Abstract
Gene duplication is an important evolutionary mechanism and no eukaryote has more duplicated gene families than the parasitic protist Trichomonas vaginalis. Iron is an essential nutrient for Trichomonas and plays a pivotal role in the establishment of infection, proliferation, and virulence. To gain insight into the role of iron in T. vaginalis gene expression and genome evolution, we screened iron-regulated genes using an oligonucleotide microarray for T. vaginalis and by comparative EST (expressed sequence tag) sequencing of cDNA libraries derived from trichomonads cultivated under iron-rich (+Fe) and iron-restricted (−Fe) conditions. Among 19,000 ESTs from both libraries, we identified 336 iron-regulated genes, of which 165 were upregulated under +Fe conditions and 171 under −Fe conditions. The microarray analysis revealed that 195 of 4,950 unique genes were differentially expressed. Of these, 117 genes were upregulated under +Fe conditions and 78 were upregulated under −Fe conditions. The results of both methods were congruent concerning the regulatory trends and the representation of gene categories. Under +Fe conditions, the expression of proteins involved in carbohydrate metabolism, particularly in the energy metabolism of hydrogenosomes, and in methionine catabolism was increased. The iron–sulfur cluster assembly machinery and certain cysteine proteases are of particular importance among the proteins upregulated under −Fe conditions. A unique feature of the T. vaginalis genome is the retention during evolution of multiple paralogous copies for a majority of all genes. Although the origins and reasons for this gene expansion remain unclear, the retention of multiple gene copies could provide an opportunity to evolve differential expression during growth in variable environmental conditions. For genes whose expression was affected by iron, we found that iron influenced the expression of only some of the paralogous copies, whereas the expression of the other paralogs was iron independent. This finding indicates a very stringent regulation of the differentially expressed paralogous genes in response to changes in the availability of exogenous nutrients and provides insight into the evolutionary rationale underlying massive paralog retention in the Trichomonas genome.
Keywords: gene duplication, iron, microarrays, EST analysis
Introduction
Gene duplications are important in evolution (Lynch and Conery 2000), and no genome has more recently duplicated genes than the parasitic flagellate Trichomonas vaginalis, the causative agent of trichomoniasis (Carlton et al. 2007; Cui et al. 2010). Trichomoniasis is the most common sexually transmitted infection of nonviral origin in humans. Although trichomoniasis is usually self-limiting in males, it causes serious health problems for female patients, including an increased risk of cervical cancer, pelvic inflammatory disease, infertility, and transmission of the HIV (Laga et al. 1993; Zhang and Begg 1994; Viikki et al. 2000; Moodley et al. 2002). Trichomoniasis during pregnancy is associated with low birth weight, the premature rupture of membranes and preterm birth (Cotch et al. 1997). The establishment of a T. vaginalis infection as well as infections by other pathogenic microorganisms is dependent on the efficient acquisition of essential nutrients such as iron, from the host environment. Trichomonads can utilize various host iron-containing proteins such as lactoferrin, transferrin, ferritin, hemoglobin, and low-molecular-weight-iron complexes (Sutak et al. 2008). Iron is required for critical housekeeping functions such as proteosynthesis, genome duplication, and energy fixation. A significant portion of the trichomonad energy metabolism takes place in hydrogenosomes, whose function is particularly dependent on iron (Gorrell 1985; Vanácová et al. 2001). These specific mitochondria-related organelles contain a number of FeS proteins that catalyze key steps in ferredoxin-linked electron transport, hydrogen production, and ATP synthesis at the level of substrate phosphorylation (Hrdy et al. 2004). The formation of FeS clusters in the catalytic centers of apoproteins is mediated by the hydrogenosomal iron–sulfur cluster (ISC) assembly machinery, which consists of approximately 10 proteins, of which cysteine desulfurase (IscS) and the molecular scaffold protein IscU are the main components (Tachezy et al. 2001; Sutak, Dolezal, et al. 2004. Iron is also required for the functions of cytosolic and nuclear FeS proteins such as Rli1p, which is a protein essential for ribosome biogenesis and function (Kispal et al. 2005; Yarunin et al. 2005). This protein is highly conserved in eukaryotes, including T. vaginalis (Smíd et al. 2008).
In addition to housekeeping functions, iron affects specific host–pathogen interactions associated with the virulence of the parasite. Experiments performed in vitro have shown that iron regulates the cytoadherence of T. vaginalis to target cells (Mundodi et al. 2006) as well as the expression of cysteine proteinases (Sommer et al. 2005; Solano-González et al. 2007; Kummer et al. 2008), ecto-ATPases and ecto-phosphatases (De Jesus et al. 2006), and it increases trichomonad resistance to complement-mediated lysis (Alderete et al. 1995). Iron-dependent enhancement of experimental infections in mice was demonstrated with the related bovine parasite Tritrichomonas foetus (Kulda et al. 1999). However, little is known about the mechanisms underlying the iron-dependent regulation of these processes. Positive iron regulation at the transcriptional level was observed for the expression of some hydrogenosomal proteins, including malic enzyme (ME) and pyruvate:ferredoxin oxidoreductase (PFOR) (Vanácová et al. 2001). A detailed study of the iron-dependent regulation of hydrogenosomal ME, which may alternatively be present on the cell surface as the adhesin (AP65–1) (Hirt et al. 2007), led to the identification of Myb recognition elements and novel Myb proteins that appear to be components of a multifarious regulatory machinery in T. vaginalis (Ong et al. 2006, 2007). Two cysteine proteases, TVCP4 and TVCP12, were recently reported to be regulated at the posttranscriptional level by an iron-responsive element/iron response protein-like system (Solano-González et al. 2007; Torres-Romero and Arroyo 2009).
We combined cDNA microarray analysis with an expressed sequence tag (EST) approach to map iron-regulated genes and to reconstruct iron-regulated pathways in T. vaginalis. Our data revealed numerous iron-responsive genes that are involved in several essential pathways, particularly in cytosolic glycolysis and extended glycolysis in hydrogenosomes, as well as genes that encode components of the FeS cluster assembly machinery. Moreover, iron affected the expression of many genes with unknown functions.
Materials and Methods
Cell Cultures
Axenic cultures of T. vaginalis strain T1 were grown in trypticase–yeast extract–maltose medium supplemented with 10% heat-inactivated horse serum, pH 6.2 (Diamond 1957). Iron-rich medium (+Fe) and iron-restricted medium (−Fe) were supplemented with 100 µM Fe-nitrilotriacetate; and 50 µM 2-2-dipyridyl (Sigma), respectively. The cells were subcultured daily in +Fe and −Fe media for 5 days prior to experiment.
cDNA Library Construction and DNA Sequencing
RNA isolation kit (Pharmacia) was used to extract the total RNA from cells grown under +Fe and −Fe conditions, and contaminating genomic DNA was digested with DNase I. PolyA+ RNA was isolated using a PolyA+ tract mRNA isolation kit (Promega). Complementary DNA was synthesized using a ZAP–cDNA synthesis kit after priming with oligo-dT. The cDNA was then directionally cloned into the EcoRI and XhoI sites of the Uni-ZAP XR vector (Stratagene). Single and well-separated plaques were cored out from agar plates and transferred to 96-well microtiter plates containing SM buffer. The phage stocks were used as templates for cDNA insert amplification with T3 and T7 primers (1 nM for each primer). The amplified products were separated in 1.5% agarose gels, and clones that yielded single polymerase chain reaction (PCR)-amplified bands were collected for sequencing. Single-pass sequencing from the 5′-end of the cDNA insert was initiated with a T3 primer using the ABI PRISM BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). The sequencing products were resolved and analysed on an ABI PRISM 377 (Applied Biosystems) or a MEGABACE DNA Sequencer (GE). The nucleotide sequences obtained were processed with the Phred/Phrap/Consed package (http://www.phrap.org/phredphrapconsed.html).
