Abstract
Prion proteins can adopt multiple different infectious strain conformations. Here we examine how the sequence of a prion protein affects its capacity to propagate specific conformations by exploiting our ability to create two distinct infectious conformations of the yeast [PSI+] prion protein Sup35p, termed Sc4 and Sc37. PNM2, a Sup35p (G58D) point mutant originally identified for its dominant interference with prion propagation, leads to rapid, recessive loss of Sc4 but does not interfere with Sc37 propagation. PNM2 destabilizes the amyloid core of Sc37 causing compensatory effects that slow prion growth but aid prion division and result in robust Sc37 propagation. In contrast, PNM2 does not affect the structure or chaperone-mediated division of Sc4, but interferes with its delivery to daughter cells. Thus, effective delivery of infectious particles during cell division is a critical and conformation-dependent step in prion inheritance.
Introduction
Infectious proteins, or prions, are a form of conformation-based inheritance, in which a prion protein aggregate binds to and catalyzes the conversion of newly-made proteins to the prion form creating stably propagating states1. Such conformation-based inheritance underlies a range of transmissible spongiform encephalopathies in mammals as well as heritable epigenetic states in fungi, of which the yeast Saccharomyces cerevisiae prion [PSI+] is arguably the best characterized2. A common feature of prion proteins is that they misfold into ordered, β-sheet rich amyloid fibers, the self-templating nature of which is thought to form the basis of prion propagation1,3. Remarkably, a single prion protein can adopt a spectrum of amyloid conformations that lead to different, heritable strain variants4–11.
The yeast prion [PSI+], which results from the aggregation of the translation termination factor Sup35p, has emerged as a powerful system for studying strain variants. Sup35p can form different amyloid conformations in vitro when polymerized at 4 °C versus 37 °C8,12,13. When introduced into yeast, these amyloid fibers form two distinct strains in vivo termed [PSI+]Sc4 and [PSI+]Sc37, respectively. (For clarity, we use [PSI+]Sc4 to describe the in vivo strain and Sc4 to describe the fiber conformation.) These strains are distinguished by the degree of Sup35p aggregation: [PSI+]Sc4 yeast exhibit a strong prion phenotype, in which the large majority of Sup35p is aggregated, while [PSI+]Sc37 yeast have a more substantial pool of soluble Sup35p and a weaker prion phenotype. Structural studies revealed that there is a dramatic expansion of the amyloid core in the Sc37 conformation relative to that found in Sc414,15. The more extensive structure in Sc37 fibers increases fiber stability and decreases the rate of prion replication by the cell’s chaperone system resulting in the weaker [PSI+]Sc37 prion phenotype16. The ability to probe the biophysical properties of these defined strain variant conformations in vitro and monitor their phenotypes in vivo makes [PSI+] an excellent system for the study of prion strain phenomena.
The precise amino acid sequence greatly biases the ability of a prion protein to adopt particular strain variants7,17–19. For example, the naturally occurring methionine/valine polymorphism at position 129 of the mammalian prion protein greatly influences disease susceptibility20. The Val129 allele does not interfere with the propagation of all prion strain variants and may even favor some, including certain iatrogenic forms of Creutzfeldt-Jakob diease19. However, the presence of even a single allele of Val129 appears to be highly protective against developing new variant Creutzfeldt-Jakob disease (nvCJD)21, which is thought to result from transmission of bovine spongiform encephalopathy (mad cow disease) to humans22,23.
This strong relationship between strain variants and the sequence of the prion protein extends to the [PSI+] prion system. A mutant of the Sup35p prion protein, which was originally identified in a screen for “Psi no more” mutants that prevented propagation of [PSI+], shows strain variant-specific effects. This mutant, termed PNM2, has a glycine to aspartate missense mutation at amino acid position 5824. Originally, the effect of PNM2 was described as causing dominant inhibition of prion propagation when co-expressed with wild type Sup35p24,25. Later studies found that PNM2 does not interfere with propagation of all [PSI+] strain variants and may enhance propagation of certain strain variants when overexpressed17.
Here, we used the PNM2 mutant and the well-defined prion strain conformations, Sc4 and Sc37, together with in-depth biophysical and in vivo analyses, to investigate how point mutations can affect prion propagation in a manner that depends on the strain variant. Our systematic characterization of fiber structure, chaperone-mediated division of fibers into seeds and delivery of seeds to daughter cells reveals a PNM2-specific defect in seed partitioning for the [PSI+]Sc4 strain variant. The findings implicate delivery of fiber seeds to daughter buds as a critical step in prion propagation.
Results
PNM2 shows strain-specific effects on [PSI+] propagation
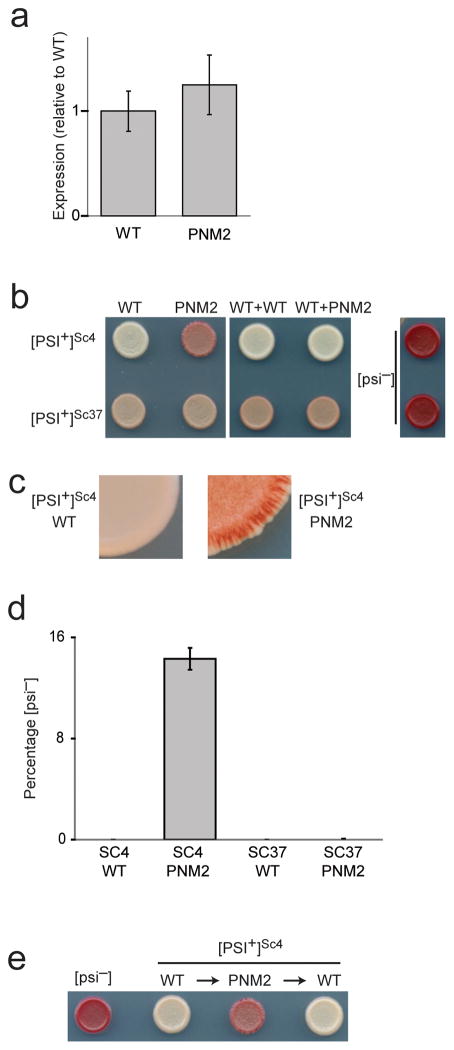
We began by characterizing the effect of the PNM2 mutation on prion propagation in [PSI+]Sc4 and [PSI+]Sc37 yeast. To allow for rapid exchange of alleles, we used a yeast background in which the genomic copy of SUP35 was deleted and replaced by a copy on a plasmid containing the counter-selectable URA3 marker. We exchanged this plasmid with one encoding wild-type Sup35p (WT) or Sup35p G58D (PNM2) and confirmed that the two proteins were expressed at similar levels (Fig. 1a). We assessed the prion phenotype with a red/white color readout that results from suppression of a nonsense mutation in an ADE1 reporter (the ade1–14 allele)26. When Sup35p is soluble ([psi−]), translation of ade1–14 terminates prematurely, resulting in accumulation of a red metabolic intermediate. When Sup35p is aggregated ([PSI+]), read-through of the nonsense mutation occurs and functional Ade1p is produced. Depending on the amount of soluble Sup35p present and perhaps the nature of the aggregates, [PSI+] strain variants have varying levels of functional Ade1p and exhibit color phenotypes ranging from pink ([PSI+]Sc37) to white ([PSI+]Sc4) (Fig. 1b). This assay also reports on the stability of prion inheritance: when a cell in a growing colony loses [PSI+], its progeny remain prion free and form a red sector (Fig. 1c).
