Abstract
We used genetic mapping to examine the genetic architecture of differences in host plant use between two species of noctuid moths, Heliothis subflexa, a specialist on Physalis spp., and its close relative, the broad generalist H. virescens. We introgressed H. subflexa chromosomes into the H. virescens background and analyzed 1,462 backcross insects. The effects of H. subflexa-origin chromosomes were small when measured as the percent variation explained in backcross populations (0.2 to 5%), but were larger when considered in relation to the interspecific difference explained (1.5 to 165%). Most significant chromosomes had effects on more than one trait, and their effects varied between years, sexes, and genetic backgrounds. Different chromosomes could produce similar phenotypes, suggesting that the same trait might be controlled by different chromosomes in different backcross populations. It appears that many loci of small effect contribute to the use of Physalis by H. subflexa. We hypothesize that behavioral changes may have paved the way for physiological adaptation to Physalis by the generalist ancestor of H. subflexa and H. virescens.
Keywords: ecologically adaptive traits, herbivore host range, plant-insect interactions, ecological genetics, complex phenotypes
Introduction
Understanding the genetic basis of ecological adaptation is a longstanding goal of evolutionary biology, but genetic studies comparable in depth to the research that informs our understanding of evolutionary ecology are still rare. Vigorous inquiry over the past fifty-plus years has resulted in a nuanced understanding of the evolutionary determinants of insect host plant use (e.g., Dethier 1954; Ehrlich and Raven 1964; Bernays and Graham 1988; Mitter et al. 1991), yet our knowledge of the genetic basis of variation in host plant use is largely limited to identification of the genes that allow insects to cope with particular plant defense compounds (e.g., Wheat et al. 2007; Scriber et al. 2008; Heidel-Fischer et al. 2010; Loxdale 2010). Happily, ongoing work in a range of systems is beginning to allow progress toward understanding the genetic architecture of host plant use (e.g., Acyrthosiphon (Hawthorne and Via 2001; Caillaud and Via 2012), Drosophila (Dekker et al. 2006; Dworkin and Jones 2009; Etges et al. 2010; Earley and Jones 2011; Wisotsky et al. 2011), and Plutella (Henniges-Janssen et al. 2011).
Host plant use is phenotypically complex, involving variation in neurosensory, physiological, morphological, and behavioral traits. In this study, we examine traits associated with host use differences between the narrow host specialist Heliothis subflexa and its close relative, the broad generalist H. virescens. In previous work, we showed that H. subflexa is behaviorally and physiologically adapted to its host plant, Physalis angulata (Oppenheim and Gould 2002a, b). In the current study, we use genetic mapping to investigate the genetic basis of interspecific differences in use of P. angulata. Meiotic recombination does not occur in female Lepidoptera (Marec 2010), so maternal-origin chromosomes are transmitted intact. This characteristic allowed us to introgress whole H. subflexa chromosomes into the H. virescens background by backcrossing interspecific hybrids to H. virescens. This approach resolves quantitative trait loci to the level of chromosome. Because heliothines have 31 small, similarly-sized (Van't Hof et al. 2008) chromosomes, each chromosome comprises only 2–5% of the genome (around 500 genes based on the estimated genome size of H. virescens) (401 Mbp; Taylor et al. 1993).
We measured several traits that contribute to the performance of larvae on P. angulata: willingness to feed, physiological adaptation, and specialized behavior. Host plant acceptance is a critical first step in host use, and has been suggested as the basis of host use differences between the specialist D. sechellia and its close relative, the generalist D. simulans (Matsuo et al. 2007). We report here on the genetic basis of interspecific differences in the willingness of larvae to feed on P. angulata. Physiological adaptation to host plant defense compounds is a well-known component of host specialization (Berenbaum and Feeny 2008), and we report on two measures of physiological adaptation: the percent change in larval weight after feeding on P. angulata, and larval assimilation efficiency (the change in larval weight per gram of P. angulata ingested). Although the genus Physalis lacks the best-known Solanaceae defense compounds (e.g., nicotinoids, capsaicinoids, steroid alkaloids) (Wink 2003), it is characterized by the presence of withanolides, a group of steroidal lactones of unusual structure (Eich 2008). Withanolides, and, more particularly physalins (withanolides found only in Physalis), have a wide range of bioactive properties, including anti-tumor, trypanocidal, and immunoregulatory effects (Chen et al. 2011). When ingested by insect herbivores, withanolides have anti-feedant effects, but do not cause acute toxicity (Ascher et al. 1987; Mareggiani et al. 2001; Bado et al. 2004).
