Abstract
Intronic DNA is a major component of eukaryotic genes and genomes and can be subject to selective constraint and have functions in gene regulation. Intron size is of particular interest given that it is thought to be the target of a variety of evolutionary forces and has been suggested to be linked ultimately to various phenotypic traits, such as powered flight. Using whole-genome analyses and comparative approaches that account for phylogenetic nonindependence, we examined interspecific variation in intron size variation in three data sets encompassing from 12 to 30 amniotes genomes and allowing for different levels of genome coverage. In addition to confirming that intron size is negatively associated with intron position and correlates with genome size, we found that on average mammals have longer introns than birds and nonavian reptiles, a trend that is correlated with the proliferation of repetitive elements in mammals. Two independent comparisons between flying and nonflying sister groups both showed a reduction of intron size in volant species, supporting an association between powered flight, or possibly the high metabolic rates associated with flight, and reduced intron/genome size. Small intron size in volant lineages is less easily explained as a neutral consequence of large effective population size. In conclusion, we found that the evolution of intron size in amniotes appears to be non-neutral, is correlated with genome size, and is likely influenced by powered flight and associated high metabolic rates.
Keywords: intron size evolution, genome size, amniotes, flight, metabolic rate
Introduction
As one of several types of noncoding DNA, introns are abundant in amniotes genomes. In most mammals, there are on average more than eight introns per gene (Roy and Gilbert 2006; Farlow et al. 2011). First discovered in protein-coding genes of viruses (Berget et al. 1977; Chow et al. 1977) and named later (Gilbert 1978), introns were initially considered nonfunctional DNA sequences because they are spliced from precursor RNAs when producing the mature messenger RNA. However, it is now well accepted that introns are not simply “junk” DNA, as they are the basis of alternative splicing, which can generate multiple proteins from a single gene; some introns also encode noncoding RNA molecules that regulate transcription.
Because of their newly discovered functions and conservation in the genome, many introns are now believed to evolve under selective constraints. The observation that many introns harbor conserved sites under purifying selection is now commonplace, and several studies have found evidence for adaptive evolution in variation segregating within introns (Parsch et al. 2010; Hayden et al. 2011; Cagliani et al. 2012), suggesting that both size and sequence may be shaped by non-neutral forces. Previous studies have found that within species, intron size varies substantially among different genes: tissue- or development-specific genes have longer introns compared with housekeeping genes, and highly expressed genes have shorter introns than lowly expressed genes (Castillo-Davis et al. 2002; Eisenberg and Levanon 2003; Urrutia and Hurst 2003; Vinogradov 2004), which could be explained by selection for economy (Castillo-Davis et al. 2002; Eisenberg and Levanon 2003; Urrutia and Hurst 2003; Pozzoli et al. 2007), mutation bias, or the “genome design” hypothesis (Vinogradov 2004, 2005, 2006), which states that the length of genomic elements is determined by their function. Even within a single gene, introns are different: first introns are generally longer than other introns (Marais et al. 2005; Gaffney and Keightley 2006; Gazave et al. 2007; Bradnam and Korf 2008), which may reflect different functional properties they possess, such as intron-mediated enhancement (IME) of heterologous gene expression (Mascarenhas et al. 1990), insertion frequency of SINE elements (Majewski and Ott 2002), or proportion of conserved elements (Keightley and Gaffney 2003; Chamary and Hurst 2004).
Moreover, intron size also varies between species, and it has been proposed that avian intron sizes, such as genome sizes, are reduced in comparison with mammals partially because of the selection pressure imposed by metabolically demanding behaviors, such as flight (Hughes and Hughes 1995), where small introns provide a slightly improved transcription efficiency or splicing accuracy (Lynch 2002). Alternatively, small introns may simply mirror reduced genomes and thus reduced cell sizes, which increase the surface to volume ratio and permit a greater rate of gas change per unit volume (Hughes and Hughes 1995), therefore beneficial for metabolically demanding behaviors. In an early study, Hughes and Hughes (1995) surveyed 111 introns homologous between humans and chickens for 31 genes and found that chicken introns are significantly smaller than those of humans. However, in a later study, Vinogradov (1999) examined 176 introns of 55 chicken–human homologous genes but failed to reveal any significant difference in intron size between these two species. Because these studies only included only one bird species (chicken), the possibility cannot be excluded that random changes occurred in chicken and that the trends observed were not bird specific but chicken specific; therefore, the role of flight in shaping the intron size variation is controversial. To overcome this concern, Waltari and Edwards (2002) studied 14 introns from 19 flighted and flightless birds and 1 nonflying relative, the American alligator; their result suggested that the evolution of intron size is consistent with neutral Brownian motion and that there was no significant correlation between intron size and metabolically costly behaviors such as flight. However, the number of introns in that study was quite small, so we still cannot rule out the influence of random effects. Thus, there is no firm conclusion regarding whether introns are smaller in avian species than in mammals and whether flight might impose selection pressures on intron sizes.
Recently, great efforts on whole genome sequencing in a larger number of species provide an opportunity to study the evolution of genomic properties in an information-rich phylogenetic context. Here, we exploited recent whole-genome data to revisit the question of intron size variation in amniotes by using a larger number of introns from more species. Our goal is to produce a better understanding of intron size variation and evolutionary forces acting on it, all the while using appropriate comparative methods (Felsenstein 1985; Harvey and Pagel 1991; Lynch 1991). Our main finding is that mammals have larger introns than birds and reptiles and that this difference is comparable to that exhibited by genome size between these two clades. Furthermore, flighted species tend to have shorter introns than their nonflying sister groups, suggesting flight or its related traits may pose selective constraints on the evolution of intron sizes.
Materials and Methods
Data Sets
We generated three different data sets in this study to serve different purposes. All genomes were downloaded from Ensembl genome browser (http://www.ensembl.org, release 59, last accessed October 3, 2012) (Flicek et al. 2011). (We also investigated a high-quality microbat genome from release 64 and achieved almost identical results. See further details in the Supplementary Material online). Data set A includes 11 species, including 9 species with published complete genomes and two prereleased bat genomes. These species are human (Homo sapiens), mouse (Mus musculus), microbat (Myotis lucifugus), megabat (Pteropus vampyrus), opossum (Monodelphis domestica), platypus (Ornithorhynchus anatinus), chicken (Gallus gallus), turkey (Meleagris gallopavo), zebra finch (Taeniopygia guttata), anole (Anolis carolinensis), and xenopus (Xenopus tropicalis). This data set allows informative comparisons between flying and nonflying species in both mammals and reptiles, and it contains a relatively small number of species to assure a large number of orthologous introns to be identified. Data set B includes 20 species with at least 6X coverage genome data to represent a high-quality data set, those are human (H. sapiens), chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), rhesus (Macaca mulatta), marmoset (Callithrix jacchus), mouse (M. musculus), rat (Rattus norvegicus), Guinea Pig (Carvia porcellus), rabbit (Oryctolagus cuniculus), cow (Bos taurus), horse (Equus caballus), dog (Canis familiaris), elephant (Loxodonta africana), opossum (Mon. domestica), chicken (G. gallus), turkey (Mel. gallopavo), zebra finch (T. guttata), anole (A. carolinensis), and xenopus (X. tropicalis). Data set C contains the two bats and eight arbitrarily chosen mammals in addition to data set B, which represents a broad phylogenetic range. These additional species are alpaca (Vicugna pacos), pig (Sus scrofa), cat (Felis catus), hedgehog (Erinaceus europaeus), shrew (Sorex araneus), lesser hedgehog tenrec (Echinops telfairi), armadillo (Dasypus novemcinctus), and wallaby (Macropus eugenii).