Functional Annotations and Sequence Analysis
BLAST tools were used to compare the assembled sequence contigs to known Trichomonas mRNAs, putative open-reading-frames from the T. vaginalis G3 genome (Carlton et al. 2007) and NCBI’s nonredundant (nr) nucleotide (E value = 10−15) and protein database. Genes were functionally annotated based on the Interpro and Gene Ontology databases. Contigs with identity greater than 60% of their length were annotated and assigned to KEGG pathways.
Putative coding regions from the EST data were collected from BLASTX alignments, and the codon usage bias was calculated. A Perl script was also written to locate poly-A tails and to search 10–35 bp upstream for putative mRNA poly-adenylation signals; the script allowed one mismatch and two mismatches from the sequence AAUAAA. Putative signal sequences and transmembrane domains in the coding regions were identified by SignalP 3.0 and TMHMM 2.0 (http://www.cbs.dtu.dk/services/), respectively. Putative Trichomonas protein kinases and peptidases were identified by sequence comparison with datasets downloaded from the KinBase (http://www.kinase.com) and the MEROPS (http://merops.sanger.ac.uk) databases, respectively.
Microarray Sample Preparation
RNA from trichomonads grown in +Fe and −Fe conditions was isolated using a QuickPrep Total RNA Extraction Kit (Amersham Biosciences) according to the manufacturer’s instructions. The total RNA was further purified using an RNeasy CleanUp Kit (Qiagen). The RNA concentration and purity were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). The integrity of the RNA was checked by agarose gel electrophoresis. The same RNA samples were used in parallel experiments for cDNA microarray analysis and quantitative real-time PCR (qRT-PCR). cDNA probes were synthesized using 2 μg of total RNA and primers labeled with Cy3 and Cy5, respectively, according to the manufacturer’s instructions (3DNA Array 900 Expression Array Detection Kit, Genisphere). Four independent RNA samples (biological replicates) from T. vaginalis strain T1 grown under +Fe and −Fe conditions were compared. A dye–swap design was used to prevent dye-associated effects on cDNA synthesis.
Microarray Analysis
A T. vaginalis customized cDNA microarray (TvArray V2.0) was implemented by the Molecular Regulation and Bioinformatics Laboratory, Chang Gung University, Taiwan. The TvArray chip contained PCR products amplified from 7,688 EST clones with an average GC content of 38.36% and an average length of 384 bp. The cDNA inserts were fabricated on GAPS™ II Coated Slides (Corning. USA). The microarrays were prehybridized in Coplin chambers using a solution containing 1% bovine serum albumine, 1% sodium dodecyl sulfate, and 3X saline sodium citrate buffer (1X SSC consists of 0.15 M NaCl with 0.015 M sodium acetate). The slides were incubated at 50°C for 30 min, washed with water and isopropanol and dried by centrifugation at 90 × g for 3 min. Hybridization with the cDNA hybridization mix and washes were performed following the protocol for the 3DNA Array 900 Expression Array Detection Kit (Genisphere). cDNA hybridizations were performed in a VersArray Hybridization Chamber (Bio-Rad) at 60°C overnight. Hybridizations with fluorescent 3DNA reagents were performed in the same chamber at 60°C for 4 h. After the final washing step, the slides were dried by centrifugation at 90 × g for 3 min and scanned using the GenePix 4200A scanner (Axon). GenePix Pro 5.1 software was used to determine the average signal intensity and the local background for each spot. The Cy3/Cy5 fluorescence ratios were Log 2 transformed and normalized by LOWESS normalization method in the TIGR microarray data analysis system version 2.19 (Saeed et al. 2003). In total, 10 independent hybridizations using samples from 4 independent cultures and RNA extractions were performed and data of the independent experiments were combined. We used MeV v4.8.1 to determine significantly regulated genes. One-class Student t test was performed and P values were based on all 1,024 possible permutations (cut-off was 0.01) and the proportion of false significant genes was set to not exceed 10 genes.
The results of all experiments are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under the array design name TvArray v1.0.
qRT-PCR Analysis
Oligonucleotide primers (supplementary table S1, Supplementary Material online) were designed using Primer Designer software v1.01 (Scientific and Education software). The primers were tested prior to qRT-PCR analysis using DNA as a template, and single amplicons of the proper size were sequenced. The RT reaction contained 1 µg of total RNA, 500 ng of Oligo(dT) 15 Primer (Invitrogen), 5 mM DTT (Invitrogen), 2 U of RNasin (Invitrogen), 10 U of SuperScriptII reverse transcriptase (Invitrogen), and 500 µM each of dATP, dCTP, dGTP, and dTTP. The reactions were incubated at 42°C for 50 min followed by 15 min at 70°C. For qRT-PCR, 1 µl of cDNA was amplified in a 25 µl reaction mixture containing each gene-specific primer at 50 nM and 12.5 µl of iQTM SYBR green Supermix (Bio-Rad). All reactions were performed in triplicate using a RotorGene 2000 Real-Time PCR cycler (Corbett Life Science). The expression levels of each gene were normalized to those of the housekeeping gene β-tubulin, expression of which is not affected by the availability of iron. Relative quantitative values were obtained by the comparative threshold cycle (2−ΔΔCT) method as described by (Livak and Schmittgen 2001).
Analysis of 5′ Untranslated Regions
An application based on the NetBeans Platform (http://platform.netbeans.org) was developed to search for MRE-like motifs in 300 bp of the upstream regions of all iron-regulated genes. We searched for the MRE eukaryotic consensus (C/T)AACG(G/T) and the specific MREs of the ME gene as described by Hsu et al. (2009): MRE1/MRE2r (A[A/T/C/G]AACGAT, CGATA, AACGATA, and TAACGA) and MRE2f (TATCGT and TATCGTC).