Behavioral adaptations can contribute to host plant adaptation (Bernays and Graham 1988), and in a previous study we found that specialized behaviors allow H. subflexa, but not H. virescens, to use the inflated calyx P. angulata as a refuge from natural enemies (Oppenheim and Gould 2002a). In the field, larvae of both species are subject to attack by specialist parasitoids, but because H. subflexa larvae enter the calyx quickly and completely, they are able to use it as a refuge. H. virescens, however, often either enter and leave repeatedly, or begin feeding before fully entering the calyx. Both behaviors leave H. virescens vulnerable to parasitoids. We used the number of holes bored in the calyx as a laboratory-based measure of this behavioral adaptation to examine its genetic basis.
Our ability to identify the loci associated with variation in the use of P. angulata is subject to all the constraints that the past two decades of mapping studies have revealed: context-dependent effects (Mackay et al. 2009), many loci of small effect whose detection depends on sample size and map resolution (Beavis 1998; Kroymann and Mitchell-Olds 2005), and pleiotropic effects of the loci involved (Edwards et al. 2006; Ehrenreich et al. 2010). Our experimental design took these factors into account: to ensure sufficient detection power, we analyzed 1,462 backcross insects; to address context-specific effects, we used a large sample size that allowed us to test whether there were interactions between genotype, sex, population, environment, and year; and we used closely related insects to reduce the amount of background genetic variation.
Materials and Methods
Study system
Heliothis subflexa and H. virescens are closely related, sharing 99% sequence similarity in the genes for which comparisons have been made (Cho et al. 1995; Fang et al. 1997). The two species are thought to have evolved about 2.5 Myr ago from a generalist ancestor (Mitter et al. 1993; Poole et al. 1993; Fang et al. 1997; Cho et al. 2008). In the laboratory, H. virescens and H. subflexa will hybridize, producing fertile F1 females and sterile F1 males (male fertility is restored after several backcross generations) (Karpenko and Proshold 1977). The sterility of the male hybrids, which is contrary to Haldane's Rule (Haldane 1922), is apparently due to deficiencies in maternally inherited mitochondria in the hybrid sperm (Miller et al. 1986).
Despite their genetic similarity and ability to hybridize, these species differ greatly in host plant use. Heliothis virescens has a very broad host range, feeding on at least 37 species in 14 plant families (Sheck and Gould 1993), while H. subflexa is narrowly specialized on plants in the genus Physalis (Laster et al. 1982); even within this genus, not all species are acceptable to H. subflexa (Bateman 2006). Heliothis virescens is not known to feed on Physalis in the field; in laboratory assays, we found 5 percent survival from neonate to 3rd instar for H. virescens on P. angulata, as compared to 55 percent survival for H. subflexa.
Insect strains and rearing
All of the insects used in these experiments originated from colonies maintained at North Carolina State University (Sheck et al. 2006). The H. virescens colony was established in 1988 using field-collected larvae from Yadkin County, North Carolina. The H. subflexa colony was established in 1997 using field-collected larvae from Orangeburg County, South Carolina. The colonies have been maintained in the laboratory at population sizes of about 250 adults. The backcross experiments described here were conducted in 2001 (when the H. virescens and H. subflexa strains had been in the laboratory for about 160 and 40 generations, respectively) and in 2007 (after about 220 and 100 generations).
Larvae were individually reared on artificial diet (Burton 1970), except during assays on Physalis. Laboratory colonies and experimental insects were maintained at 23°C and 50–70% relative humidity on a 16:8 h light-dark cycle. In 2007, some eggs and larvae were held at 5°C for 2–5 days to slow their development to coincide with experimental resources.
