regulation tend to have longer 5′-UTRs, leading to lower initiation probabilities (Lin and Li 2012).
In summary, we find that shorter genes have higher 5′-mRNA folding energies and codon bias, suggesting that shorter genes both initiate and elongate faster than longer genes. Both of these trends hold across a diverse set of eukaryotic taxa. Because faster elongation leads to lower ribosome densities, the elevated ribosome densities of short eukaryotic genes is a result of initiation rates, rather than elongation rates.
Materials and Methods
Data Sets
Coding sequences with 4-bp upstream data for most species were downloaded from ensembl genomes servers (http://www.ensemblgenomes.org, last accessed March 25, 2011). The coding sequences of Yarrowia lipolytica with 1,000 bp upstream sequences and 300 bp downstream sequences were downloaded from Génolevures (Sherman et al. 2009) (www.genolevures.org/yali.html, last accessed March 25, 2011). All the coding sequences were preprocessed, so that sequences whose length is not a multiple of 3, those with premature stop codons, or a continuous string of more than three ambiguous “N” symbols are discarded. We only considered coding sequences at least 42 nucleotides long. The complete list of species used in this study is listed in supplementary table S1, Supplementary Material online.
We identified ribosomal genes for the purpose of computing CAI from one of three sources: 1) the ribosomal gene sequences for 24 species were downloaded from the HOGENOMDNA (Penel et al. 2009) database (http://pbil.univ-lyon1.fr/databases/hogenom/acceuil.php, last accessed February 1, 2011).
Orthologous groups of ribosomal genes from the HOGENOM database are listed in supplementary table S6, Supplementary Material online. 2) The ribosomal genes for 64 species were obtained from Orthologous MAtrix Project (Altenhoff et al. 2011) (http://omabrowser.org, last accessed March 25, 2011). We used Saccharomyces cerevisiae as our genome of reference and obtained orthologs of its ribosomal genes. The OMA orthologous groups and organism-specific ribosomal genes are listed in supplementary table S7, Supplementary Material online. 3) The ribosomal genes for Y. lipolytica were obtained by performing a protein blast search against the ribosomal gene coding sequences for S. cerevisiae and taking the top hit for each gene provided it has an E value <10−5. The number of identified ribosomal genes per species in our data set ranged from 19 to 184 genes with a median value of 44.
Calculating 5′-mRNA Folding Free Energy
To get an estimate of the translation initiation rates, we used the program RNAfold from Vienna RNA package (Hofacker et al. 1994) to calculate the mRNA folding energy from base −4 to 37 for each gene. For each species, we calculated the 5′-folding energy and length of every gene and then obtained the Spearman rank correlation coefficient and a two-tailed P value using the function spearmanr in the SciPy (Jones et al. 2001) package of Python (Van Rossum and Drake 2001). We chose 0.05 as the significance level.
We then counted the number of species in which the 5′ free energy has a negative Spearman rank correlation with sequence length and also the number of species in which the correlations are significant. We calculated a two-tailed P value to assess whether there is an overall trend in the direction of rank correlation between 5′-mRNA folding energy and coding sequence length.
Calculating 5′-CAI
To obtain an estimate of the translation early elongation rates, we calculated the CAI (Sharp and Li 1987) for the first 50 codons of each gene. The 5′-CAI of a gene is defined as the geometric mean of the relative adaptiveness values of all the considered codons in a particular gene. The relative adaptiveness values of each codon are defined as ratio of occurrences of the codon to occurrences of the most abundant synonymous codon, using the ribosomal gene sequences from each species. In the above calculations, we removed coding sequences less than 51 codons long. Alternatively, for these short sequences, we also calculated 5′-CAI using the whole sequence and obtained the same qualitative results (supplementary table S5, Supplementary Material online).
Supplementary Material
Supplementary figure S1 and tables S1–S8 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank two anonymous referees for constructive comments. This work was supported by the Burroughs Wellcome Fund, the David and Lucile Packard Foundation, the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, and grant D12AP00025 from the U.S. Department of the Interior and Defense Advanced Research Projects Agency to J.B.P. and by the Penn Genome Frontiers Institute to Y.D.
