Abstract
In both vertebrates and insects, the conservation of local gene order among distantly related species (microsynteny) is higher than expected in the presence of highly conserved noncoding elements (HCNEs). Dense clusters of HCNEs, or HCNE peaks, have been proposed to mediate the regulation of sometimes distantly located genes, which are central for the developmental program of the organism. Thus, the regions encompassing HCNE peaks and their targets in different species would form genomic regulatory domains (GRDs), which should presumably enjoy an enhanced stability over evolutionary time. By leveraging genome rearrangement information from nine Drosophila species and using gene functional and phenotypic information, we performed a comprehensive characterization of the organization of microsynteny blocks harboring HCNE peaks and provide a functional portrait of the putative HCNE targets that reside therein. We found that Drosophila HCNE peaks tend to colocalize more often than expected and to be evenly distributed across chromosomal elements. Putative HCNE peak targets are characterized by a tight association with particular promoter motifs, higher incidence of severe mutant phenotypes, and evidence of a more precise regulation of gene expression during important developmental transitions. As for their physical organization, ∼65% of these putative targets are separated by a median of two genes from their nearest HCNE peaks. These observations represent the first functional portrait of this euchromatic fraction of the Drosophila genome with distinctive evolutionary dynamics, which will facilitate future experimental studies on the interactions between HCNE peaks and their targets in a genetically tractable system such as Drosophila melanogaster.
Keywords: genome organization, microsynteny, functional constraints, HCNEs, Drosophila
Microsynteny and Genomic Regulatory Domains
The two main mechanisms considered to underlie microsynteny in metazoans are the presence of fragile regions at the edges of chromosomal rearrangements (Pevzner and Tesler 2003) and the presence of constraints that influence where breakpoints cannot be accommodated (Hurst et al. 2004). Studies in the genus Drosophila indicate that although fragile regions seem to have prevailed in shaping gene organization over time, constraints have prevented the occurrence of chromosomal breakpoints in at least ∼15% of the intergenic regions (von Grotthuss et al. 2010). Analysis of the top 1% largest microsynteny blocks, or ultraconserved regions (UCRs), revealed that the genomic feature most prominently found was an unusually high presence of nucleotide sequences that are 98% identical over at least 50 bp in all pairwise species comparisons, which are otherwise known as highly conserved noncoding elements (HCNEs) (Engstrom et al. 2007; von Grotthuss et al. 2010).
Experimental evidence has revealed a role for HCNEs in regulating gene expression, sometimes over long distances (de la Calle-Mustienes et al. 2005; Shin and Cho 2005; Woolfe et al. 2005; Pennacchio et al. 2006). In addition, dense clusters of HCNEs (HCNEs peaks hereafter) have been found around genes regulating development (Lindblad-Toh et al. 2005). Because HCNE peaks predominantly reside in microsynteny blocks that contain developmental regulatory genes both in vertebrates and insects (Engstrom et al. 2007; Kikuta et al. 2007), it has been proposed that the pressure to maintain these regulatory interactions would be the major constraint that explains microsynteny. Perturbation of these interactions might result in gene misexpression with a corresponding fitness cost (Goode et al. 2005; Spitz et al. 2005; Jeong et al. 2006).
Microsynteny blocks harboring 164 HCNE peaks have been documented in comparative analyses of the genus Drosophila (Engstrom et al. 2007). Some of these same microsynteny blocks were found to include genes involved in development as inferred by their Gene Ontology (GO) term annotation. These genes preferentially possess initiator-type (Inr) core promoters, thus reinforcing the notion that HCNEs might be selectively targeting a particular class of genes and that the microsynteny blocks where HCNEs reside encompass genomic regulatory domains (GRDs). However, this early characterization of microsynteny blocks harboring putative GRDs was hampered for several reasons. First, the physical limits of the microsynteny blocks could not be precisely delineated because several Drosophila lineages that have accumulated multiple chromosomal breakpoints were not included. In addition, the gene expression profile used in the study was confined to embryogenesis in Drosophila melanogaster (Manak et al. 2006), and the GO annotation was incomplete; other types of phenotypic information such as essentiality were not considered. Therefore, a precise characterization of the microsynteny blocks that encompass putative GRDs in the genus Drosophila is still lacking.