Figure 1. Characterization of in vivo prion phenotypes.
(a) Expression levels of WT or PNM2 expressed from a plasmid in a [PSI+]Sc4 background. Expression levels were quantitated by immunoblotting with a polyclonal anti-SupNM antibody. Values represent the mean ± s.d. for three experiments. (b) Representative in vivo prion phenotypes of yeast spotted on low adenine media. WT Sup35p was replaced with (left panel), or was co-expressed with (middle panel) either WT or PNM2. (c) Enlarged view of the edge of the yeast spot. Presence of red sectors in [PSI+]Sc4 PNM2 indicates a loss of [PSI+]. (d) Quantification of loss of [PSI+] as determined by counting the number of [psi−] colonies after growing for 24 hours in YEPD. Values represent the mean ± s.d. for three experiments. (e) Representative in vivo prion phenotypes of yeast spotted on low adenine media. WT Sup35p was replaced with PNM2, which was subsequently replaced with WT through plasmid exchanges.
Using the [PSI+]Sc4 and [PSI+]Sc37 yeast backgrounds, we found that PNM2 has dramatic, strain variant-specific effects on prion propagation. When expressed as the sole copy of Sup35p, PNM2 strongly compromised propagation of [PSI+]Sc4, apparent by the continuous generation of prion-free ([psi−]) states (Fig. 1b,c,d). This effect is similar to the originally reported description of the PNM2 phenotype24,25, though there are differences between studies: PNM2 is recessive in our strain background and thus does not strongly interfere with [PSI+]Sc4 propagation when co-expressed with WT (Fig. 1b). Thus, we focused on analyzing effects of prion propagation when PNM2 is expressed as the sole copy of Sup35p.
In contrast to its dramatic effect on [PSI+]Sc4 propagation, PNM2 had a much weaker effect on propagation of the [PSI+]Sc37 strain variant, with at most a mild darkening of the color phenotype and no discernable increase in prion loss (Fig. 1b,d). This result was surprising given that previous studies indicate that the PNM2 mutation is located in a region that is structured in Sc37 but not in Sc415, leading to the expectation that the PNM2 mutation would preferentially interfere with propagation of the Sc37 conformation.
We first asked whether the defect in stable inheritance seen in [PSI+]Sc4 yeast could arise from the PNM2 mutation forcing an irreversible structural change that results in an amyloid conformation that is poorly propagated. We carried out a series of plasmid exchanges, replacing the WT plasmid with the PNM2 plasmid, then exchanging it back with the WT plasmid. Reintroduction of the WT plasmid fully restored the original [PSI+]Sc4 phenotype (Fig. 1e), indicating that the PNM2 Sc4 fibers retained the structural information necessary to template WT to the characteristic Sc4 amyloid conformation.
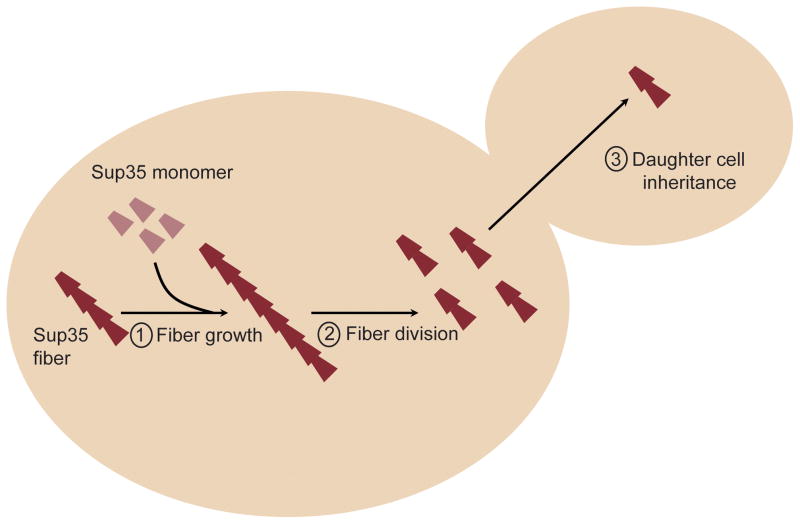
To investigate what accounts for the negative impact of PNM2 on [PSI+]Sc4, but not on [PSI+]Sc37, propagation, we systematically tested the parameters that affect each step in the prion replication cycle (Fig. 2): fiber growth; fiber division, of which fiber stability and ability to interact with the in vivo chaperone prion-replication machinery are key determinants; and delivery of prion particles to daughter cells during cell division.
Figure 2.
Schematic of the prion propagation cycle that includes (1) fiber growth, (2) chaperone mediated division, and (3) partitioning to daughter cells.