Results and Discussion
Microarray Analysis
To investigate changes in gene expression caused by iron availability, trichomonads were grown in media supplemented with 100 µM Fe-nitrilotriacetate (iron-rich conditions, +Fe) or with the iron chelator 2,2-dipyridyl (50 µM; iron-restricted conditions, −Fe). To estimate timeframe for cultivation prior to RNA isolation, we monitored enzymatic activities of PFOR and ME (Rasoloson et al. 2002) as their expression is known to be affected by iron availability in trichomonads (Vanácová et al. 2001). The activities of both enzymes decreased during 24 h cultivation under −Fe conditions and then remained stable (supplementary fig. S1, Supplementary Material online). Thus, to assure ultimate effect of iron on trichomonads, we cultivated the cells for 5 days under +Fe/−Fe conditions prior to each experiment. The total RNA was then isolated from both treatment groups and hybridized onto DNA microarray slides containing PCR products amplified from 7,688 EST clones representing 4,950 unique T. vaginalis genes. A direct pairwise comparison strategy was used to identify differentially expressed genes. Altogether, 10 independent hybridizations were performed using samples from four independent experiments. Genes whose expression ratios changed by a factor of at least 1.3 and had P values lower than 0.01 were considered to be significantly regulated by iron. In total, 195 genes met this criterion that represent approximately 4% of unique genes; 117 and 78 genes were upregulated in cells cultivated under +Fe and −Fe conditions, respectively (fig. 1 and supplementary table S2a, Supplementary Material online). The distribution of genes into fold change categories is given in supplementary table S2b, Supplementary Material online. The complete dataset including genes expression of which was not affected by iron was deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under the array design name TvArray v1.0.
Fig. 1.—
Heatmap visualization of iron-regulated genes based on microarray analysis. Results of 10 experiments are given in columns A–J. (A) genes upregulated in +Fe conditions; (B) genes upregulated in −Fe conditions.
To verify the cut-off limit and to validate the microarray data, we selected 15 genes for qRT-PCR analyses. The selection included genes that were not affected by iron (ferredoxin 6, IscS-2), genes with moderate changes in expression (−1.42- to 1.67-fold change) and genes with a greater than 2-fold change in expression (hydrogenase, alcohol dehydrogenase, and PFOR) according to the microarray data (table 1). In each instance, the qRT-PCR confirmed regulation trends observed in the microarray analysis. The ratios between paired samples (fold change) determined by qRT-PCR were, however, greater than the fold change obtained for the same gene by microarray analysis. These results are consistent with those of Yuen et al. (2002) and Dallas et al. (2005), who showed that microarray analysis underestimates the changes in gene expression compared with the more qRT-PCR assay, although correlations between microarray and qRT-PCR data are generally strong. The list of genes that met the cut-off limit 1.3 in our study included majority of genes coding for proteins that were showed previously to be affected by iron on protein level in T. vaginalis and related parasite T. foetus such as PFOR, hydrogenosomal ME, hydrogenase, ferredoxin, cytosolic malate dehydrogenase, and cysteine protease (Vanácová et al. 2001; De Jesus et al. 2007; Dolezal et al. 2007) that further supports validity of the microarray analysis.
Table 1.
Fold Changes Detected by qRT-PCR in Comparison with Results of Microarray Analysis
| TrichDB 1.2 Accession No. | Annotation | Microarrays Upregulation Rate* | Fold Change Detected by qRT-PCR** |
|---|---|---|---|
| TVAG_239660 | IscS-2 | NSC | NSC |
| TVAG_251200 | Ferredoxin 6 | NSC | NSC |
| TVAG_129940 | IBP39 | 1.11 | 1.88 |
| TVAG_348330 | Glycogen phosphorylase | 1.24 | 1.95 |
| TVAG_281070 | Phosphofructokinase | 1.31 | 4.62 |
| TVAG_292710 | Ferredoxin 4 | 1.33 | 2.89 |
| TVAG_104250 | Hmp-35-2 | 1.56 | 2.28 |
| TVAG_238830 | Malic enzyme B | 1.67 | 2.51 |
| TVAG_198110 | PFO A | 4.01 | 24.96 |
| TVAG_037570 | Iron hydrogenase 64 kDa | 2.48 | 109.64 |
| TVAG_422780 | Alcohol dehydrogenase | 2.57 | 31.44 |
| TVAG_165030 | Malate dehydrogenase | −1.33 | −1.62 |
| TVAG_381311 | Lactate dehydrogenase | −1.81 | −5.02 |
| TVAG_361540 | IscA-2 | −1.36 | −2.26 |
| TVAG_008840 | NfU-2 | −1.42 | −1.47 |
Note.—NSC, no significant change of gene expression.
*P ≤ 0.01.
**P ≤ 0.05.
Comparative EST Analysis
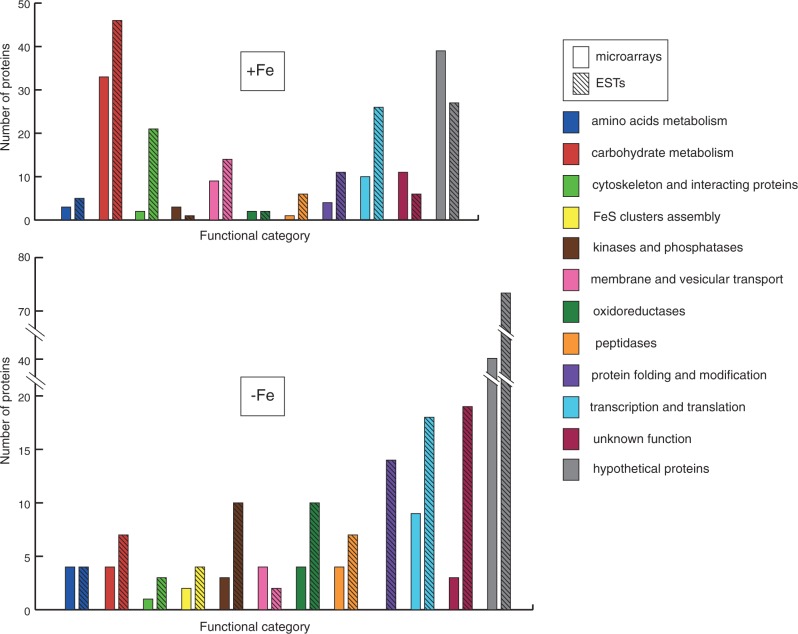
As a second technique to identify differentially expressed genes, we employed a comparative EST approach (Lee et al. 1995). 10,042 and 9,032 ESTs were sequenced from two distinct cDNA libraries that were derived from trichomonads grown under +Fe and −Fe conditions, respectively. The relative frequency (RF) of an EST was calculated as the number of ESTs per 10,000 clones. The upregulation index was calculated as the difference between the RF under +Fe conditions and the RF under −Fe conditions. The distribution of unique genes to upregulation index categories is summarized in supplementary table S2d, Supplementary Material online. The upregulation index values ranged from 1 to 70, which was considerably higher than the range of the fold change based on microarray analysis. Consequently we used higher cut-off limit: a gene was considered to be significantly upregulated if the upregulation index was greater than or equal to five. This criterion met 336 genes (∼5%) out of a total number of 6,381 unique genes that were generated by the assembly of the ESTs. Of those genes, 165 were upregulated under +Fe conditions, and 171 were upregulated under −Fe conditions (supplementary table S2c, Supplementary Material online). A representation of the gene categories that were affected by iron availability is shown in figure 2.
Fig. 2.—
Classification of iron-regulated genes by functional category. Numbers of genes that were upregulated under iron-rich (+Fe) and iron-restricted (−Fe) conditions. Results of microarray and EST analysis are represented by open and hatched bars, respectively. Color code of functional categories is given in the legend.