Genome Size
Data on genome size were retrieved from the Animal Genome Size Database (http://www.genomesize.com, last accessed October 3, 2012).
Identification of Orthologous Introns
Intron size and position information were downloaded from Ensembl genome browser (release 59) for each species under study. To identify orthologous introns, we first defined orthologous genes. For data set A, we downloaded peptide sets for the 11 species mentioned above to perform blastp search using the Basic Local Alignment Search Tool (BLAST) suite (Altschul et al. 1990) for each pair of species and used the “reciprocal best hit” method to define orthologous genes. For data sets B and C, we avoided the above method due to computing power limit; instead, we downloaded orthologous genes from Ensembl BioMart, requiring one-to-one orthology type. If a gene had more than one splicing form, only the longest one was used. Then, we denoted human (H. sapiens) genes as query and aligned to them corresponding orthologous genes from other species by performing a 1-to-1 BLASTP. Next, intron positions were mapped to the alignment, and orthologous introns were defined if their positions are within three amino acids in the alignment. Finally, only introns larger than 20 bp were considered to reduce the annotation uncertainty on short introns (Brawand et al. 2011).
Phylogenetic Tree Construction
The phylogenetic tree was downloaded from Ensembl with manual removal of unused species. To construct species trees and to estimate branch lengths, autosomal regions with refSeq annotations were used to create multiple-species alignments. The program phyloFit was applied to generate the tree and branch length, after adjusting the frequencies of the alignment back to a genome-wide GC percent of 0.41.
Ancestral State Reconstruction
To study differences in intron size between mammals and reptiles, we compared the intron size of ancestors of each group. To reconstruct ancestral intron sizes, we used the R package “Analysis of Phylogenetics and Evolution” (ape) (Paradis et al. 2004) to reconstruct ancestral states. For continuous traits such as intron size, a Brownian motion model was assumed. Using custom python scripts, both maximum likelihood (ML) (Schluter 1997) and phylogenetically independent contrast (PIC) method (Felsenstein 1985) were used to fit the model to yield ancestral values for each intron.
Phylogenetically Corrected Tests
To account for the phylogenetic signal between two phylogenetic groups in a comparison, we used phylogenetic generalized least squares (PGLS) method (Martins and Hansen 1997; Cunningham et al. 1998), which is a powerful tool to estimate unknown parameters in a linear regression (LR) model when the observations have a certain degree of correlation (Butler and King 2004). The R package “Linear and Nonlinear Mixed Effects Models” (nlme) (http://cran.r-project.org/web/packages/nlme/index.html, last accessed October 3, 2012) was used to conduct PGLS-based tests. In terms of comparing two phylogenetic groups, we assumed that the trait evolves by Brownian motion and added a binary dummy variable to distinguish two groups in the comparison (e.g., 1 for one group and 0 for another group) and constructed a regression model. If the slope coefficient in the regression model deviated significantly from 0, those groups in the comparison are significantly different.
Binomial Test for Phylogenetic Correction
We assumed that after the separation of mammals and reptiles/birds, introns evolve neutrally on each branch. Then for a given orthologous intron, the probability that it is larger in mammals than in reptiles (including birds) should be 0.5, thus the total number of larger orthologous introns in mammals compared with that in reptiles/birds should follow the binomial distribution with P = 0.5. Significant deviations from this distribution will suggest a violation of the null hypothesis and could indicate non-neutral evolution.
Permutation Test
To confirm that the intron size contraction we found in volant species is not due to random effects, because one could conceive of flying and nonflying groups species as having a 50:50 chance of having “small” or “large” introns, we developed a permutation test. Treating mammals and reptiles separately, we first permuted the distribution of intron sizes across all the species for each intron within each clade. We then counted the number of introns that are smaller in flyers when compared with their nonflying sister group. This process was repeated 1,000 times, and we recorded the number of permutations that are as extreme as the observed numbers to calculate the P value.
Phylogenetically Corrected Correlation
To test the correlation between two traits, such as intron size and genome size, we constructed a simple regression model yi = α + βxi + εi, where yi is the dependent variable and xi is the independent variable. To account for the evolutionary nonindependence of trait data, we used the program BayesTraits (http://www.evolution.rdg.ac.uk, last accessed October 3, 2012), which integrates PGLS in a Bayesian framework (Pagel 1999). A Markov chain Monte Carlo (MCMC) algorithm is applied in BayesTraits to produce posterior distributions of regression parameters. Before MCMC analysis, we used ML to decide whether phylogenetic correction is necessary by estimating the phylogenetic signal λ, which indicates whether species are not independent for a given phylogenetic tree and trait. If λ = 1, the trait is evolving as expected by a random walk model, whereas λ = 0 means a trait is evolving among species as if they were independent and no phylogenetic correction is needed. Then the MCMC was run for 5,050,000 iterations with a burn in of 50,000 and a sample period of 1,000. We manually controlled the rate deviation, which determines the boldness of the proposal procedure of the MCMC, to be consistent with acceptance rates ranging between 0.2 and 0.4 (proportion of proposals accepted). To assess the significance of correlations, we compared the proportion of the posterior distribution of slope parameters (β) that crossed 0 (the null model), as suggested elsewhere (Organ et al. 2007). We also used BayesTraits to test the hypothesis that smaller intron size and flight could be correlated when treated as binary traits. For these tests, we used an ML framework with 50 iterations. We first ran the data with all parameters and ancestors unconstrained and then with the common ancestor of birds and Anolis and of bats, horse, cat, and dog constrained to be flightless, forcing the characters to change to flighted and small introns on the appropriate branches.
Repetitive Elements
The repetitive element (RE) data were retrieved from Ensembl. By comparing repeat masked genomic sequences to raw sequences, we obtained the position and length information for REs.