Comparative analysis of nine Drosophila species that accumulate ∼380 million years of divergence allowed the mapping of 145 HCNE peaks in 123 of the 2,683 microsynteny blocks that form the Drosophila genome (von Grotthuss et al. 2010). Further, transcriptome information from 30 developmental stages (Graveley et al. 2011), and extended functional annotation and mutant phenotypic information are currently available allowing for a more refined characterization. Here, we use these more comprehensive data sets to examine the structural hallmarks of these 123 microsynteny blocks, perform a thorough characterization of the phenotypic attributes of putative HCNE targets residing in these regions, and inspect the physical relationship between these putative targets and HCNE peaks.
Genomic Organization of Microsynteny Blocks Encompassing Conserved GRDs
The frequency of HCNEs per sequence length unit is known to increase with the size of the microsynteny blocks, in stark contrast to peaks of stretches of coding DNA (Engstrom et al. 2007). Early estimates also indicated that only 12% of the microsynteny blocks spanning HCNE peaks were of small size (<3 protein-coding genes). Collectively, these observations suggested a positive correlation between the size of the microsynteny blocks, which can be thought of as a proxy for genome stability, and the presence of HCNE peaks. To analyze this with a more comprehensive data set, we retrieved the coordinates and gene content of the 123 microsynteny blocks delineated based on comparative information of nine Drosophila species and known to harbor HCNE peaks (von Grotthuss et al. 2010). These 123 microsynteny blocks include 1,321 protein-coding genes inferred to be present in the ancestor of the species analyzed. Four Drosophila species not previously included in the previous analysis (D. erecta, D. yakuba, D. willistoni, and D. grimshawi) were included here. They represent lineages that have accumulated ∼40% of the total number of chromosomal breakpoints (von Grotthuss et al. 2010). This increase in the number of chromosomal breakpoints did not result in a substantial difference in the proportion of microsynteny blocks that were small; eight (or 6.5%) microsynteny blocks were found to include <3 protein-coding genes (supplementary data set S1, Supplementary Material online). In accordance with this, the 123 microsynteny blocks spanning HCNE peaks were of a larger than expected size when tested against a random distribution of HCNEs across the genome (Monte Carlo simulations, P < 0.01). This result was robust regardless of the unit used to measure the size of the microsynteny blocks (table 1). We also examined whether the larger size trend holds when the UCRs are excluded because unlike these, other microsynteny blocks harboring HCNE peaks can have a much more limited size (supplementary data set S1, Supplementary Material online). We found this to be the case (Monte Carlo simulations, P < 0.01; table 1). Therefore, the association between HCNE density and genome stability over evolutionary time, using the size of microsynteny blocks as a proxy, is a general property of the fraction of the genome represented by all microsynteny blocks harboring HCNE peaks and not exclusive of UCRs.
Table 1.
Average Size of the Microsynteny Blocks Encompassing HCNE Peaks
| Size Unit |
|||
|---|---|---|---|
| IGAs | Genes | kb | |
| All 123 MB | |||
| Observeda | 10.24b | 11.33b | 208.74 |
| Expectedc | 5.74–8.30 | 6.76–9.58 | 63.89–106.22 |
| Excluding UCRs | |||
| Observeda | 8.18b | 9.15b | 186.33 |
| Expectedc | 4.82–6.74 | 5.76–7.94 | 51.54–89.32 |
Note.—MB, microsynteny block; IGA, independent gene anchor. IGA refers to physically related genes, for example, overlapping or nested, which are counted only once.
aCalculated based on data from von Grotthuss et al. (2010).
bP < 0.01.
cUsing Monte Carlo simulations, 0.5th–99.5th percentiles of the distribution were obtained.