Major Iron-Regulated Pathways
Glycolysis
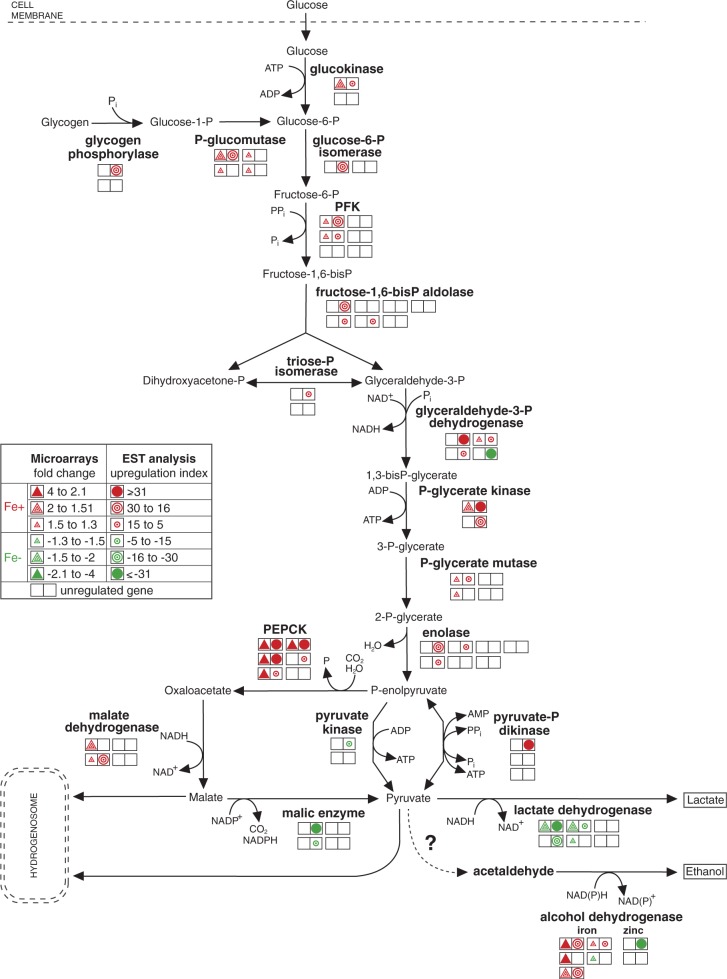
The energy metabolism of T. vaginalis relies on fermentative carbohydrate catabolism in the cytosol that is extended to malate or pyruvate degradation in the hydrogenosome (Müller et al. 2012). At least one (but usually several) of multiple gene copies encoding glycolytic proteins showed significant iron-dependent regulation (fig. 3 and supplementary table S3, Supplementary Material online), with similar results obtained from both methods (microarray and EST analysis). The enzymes that supply the glycolytic pathway with substrates (glucokinase, glycogen phosphorylase, and phosphoglucomutase) and the successive glycolytic enzymes were significantly upregulated under +Fe conditions. A striking exception was the upregulation of one of the four detected glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes (TVAG_366380) under −Fe conditions according to the EST analysis, as the other three GAPDH genes were upregulated under +Fe conditions (fig. 3 and supplementary table S3, Supplementary Material online). It is tempting to speculate that GAPDH TVAG_366380 gene copy encodes protein with the function distinct from other paralogs. Multifunctional character of GAPDH was reported in macrophages where GAPDH serves as a surface receptor that is upregulated upon iron depletion (Rawat et al. 2012). Interestingly, T. vaginalis possesses two types of phosphofructokinase (PFK) (Liapounova et al. 2006) Inorganic pyrophosphate (PPi)-dependent PFK catalyzes reversible phosphorylation of fructose-6-phosphate in the cytosol (Mertens et al. 1998). More recently, putative ATP-dependent PFK has been identified in proteome of hydrogenosomes (Rada et al. 2011). Two of the three genes that encode the PPi-dependent PFK and one of two genes for the ATP-dependent PFK were upregulated under +Fe conditions (supplementary table S3, Supplementary Material online). The most significant changes in iron-dependent gene expression were associated with the pathways that follow the conversion of phosphoenolpyruvate (PEP), the branch-point of carbohydrate metabolism. The enzymes that catalyze the formation of malate from PEP via oxaloacetate (PEP carboxykinase and malate dehydrogenase) were considerably upregulated under +Fe conditions (fig. 3). In contrast, the pathway that converts malate to lactate was upregulated under −Fe conditions. This pathway involves cytosolic NADP-dependent ME and NADH-dependent lactate dehydrogenase (fig. 3). These findings indicate that under +Fe conditions, malate preferentially enters the hydrogenosome and serves as a substrate for hydrogenosomal energy metabolism. However, under −Fe, when hydrogenosomal metabolism is ceased, malate is metabolized in the cytosol via pyruvate to lactate. Thus, the ability to switch between hydrogenosomal and cytosolic malate metabolism seems to be important for the ability of trichomonads to quickly adapt to changes in iron availability in their environment. Similar changes in carbohydrate metabolism were reported in T. vaginalis that had impaired hydrogenosomal metabolism because of the induction of metronidazole resistance (Kulda et al. 1993).
Fig. 3.—
Iron-dependent regulation of genes encoding enzymes in glycolytic pathway. Each double square represents a single gene copy. Triangles represent the results of the microarray analysis, and circles represent the results of the EST analysis. Upregulation under +Fe and −Fe conditions is indicated by red and green colors, respectively. The empty squares represent detected, but unregulated gene copies.
Besides lactate, T. vaginalis also produces low amount of ethanol (Cerkasovová et al. 1986). In our dataset, we identified two types of alcohol dehydrogenases (ADHs) that differ in the metal ion present in the active site of the enzyme. Notably, four of five genes coding for the iron-containing ADH were upregulated under +Fe conditions, whereas one gene was upregulated under −Fe conditions together with one of two genes coding for the zinc-containing ADH (fig. 3). In the related organism T. foetus, acetaldehyde that is reduced by ADH to ethanol is formed from pyruvate by the enzyme pyruvate decarboxylase (Sutak, Tachezy, et al. 2004). However, pyruvate decarboxylase activity was not detected in T. vaginalis, and the gene encoding this enzyme was not identified in the genome (Carlton et al. 2007). Thus, the pathway responsible for the formation of acetaldehyde remains unclear.