PNM2 disrupts Sc37 growth rate and structural stability
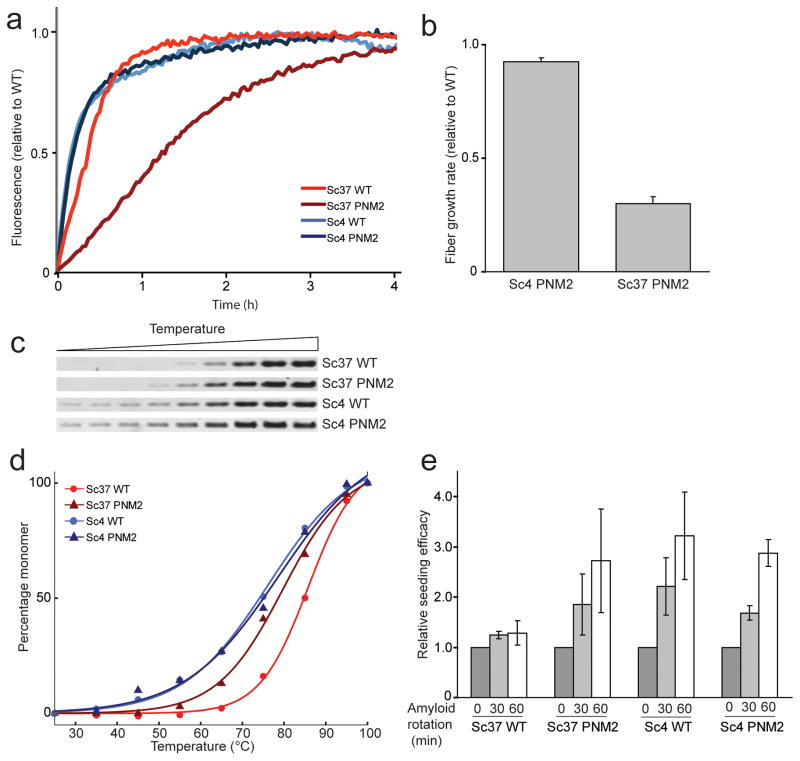
To determine whether changes in fiber growth rates account for the observed phenotypic differences, we compared in vitro growth rates of WT and PNM2 fibers. While earlier studies found that PNM2 fibers have a partial growth defect, the nature of the prion strain variants in these studies was not well defined27. For our studies, we used a fragment of Sup35p (SupNM; residues 1–254, containing the Q/N-rich N-terminal and highly charged middle domains of Sup35p) that is necessary and sufficient to support prion propagation28,29. The seeded polymerization rates of soluble SupNM were monitored by measuring the increase in fluorescence intensity of Thioflavin T. We found that when seeded, PNM2 SupNM polymerizes into the Sc37 conformation at a rate slower than that observed with WT SupNM (Fig. 3). Since this change in growth rate would be predicted to weaken the prion phenotype, other mechanisms must account for the similar phenotype of PNM2 and WT in the Sc37 conformation.
Figure 3.
Characterization of the physical properties of fibers formed in vitro.
(a) A representative experiment monitoring the relative growth rates of WT and PNM2 SupNM. Polymerization of SupNM was performed with 5% (w/w) WT seed of the specified conformation, and the rate of addition of SupNM monomers was monitored by Thioflavin T fluorescence. Data were normalized to initial and final intensities. (b) The growth rates of Sc4 and Sc37 PNM2 SupNM normalized to those of WT SupNM polymerized on the relevant seed. Initial time points were fitted to a line and the slope (initial growth rate) was calculated. Values represent the mean ± s.d. for three experiments. (c,d) Thermal stability of WT and PNM2 fibers in Sc4 and Sc37 conformations. WT and PNM2 SupNM fibers in the Sc4 or Sc37 conformations were incubated at increasing temperatures, and samples were subjected to SDS-PAGE. Band intensities (susceptibility of aggregates to thermal solubilization) were plotted against temperature and fitted to a sigmoidal function. (e) Relative seeding efficacy of WT or PNM2 fibers in the Sc4 or Sc37 conformation before and after stirring for 30 or 60 min. Seeding efficacy was determined by monitoring the initial fiber growth rates of polymerization reactions using stirred samples as seeds. Values represent mean ± s.e.m. for three experiments.
A PNM2-induced change in amyloid structure could explain the change in Sc37 fiber growth rate as well as provide insight into prion division rates in vivo16. Therefore, we used a range of biophysical techniques, including hydrogen/deuterium (H/D) exchange NMR, fiber thermal denaturation, and propensity for shearing to produce new fiber seeds, to analyze the structure and stability of PNM2 SupNM fibers. Shearing efficacy was determined by the ability of long fibers fragmented via stirring to seed monomer growth as measured from the initial polymerization rates of such reactions.
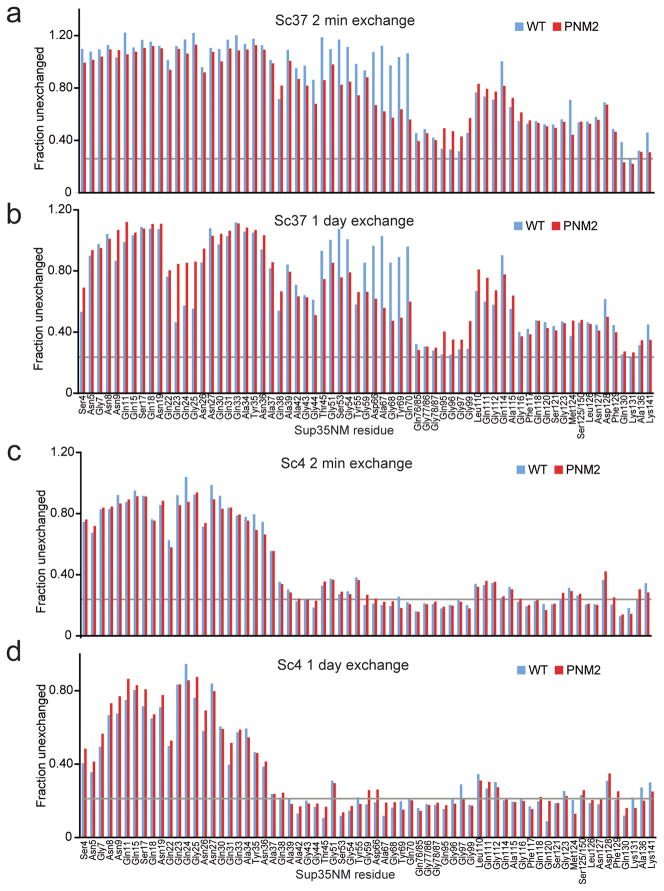
For the H/D exchange experiments, we monitored the extent of backbone amide exchange for both WT and PNM2 fibers at two different time points: after a short exchange period of 2 minutes and after a more extensive exchange period of 1 day. Based on previous assignments15, we measured exchange for 132 residues, including extensive probes throughout the amyloid core of both the Sc4 and Sc37 conformations.