Plants
Although many species of Physalis will support H. subflexa development, larvae do particularly well on P. angulata (Bateman 2006), and we used this species (referred to hereafter as Physalis) for all experiments. In the 2001 experiments, seeds from multiple plants were used. In 2007, all plants were from the seeds of a single founder plant. See Supplementary Materials for plant source and cultivation information.
Backcross matings
Experiments were conducted on seven backcross families (Figure 1). Because F1 males are sterile, all backcrosses involved F1 females. Each backcross involved a grandparental mating of an H. virescens female to an H. subflexa male. An F1 female was then backcrossed to an H. virescens male, and their progeny (referred to as VS) were used in the experiments. Backcrosses were done in this direction to isolate the H. subflexa-origin chromosomes involved in performance on Physalis.
Figure 1.
Single pair matings in 2001 and 2007. Sex chromosome states are shown only once, but are the same in all seven families. WS and ZS are the female and male sex chromosomes from Heliothis subflexa; WV and ZV are the female and male sex chromosomes from H. virescens.
We tested two backcross families in 2001 (VS01A and VS01B) and five in 2007 (VS07A, VS07B, VS07C1, VS07C2, and VS07C3). Offspring from the five VS07 families were paternal cousins, and two families (VS07C1 and VS07A) were half-siblings.
Measurement of larval phenotypes
The performance of H. virescens, H. subflexa, their F1 progeny, and VS backcross individuals on Physalis was evaluated by allowing each larva to feed on a single Physalis fruit for 48 hours. Newly-hatched larvae were reared on artificial diet and checked twice daily to determine developmental stage. Larvae were assayed 4–8 hours after molting to 2nd instar. We used 2nd instars because testing this stage provides good discrimination between H. virescens and H. subflexa phenotypes but does not result in unacceptably high mortality (SJO, personal observation).
Larvae were presented with Physalis fruits that were still within their calyces; to feed on the fruits, larvae had first to bore an entry hole through the calyx. At the beginning of each assay, we recorded: larval weight (mg); fruit weight (g); larval age (days); larval rearing temperature (23°C or 5°C; if the latter, number of days at 5°C); identity of plant providing fruit; and time of day. At the conclusion of each assay, we recorded: larval weight, fruit weight, assay duration (hours), occurrence of larval feeding* (judged by damage to fruit, recorded as 0 or 1), and the number of holes in the calyx* (determined by visual inspection of calyces). From these data, we calculated: change in larval weight (larval end weight – larval start weight); change in fruit weight (fruit start weight – fruit end weight); percent change in fruit weight and percent change in larval weight* (weight change ÷ start weight); and assimilation efficiency* (change in larval weight ÷ change in fruit weight). (The four traits marked with an asterick were used in genetic mapping).
After the assay, larvae were maintained on artificial diet. Sex was determined at the pupal stage. Insects were inspected daily, and usually held until adult emergence before freezing at −80°C. When inspection indicated that an insect might not survive to adulthood, that insect was frozen immediately to maintain the largest possible population for molecular analysis. Table 1 gives sample sizes for each year and population.
Table 1.
The number of backcross and control insects assayed each year.
| 2001 | 2007 | ||||
|---|---|---|---|---|---|
| Type | Population | N | Population | N | Total |
| Backcross | VS07 | 1157 | 1462 | ||
| VS01 | 291 | ♀VS07 | 648 | ||
| ♀VS01 | 160 | ♂VS07 | 509 | ||
| ♂VS01 | 145 | VS07A | 242 | ||
| VS01A | 180 | VS07B | 211 | ||
| VS01B | 125 | VS07C1 | 219 | ||
| VS07C2 | 243 | ||||
| VS07C3 | 242 | ||||
| Control | H. subflexa | 98 | H. subflexa | 97 | 195 |
| H. virescens | 119 | H. virescens | 130 | 249 | |
To control for differences among dates, assays of H. subflexa and H. virescens controls were conducted on multiple dates, simultaneous with backcross and F1 assays. The effect of holding larvae at 5°C for 2–5 days was controlled for by exposing H. subflexa, H. virescens, and larvae from multiple backcross families to this regime.