Results
Data Set Summary
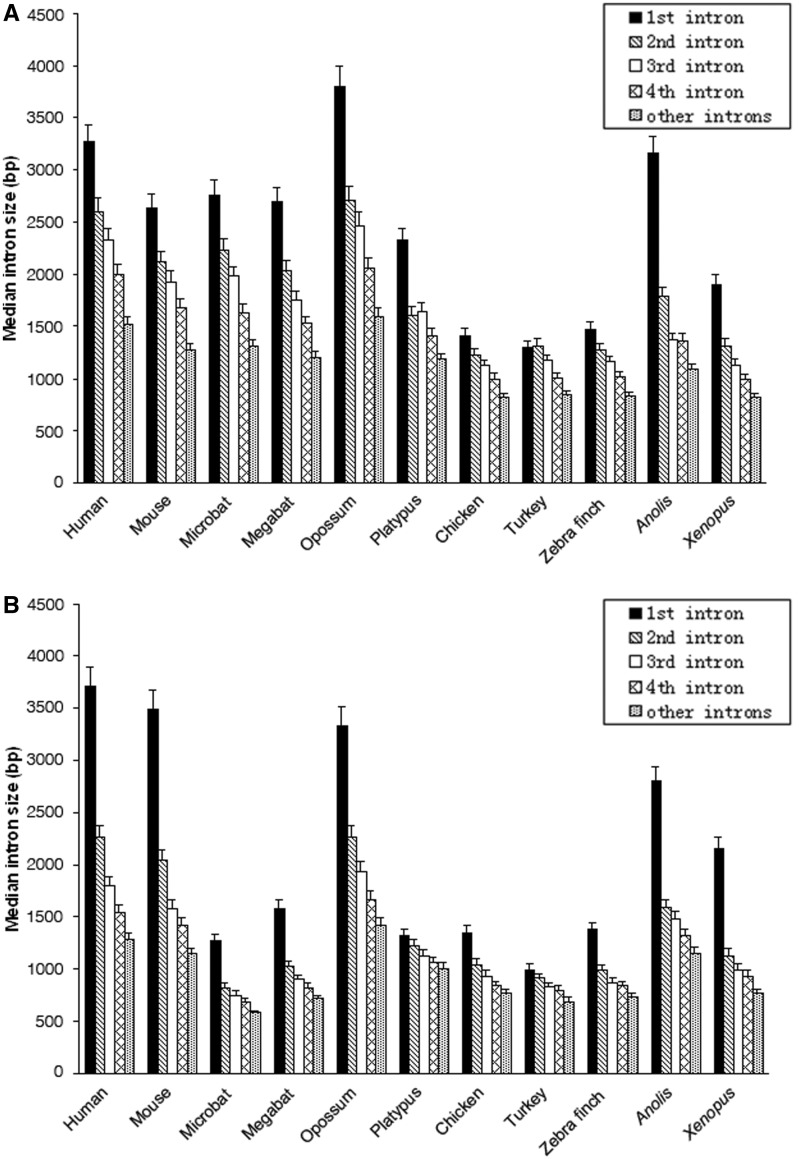
In this analysis, we built three nonexclusive data sets with different number of species and thus representing different phylogenetic depth. In our study, data sets with sparse phylogenetic sampling maximize the number of identified orthologous introns, which could avoid the possibility of drawing conclusions based on a small number of introns. Meanwhile, data sets with deeper phylogenetic coverage give us a broad picture of intron size evolution and avert biased results by focusing on few species. Throughout we used data from Ensembl release 59, but we also performed analyses using a recently released high-quality microbat (Myo. lucifugus) genome but found few differences from our initial analyses (see the Supplementary Material online), so we report results using data from Ensembl 59). Using a reciprocal-best-hit approach, we identified 12,506 homologous introns in 11 selected species, which are designated as data set A; and we also exploited the protein ortholog annotation from Ensembl to identify 562 and 98 homologous introns in data sets that we designate B and C, respectively. These introns belong to 2,300, 367, and 67 genes (see Materials and Methods). The small number of introns identified in the latter two data sets was probably due to stringent filters in our method (to pass the filters, introns were required to occur within coding regions, which in turn had to have orthologs in each species that had to occur at orthologous sites in all species); therefore, when more species are used, the probability of changes in exon–intron structure occur, ruling out inclusion in our study. To test this, we relaxed constraints in data set C by requiring orthologous introns presented in bats, reptiles and could be missing in at most one other species, which resulted in 1,070 introns. However, the pattern is very similar to what we observed for the small number of introns (data not shown), so we are convinced that even though data sets B and C contain a small number of introns, analyses based on them are representative. Alternatively, including more incompletely annotated genomes, as in data set B, could also lead to a small number of orthologous regions in all species. Because we used different methods to identify orthologous introns, it is important to determine whether results generated by different methods are consistent. The comparisons of median size of introns in eight species represented in all three data sets showed that data set A is significantly correlated with data sets B and C (P < 0.01), suggesting these two methods are consistent. Data sets B and C are also closely correlated (P < 0.001), which implies that little bias was introduced when we used fewer introns as a result of more species considered. Similar to previous studies on metazoans, we found that the first intron of the amniote genomes we studied was significantly larger than the other introns (fig. 1), presumably due to harboring more functional sequences than other introns (Marais et al. 2005; Gaffney and Keightley 2006; Gazave et al. 2007; Bradnam and Korf 2008).
Fig. 1.—
Distribution of intron median size in 11 species used in data set A. “Other introns” include all other introns after the fourth intron. (A) Introns identified in data set A. (B) Introns from genes with at least five introns in each species.