Hydrogenosomal Energy Metabolism
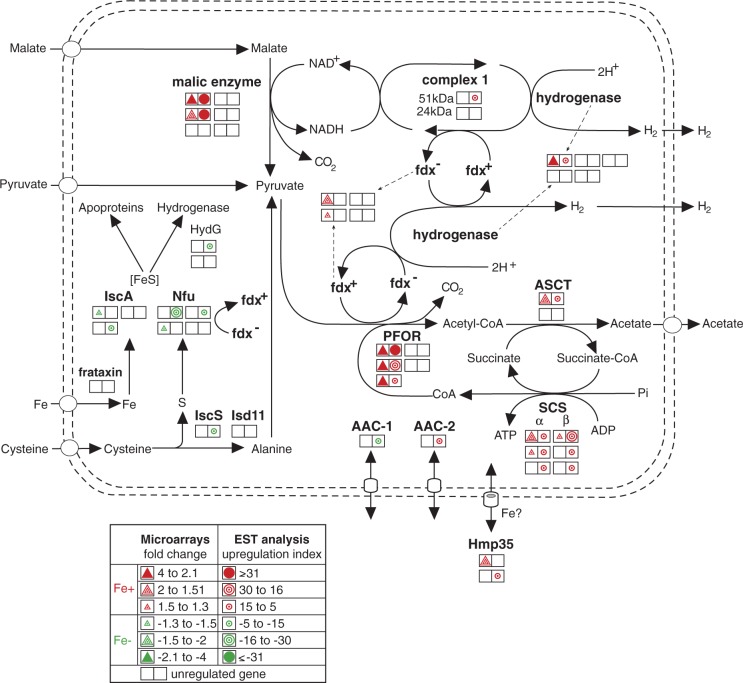
Iron increased the transcription of all critical enzymes in hydrogenosomal carbohydrate catabolism; at least one copy of each gene was significantly upregulated (fig. 4 and supplementary table S3, Supplementary Material online). PFOR and ME, which are the enzymes that catalyze the oxidative decarboxylation of the hydrogenosomal substrates pyruvate and malate, respectively, were the most highly upregulated enzymes of the pathway. Three genes encoding PFOR (PFOR-A, BI and BII) displayed the highest upregulation observed in the microarray analysis (4.01, 3.71, and 3.64, respectively), and also had high upregulation scores determined by the EST analysis (32, 18, and 9, respectively) (supplementary table S2a and c, Supplementary Material online). The gene coding for ME-H was the most highly upregulated according to the EST analysis, with an upregulation index value of 63 (supplementary table S2c, Supplementary Material online). The re-oxidation of NADH resulting from ME activity is mediated by heterodimeric NADH dehydrogenase (remnant of respiratory complex I). The 51 kDa subunit of this enzyme was significantly upregulated under +Fe conditions according to EST analysis, but the upregulation of the 24 kDa subunit did not reach the cut-off limit. Electrons generated by PFOR are transferred via ferredoxin to the hydrogenase responsible for the synthesis of molecular hydrogen. The genes that code for ferredoxin-1 and the 64 kDa [Fe]-hydrogenase were upregulated under +Fe conditions. Finally, the acetyl-CoA that is generated by PFOR activity is used in two steps of ATP synthesis: the reactions catalyzed by acetate:succinate-CoA transferase (ASCT) and heterodimeric succinyl-CoA synthase (SCS). One of the four ASCT genes and all of the genes that encode SCS subunits showed significant upregulation under +Fe conditions.
Fig. 4.—
Iron-dependent regulation of hydrogenosomal metabolism. Each double square represents a single gene copy. Triangles represent the results of the microarray analysis, and circles represent the results of the EST analysis. Upregulation under +Fe and −Fe conditions is indicated by red and green colors, respectively. The empty squares represent detected, but unregulated gene copies.
FeS Cluster Assembly
The biosynthesis of the ISCs that are necessary for the maturation of FeS proteins is an indispensable process that occurs in the hydrogenosomes of T. vaginalis. In contrast to the components of energy metabolism, the ISC assembly machinery appeared to be upregulated under −Fe conditions (fig. 4 and supplementary table S3, Supplementary Material online). Our screen detected genes that encode two paralogs of the scaffold protein IscA, three Nfu paralogs, and the cysteine desulfurase IscS-2. Frataxin was previously shown to be upregulated under −Fe conditions using a nuclear run-on assay (Dolezal et al. 2007). This trend was also observed in our analysis, but the cut off limit was not reached (EST upregulation index −2). Interestingly, −Fe conditions also caused a significant increase in the expression of the hydrogenase maturase (Hyd–G) that is required for the assembly of the hydrogenase-specific H cluster (Putz et al. 2006). Altogether, these results suggest a common regulatory mechanism for the genes that encode the multiple components of the ISC assembly machinery that are upregulated under −Fe conditions. This regulation might be related to the sensing of an increased cellular demand for FeS cluster synthesis during −Fe conditions.
Hydrogenosomal Membrane Proteins
Hydrogenosomes are surrounded by two membranes that possess a specific set of proteins that facilitate the exchange of metabolites between the cytosol and organelles. We detected four hydrogenosomal membrane proteins [two Hmp35 and two ADP/ATP carriers (AACs)] that were differentially expressed in response to iron availability (supplementary table S3, Supplementary Material online). Hmp35, which is a unique form of a β-barrel protein that is localized to the outer hydrogenosomal membrane, was significantly upregulated under +Fe conditions. Hmp35 is a cysteine-rich protein with the cysteine residues clustered near the C terminus, where they form a metal-binding motif (Dyall et al. 2003; Rada et al. 2011). These structural properties, together with the observed upregulation of the protein under +Fe conditions, allow us to speculate that Hmp35 may be involved in iron transport. Five members of the mitochondrial carrier protein family (MCF) that serve as AACs in the inner hydrogenosomal membrane have been identified (Dyall et al. 2000; Rada et al. 2011). In this study, two of these proteins showed significant iron-dependent regulation: AAC-1 was upregulated under −Fe conditions, whereas AAC-2 was upregulated under +Fe conditions (fig. 4 and supplementary table S3, Supplementary Material online). As described earlier, active hydrogenosomal energy metabolism is dependent on iron. Under these conditions, a portion of the ATP synthesized in the hydrogenosomes is directly used by processes such as ISC assembly and HSP-70–dependent protein transport and maturation. Some ATP could also be exported by AACs to the cytosol in exchange for ADP as in mitochondria. However, ATP is also required for hydrogenosomal functions under iron-limiting conditions when the expression of enzymes necessary for ATP synthesis is ceased. Thus, ATP might be imported from the cytosol by AACs. The expression of AAC-1 is increased under −Fe conditions, which renders it the most likely candidate for the transportation of ATP into hydrogenosomes.
Amino Acid Metabolism
The arginine dihydrolase pathway contributes to energy metabolism in T. vaginalis (Yarlett et al. 1996). Two components of the pathway, ornithine carbamoyltransferase and carbamate kinase, were significantly upregulated under +Fe conditions (supplementary table S3, Supplementary Material online), whereas arginine deiminase, which was recently shown to be localized in hydrogenosomes (Morada et al. 2011), was not regulated by iron availability.