The H/D exchange experiments revealed that the PNM2 mutation induced specific, localized structural defects in the Sc37 conformation. Specifically, we observed a region of increased exchange in residues proximal to the site of the PNM2 mutation for both early and late exchange time points (Fig. 4a,b). Consistent with this result, we observed a decrease in the thermal stability of PNM2 SupNM Sc37 fibers (Tm = 80 °C ± 2) compared to WT SupNM Sc37 fibers (Tm = 86 °C ± 2) (Fig. 3c,d). Additionally, when subjected to shearing forces, PNM2 SupNM Sc37 fibers fragmented more than WT SupNM Sc37 fibers (Fig. 3e). These results suggest that the PNM2 mutation in the Sc37 conformation causes a localized structural destabilization with a resulting decrease in fiber stability.
Figure 4.
H/D exchange of WT and PNM2 SupNM fibers. Intensities for assigned and unambiguous peaks corresponding to residues 1–141 were plotted as the fraction of the non-exchanged intensity after 2 min (a, c) and 1 day (b, d) of exchange for both WT (blue) and PNM2 (red) fibers in the Sc37 (a, b) and Sc4 (c, d) conformations. Unassigned and ambiguous residues are not displayed. The gray line represents the estimated minimum peak intensity following complete exchange.
The affects of the PNM2 mutation on Sc37 fibers may have opposing consequences for prion propagation. The localized structural defect would be expected to enhance the rate of prion division in vivo, causing an increase in the rate of generation of free fiber ends and subsequently strengthening the prion phenotype15,16. In contrast, the decrease in fiber growth would be expected to reduce monomer addition onto fibers and weaken the prion phenotype. Therefore, we hypothesize that the modest overall effect of the PNM2 mutation on the [PSI+]Sc37 phenotype is the result of compensatory changes.
PNM2 does not affect Sc4 growth, stability or structure
To determine the mechanism by which the PNM2 mutation interferes with [PSI+]Sc4 propagation, we carried out a set of experiments on Sc4 fibers analogous to those done on the Sc37 conformation. When seeded by Sc4, PNM2 SupNM polymerizes into the Sc4 conformation at a rate similar to WT SupNM indicating that the negative effect of the PNM2 mutation on [PSI+]Sc4 propagation cannot be explained by a decrease in fiber growth rate (Fig. 3a,b). The Sc4 fiber structure was also unaffected by the PNM2 mutation: H/D exchange experiments showed no appreciable differences between WT SupNM and PNM2 SupNM fibers in the Sc4 conformation (Fig. 4c,d). Both the regions of protection and the extent of protection for all residues was virtually indistinguishable between the two samples for both short and long exchange times, suggesting that the structures were stable as well as very similar. Consistent with the above results, the melting temperatures of WT SupNM and PNM2 SupNM in the Sc4 conformation were comparable (77 °C ± 2 and 77 °C ± 5, respectively) (Fig. 3c,d). Lastly, when subjected to shearing forces, both PNM2 SupNM and WT SupNM fragmented to a similar extent (Fig. 3e).
Taken together, these results argue that PNM2 SupNM is able to polymerize into fibers with growth, structure and stability indistinguishable from that of WT Sc4 fibers, and that the defect in [PSI+]Sc4 propagation cannot be explained by the biophysical properties of the fibers.
PNM2 does not affect Sc4-chaperone interactions
Prion propagation is critically dependent on the host chaperone machinery, and in particular on Hsp104p26. A growing body of evidence indicates that Hsp104p acts in concert with Ssa1p (Hsp70) and Sis1p (Hsp40) to divide prion particles and therefore facilitate prion replication30–33. Specifically, Ssa1p and Sis1p bind to prion particles and deliver them to Hsp104p, which is thought to extract prion particles from fibers in an ATP-dependent process that involves translocation of the extracted monomers through its axial pore34. In addition to its role in propagation, Hsp104p also causes curing of the prion (i.e., conversion from [PSI+] to [psi−]) when expressed at high levels via a mechanism separate from fiber division. Therefore, the overall effect of Hsp104p is a balance between fiber division necessary for propagation at low levels and curing at high levels26. Since a fiber’s differential susceptibility to fragmentation and curing by Hsp104p can directly affect its propagation, we sought to evaluate whether changes in chaperone-prion interactions cause the PNM2 phenotype.