We next explored the genome distribution of HCNE peaks; 14.6% (18/123) of the microsynteny blocks contained more than one HCNE peak. This colocalization of HCNE peaks in the same microsynteny block was significantly higher than expected by chance, because Monte Carlo simulations indicated that a random distribution would result in 145 HCNE peaks being scattered over 131–144 microsynteny blocks, with only 1–13 of these harboring more than one HCNE peak (0.5th–99.5th percentiles). Further, HCNE peaks were found to be randomly distributed along the main chromosomal elements that make up the Drosophila genome (Muller's elements A–E; Muller 1940) (supplementary table S1, Supplementary Material online). Together, these results indicate that HCNE peaks are located in genomic regions that are more refractory to disruption by chromosomal rearrangements, tend to colocalize more often than expected by chance, and are evenly distributed across Drosophila chromosomes.
Functional Attributes of HCNE Targets
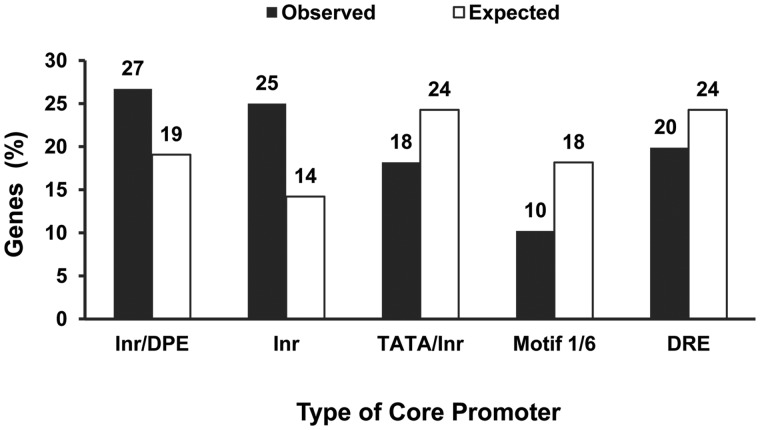
Two proxies have been used to identify the genes that are regulated by HCNE peaks (Engstrom et al. 2007). The first is their association with diagnostic GO terms related to the regulation of developmental processes (i.e., with regulation of transcription, GO:0006355 and/or multicellular organismal development, GO:0007275). The second is the presence of specific types of core promoters that might be selectively regulated by HCNE enhancers. Approximately 95% of developmental regulators present in microsynteny blocks harboring HCNE peaks contained some kind of Inr motif: Inr alone; Inr/DPE (initiator followed by a downstream promoter element); or TATA/Inr (TATA box followed by initiator) (Engstrom et al. 2007). We inspected genes encompassed in the 123 microsynteny blocks harboring HCNE peaks and found reliable core promoter predictions for ∼42.1% (556/1,321) of genes, which were distributed across all but one of the microsynteny blocks (see Materials and Methods and supplementary data set S2, Supplementary Material online). Furthermore, ∼20.5% (271/1,321) of the genes were found to be annotated with at least one diagnostic GO term. The new data included in this analysis led to a redistribution of the proportions of genes found both with specific promoter types and with the regulation of development GO terms, compared with the previous analysis (supplementary fig. S1a and b, Supplementary Material online).
To better characterize the genes that are most likely to be targeted by HCNE peaks, we further examined 176 genes that had both core promoter predictions and were associated with diagnostic GO terms (supplementary table S2, Supplementary Material online). Genes with core promoters spanning the motifs Inr alone and Inr/DPE were more tightly associated (∼10.8% and ∼7.6%, respectively, more than expected by chance) with diagnostic GO terms than other types of promoter motifs (randomization test of goodness-of-fit, Padjusted = 6.0 × 10−5; table 2; fig. 1). This led us to group genes with these two types of promoters in downstream analyses. Importantly, and unlike in a previous study (Engstrom et al. 2007), for our downstream analyses, we considered genes associated with at least one diagnostic GO term and did not limit our analyses to those associated with both, that is, developmental regulator genes. There are two other patterns of expression that are also compatible with regulation mediated by HCNEs. First, genes involved in developmental and differentiation tasks that are not transcription factors, for example, those encoding ligands and protein receptors, can have comparable complex regulatory inputs as inferred by the size of the downstream and upstream regions where their regulatory sequences reside (Nelson et al. 2004). Second, genes associated with only one diagnostic GO term often showed a pattern of expression developmentally regulated during the life cycle (supplementary fig. S2, Supplementary Material online), which was further supported by the observation that ∼43% of these genes exhibited precise spatial expression in the embryo (GO:0006355: 11/29; GO:0007275: 67/151; supplementary data set S2, Supplementary Material online). Using these criteria, we considered genes with core promoter motif Inr alone or Inr/DPE and associated with at least one diagnostic GO term as the best candidates to be targets of HCNE peaks. On the basis of this definition, we found at least one putative target gene in 63 of the 123 microsynteny blocks harboring HCNE peaks.