Reptiles (Including Birds) Have Smaller Introns Compared with Mammals
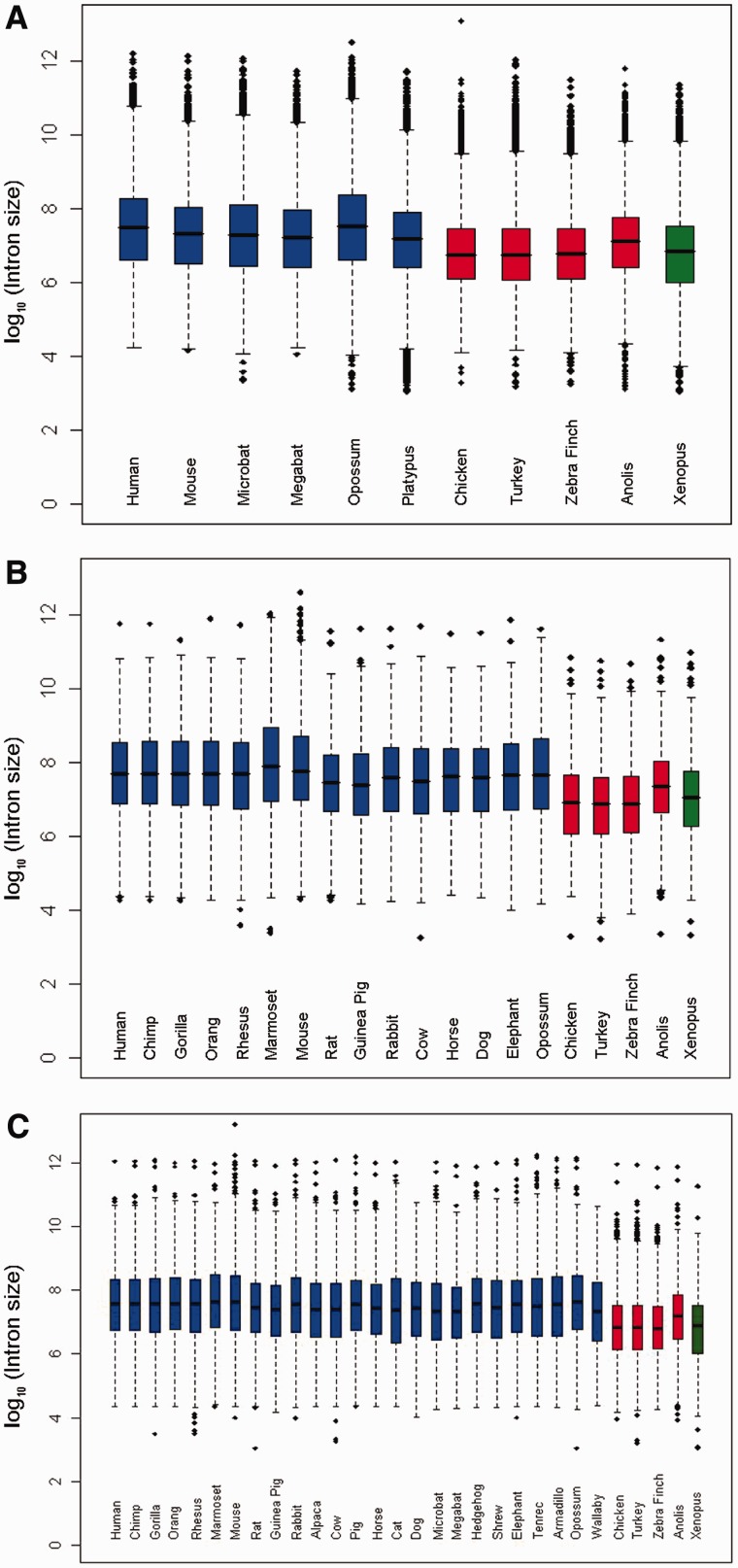
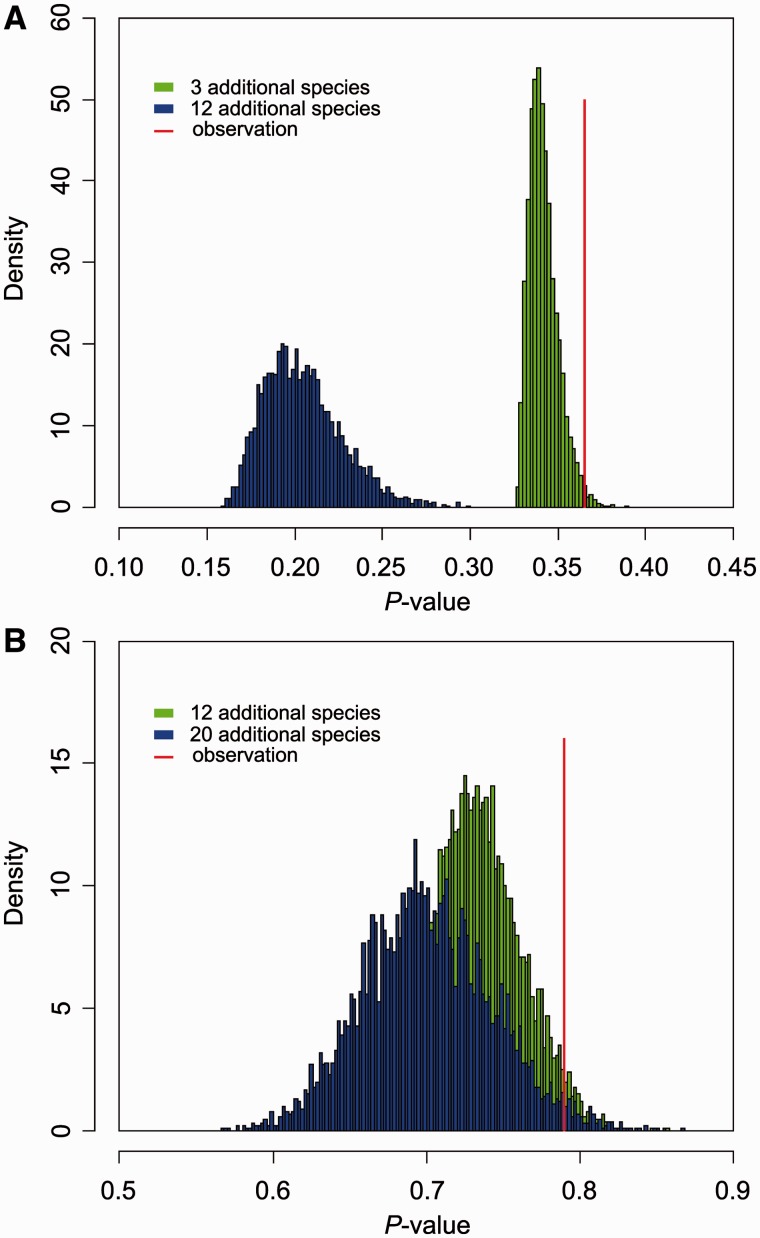
Mammals and reptiles/birds differ in many genomic characteristics, such as genome size and the proportion of REs. Here, we compared the intron size between these two sister groups, and we found for all three data sets, reptiles (including birds) have smaller introns compared with mammals (fig. 2). To understand whether these differences in intron size are statistically significant or simply random fluctuation, we performed t-tests on the median intron size of these species within a PGLS framework that accounts for nonindependence among data points introduced by shared evolutionary history. In these analyses, no significant P value was found for introns either categorized by position or as a whole (data not shown), suggesting that this apparent pattern is not strong in a phylogenetic context. However, the small sample size of reptiles in our data set (only four species included in our analysis) could affect the power of our test because of the resulting small degrees of freedom. To explore this possibility, we constructed several large species trees by adding different number of birds to our existing trees, based on tree topologies and branch lengths from recent phylogenetic surveys (Hackett et al. 2008). Then, we randomly assigned intron sizes for these additional bird species from a normal distribution with parameters estimated from three known birds (chicken, turkey, and zebra finch). Overall, we created four simulated data sets, two derived from data set A (A03, which has 3 newly added birds, and A12, which has 12 newly added birds), and the other two derived from data set B (B12, which contains 12 newly added birds, and B20, which contains 20 newly added birds). We next repeated the above PGLS analysis 5,000 times, and the result demonstrated that smaller P values were produced as sample size became larger (fig. 3), which suggests that the PGLS-based t-test is heavily affected by the number of species used and has low statistical power if that number is small. Therefore, we used a binomial test (see Materials and Methods) to overcome the confounding phylogenetic effect. To test this hypothesis, we reconstructed the intron size for the common ancestor of mammals and that of reptiles, by both ML method and the PIC method. In data set A, 8,728 of 12,506 (∼70%) introns are longer in the mammalian ancestor compared with the reptile ancestor (P < 0.001) using ML reconstruction and 8,974 of 12,506 (∼72%, P < 0.001) for PIC reconstruction. Similar results are found in data sets B and C with all P values <0.001. These results suggest that reptiles have smaller introns compared with mammals and that this contraction is consistent in direction across large numbers of introns, implying the action of non-neutral or genome-wide forces.