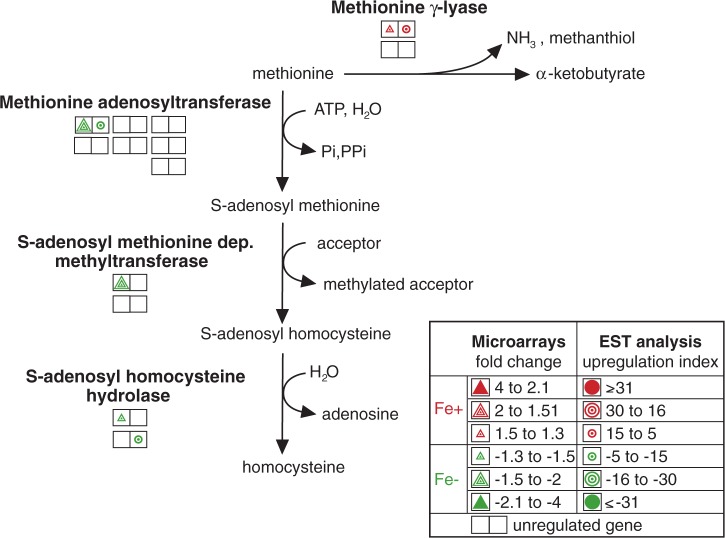
Methionine can be directly degraded to α-ketobutyrate, ammonia, and thiols or converted to homocysteine. These two metabolic pathways appear to be regulated by iron in opposing manners. Methionine degradation is catalyzed by the unique enzyme methionine γ-lyase (McKie et al. 1998). We found that one of the two genes that encode this enzyme was upregulated under +Fe conditions. In contrast, genes that encode three components of the homocysteine-forming pathway (methionine adenosyltransferase, S-adenosyl methionine-dependent methyltransferase, and S-adenosyl homocysteine hydrolase) appear to be upregulated under −Fe conditions (fig. 5 and supplementary table S3, Supplementary Material online).
Fig. 5.—
Iron-regulated genes involved in methionine metabolism. Each double square represents a single gene copy. Triangles represent the results of the microarray analysis, and circles represent the results of the EST analysis. Upregulation under +Fe and −Fe conditions is indicated by red and green colors, respectively. The empty squares represent detected, but unregulated gene copies.
Proteases
Multiple proteinases have been found in T. vaginalis (Carlton et al. 2007), and many of them have been implicated in the virulence of the parasite (Dailey et al. 1990; Ramon-Luing et al. 2010). The expression of 16 genes coding for cysteine proteases and metallopeptidases was found to be regulated by iron, and 9 of these genes were upregulated under −Fe conditions (supplementary table S3, Supplementary Material online). De Jesus et al. (2007) used comparative proteomics to show that CP3 and the legumain-like cysteine proteinase-1(LEGU1) are downregulated under −Fe conditions, and their results are consistent with upregulation of these proteins that we observed with EST under +Fe conditions. Kummer et al. (2008) isolated an extracellular protein fraction from T. vaginalis that they called CP30 and that contained CP2, CP3, CP4, and CPT, and they demonstrated that trichomonads grown under −Fe conditions had increased CP30 fraction protease activity. These results are consistent with the increased transcription of CP2 observed in our experiments (supplementary table S3, Supplementary Material online), whereas opposite trend we obtained for CP3 and CPT (supplementary table S3, Supplementary Material online).
Regulation of Transcription
Proteins that are involved in the regulation of transcription and translation, including ribosomal proteins and proteins that affect DNA metabolism, constitute approximately 10% of the iron-regulated genes (fig. 2 and supplementary table S3, Supplementary Material online). Of particular interest are the Myb-like transcription factors, which are exceptionally abundant in the T. vaginalis genome and have been suggested to be major regulators of gene transcription (Hsu et al. 2009). In our dataset, we detected the expression of 97 Myb-like DNA-binding proteins, five of which were significantly regulated by iron. One gene was upregulated under +Fe conditions, and four genes were upregulated under −Fe conditions (supplementary table S3, Supplementary Material online). To date, four T. vaginalis Myb-like proteins have been characterized (Ong et al. 2006, 2007; Hsu et al. 2009; Smith et al. 2011). It was demonstrated that the temporal and iron-inducible transcription of the ME is regulated by the synergistic or antagonistic actions of three proteins that can selectively bind to two discrete Myb protein recognition sites in the 5′-untranslated regions (5’-UTR) of the ME gene. Myb1 and Myb2, that play the roles of a repressor and an activator, respectively, are able to bind to both sites with affinities that differ over time and according to iron availability (Ong et al. 2006, 2007). Myb3 activates basal and prolonged iron-inducible transcription by binding solely to the MRE1 site (Hsu et al. 2009). In the T. vaginalis genome, there are three genes that encode Myb-3-like proteins, and all three products contain conserved base-contacting amino acids. Hsu et al. (2009) showed that the gene copy TVAG_475500 is upregulated under +Fe conditions. In our screen, we detected a second copy (TVAG_252420) that was upregulated under −Fe conditions. We also identified a Myb2 gene; however, under our experimental conditions, Myb2 was not regulated by iron, which contradicts the findings of Ong et al. (2007).
Because MRE sites were demonstrated to be essential for the iron-dependent regulation of ME, we searched 300 bp of the 5′-UTR of all iron-regulated genes detected in our screen for the presence of MRE motifs. We found numerous genes that had upstream sequences containing the MRE eukaryotic consensus sequence (C/T)AACG(G/T) or specific MRE-like sites similar to those found in the ME gene (supplementary table S4, Supplementary Material online). The genes that contain both MRE1/MRE2r and MRE2f in their upstream regions, and that have the motifs in the same order as in ME upstream region, are listed in table 2. Interestingly, the 5′-UTR of the genes that encode ADH, hypothetical protein TVAG_383310 and thioredoxin reductase contain both elements at positions similar to those in the 5′-UTR sequence of the ME gene (table 2), which suggests a similar multifarious Myb-mediated regulation of transcription.
Table 2.
Iron-Regulated Genes That Contain Both MRE1/MRE2r and MRE2f in Their Upstream Regions
| TrichDB 1.2 Accession No. | Annotation | MRE1/MRE2r | MRE2f |
|---|---|---|---|
| +Fe | ANAACGATA | TATCGT | |
|
−111/−103 |
−59/−54 |
||
| TVAG_422780 | Alcohol dehydrogenase | −136/−129 |
−37/−32 |
| CGATA | TATCGT | ||
|
−107/−103 |
−59/−54 |
||
| TVAG_035180 | Arp2/3 complex subunit | −300/−296 | −45/−40 |
| TVAG_218790 | Conserved hypothetical protein | −230/−226 | −110/−105 |
| TVAG_405900 | Phosphoglucomutase | −286/−282 | −199/−194 |
| TAACGA | TATCGTC | ||
|
−110/−105 |
−59/−53 |
||
| TVAG_072120 | Conserved hypothetical protein | −173/−168 | −19/−13 |
| −Fe | CGATA | TATCGT | |
|
−107/−103 |
−59/−54 |
||
| TVAG_383370 | Conserved hypothetical protein | −114/−110 | −81/−76 |
| TVAG_474980 | Thioredoxin reductase | −151/−147 | −107/−102 |
Note.—The numbers indicate position of the sequence motifs of translation start site. Values in bold indicate position of the motifs in the 5′-UTR of ME (Ong et al. 2006).