Table 2.
Relationship between Different Functional Features and Protein-Coding Genes Predicted to Have a Particular Type of Core Promoter
| Number of Genes Associated/Not Associated |
|||||
|---|---|---|---|---|---|
| Genomic Features | Inr Only and Inr/DPE | TATA/Inr | Motif 1/6 | DRE | Padja |
| Diagnostic GO terms (GO:0006355, GO:0007275) | 91/94 | 32/103 | 18/83 | 35/100 | 6.0 × 10−5 |
| Severe detrimental mutant phenotype | 81/34 | 29/18 | 26/21 | 43/20 | 1.6 × 10−2 |
| Expression profilesb | |||||
| >80th percentile (E, P) | 172/13 | 129/6 | 96/5 | 125/10 | 6.9 × 10−1 |
| >95th percentile (E, P) | 165/20 | 104/31 | 78/23 | 99/36 | 5.7 × 10−3 |
| >80th percentile (E, P) and <40th percentile (L, AM, AF) | 148/37 | 95/40 | 75/26 | 97/38 | 2.4 × 10−1 |
| >95th percentile (E, P) and <40th percentile (L, AM, AF) | 145/40 | 86/49 | 66/35 | 83/52 | 8.4 × 10−3 |
Note.—Inr/DPE, initiator followed by downstream promoter element; TATA/Inr, TATA box followed by initiator; Motif 1/6, Motif 1 followed by Motif 6; DRE, DNA replication element binding factor.
aA randomization test of goodness-of-fit was performed for each genomic feature; P ≥ χ2 with 3 degrees of freedom (100,000 simulations). Subsequently, the Benjamini–Hochberg (Benjamini and Hochberg 1995) correction for multiple testing was applied.
bHigher than a given percentile in at least one timepoint during embryogenesis (E), pupa stage (P), or both and, if it is the case, lower than a given percentile in at least one timepoint in at least two stages among larva (L), adult male (AM), and adult female (AF). Genes examined: with Inr only or Inr/DPE promoter types, 185; with TATA/Inr promoter type, 135; with Motif 1/6 promoter type, 101; and with DRE promoter type, 135.
Fig. 1.—
Association between the type of predicted core promoter and diagnostic GO terms that denote involvement in developmental tasks. Unlike protein-coding genes predicted to have core promoters TATA/Inr, Motif 1/6, or DRE, those with core promoters Inr/DPE and Inr alone show a significant association with diagnostic GO terms (table 2 and supplementary table S2, Supplementary Material online).