Literature Cited
- Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–D294. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiol Mol Biol Rev. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Boas FE, Brown PO, Herschlag D. Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res. 2005;33:2421–2432. doi: 10.1093/nar/gki331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettany AJ, et al. 5′-secondary structure formation, in contrast to a short string of non-preferred codons, inhibits the translation of the pyruvate kinase mRNA in yeast. Yeast. 1989;5:187–198. doi: 10.1002/yea.320050308. [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot. 2005;96:647–660. doi: 10.1093/aob/mci217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capon F, et al. A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum Mol Genet. 2004;13:2361–2368. doi: 10.1093/hmg/ddh273. [DOI] [PubMed] [Google Scholar]
- Cataldo L, Mastrangelo MA, Kleene KC. A quantitative sucrose gradient analysis of the translational activity of 18 mRNA species in testes from adult mice. Mol Hum Reprod. 1999;5:206–213. doi: 10.1093/molehr/5.3.206. [DOI] [PubMed] [Google Scholar]
- Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- Duret L. Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev. 2002;12:640–649. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. Synonymous codon bias is related to gene length in Escherichia coli: selection for translational accuracy? Mol Biol Evol. 1996;13:864–872. doi: 10.1093/oxfordjournals.molbev.a025646. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Bulmer M. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 1993;21:4599–4603. doi: 10.1093/nar/21.19.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- Hirsh AE, Fraser HB, Wall DP. Adjusting for selection on synonymous sites in estimates of evolutionary distance. Mol Biol Evol. 2005;22:174–177. doi: 10.1093/molbev/msh265. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, et al. Fast folding and comparison of RNA secondary structures. Monatshefte Chem. 1994;125:167–188. [Google Scholar]
- Hurowitz EH, Brown PO. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Pearu P. SciPy: open source scientific tools for Python. 2001 [Google Scholar]
- Keller TE, Mis SD, Jia KE, Wilke CO. Reduced mRNA secondary-structure stability near the start codon indicates functional genes in prokaryotes. Genome Biol Evol. 2012;4:80–88. doi: 10.1093/gbe/evr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, et al. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell. 2007;26:145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacsina JR, LaMonte G, Nicchitta CV, Chi JT. Polysome profiling of the malaria parasite plasmodium falciparum. Mol Biochem Parasitol. 2011;179:42–46. doi: 10.1016/j.molbiopara.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Oh E, Weissman JS. The anti-shine-dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Li WH. Evolution of 5′ untranslated region length and gene expression reprogramming in yeasts. Mol Biol Evol. 2012;29:81–89. doi: 10.1093/molbev/msr143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Oh E, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penel S, et al. Databases of homologous gene families for comparative genomics. BMC Bioinformatics. 2009;10(Suppl 6):S3. doi: 10.1186/1471-2105-10-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Ahn S, Speed TP, Rubin GM. Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome Biol. 2007;8:R63. doi: 10.1186/gb-2007-8-4-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao YS, et al. Selection for the compactness of highly expressed genes in Gallus gallus. Biol Direct. 2010;5:35. doi: 10.1186/1745-6150-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuveni S, Meilijson I, Kupiec M, Ruppin E, Tuller T. Genome-scale analysis of translation elongation with a ribosome flow model. PLoS Comput Biol. 2011;7:e1002127. doi: 10.1371/journal.pcbi.1002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA, Hartl DL. Population genetics of polymorphism and divergence. Genetics. 1992;132:1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Gilchrist MA. Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLoS Genet. 2010;6:e1001128. doi: 10.1371/journal.pgen.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Gilchrist MA. Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift. Proc Natl Acad Sci U S A. 2011;108:10231–10236. doi: 10.1073/pnas.1016719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Averof M, Lloyd AT, Matassi G, Peden JF. DNA sequence evolution: the sounds of silence. Philos Trans R Soc Lond B Biol Sci. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Emery LR, Zeng K. Forces that influence the evolution of codon bias. Philos Trans R Soc Lond B Biol Sci. 2010;365:1203–1212. doi: 10.1098/rstb.2009.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LX, Basilion JP, Stanton VP., Jr Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc Natl Acad Sci U S A. 1999;96:7871–7876. doi: 10.1073/pnas.96.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DJ, et al. Genolevures: protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res. 2009;37:D550–D554. doi: 10.1093/nar/gkn859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci U S A. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Van Rossum G, Drake F. VA: PythonLabs; 2001. Python reference manual. [cited March 2011]. Available at http://www.python.org. [Google Scholar]
- Zhou T, Wilke CO. Reduced stability of mRNA secondary structure near the translation-initiation site in dsDNA viruses. BMC Evol Biol. 2011;11:59. doi: 10.1186/1471-2148-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]