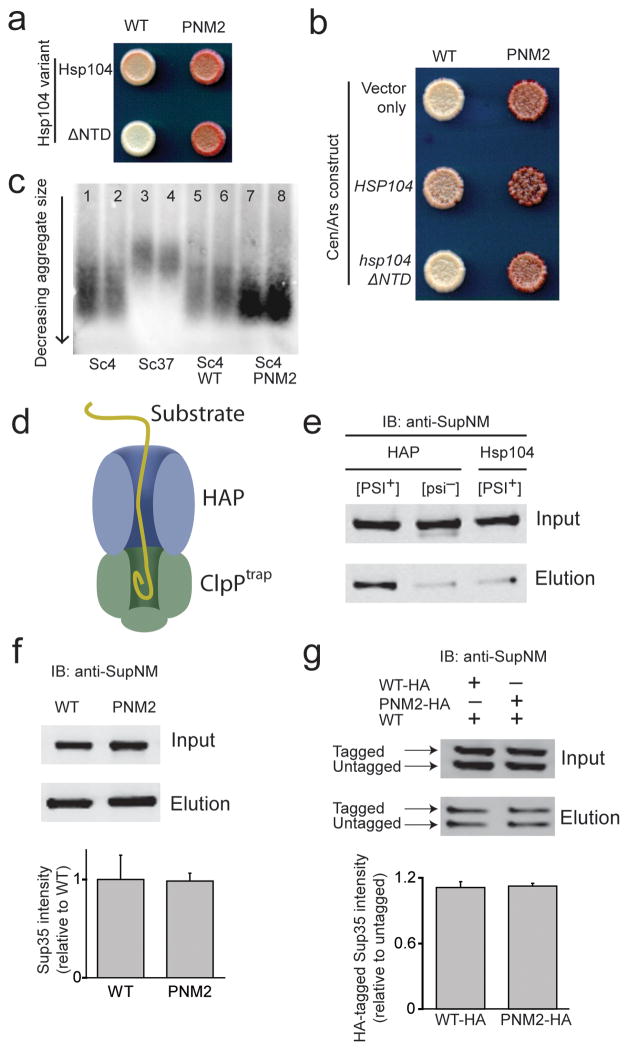
One possible explanation for the interference in [PSI+]Sc4 propagation by the PNM2 mutation is that it causes loss of [PSI+] through increased susceptibility to curing by Hsp104p. To explore this possibility, we used a truncated variant of Hsp104p that is missing its N-terminal domain, termed Hsp104ΔNTD35. When this variant is expressed from the endogenous HSP104 locus in the genome, it is fully active in [PSI+] propagation and can mediate thermotolerance, but does not cure [PSI+] with overexpression35 (Supplementary Fig. 1a). We found that the defect in [PSI+] propagation in PNM2 is observed even in the strain expressing Hsp104ΔNTD establishing that the sectoring seen in PNM2 is not due to curing via the mechanism active with overexpression of Hsp104p (Fig. 5a).
Figure 5.
PNM2 fibers in the Sc4 conformation interact with the in vivo chaperone machinery. (a) HSP104 (top row) and hsp104ΔNTD (bottom row) were expressed in [PSI+]Sc4 yeast expressing either WT (left) or PNM2 (right) Sup35p and spotted onto low adenine media. (b) HSP104 and hsp104ΔNTD were expressed at increased levels along with endogenous Hsp104p in [PSI+]Sc4 yeast expressing WT or PNM2 Sup35p. “Vector only” denotes transformation with an empty CEN/ARS plasmid. (c) SDD-AGE analysis of prion particle size in duplicate from lysates of [PSI+]Sc4 (lanes 1–2 and 5–6), [PSI+]Sc37 (lanes 3–4) and PNM2 [PSI+]Sc4 (lanes 7–8). SUP35 is genomic in lanes 1–4 and on a plasmid in lanes 5–8. Yeast express wild type HSP104 in all lanes. (d) Schematic of the in vivo HAP/ClpPtrap reaction. (e) Representative blot of the ClpPtrap affinity purification. WT Sup35p and ClpPtrap were expressed in backgrounds that were [PSI+]Sc4 or [psi−] expressing HAP or Hsp104p. (f) Monitoring the translocation of PNM2. HAP and ClpPtrap were expressed in [PSI+]Sc4 cells that also expressed WT or PNM2 Sup35p. Intensities of Sup35p elution signal from western blot (top panel) were normalized for input signal (bottom panel). (g) Monitoring translocation of PNM2 when co-expressed with WT. As in (f), but untagged WT Sup35p was also expressed in cells expressing HA-tagged WT or PNM2 Sup35p. The higher molecular weight band corresponds to HA-tagged Sup35p. Intensities of tagged-Sup35p elution signals were normalized for input signal and for untagged-Sup35p elution signal. For Sup35 intensity, values are expressed as mean ± s.e.m for three experiments.
An alternate hypothesis is that the PNM2 mutation causes increased susceptibility to the fiber division activity of Hsp104p leading to fiber resolubilization. To test this, we expressed HSP104 and hsp104ΔNTD from a CEN/ARS plasmid that leads to a 2 to 3-fold increase in expression. At this expression level, wild type Hsp104p destabilized [PSI+] while Hsp104ΔNTD did not (Fig. 5b) indicating that the destabilization must be due to the curing activity of Hsp104p and not over-fragmentation. Importantly, we find that overexpression of Hsp104ΔNTD did not enhance PNM2 sectoring arguing that PNM2 fibers are not overly susceptible to fragmentation and resolubilization by Hsp104p (Fig. 5b, Supplementary Fig. 1b). These results are distinct from those of Serio and coworkers (NSMB This Issue), who found that the PNM2 mutation can lead to fiber destabilization and enhanced Hsp104p-mediated fiber dissolution. The differences between the two studies likely results from differences in the prion strain conformations that were examined as Serio et al. observe dominant curing of [PSI+] when WT and PNM2 Sup35p are coexpressed whereas we find that the PNM2 phenotype is recessive in Sc4. Additionally, the fact that the PNM2 mutation destabilizes the fibers in those studies suggest that residue 58 is within the amyloid core of their strain conformation. Indeed we have generated examples of prion strain conformation that, like the Sc4 conformation lead to “strong” prion phenotypes, but nonetheless have amyloid cores that extend across residue 58 (unpublished data).
In an independent assay of Hsp104p activity, we looked at the size distribution of prion fibers by semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE). Previous studies have shown that changes in Hsp104p activity affect aggregate size36 and that Sc37 fibers, which are severed less efficiently by Hsp104p in vivo, are larger than those in the Sc4 conformation16. We examined the relative sizes of Sc37 and Sc4 fibers and found that PNM2 and WT Sc4 fibers have the same size distribution (Fig. 5c). This finding is in agreement with our Hsp104ΔNTD data and indicates that PNM2 and WT Sc4 interact equally with Hsp104p.