For many genes, there are now data available on associated mutant phenotypes (Tweedie et al. 2009) and time series expression profiles (Graveley et al. 2011). We evaluated how this new functional information could also refine current proxies for the identification of targets of HCNE peaks. With the mutant phenotype data, we hypothesized that these targets would be more likely to have a severe detrimental mutant phenotype, signifying a functional role during development. Therefore, there would be a tighter association between genes spanning core promoter types Inr alone or Inr/DPE and severe phenotypes (lethal, sterile, or both) than between those same genes and nonsevere phenotypes (viable, fertile, or both), when compared with genes with other core promoters. We could confirm severe and nonsevere detrimental phenotypes for 180 and 93 genes (of 556), respectively. We found a significantly higher ratio of severe to nonsevere detrimental phenotypes for genes with core promoters type Inr alone or Inr/DPE (∼2.4-fold) than for genes with types DRE (DNA replication element binding factor; ∼2.1-fold), TATA/Inr (∼1.6-fold), and Motif 1/6 (Motif 1 followed by Motif 6; ∼1.2-fold) (randomization test of goodness-of-fit, Padjusted = 1.6 × 10−2; table 2). We further examined this relationship between mutant phenotypes and different types of core promoters by focusing on the genes associated with the diagnostic GO terms that reflect a specific, essential role within the developmental program of Drosophila and thus presumably those genes likely regulated by HCNE peaks. We found that the mutant phenotypes of this refined group of genes were less likely to be severe when they had core promoters types TATA/Inr, Motif 1/6, and DRE (i.e., likely non-HCNE targets), compared with genes with core promoter types Inr only or Inr/DPE (two-tailed Fisher's exact test, FET; Inr only or Inr/DPE, Padjusted = 1.5 × 10−8; e.g., Motif 1/6, Padjusted = 1.3 × 10−3, the closest one; table 3). Therefore, when we confined our analysis to the best candidate genes to be targets of HCNE peaks, we found that they are more likely to be essential compared with genes with other core promoter types.
Table 3.
Association between Genes with and without Diagnostic GO Terms and Functional Features for Protein-Coding Genes Predicted to Have Different Types of Core Promoters
| Genes with/without Diagnostic GO Terms (Padja) |
||||
|---|---|---|---|---|
| Alternative Proxy | Inr Only and Inr/DPE | TATA/Inr | Motif 1/6 | DRE |
| Severe detrimental mutant phenotype | 70/9 (1.5 × 10−8) | 22/5 (8.2 × 10−3) | 13/3 (2.5 × 10−2) | 26/2 (1.3 × 10−3) |
| Expression during developmentb | ||||
| >80th percentile (E, P) | 88/84 (9.6 × 10−2) | 31/98 (1) | 18/78 (7.9 × 10−2) | 35/90 (6.1 × 10−1) |
| >95th percentile (E, P) | 87/78 (1.8 × 10−2) | 28/76 (1.7 × 10−1) | 18/60 (3.6 × 10−2) | 31/68 (2.1 × 10−2) |
| >80th percentile (E, P) and <40th percentile (L, AM, AF) | 87/61 (3.2 × 10−7) | 28/67 (2.5 × 10−2) | 17/58 (2.5 × 10−2) | 31/66 (4.9 × 10−2) |
| >95th percentile (E, P) and <40th percentile (L, AM, AF) | 86/59 (3.2 × 10−7) | 26/60 (3.2 × 10−2) | 17/49 (7.0 × 10−3) | 29/54 (1.2 × 10−2) |
Note.—Inr/DPE, initiator followed by downstream promoter element; TATA/Inr, TATA box followed by initiator; Motif 1/6, Motif 1 followed by Motif 6; DRE, DNA replication element binding factor.
aTwo-tailed Fisher's exact test; the Benjamini–Hochberg correction (Benjamini and Hochberg 1995) was applied to correct for multiple testing. For mutant phenotypes, the total number of genes for the two categories (severe vs. nonsevere) is 81/34 (Inr only and Inr/DPE), 29/18 (TATA/Inr), 43/20 (Motif 1/6), and 26/21 (DRE). For expression data, the total number of genes for the two categories is 91/94 (Inr only and Inr/DPE), 32/103 (TATA/Inr), 18/83 (Motif 1/6), and 35/100 (DRE).
bHigher than a given percentile in at least one timepoint during embryogenesis (E), pupa stage (P), or both and, if it is the case, lower than a given percentile in at least one timepoint in at least two stages among larva (L), adult male (AM), and adult female (AF).