Fig. 2.—
Intron size distributions in different data sets. Boxplot is used to display the logarithmized size distribution of introns in each data set. Species names in black represent mammals, names in red represent reptiles/birds, and names in dark green represent amphibians. (A) Data set A; (B) data set B; and (C) data set C.
Fig. 3.—
The influence of greater taxon sampling on the significance of PGLS-based t-tests. We generated four larger phylogenetic trees with more bird species (A03 and A12 derived from data set A and B12 and B23 derived from data set B). Then we used the median size of a specific intron class in each species as node values in a phylogenetic tree and performed PGLS analysis. For newly added bird species, node values were generated by normal distribution (see text for details). To get a hypothetical distribution, this procedure was repeated 5,000 times. In each diagram, the red line denotes the P value from PGLS analysis in the original data set, and the blue and green bars denote the 5,000-time simulation of such P value in two simulated data sets derived from a same original data set. (A) Simulation based on the median size of first introns in data set A. (B) Simulation based on the median size of first introns in data set B.
Volant Species Have Smaller Introns Compared with Nonflying Relatives
We used large-scale data sets to study whether there was relationship between flight and intron size by comparing intron sizes in flying species and nonflying sister lineages in both mammals and birds. In mammals, we compared bats with their sister clade on our consensus phylogenetic tree; here, in data set A, bats were compared with humans and mice, whereas in data set C, bats were compared with horses, cats, and dogs. Figure 2 reveals that in general, flying species have shorter introns than their flightless close relatives. To diminish the influence of correlations imposed by phylogeny, we reconstructed the value for intron lengths in the common ancestor of the two bats and that of their sister group by the ML method. A total of 7,877 of 12,506 (63%) introns in data set A and 69 of 98 (70%) introns in data set C are smaller in the common ancestor of the two bats we studied than in the common ancestor of close mammalian relatives (P < 0.001, fig. 4). In addition, we also used permutation-based tests to exclude the possibility of random effect. For each intron, we permuted the intron size distribution across mammals. Then we counted the number of introns that are smaller in bats, in the same way as described above, repeating this process 1,000 times. We recorded the number of runs that have as many smaller introns in bats as observed in our data (observation). We found that the pattern of a large number of small introns in bats is unlikely to be caused by random effects (P < 0.001 and P = 0.002 for data sets A and C, respectively). In reptiles/birds, comparisons between the three birds (chicken, turkey, and zebra finch) and the green anole were conducted and we observed a similar pattern. As with the mammals, significantly more avian introns are smaller than their anole orthologs (7,552 of 12,506 [60%] introns in data set A, 361 of 562 [64%] introns in data set B, and 59 of 98 [60%] introns in data set C, P < 0.001). Again, permutation tests within Reptilia confirmed the nonrandomness of this pattern (P < 0.001 for all three data sets). Similar results were obtained when using PIC to reconstruct ancestral values for intron length or when using mean size for each group in the comparison. Thus, we found a convergent pattern in mammals and reptiles/birds that flying species have smaller introns than flightless species closely related to them.
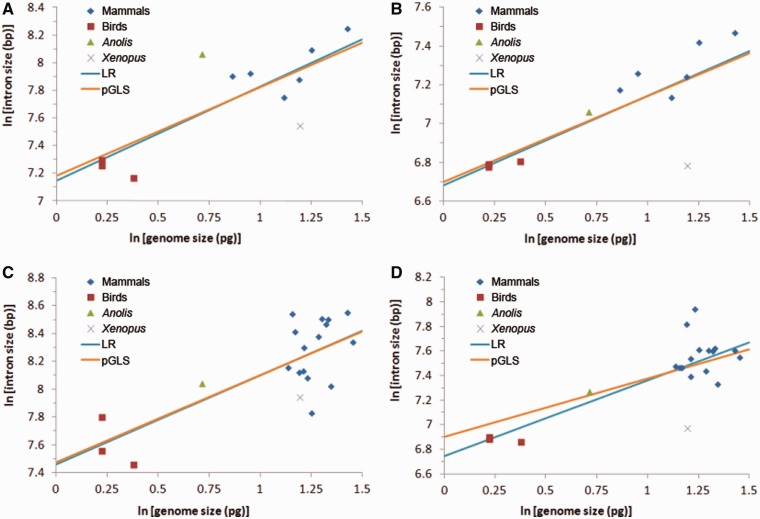
Fig. 4.—
Correlation between genome size and intron size. Light-blue lines indicate regression lines derived from normal linear regression model; and brown lines indicate regression lines derived from PGLS model, which accounts for nonindependence among data points. (A) Median size of first introns in data set A; (B) median size of other introns (introns except first introns) in data set A; (C) median size of first introns in data set B; and (D) median size of other introns (introns except first introns) in data set B.
Intron Size Variation Is Correlated with Genome Size Variation
We have shown that mammalian introns are longer than their orthologs in Reptilia. Because previous studies showed that genome size is smaller in avian species compared with other amniotes (Hughes and Hughes 1995; Hughes 1999; Organ et al. 2007), it is interesting to determine whether intron size and genome size are correlated. Because first introns are larger and functionally distinct from other introns, we treated them separately, and data set C was excluded due to the small number of first introns in it. We found a significant correlation between genome size and median intron size (fig. 4a–d). Under the normal LR model, genome size explains 62% and 57% of the variation of first introns in data sets A and B (P < 0.005), and for other introns, genome size explains 58% and 60% of the variation in data sets A and B. Because data points are nonindependent due to shared ancestry, we used the statistical package BayesTraits, which incorporates a Bayesian framework, to account for the phylogenetic signal and build a PGLS model. Again, genome size showed strong correlation with both first introns and other introns and explained 52% and 43% of the variation for the first introns and 57% and 32% for other introns in data sets A and B, respectively (P < 0.05 for all correlations). However, we did not find such correlation between genome size and exon size, presumably because exon size is more conserved than intron size (data not shown). These patterns are consistent with the notion that exons are under strong purifying selection with respect to length because indels are generally deleterious, even when preserving the reading frame.