Conclusions
The 160 Mb T. vaginalis genome is the largest protozoan genome that has been sequenced thus far (Carlton et al. 2007). A unique feature of the Trichomonas genome among all sequenced eukaryotes is the massive expansion of many gene families and the retention of multiple copies for almost all genes (Cui et al. 2010). The mechanistic origins and biological functions of these multiple gene copies are yet unknown, but it is clear that they provided abundant starting material for evolutionary innovation while also contributing to the genetic robustness of the organism (Li et al. 2010). Indeed, because the T. vaginalis genome appears to be haploid, genetic redundancy might be particularly important to buffer gene mutations. Moreover, the differential expression of paralogous copies could be necessary for optimal growth in response to various environmental conditions (Giaever et al. 2002). We demonstrated that iron, a critical nutrient for T. vaginalis, has broad effects on the parasite’s trancriptome, which is consistent with the observation that iron modulates trichomonad growth, metabolic fluxes, and virulence phenotypes such as cytoadherence (Mundodi et al. 2006). However, we found that in most cases iron regulated the expression of a single gene or a portion of the gene copies, whereas the expression of other paralogous copies of the gene was iron independent.
The strongest iron-dependent upregulation we observed was for ME and PFOR, enzymes that play critical roles in hydrogenosomal energy metabolism and that are known to exhibit iron-dependent changes in protein expression and enzyme activity (Vanacova et al. 2001; Leitsch et al. 2009. However, we also found that there were four gene copies for ME, and two for PFOR, whose expression was not iron dependent. Of all major enzymes involved in hydrogenosomal energy metabolism, only SCS, which mediates the final step of ATP synthesis, appears to be an exception in that the expression of all copies of both SCS subunits was iron dependent. Further studies of expression profiles for cells grown under various conditions such as the absence of external glucose, temperature or oxygen stress, the induction of the amoeboid form or comparison with nonpathogenic forms should lead to further insights as to how individual copies among the myriad trichomonad gene families have come to fall under differential iron regulation during evolution.
Supplementary Material
Supplementary figure S1 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank M. Marcinčiková for technical support and J. Horváth for the development of the sequence motif searching software. This work was supported by the Czech Ministry of Education grants [MSM0021620858, LC07032] to J.T. and the Charles University GAUK 179/2006/B-BIO/PrF to L.Š. The T. vaginalis EST sequencing project was supported by the Chang Gung Memorial Hospital research grant [SMRPD33002] to P.T.
Literature Cited
- Alderete JF, Provenzano D, Lehker MW. Iron mediates Trichomonas vaginalis resistance to complement lysis. Microb Pathog. 1995;19:93–103. doi: 10.1006/mpat.1995.0049. [DOI] [PubMed] [Google Scholar]
- Carlton JM, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerkasovová A, Novák J, Cerkasov J, Kulda J, Tachezy J. Metabolic properties of Trichomonas vaginalis resistant to metronidazole under anaerobic conditions. In: Kulda J, Cerkasov J, editors. Trichomonads and trichomoniasis. Vol. 2. Prague: Acta Universitatis Carolinae Biologica; 1986. pp. 505–512. [Google Scholar]
- Cotch MF, et al. Trichomonas vaginalis associated with low birth weight and preterm delivery. The Vaginal Infections and Prematurity Study Group. Sex Transm Dis. 1997;24:353–360. doi: 10.1097/00007435-199707000-00008. [DOI] [PubMed] [Google Scholar]
- Cui J, Das S, Smith TF, Samuelson J. Trichomonas transmembrane cyclases result from massive gene duplication and concomitant development of pseudogenes. PLoS Negl Trop Dis. 2010;4:e782. doi: 10.1371/journal.pntd.0000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey DC, Chang TH, Alderete JF. Characterization of Trichomonas vaginalis haemolysis. Parasitology. 1990;101(Pt 2):171–175. doi: 10.1017/s0031182000063204. [DOI] [PubMed] [Google Scholar]
- Dallas PB, et al. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR—how well do they correlate? BMC Genomics. 2005;6:59. doi: 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jesus JB, et al. Iron modulates ecto-phosphohydrolase activities in pathogenic trichomonads. Parasitol Int. 2006;55:285–290. doi: 10.1016/j.parint.2006.08.002. [DOI] [PubMed] [Google Scholar]
- De Jesus JB, et al. A further proteomic study on the effect of iron in the human pathogen Trichomonas vaginalis. Proteomics. 2007;7:1961–1972. doi: 10.1002/pmic.200600797. [DOI] [PubMed] [Google Scholar]
- Diamond LS. The establishment of various trichomonads of animals and man in axenic cultures. J Parasitol. 1957;43:488–490. [PubMed] [Google Scholar]
- Dolezal P, et al. Frataxin, a conserved mitochondrial protein, in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2007;6:1431–1438. doi: 10.1128/EC.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, et al. Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol Cell Biol. 2000;20:2488–2497. doi: 10.1128/mcb.20.7.2488-2497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, et al. Trichomonas vaginalis Hmp35, a putative pore-forming hydrogenosomal membrane protein, can form a complex in yeast mitochondria. J Biol Chem. 2003;278:30548–39561. doi: 10.1074/jbc.M304032200. [DOI] [PubMed] [Google Scholar]
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gorrell TE. Effect of culture medium iron content on the biochemical composition and metabolism of Trichomonas vaginalis. J Bacteriol. 1985;161:1228–1230. doi: 10.1128/jb.161.3.1228-1230.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt RP, Noel CJ, Sicheritz-Ponten T, Tachezy J, Fiori PL. Trichomonas vaginalis surface proteins: a view from the genome. Trends Parasitol. 2007;23:540–547. doi: 10.1016/j.pt.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Hrdy I, et al. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Hsu HM, Ong SJ, Lee MC, Tai JH. Transcriptional regulation of an iron-inducible gene by differential and alternate promoter entries of multiple Myb proteins in the protozoan parasite Trichomonas vaginalis. Eukaryot Cell. 2009;8:362–372. doi: 10.1128/EC.00317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulda J, Poislova M, Suchan P, Tachezy J. Iron enhancement of experimental infection of mice by Tritrichomonas foetus. Parasitol Res. 1999;85:692–699. doi: 10.1007/s004360050617. [DOI] [PubMed] [Google Scholar]
- Kulda J, Tachezy J, Cerkasovova A. In vitro induced anaerobic resistance to metronidazole in Trichomonas vaginalis. J Eukaryot Microbiol. 1993;40:262–269. doi: 10.1111/j.1550-7408.1993.tb04915.x. [DOI] [PubMed] [Google Scholar]
- Kummer S, et al. Induction of human host cell apoptosis by Trichomonas vaginalis cysteine proteases is modulated by parasite exposure to iron. Microb Pathog. 2008;44:197–203. doi: 10.1016/j.micpath.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Laga M, et al. Non-ulcerative sexually transmitted diseases as risk factors for HIV-1 transmission in women: results from a cohort study. AIDS. 1993;7:95–102. doi: 10.1097/00002030-199301000-00015. [DOI] [PubMed] [Google Scholar]