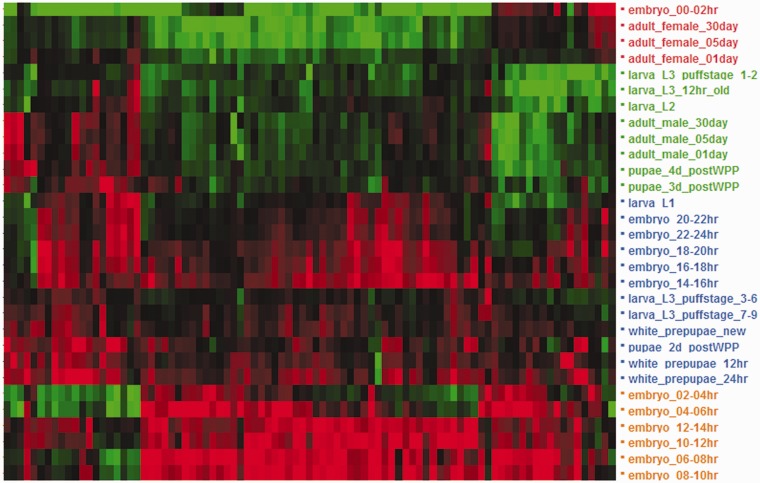
We next examined expression profiles during the Drosophila life cycle. We extracted expression levels across 30 timepoints within five stages (embryo, larva, pupa, adult male, and adult female) and assessed the relationship between expression profiles and particular types of promoters. Genes with Inr only or Inr/DPE promoter types showed the strongest association with expression levels above the 95th percentile in at least one timepoint within embryogenesis, pupa stage, or both (randomization test of goodness-of-fit, Padjusted = 5.7 × 10−3; fig. 2 and table 2). Thus, ∼89.2% (165/185) of the genes with core promoter type Inr alone or Inr/DPE, as opposed to a ∼77.2% (78/101) among genes with core promoter type Motif 1/6, the closest one, exhibit a developmentally specific expression profile. When this demanding expression threshold is relaxed (80th percentile), no differences were found across genes with different promoters (Padjusted = 6.9 × 10−1; table 2). Further, coupling the requirement of being expressed above the 95th percentile in at least one timepoint within embryogenesis, pupa stage, or both with a limited expression (i.e., <40th percentile) in at least two timepoints among the other three stages (larva, adult male, and adult female) was of similar significance to expression levels above the 95th percentile in at least one timepoint within embryogenesis, pupa stage, or both (table 2). This result stresses that the most robust pattern associated with the expression profile of genes with core promoter type Inr alone or Inr/DPE is their high expression level during key developmental transitions in Drosophila regardless of their magnitude of expression in other life stages. However, if we analyze only the genes associated with the diagnostic GO terms, being expressed above the 95th percentile in at least one timepoint within embryogenesis, pupa stage, or both, coupled with limited expression (<40th percentile) in at least one timepoint within at least two of the other three stages (larva, adult male, and adult female), is now more commonly found in genes with promoter type Inr only or Inr/DPE than in genes with different promoter motifs. Under this combination of requirements, genes with promoter type Inr only or Inr/DPE are clearly more tightly associated with this temporal expression profile (FET; Inr only or Inr/DPE, Padjusted = 3.2 × 10−7; e.g., Motif 1/6, Padjusted = 7.0 × 10−3, the closest one; table 3). This pattern now does hold when the expression threshold during key developmental transitions is decreased from the 95th percentile to the 80th percentile. This result substantiates that genes with promoter type Inr only or Inr/DPE, and also associated with diagnostic GO terms, are more often developmentally regulated than genes with those same GO terms but that posses a different type of promoter.
Fig. 2.—
Two-way hierarchical clustering of the levels of expression for genes with core promoter type Inr alone or Inr/DPE, and associated with either one or both diagnostic GO terms, across 30 timepoints of the life cycle of D. melanogaster. Red, overexpession; green, underexpression. High levels of expression are common at some point during embryogenesis, and intermediate to high levels of expression are often observed during the larval–pupal transition. A more reduced number of genes show additionally substantial expression in adult males and females.
In sum, the best candidate genes to be targets of HCNE peaks present in the 123 microsynteny blocks analyzed are characterized by an expression profile more markedly regulated throughout development and by a tendency to show severe mutant phenotypes as opposed to the rest of genes considered. These patterns agree well with the role thought to be played by genes targeted by HCNEs peaks during key developmental transitions although this is not incompatible with being expressed in other life stages.