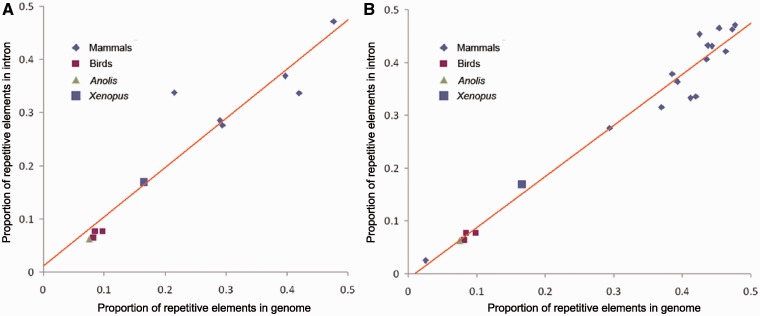
Because most of the genome size variation among amniotes is due to variation in the abundance of REs (Ohno 1970; Cavalier-Smith 1985; Pagel and Johnstone 1992), we also examined whether intron size variation correlates with the proportion of REs among species or, stated differently, whether the proportion of REs is similar between intronic regions and whole genomes among species. Our result showed a significant correlation between genomic and intronic RE proportion (fig. 5, R2 = 0.88 in data set A, R2 = 0.97 in data set B, P < 0.001 for both correlations). These results confirm that intron size and genome size in amniotes are correlated and suggest that REs may be a common driver of both.
Fig. 5.—
Correlations between the proportion of repetitive elements in introns and genomes. Brown lines indicate regression lines from normal linear regression model. (A) Data from data set A and (B) data from data set B.
Discussion
Although the underlying mechanisms are poorly understood, genome size has been shown to be related to various phenotypic traits (Petrov 2001), such as cellular and nuclear sizes (Cavalier-Smith 1982; Gregory and Hebert 1999), the rate of cell division, transcriptional process, and cellular respiration (Kozlowski et al. 2003), duration of mitosis and meiosis (Bennett 1987), weediness in plants (Neal Stewart et al. 2009; Lavergne et al. 2010), embryonic development time (Jockush 1997), morphological complexity in the brains (Roth et al. 1994), and response to CO2 (Jasienski and Bazzaz 1995). It has also been proposed that in warm-blooded amniotes, genome size may be under physiological constraints (Waltari and Edwards 2002), which favor smaller cells and thus larger surface area to volume ratios with an attendant greater ability for gas exchange to maintain a high metabolic rates (Szarski 1983; Hughes and Hughes 1995; Organ et al. 2007). Similarly, small genomes and thus small introns are thought to be favored in volant lineages due to the demands of powered flight (Hughes and Hughes 1995; Hughes 1999), which require high metabolic rates that can be facilitated by small cells with more efficient gas exchange. In support of this claim, several studies found smaller genomes in birds and bats compared with other eutherian mammals (Hughes and Hughes 1995; Van den Bussche et al. 1995), and hummingbirds, which engage in very energy-intensive maneuvers such as hovering flight, have the smallest genomes among birds studied thus far (Gregory et al. 2009).
However, Organ et al. (2007) studied the origin of avian genome size by reconstructing ancestral genomes in extant and extinct amniotes and suggested the reduction of genome size occurred along the lineage leading to basal and theropod dinosaurs, long before the origin of birds and powered flight (Organ et al. 2007). Consistent with this pattern, our analysis showed that birds and reptiles together have smaller introns compared with mammals but that within reptiles and mammals, intron size in flighted lineages is smaller than in close relatives that do not fly, suggesting a possible correlation between intron size/genome size and flight ability. Similar to Organ et al. (2007), we suggest that although genome size reduction in reptiles may have occurred before the origin of powered flight in birds and bats, flight nonetheless further reduced genome size in these lineages, leading to further reductions in of intron sizes, likely through biased deletion or ultimately through reduction of cell volume (Johnson 2004). Additional paleogenomics studies have confirmed smaller genomes in other volant reptile lineages, such as pterosaurs (Organ and Shedlock 2009).
Although we have found some evidence for a role of flight in reducing intron size in amniotes, it is reasonable to wonder whether the one or two evolutionary events in which these changes took place (on the one or two branches of the trees in our three data sets leading to flight from flightless ancestors) constitute a statistically significant association, given our tree, branch lengths and the distribution of character states among taxa. To investigate this, we ran a simple test of the hypothesis that the binary traits of flight and smaller intron size are significantly associated using BayesTraits (Pagel 1994; Barker and Pagel 2005). In our test, we scored states for both flightless and large introns as “0” and volant and small introns as “1.“ Using the ML mode and leaving all rate parameters between states unconstrained, we found that a model in which flight and small introns were associated was a slightly better explanation of the data than a model in which they were independent in two of three data sets (P = 0.09 in data sets A and B and P = 0.29 in data set C, χ2 test). In the dependent model, the probability that the common ancestors of bats and Zooamata, which comprised the horse–dog–cat clade (Waddell et al. 1999; Benton et al. 2009), or of birds and Anolis arose was flightless and had large introns was surprisingly and perhaps unrealistically small [P(0,0) = 0.1804 or 0.0735 for the Anolis–bird ancestor or the bat–Zooamata ancestor, respectively]. We expect, for example, the ancestor of birds and lizards to have been flightless based on the fossil record. The same was true for the uncorrelated model (P[0] = 0.3946 or 0.1498 for Anolis–bird and bat–Zooamata ancestors). This result may have arisen because the ML estimates of the transition rates from flightless to volant or from large to small introns (rates q12 and q13 in the model) were very small, presumably because the number of transitions from flightless to volant (0→1) was small. To create a more realistic model, we first used the largest data set, data set C, and constrained q12 and q13 to be higher, varying the rate from 10 to 100. Under these scenarios, the probability that the common ancestor at the branch leading to bats or birds arose was flightless and had large introns in the dependent model was higher [P(0,0) = 0.3287 or 0.3076 for q12 = q13 = 100]. In this more realistic case, the difference in log likelihood between the dependent and independent models was even greater (P = 2.5 × 10−5, χ2 test, d.f. = 4) than when transition rates were unconstrained, supporting the hypothesis that just two transitions to flight and small intron size is indeed statistically significant in a likelihood framework. We also confirmed biological intuition by finding that the likelihood of dependent models in which the ancestor of birds and Anolis or bats and Zooamata was forced to be flightless was significantly higher than models in which that ancestor was volant (P = 0.004, χ2 test, d.f. = 2). Additionally, we found that the dependent model in which these ancestors were forced to be flightless with large introns was a much better explanation of the character data than was the independent model (P = 0.0007, χ2 test, d.f. = 4). All these results strongly support a model in which flight and small genomes are correlated, if not related causally, given two origins of powered flight among extant amniotes. This analysis does not include extinct lineages such as pterosaurs, which we now infer to have small genomes (Organ and Shedlock 2009) and could constitute a third origin of the genomic syndrome associated with powered flight.