Finally, to more directly look at interactions between Hsp104p and Sc4 fibers, we used a system developed to monitor the flux of substrates through the axial pore of Hsp104p (Fig. 5d). Tessarz et al.34 and Tipton et al.33 engineered derivatives of Hsp104p (HAP and 4BAP, respectively) that interact with ClpP, a bacterial protein that forms a proteolytic chamber. When used in conjunction with a catalytically dead mutant of ClpP (ClpPtrap), substrates of Hsp104p are translocated through HAP and trapped in the ClpP chamber. Thus, following its induction in HAP-expressing yeast, ClpPtrap was affinity purified to identify Hsp104p substrates. Given that translocation through Hsp104p is likely to be the terminal event in an interaction between Sup35p and the chaperone machinery, defects in any aspect of this interaction would be expected to yield a difference in the flux of Sup35p through Hsp104p.
Previous studies have shown that Sup35p is delivered to ClpPtrap in a HAP-dependent manner, and HAP only translocates Sup35p into ClpPtrap in [PSI+] cells33,34. Moreover, substantially less Sup35p is trapped in the weak [PSI+]Sc37 strain than in the strong, and more efficiently divided16, [PSI+]Sc4 strain33. We both confirmed that WT Sup35p was trapped in a [PSI+]-dependent manner (Fig. 5e), and found that the flux of PNM2 through Hsp104 was similar to WT when expressed as either the sole source of Sup35p or co-expressed with WT (Fig. 5f,g). Therefore, in agreement with our Hsp104ΔNTD and SDD-AGE data, the HAP/ClpPtrap system shows that PNM2 Sc4 fibers interact normally with the Hsp104p chaperone machinery necessary for propagation.
PNM2 causes a defect in Sc4 propagon partitioning
We investigated whether the PNM2 mutation causes a defect in delivery of [PSI+]Sc4 infectious particles (propagons) to daughter cells during cell division using a method developed by Cox, Ness, and Tuite37. Briefly, corresponding pairs of mother and daughter cells are separated and grown in media containing guanidine, which reversibly inhibits Hsp104p38–40, such that the number of propagons remain fixed. As the cells divide, a point is reached at which there is no more than one propagon per cell. When Hsp104p function is restored by removal of guanidine, the number of [PSI+] colonies that arise from the mother versus the daughter reports on the relative number of propagons in each mother/daughter pair. Although this assay does not yield a quantitative measure of the absolute number of prion particles, it allows monitoring of relative propagon numbers and loss of [PSI+] by one of the partners.
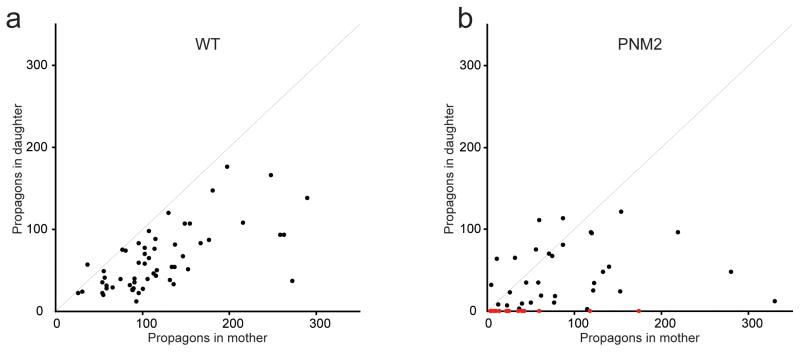
Using the above assay for [PSI+]Sc4 cells expressing WT or PNM2 Sup35p, we found evidence of partitioning dysregulation in PNM2 [PSI+]Sc4 cells (Fig. 6a,b). The mild bias in the distribution of propagons toward the mother and positive correlation between the number of propagons in the mother and daughter WT [PSI+]Sc4 cells seen previously37,41 was lost in PNM2 [PSI+]Sc4 cells. Strikingly, for a subset of pairs, the mother contained a large number of propagons while the daughter did not inherit any. In contrast, we did not observe any pairs in which the daughter had propagons, but the mother did not. The failure of propagons to partition into daughter cells accounts for the rapid appearance of red [psi−] sectors in PNM2 [PSI+]Sc4 yeast colonies.
Figure 6. PNM2 in the Sc4 conformation shows a defect in partitioning.
The number of propagons in the daughter was plotted against number of propagons in the mothers for both WT (a) and PNM2 (b) [PSI+]Sc4 backgrounds. Red dots represent mother/daughter pairs in which the mother contained propagons, but the daughter did not.
Discussion
Our studies point to a critical role for prion conformation in modulating the effects of changes in the sequence of a prion protein on prion propagation. In the case of [PSI+]Sc37, the PNM2 mutation affects the structural and physical characteristics of the amyloid fibers in a manner that has compensatory effects on prion propagation resulting in a mild effect on prion propagation in vivo. A different picture, however, emerges from studying PNM2 in the context of [PSI+]Sc4, where the mutation does not affect the amyloid structure or chaperone-mediated fiber division, but has a striking negative effect on propagation. The effect of PNM2 on [PSI+]Sc4 points to a critical role for the least understood step of [PSI+] propagation: the partitioning of prion particles. In PNM2 [PSI+]Sc4 yeast, we observed a pronounced defect in the delivery of aggregates to daughter cells. This finding highlights the question of how [PSI+] prion particles are delivered to daughter cells.
Past studies indicate that the observed mother bias in the allocation of [PSI+] particles is roughly consistent with differences in cell volume between the mother and the daughter, suggestive of partitioning by passive diffusion41. Other studies point to the potential for more active mechanisms in the segregation of protein aggregates, including the active recruitment of cytosolic aggregates to specific loci42,43. Because the PNM2 mutation lies outside of the amyloid core of the Sc4 conformation, it could disrupt interactions between this region of the prion protein and host factors responsible for prion segregation.
In support of a role for host factors in prion particle partitioning, we carried out an unbiased genetic screen for high copy suppressors of the PNM2 phenotype, in which we identified multiple independent isolates of two genes, LSB3 and ART5, that when overexpressed minimized sectoring in PNM2 [PSI+]Sc4 yeast (Supplementary Fig. 2a,b, Supplementary Methods). Lsb3p interacts with Las17p as well as the Sla1p/End3p/Pan1p complex, both factors involved in the regulation of actin dynamics44–46. The actin cytoskeleton has previously been implicated both in transport of general protein aggregates between mother and daughter cells42 as well as in the regulation of prion formation47,48. Art5p is primarily uncharacterized, but has proposed activity in the regulation of endocytosis49,50. While a full characterization of the role of these proteins in prion biology is beyond the scope of this study, it will be interesting to determine how prion particles are affected by actin-based transport and if endocytic pathways play a role in prion delivery to daughter cells.