Internal Organization of Microsynteny Blocks Encompassing Conserved GRDs
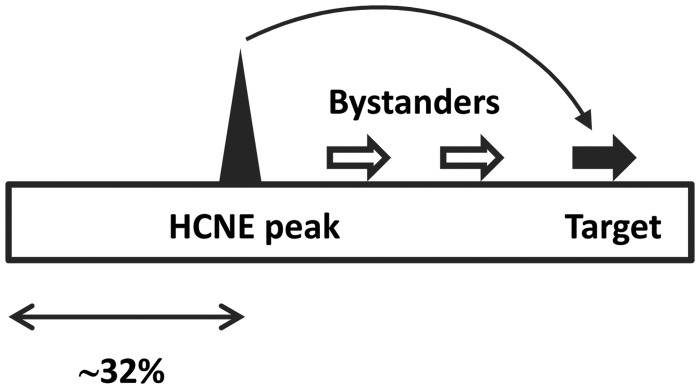
We next evaluated the location of HCNE peaks within microsynteny blocks. The median distance between HCNE peaks and the closest outermost markers equaled 32% of the size of the microsynteny blocks examined (supplementary fig. S3, Supplementary Material online), in good agreement with previous estimates (Engstrom et al. 2007). Using D. melanogaster as a reference, we find that HCNE peaks are separated from their putative targets by ∼65 ± ∼55/∼53 kb (average ± standard deviation [SD]/median) or ∼32 ± ∼42/∼26% of the size of the microsynteny block where both reside. Approximately 3 ± ∼5/∼2 intervening protein-coding genes were found between HCNE peaks and their putative target genes, although for ∼37% of these targets there was no intervening protein-coding gene (fig. 3 and supplementary fig. S4, Supplementary Material online).
Fig. 3.—
Canonical view of the internal organization of microsynteny blocks that harbor GRDs in the genus Drosophila. Below, median distance between the HCNE peak and the closest outermost marker expressed as a fraction of the total length of the microsynteny block. The distance between the HCNE peak and a putative target gene is indicated as the median number of intervening, or bystander, genes.
We also explored the patterns of colocalization between HCNE peaks and their putative targets on individual microsynteny blocks. We found no correlation between them (Pearson’s correlation coefficient, r = −0.0054; >1 putative target and >1 HCNE peak on the same microsynteny block occurred in 4 of 63 cases). In fact, Monte Carlo simulations showed that finding a correlation equal or lower than that observed was unlikely (P = 0.0039). This raises the possibility that a single HCNE peak might regulate more than one putative target gene in some microsynteny blocks, whereas in others, several HCNE peaks might cooperatively regulate a single target. We found 18 and 3 cases out of 63 following these tentative organizational patterns, respectively.
With the criteria used, we have extracted the main patterns of the architecture of genomic regions with gene arrangement conserved across species harboring HCNE peaks and their putative targets. Nevertheless, these emerging patterns are not based on the 123 microsynteny blocks, because in 49% (60/123) of them, we did not find a reliable prediction for the presence of at least one putative target. This can result from two nonmutually exclusive possibilities. The first is that a large fraction of genes associated with one or both of the proxies used for the identification of targets of HCNE peaks still remain to be characterized. The second is that the regulation of HCNE peaks may not be as restrictive as previously presumed and may involve genes with core promoter motifs other than Inr only and Inr/DPE. The use of genome engineering techniques (Spitz et al. 2005; Diaz-Castillo et al. 2012) to perturb the interactions between HCNE peaks and their putative targets, coupled with expression assays (Woolfe et al. 2005), will help to more precisely dissect the role of HCNE peaks in impacting gene function within the context of chromosome repatterning. This kind of experimental framework will also generate valuable information that can be used to develop improved computational pipelines for the identification of targets of HCNE peaks, especially in regions where none have been predicted. On the whole, this integrative approach will enable to generate an ultimate portrait of the structural and functional organization of this fraction of the Drosophila euchromatin with distinctive evolutionary dynamics.