An alternative explanation for genome and intron size variation in amniotes is suggested by theories of neutral processes and their effect on genome architecture (Lynch 2007). For example, Lynch and Conery (2003) studied 43 eukaryotic species and suggested that changes of genome complexity and/or genomic characteristics passively respond to long-term changes in population size. Based on their hypothesis, the contraction of genomes and introns that we observe in birds and bats is the result of their larger effective population sizes relative to close nonflying relatives, thereby allowing selection for smaller genome size to proceed more efficiently than in small populations. However, several lines of evidence suggest that the influence of effective population size in genome/intron size variation might not be enough to explain the pattern we observed in amniotes. First, human and mouse genomes are similar in size (3.5 pg vs. 3.29 pg), but the estimated effective population size of mice is at least 10-fold larger than in humans (Eyre-Walker et al. 2002; Halligan et al. 2010). Second, the majority of estimates of effective population sizes of birds are generally an order of magnitude smaller than 106 (Jennings 2005; Lynch 2007; Lanfear et al. 2010) and are on par with those of rodents (Eyre-Walker et al. 2002; Halligan et al. 2010), but avian genomes are significantly reduced in comparison with rodent genomes. Third, in the work by Lynch and Conery, only two amniotes (H. sapiens and M. musculus) were used in the regression analysis including intron size: this small number could introduce bias, and conclusions based on such a data set cannot easily be extrapolated to amniotes as a whole. Furthermore, in their analysis, the product of effective population size (Ne) and per site mutation rate (μ) is larger in humans than in mice (fig. 1A in their article), which contradicts the well-accepted result that mice have much larger genetic diversities than do humans. Hence, although the effective population size hypothesis may be generally true across broader phylogenetic groups, it does not seem capable of explaining phylogenetically local variation of genome characteristics in amniotes such as we observe here. There are certainly other neutral processes that could explain smaller genomes in birds, such as the fixation of mechanisms that yield a biased spectrum of deletions during replication. Such processes may or may not have fitness effects on lineages that bear them. If, however, smaller genomes do confer a physiological advantage to those lineages, it seems more plausible to us that genome reduction in birds and bats is not a neutral process.
Overall, our study demonstrates a complex pattern of intron size evolution suggesting that forces of mutation and natural selection vary among introns within a gene and between species. Although our study is consistent with an influence of powered flight on genome and intron size, additional studies clarifying the mechanism linking these traits are needed. We believe that our understanding of introns will increase with the addition of new amniote genomes, particularly those of reptiles, which are still underrepresented in the databases (Castoe et al. 2011; St John et al. 2012).
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Drs Chris Organ, Andrew Meade, Charlie Nunn, and Luke Matthews for help on BayesTraits and PGLS analysis. They also thank Dr Rachel Mueller and Dr Niclas Backström, Dr Eugene Koonin, and two anonymous reviewers for helpful comments on this manuscript. This work was supported by the Department of Human Evolutionary Biology, Harvard University and a grant from the National Science Foundation to S.V.E. (DEB-0743616).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barker D, Pagel M. Predicting functional gene links from phylogenetic-statistical analyses of whole genomes. PLoS Comput Biol. 2005;1:e3. doi: 10.1371/journal.pcbi.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD. Ordered disposition of parental genomes and individual chromosomes in reconstructed plant nuclei, and their implications. Somat Cell Mol Genet. 1987;13:463–466. doi: 10.1007/BF01534949. [DOI] [PubMed] [Google Scholar]
- Benton M, Donoghue P, Asher R. Calibrating and constraining molecular clocks. In: Hedges S, Kumar S, editors. The timetree of life. New York: Oxford University Press; 2009. pp. 35–86. [Google Scholar]
- Berget SM, Moore C, Sharp PA. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, Korf I. Longer first introns are a general property of eukaryotic gene structure. PLoS One. 2008;3:e3093. doi: 10.1371/journal.pone.0003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Butler MA, King AA. Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am Nat. 2004;164:13. doi: 10.1086/426002. [DOI] [PubMed] [Google Scholar]
- Cagliani R, et al. Variants in SNAP25 are targets of natural selection and influence verbal performances in women. Cell Mol Life Sci. 2012;69:1705–1715. doi: 10.1007/s00018-011-0896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Castoe TA, et al. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 2011;12:406. doi: 10.1186/gb-2011-12-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Skeletal DNA and the evolution of genome size. Annu Rev Biophys Bioeng. 1982;11:273–302. doi: 10.1146/annurev.bb.11.060182.001421. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The evolution of genome size. Chichester (United Kingdom): John Wiley & Sons; 1985. [Google Scholar]
- Chamary JV, Hurst LD. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Mol Biol Evol. 2004;21:1014–1023. doi: 10.1093/molbev/msh087. [DOI] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Cunningham CW, Omland KE, Oakley TH. Reconstructing ancestral character states: a critical reappraisal. Trends Ecol Evol. 1998;13:361–366. doi: 10.1016/s0169-5347(98)01382-2. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD, Smith NG, Gaffney D. Quantifying the slightly deleterious mutation model of molecular evolution. Mol Biol Evol. 2002;19:2142–2149. doi: 10.1093/oxfordjournals.molbev.a004039. [DOI] [PubMed] [Google Scholar]
- Farlow A, Meduri E, Schlotterer C. DNA double-strand break repair and the evolution of intron density. Trends Genet. 2011;27:1–6. doi: 10.1016/j.tig.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Flicek P, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, Keightley PD. Genomic selective constraints in murid noncoding DNA. PLoS Genet. 2006;2:e204. doi: 10.1371/journal.pgen.0020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Marques-Bonet T, Fernando O, Charlesworth B, Navarro A. Patterns and rates of intron divergence between humans and chimpanzees. Genome Biol. 2007;8:R21. doi: 10.1186/gb-2007-8-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Gregory TR, Andrews CB, McGuire JA, Witt CC. The smallest avian genomes are found in hummingbirds. Proc Biol Sci R Soc. 2009;276:3753–3757. doi: 10.1098/rspb.2009.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR, Hebert PD. The modulation of DNA content: proximate causes and ultimate consequences. Genome Res. 1999;9:317–324. [PubMed] [Google Scholar]
- Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Halligan DL, Oliver F, Eyre-Walker A, Harr B, Keightley PD. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 2010;6:e1000825. doi: 10.1371/journal.pgen.1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PH, Pagel M. New York: Oxford University Press; 1991. The comparative method in evolutionary biology. [Google Scholar]
- Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Adaptive evolution of genes and genomes. Oxford: Oxford University Press; 1999. [Google Scholar]
- Hughes AL, Hughes MK. Small genomes for better flyers. Nature. 1995;377:391. doi: 10.1038/377391a0. [DOI] [PubMed] [Google Scholar]
- Jasienski M, Bazzaz FA. Genome size and high CO2. Nature. 1995;376:2. [Google Scholar]
- Jennings WB, Edwards SV. Speciational history of Australian grass finches (Poephila) inferred from thirty gene trees. Evolution. 2005;59:5. [PubMed] [Google Scholar]
- Jockush EL. An evolutionary correlate of genome size change in plethodontid salamanders. Proc R Soc Lond B Biol Sci. 1997;264:8. [Google Scholar]
- Johnson KP. Deletion bias in avian introns over evolutionary timescales. Mol Biol Evol. 2004;21:599–602. doi: 10.1093/molbev/msh051. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Gaffney DJ. Functional constraints and frequency of deleterious mutations in noncoding DNA of rodents. Proc Natl Acad Sci U S A. 2003;100:13402–13406. doi: 10.1073/pnas.2233252100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski J, Konarzewski M, Gawelczyk AT. Cell size as a link between noncoding DNA and metabolic rate scaling. Proc Natl Acad Sci U S A. 2003;100:14080–14085. doi: 10.1073/pnas.2334605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Ho SY, Love D, Bromham L. Mutation rate is linked to diversification in birds. Proc Natl Acad Sci U S A. 2010;107:20423–20428. doi: 10.1073/pnas.1007888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne S, Muenke NJ, Molofsky J. Genome size reduction can trigger rapid phenotypic evolution in invasive plants. Ann Bot. 2010;105:8. doi: 10.1093/aob/mcp271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Methods for the analysis of comparative data in evolutionary biology. Evolution. 1991;45:1065–1080. doi: 10.1111/j.1558-5646.1991.tb04375.x. [DOI] [PubMed] [Google Scholar]
- Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci U S A. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Sunderland (MA): Sinauer Associates; 2007. The origins of genome architecture. [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Nouvellet P, Keightley PD, Charlesworth B. Intron size and exon evolution in Drosophila. Genetics. 2005;170:481–485. doi: 10.1534/genetics.104.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins EP, Hansen TF. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am Nat. 1997;149:22. [Google Scholar]
- Mascarenhas D, Mettler IJ, Pierce DA, Lowe HW. Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol Biol. 1990;15:913–920. doi: 10.1007/BF00039430. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin: Springer; 1970. [Google Scholar]
- Organ CL, Shedlock AM. Palaeogenomics of pterosaurs and the evolution of small genome size in flying vertebrates. Biol Lett. 2009;5:47–50. doi: 10.1098/rsbl.2008.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ CL, Shedlock AM, Meade A, Pagel M, Edwards SV. Origin of avian genome size and structure in non-avian dinosaurs. Nature. 2007;446:180–184. doi: 10.1038/nature05621. [DOI] [PubMed] [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies: a general method for comparative analysis of discrete characters. Proc R Soc Lond B. 1994;255:37–45. [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pagel M, Johnstone RA. Variation across species in the size of the nuclear genome supports the junk-DNA explanation for the C-value paradox. Proc Biol Sci. 1992;249:119–124. doi: 10.1098/rspb.1992.0093. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Parsch J, Novozhilov S, Saminadin-Peter SS, Wong KM, Andolfatto P. On the utility of short intron sequences as a reference for the detection of positive and negative selection in Drosophila. Mol Biol Evol. 2010;27:1226–1234. doi: 10.1093/molbev/msq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA. Evolution of genome size: new approaches to an old problem. Trends Genet. 2001;17:23–28. doi: 10.1016/s0168-9525(00)02157-0. [DOI] [PubMed] [Google Scholar]
- Pozzoli U, et al. Intron size in mammals: complexity comes to terms with economy. Trends Genet. 2007;23:20–24. doi: 10.1016/j.tig.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Roth G, Blanke J, Wake DB. Cell size predicts morphological complexity in the brains of frogs and salamanders. Proc Natl Acad Sci U S A. 1994;91:4796–4800. doi: 10.1073/pnas.91.11.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers AO, Ludwig D. Likelihood of ancestor states inadaptive radiation. Evolution. 1997;51:1699–1711. doi: 10.1111/j.1558-5646.1997.tb05095.x. [DOI] [PubMed] [Google Scholar]
- Stewart CN, Jr, et al. Evolution of weediness and invasiveness: charting the course for weed genomics. Weed Sci. 2009;57:12. [Google Scholar]
- St John JA, et al. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 2012;13:415. doi: 10.1186/gb-2012-13-1-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarski H. Cell size and the concept of wasteful and frugal evolutionary strategies. J Theor Biol. 1983;105:201–209. doi: 10.1016/s0022-5193(83)80002-2. [DOI] [PubMed] [Google Scholar]
- Urrutia AO, Hurst LD. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bussche RA, Longmire JL, Baker RJ. How bats achieve a small C-value: frequency of repetitive DNA in Macrotus. Mamm Genome. 1995;6:521–525. doi: 10.1007/BF00356168. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. Intron-genome size relationship on a large evolutionary scale. J Mol Evol. 1999;49:376–384. doi: 10.1007/pl00006561. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. Compactness of human housekeeping genes: selection for economy or genomic design? Trends Genet. 2004;20:248–253. doi: 10.1016/j.tig.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. Noncoding DNA, isochores and gene expression: nucleosome formation potential. Nucleic Acids Res. 2005;33:559–563. doi: 10.1093/nar/gki184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. “Genome design” model: evidence from conserved intronic sequence in human-mouse comparison. Genome Res. 2006;16:347–354. doi: 10.1101/gr.4318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell PJ, Okada N, Hasegawa M. Towards resolving the interordinal relationships of placental mammals. Syst Biol. 1999;48:1–5. [PubMed] [Google Scholar]
- Waltari E, Edwards SV. Evolutionary dynamics of intron size, genome size, and physiological correlates in archosaurs. Am Nat. 2002;160:539–552. doi: 10.1086/342079. [DOI] [PubMed] [Google Scholar]