Recent studies have suggested that size selection plays a vital role in prion particle transmission to daughter cells51. In this “size-based” model, the small prion particles, which are preferentially propagated to daughter cells, might be the species primarily affected by the PNM2 mutation such that these transmission-competent particles are either not delivered to daughter buds or actively retained in the mother cell. These possibilities may be related to the more general mechanism in which the cell specifically recognizes and concentrates misfolded protein aggregates42,43. Going forward, an understanding of the relationship between the cellular machinery for segregation of general protein aggregates and prion transmission is essential.
Our studies illustrate the importance of delivery of infectious proteins to daughter cells as a critical step in the [PSI+] propagation cycle. In the future, we anticipate that the [PSI+] system will prove useful for studying how cells survey, partition, and sequester protein aggregates. The PNM2 mutation in the Sc4 conformation should provide a critical tool for such studies, as it exhibits a distinct partitioning defect that is not confounded by changes in amyloid structure or chaperone-mediated fiber division.
Materials and Methods
Strains and plasmids
All strains were converted to [PSI+] by infection with in vitro formed Sc4 or Sc37 fibers. See Supplementary Information Tables 1 and 2 for full list of strains and plasmids.
Fiber preparation
Fibers were produced as described previously, using bacterially produced pure SupNM proteins carboxy-terminally tagged with 7x-histidine52.
In vivo yeast prion propagation characterization
YJW1110 and YJW1111 were transformed with a LEU2-marked plasmid expressing WT or PNM2 from the endogenous promoter (pRS315 WT Sup35p or pRS315 PNM2 Sup35p, respectively). The resulting transformants were selected on media lacking uracil and leucine (SD–Ura–Leu), then subsequently passaged on YEPD, then 5-FOA, then 1/4 YEPD. For the swap back experiment, the [PSI+]Sc4 yeast background containing the pRS315 PNM2 Sup35p plasmid was transformed with the original pRS316 WT Sup35p plasmid. Resulting transformants were selected on SD–Ura–Leu. After passaging several times on SD–Ura, colonies that required leucine for growth were identified. The [PSI+] phenotype was determined by observing the color on low adenine media (1/4 YEPD). The degree of sectoring, or loss of [PSI+], was determined by growing cultures in YEPD liquid media for 24 hours at 30 °C and plating onto 1/4 YEPD plates at a density of ~400 colonies per plate. The number of [PSI+] and [psi−] colonies were then counted.
In vitro analysis of the physical properties of strain conformations
Fiber growth rates15, thermal stabilities8, and susceptibility to shearing16 were all determined as described previously.
H/D exchange NMR
Uniformly 15N labeled SupNM was expressed in E. coli and purified as previously described15. 15N-SupNM seeded fibers of each strain were made as described15 and concentrated to 1/25th of their original volume. The fibers were then diluted 1:10 into D2O buffer at pH 7.0 to begin the exchange. After the desired time, exchange was quenched by adjusting the pH to 2.5, and fibers were centrifuged at 100,000g for 25 min. The pellet was washed once with 5mM DCl in D2O, then centrifuged again at 100,000g for 20 min. The pellet was frozen, freeze-dried and stored at −80 °C until NMR acquisition. The NMR spectra were acquired as described previously15. Estimated minimum peak intensity was calculated by averaging the intensity of a set of fully exchanged residues.
In vivo analysis of Hsp104p activity using the variant Hsp104ΔNTD
The Hsp104p variant Hsp104ΔNTD, first constructed by Hung and Masison35, has the nucleotides corresponding to the N-terminal 146 residues deleted. hsp104ΔNTD was amplified and fused to the NAT cassette by PCR, and inserted into the HSP104 genomic locus of YJW1110 by homologous recombination, resulting in strain YJW1667. Plasmids pRS399 Hsp104p and pRS399 Hsp104ΔNTD35 were used to mildly (2–3 fold) overexpress Hsp104p or Hsp104ΔNTD. pRS399 is a pRS315 plasmid where the LEU2 marker has been replaced by a KanR cassette. Plasmids were transformed into YJW1110 and plated on 1/4 YEPD containing the antibiotic G418 for visualization of the [PSI+] phenotype.
Semi-Denaturing Detergent-Agarose Gel Electrophoresis (SDD-AGE)
Relative aggregate size was determined by SDD-AGE as previously described53,54. Sup35p was probed for by western blotting using polyclonal anti-SupNM.
ClpPtrap Affinity Purification Experiments
The ClpPtrap affinity purification experiments were performed as described previously34. Briefly, YJW1112, YJW1113, YJW1114, or YJW1115 that had been transformed with pRS313 WT Sup35p, pRS313 PNM2 Sup35p, pRS313 WT Sup35HA, or pRS313PNM2 Sup35HA were grown in SD media with 50 μM CuSO4 from OD600 = 0.1 to 1 and lysed at 4 °C by bead beating in IP buffer (50 mM Tris/Cl pH 8.0, 1 M NaCl, 2 mM EDTA, 0.5% (v/v) Triton X-100, 5 mM β-mercaptoethanol, + Roche Complete Protease Inhibitor Cocktail). The lysate was cleared by centrifugation at 15,000g for 15 min, incubated with Streptavidin Sepharose (GE Healthcare) at RT for 1 hr, washed with 40 column volumes of IP buffer, and eluted in IP buffer + 150 mM NaCl and 4 mM biotin. The eluate was subjected to SDS-PAGE and Sup35p was probed for by western blotting using polyclonal anti-SupNM.