- Lee NH, et al. Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci U S A. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D, et al. Trichomonas vaginalis: metronidazole and other nitroimidazole drugs are reduced by the flavin enzyme thioredoxin reductase and disrupt the cellular redox system. Implications for nitroimidazole toxicity and resistance. Mol Microbiol. 2009;72:518–536. doi: 10.1111/j.1365-2958.2009.06675.x. [DOI] [PubMed] [Google Scholar]
- Li J, Yuan Z, Zhang Z. The cellular robustness by genetic redundancy in budding yeast. PLoS Genet. 2010;6:e1001187. doi: 10.1371/journal.pgen.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liapounova NA, et al. Reconstructing the mosaic glycolytic pathway of the anaerobic eukaryote Monocercomonoides. Eukaryot Cell. 2006;12:2138–2146. doi: 10.1128/EC.00258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- McKie AE, Edlind T, Walker J, Mottram JC, Coombs GH. The primitive protozoon Trichomonas vaginalis contains two methionine gamma-lyase genes that encode members of the gamma-family of pyridoxal 5’-phosphate-dependent enzymes. J Biol Chem. 1998;273:5549–5556. doi: 10.1074/jbc.273.10.5549. [DOI] [PubMed] [Google Scholar]
- Mertens E, et al. The pyrophosphate-dependent phosphofructokinase of the protist, Trichomonas vaginalis, and the evolutionary relationships of protist phosphofructokinases. J Mol Evol. 1998;47(6):739–750. doi: 10.1007/pl00006433. [DOI] [PubMed] [Google Scholar]
- Moodley P, Wilkinson D, Connolly C, Moodley J, Sturm AW. Trichomonas vaginalis is associated with pelvic inflammatory disease in women infected with human immunodeficiency virus. Clin Infect Dis. 2002;34:519–522. doi: 10.1086/338399. [DOI] [PubMed] [Google Scholar]
- Morada M, et al. Hydrogenosome-localization of arginine deiminase in Trichomonas vaginalis. Mol Biochem Parasitol. 2011;176:51–54. doi: 10.1016/j.molbiopara.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundodi V, Kucknoor AS, Chang TH, Alderete JF. A novel surface protein of Trichomonas vaginalis is regulated independently by low iron and contact with vaginal epithelial cells. BMC Microbiol. 2006;6:6. doi: 10.1186/1471-2180-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SJ, Hsu HM, Liu HW, Chu CH, Tai JH. Activation of multifarious transcription of an adhesion protein ap65-1 gene by a novel Myb2 protein in the protozoan parasite Trichomonas vaginalis. J Biol Chem. 2007;282:6716–6725. doi: 10.1074/jbc.M610484200. [DOI] [PubMed] [Google Scholar]
- Ong SJ, Hsu HM, Liu HW, Chu CH, Tai JH. Multifarious transcriptional regulation of adhesion protein gene ap65-1 by a novel Myb1 protein in the protozoan parasite Trichomonas vaginalis. Eukaryot Cell. 2006;5:391–399. doi: 10.1128/EC.5.2.391-399.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz S, et al. Fe-hydrogenase maturases in the hydrogenosomes of Trichomonas vaginalis. Eukaryot Cell. 2006;5:579–586. doi: 10.1128/EC.5.3.579-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada P, et al. The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis. PloS One. 2011;6:e24428. doi: 10.1371/journal.pone.0024428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon-Luing LA, et al. Immunoproteomics of the active degradome to identify biomarkers for Trichomonas vaginalis. Proteomics. 2010;10:435–444. doi: 10.1002/pmic.200900479. [DOI] [PubMed] [Google Scholar]
- Rasoloson D, et al. Mechanisms of in vitro development of resistance to metronidazole in Trichomonas vaginalis. Microbiology. 2002;148(Pt 8):2467–2477. doi: 10.1099/00221287-148-8-2467. [DOI] [PubMed] [Google Scholar]
- Rawat P, Kumar S, Sheokand N, Raje CI, Raje M. The multifunctional glycolytic protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a novel macrophage lactoferrin receptor. Biochem Cell Biol. 2012;90(3):329–338. doi: 10.1139/o11-058. [DOI] [PubMed] [Google Scholar]
- Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Smith AJ, et al. Novel core promoter elements and a cognate transcription factor in the divergent unicellular eukaryote Trichomonas vaginalis. Mol Cell Biol. 2011;31:1444–1458. doi: 10.1128/MCB.00745-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smíd O, et al. Reductive evolution of the mitochondrial processing peptidases of the unicellular parasites Trichomonas vaginalis and Giardia intestinalis. PLoS Pathogens. 2008;4:e1000243. doi: 10.1371/journal.ppat.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano-González E, et al. The trichomonad cysteine proteinase TVCP4 transcript contains an iron-responsive element. FEBS Lett. 2007;581:2919–2928. doi: 10.1016/j.febslet.2007.05.056. [DOI] [PubMed] [Google Scholar]
- Sommer U, et al. Identification of Trichomonas vaginalis cysteine proteases that induce apoptosis in human vaginal epithelial cells. J Biol Chem. 2005;280:23853–23860. doi: 10.1074/jbc.M501752200. [DOI] [PubMed] [Google Scholar]
- Sutak R, Dolezal P, et al. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc Natl Acad Sci U S A. 2004;101:10368–10373. doi: 10.1073/pnas.0401319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutak R, Lesuisse E, Tachezy J, Richardson DR. Crusade for iron: iron uptake in unicellular eukaryotes and its significance for virulence. Trends Microbiol. 2008;16:261–268. doi: 10.1016/j.tim.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Sutak R, Tachezy J, Kulda J, Hrdy I. Pyruvate decarboxylase, the target for omeprazole in metronidazole-resistant and iron-restricted Tritrichomonas foetus. Antimicrob Agents Chemother. 2004;48:2185–2189. doi: 10.1128/AAC.48.6.2185-2189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy J, Sánchez LB, Müller M. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol Biol Evol. 2001;18:1919–1928. doi: 10.1093/oxfordjournals.molbev.a003732. [DOI] [PubMed] [Google Scholar]
- Torres-Romero JC, Arroyo R. Responsiveness of Trichomonas vaginalis to iron concentrations: evidence for a post-transcriptional iron regulation by an IRE/IRP-like system. Infect Genet Evol. 2009;9:1065–1074. doi: 10.1016/j.meegid.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Vanácová S, et al. Iron-induced changes in pyruvate metabolism of Tritrichomonas foetus and involvement of iron in expression of hydrogenosomal proteins. Microbiology. 2001;147:53–62. doi: 10.1099/00221287-147-1-53. [DOI] [PubMed] [Google Scholar]
- Viikki M, Pukkala E, Nieminen P, Hakama M. Gynaecological infections as risk determinants of subsequent cervical neoplasia. Acta Oncol. 2000;39:71–75. doi: 10.1080/028418600431003. [DOI] [PubMed] [Google Scholar]
- Yarlett N, Martinez MP, Moharrami MA, Tachezy J. The contribution of the arginine dihydrolase pathway to energy metabolism by Trichomonas vaginalis. Mol Biochem Parasitol. 1996;78:117–125. doi: 10.1016/s0166-6851(96)02616-3. [DOI] [PubMed] [Google Scholar]
- Yarunin A, et al. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen T, Wurmbach E, Pfeffer RL, Ebersole BJ, Sealfon SC. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 2002;30:e48. doi: 10.1093/nar/30.10.e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZF, Begg CB. Is Trichomonas vaginalis a cause of cervical neoplasia? Results from a combined analysis of 24 studies. Int J Epidemiol. 1994;23:682–690. doi: 10.1093/ije/23.4.682. [DOI] [PubMed] [Google Scholar]