Materials and Methods
Synteny Information and HCNE Peaks
We used synteny maps constructed under the requirement of conservation of gene order but not orientation (GO synteny definition in von Grotthuss et al. 2010) across the species D. ananassae, D. erecta, D. grimshawi, D. melanogaster, D. mojavensis, D. pseudoobscura, D. virilis, D. willistoni, and D. yakuba. Coordinates of the 145 HCNEs peaks in the D. melanogaster genome (release 4.3) were retrieved from Engstrom et al. (2007).
Promoter Analysis
Core promoter predictions for transcripts associated with protein-coding genes in microsynteny blocks were performed with McPromoter using the most stringent parameter values (Ohler 2006). We inspected 500 nt upstream of the 5' untranslated region (5'-UTR) start of each transcripts as annotated in FlyBase (Tweedie et al. 2009). In the absence of an annotated 5’-UTR, a stretch of DNA of equal length upstream of the first annotated nucleotide was examined.
Per Gene Phenotypic and Functional Data
Expression profiles during the life cycle of D. melanogaster (Graveley et al. 2011) were retrieved from FlyBase (Tweedie et al. 2009). Reads per kilobase of exon model per million mapped reads across 30 timepoints were log transformed, their relative level of expression per life stage quantified relative to each gene, and compared by unsupervised hierarchical clustering using Ward’s minimum variances as a distance metric. The first principal component was used to assist in the ordering of expression levels. Spatial patterns of expression were obtained from FlyExpress (Kumar et al. 2011). Severe (lethal and/or sterile alleles) and nonsevere (viable and/or fertile alleles) phenotypes were obtained from FlyBase (Tweedie et al. 2009). GO annotations for the category biological process were retrieved from modMine (http://intermine.modencode.org/; last accessed February 2012).
Statistical Analyses
With the exception of the Monte Carlo simulations, all statistical analyses were performed using SAS 9.2 (SAS Institute).
Supplementary Material
Supplementary figures S1–S4, tables S1 and S2, and data sets S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Carlos Díaz-Castillo for technical help. This work was supported by a grant from the National Science Foundation [MCB-1157876] to J.M.R. V.S. is grateful to the Bridges to the Baccalaureate Program supported by a National Institute of Health grant [R25-GM056647] award to Luis Mota-Bravo.
Literature Cited
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- de la Calle-Mustienes E, et al. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Castillo C, Xia XQ, Ranz JM. Evaluation of the role of functional constraints on the integrity of an ultraconserved region in the genus Drosophila. PLoS Genet. 2012;8:e1002475. doi: 10.1371/journal.pgen.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode DK, Snell P, Smith SF, Cooke JE, Elgar G. Highly conserved regulatory elements around the SHH gene may contribute to the maintenance of conserved synteny across human chromosome 7q36.3. Genomics. 2005;86:172–181. doi: 10.1016/j.ygeno.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- Kikuta H, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, et al. FlyExpress: visual mining of spatiotemporal patterns for genes and publications in Drosophila embryogenesis. Bioinformatics. 2011;27:3319–3320. doi: 10.1093/bioinformatics/btr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Manak JR, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Bearings of the Drosophila work on systematics. In: Huxley J, editor. The new systematics. Oxford: Clarendon Press; 1940. pp. 185–268. [Google Scholar]
- Nelson CE, Hersh BM, Carroll SB. The regulatory content of intergenic DNA shapes genome architecture. Genome Biol. 2004;5:R25. doi: 10.1186/gb-2004-5-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006;34:5943–5950. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci U S A. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JM, Cho DH. PDB-Ligand: a ligand database based on PDB for the automated and customized classification of ligand-binding structures. Nucleic Acids Res. 2005;33:D238–D241. doi: 10.1093/nar/gki059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- Tweedie S, et al. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grotthuss M, Ashburner M, Ranz JM. Fragile regions and not functional constraints predominate in shaping gene organization in the genus Drosophila. Genome Res. 2010;20:1084–1096. doi: 10.1101/gr.103713.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]