Mother/Daughter propagon counting
The propagons in mothers and daughters were counted as described previously37. Briefly, mothers and daughters of YJW1110 (containing pRS315 WT Sup35p or pRS315 PNM2 Sup35p as the sole source of Sup35p) were separated by micromanipulation onto YEPD plates containing 3mM Guanidine HCl. After growing at 30 °C for about 40 hours, whole colonies were isolated using a cut pipette tip, resuspended in a small volume of H2O, and plated onto SD–Ade + 5% (w/v) YEPD. After growing at 30 °C for 10 to 14 days, the number of [PSI+] colonies was counted. For the WT samples, [PSI+] colonies were easily distinguished from [psi−] and Ade revertants by color. The PNM2 samples contained background indistinguishable from true [PSI+] colonies by color. We estimated the average background by repeating the experiment using a [psi−] PNM2 strain created by curing the [PSI+]Sc4 PNM2 strain by successive passaging on YEPD containing 3 mM guanidine HCl. The average background of ~25 colonies per sample was subtracted for all counted values.
Supplementary Material
Acknowledgments
We would like to thank B. Bukau, A. Mogk, and P. Tessarz for reagents; M. Kelly for NMR assistance; D. Cameron, S. Collins, C. Gross, D. Mullins, G. Narlikar, M. Tanaka, and K. Tipton for helpful discussions; and O. Brandman, C. Foo, A. Frost, N. Ingolia, E. Oh, E. Quan, and E. Rodriguez for critical reading of the manuscript. This work was funded by the Howard Hughes Medical Institute (J.S.W.), the National Science Foundation Graduate Research Fellowship program (K.J.V), the Hertz Foundation Peter Strauss Fellowship (M.H.S.), and the National Institutes of Health.
Footnotes
Author Contributions
KJV, MHS and JSW designed this study and wrote the manuscript. JSW supervised this work. KJV and MHS performed the majority of the experiments. BHT designed the H/X NMR experiments and acquired and analyzed the NMR data.
References
- 1.Caughey B, Baron GS, Chesebro B, Jeffrey M. Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem. 2009;78:177–204. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 4.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 6.Santoso A, Chien P, Osherovich L. Molecular basis of a yeast prion species barrier. Cell. 2000 doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 7.Morales R, Abid K, Soto C. The prion strain phenomenon: molecular basis and unprecedented features. Biochim Biophys Acta. 2007;1772:681–691. doi: 10.1016/j.bbadis.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 9.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99 (Suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 12.Chien P, Weissman J. Conformational diversity in a yeast prion dictates its seeding specificity. Nature. 2001 doi: 10.1038/35065632. [DOI] [PubMed] [Google Scholar]
- 13.Sparrer HE, Santoso A, Szoka FC, Weissman JS. Evidence for the prion hypothesis: induction of the yeast [PSI+] factor by in vitro- converted Sup35 protein. Science. 2000;289:595–599. doi: 10.1126/science.289.5479.595. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 17.Derkatch IL, Bradley ME, Zhou P, Liebman SW. The PNM2 mutation in the prion protein domain of SUP35 has distinct effects on different variants of the [PSI+] prion in yeast. Curr Genet. 1999;35:59–67. doi: 10.1007/s002940050433. [DOI] [PubMed] [Google Scholar]
- 18.Green KM, et al. The elk PRNP codon 132 polymorphism controls cervid and scrapie prion propagation. J Gen Virol. 2008;89:598–608. doi: 10.1099/vir.0.83168-0. [DOI] [PubMed] [Google Scholar]
- 19.Takemura K, Kahdre M, Joseph D, Yousef A, Sreevatsan S. An overview of transmissible spongiform encephalopathies. Anim Health Res Rev. 2004;5:103–124. doi: 10.1079/ahr200494. [DOI] [PubMed] [Google Scholar]
- 20.Zeidler M, Stewart G, Cousens SN, Estibeiro K, Will RG. Codon 129 genotype and new variant CJD. Lancet. 1997;350:668. doi: 10.1016/s0140-6736(05)63366-1. [DOI] [PubMed] [Google Scholar]
- 21.Mead S, et al. Creutzfeldt-Jakob disease, prion protein gene codon 129VV, and a novel PrPSc type in a young British woman. Arch Neurol. 2007;64:1780–1784. doi: 10.1001/archneur.64.12.1780. [DOI] [PubMed] [Google Scholar]
- 22.Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 23.Hill AF, et al. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–450. 526. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 24.Doel SM, McCready SJ, Nierras CR, Cox BS. The dominant PNM2-mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young CSH, Cox BS. Extrachromosomal elements in a super-suppression system of yeast: I. A nuclear gene controlling the inheritance of the extrachromosomal elements. Heredity. 1971;26:413–422. [Google Scholar]
- 26.Chernoff Y, Lindquist S, Ono B, Inge-Vechtomov S, Liebman S. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 27.Kochneva-Pervukhova NV, et al. Mechanism of inhibition of Psi+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. Embo J. 1998;17:5805–5810. doi: 10.1093/emboj/17.19.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 29.King CY, et al. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci U S A. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci U S A. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–832. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- 32.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. Embo J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 35.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 37.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 39.Grimminger V, Richter K, Imhof A, Buchner J, Walter S. The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J Biol Chem. 2004;279:7378–7383. doi: 10.1074/jbc.M312403200. [DOI] [PubMed] [Google Scholar]
- 40.Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- 41.Byrne LJ, et al. The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae. PLoS ONE. 2009;4:e4670. doi: 10.1371/journal.pone.0004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B, et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 43.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robertson AS, et al. The WASP homologue Las17 activates the novel actin-regulatory activity of Ysc84 to promote endocytosis in yeast. Molecular Biology of the Cell. 2009;20:1618–1628. doi: 10.1091/mbc.E08-09-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tonikian R, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madania A, et al. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Molecular Biology of the Cell. 1999;10:3521–3538. doi: 10.1091/mbc.10.10.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganusova EE, et al. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailleul P, Newnam G, Steenbergen J, Chernoff Y. Genetic Study of Interactions Between the Cytoskeletal Assembly Protein Sla1 and Prion-Forming Domain of the Release Factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics. 1999;153:81. doi: 10.1093/genetics/153.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin CH, Macgurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 50.Nikko E, Pelham HRB. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Derdowski A, Sindi SS, Klaips CL, Disalvo S, Serio TR. A size threshold limits prion transmission and establishes phenotypic diversity. Science. 2010;330:680–683. doi: 10.1126/science.1197785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Halfmann R, Lindquist S. Screening for Amyloid Aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. J Vis Exp. 2008:e838. doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Meth Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.