While it was not possible to test every insect population on fruits from each of the fifty plants used in 2007, the effects of fruit source were tested by simultaneously assaying H. virescens, H. subflexa, and two or more backcross families on fruits from each of eight different plants. Furthermore, fruits from each of the fifty plants were used in assays of at least two backcross families, allowing us to detect any gross anomalies due to fruit source. Variation in fruit size and maturity can affect larval performance (Bateman 2006), so only fruits of similar size (range: 1.7 to 2.8 g) and stage of ripeness were used.
AFLP Markers
We extracted DNA from frozen adults, larvae, and pupae, using the Qiagen (Chatsworth, CA) DNeasy 96 extraction kit. After extraction, DNA was prepared for AFLP mapping using a modified version of the procedure described by Vos et al. (Vos et al. 1995). Selective amplification was carried out using 19 different primer pairs, and the resulting fragments visualized using fluorescently labeled primers. Fragments were separated by capillary electrophoresis on a CEQ 8000 (Beckman-Coulter, Fullerton, CA), following the recommended protocol. The resulting electropherograms were first analyzed with the CEQ AFLP software (version 9). Final scoring of all fragments was done manually to ensure that all legitimate peaks were included, and spurious peaks excluded. See Supplementary Materials for detailed methods.
Gene-Based Markers
AFLP fragment homology is generally very high between different populations within a species (Althoff et al. 2007). However, to confirm that apparently homologous linkage groups did indeed represent the same chromosomes, we used gene-based markers as anchors for linkage groups. We used eleven codominant markers developed in the Gould lab (Gould et al. 2010), and generated 14 new ones (their development is described in Supplementary Materials). Twenty-five primer pairs were used to produce gene-based markers (see Supplementary Table 1 for primer and nucleotide sequence information for all gene-based markers). We amplified DNA with these 25 primer pairs in 176 backcross individuals from five VS families, and added the resulting marker phenotypes to our AFLP-based linkage map.
Linkage Mapping
For mapping, we used AFLP markers that were absent in H. virescens, present in H. subflexa, present in F1 mothers, and segregating approximately 1:1 in the VS backcross. All grandparental crosses were of an H. virescens female to an H. subflexa male, so all F1 mothers had a W sex chromosome from H. virescens and a Z sex chromosome from H. subflexa. Backcrossing to H. virescens resulted in female progeny with both sex chromosomes from H. virescens and male progeny with one Z chromosome from H. virescens and one from H. subflexa (Figure 1). Thus, the effects of the H. subflexa sex chromosomes could not be determined, because they were either confounded with overall differences between the sexes (the Z chromosome, present in all males and absent in all females) or universally absent (the W chromosome). We did, however, examine differences between the sexes in the effects of H. subflexa-origin autosomes.
We used the program JoinMap (version 3.0) to sort our AFLP and gene-based markers into linkage groups. We made separate maps for each data set, using LOD threshold ≥ 10 and recombination ≤ 0.5. Because no recombination occurred in F1 females, linkage between markers on the same chromosome should be complete, and the level of recombination between them should be zero. In practice, however, missing data and errors in determining marker genotypes combine to reduce the association between markers. Thus, small departures from the ideal values are treated as experimental error.
Mapping
i. Statistical analysis
We used permutation to determine empirical significance thresholds for chromosome-phenotype association in a model that included all chromosomes as the independent variables and a given phenotypic trait as the dependent variable (Churchill and Doerge 1994). To determine genome-wide significance thresholds at an experiment-wise error rate of α = 0.05 or α = 0.1, we randomly permuted the phenotype values among chromosomal genotypes. We performed 1000 permutations for each phenotypic variable, and recorded the maximum F statistic generated in each replicate. The resultant population of F statistics was sorted from lowest to highest, and the 900th and 950th greatest F statistics (corresponding to α = 0.1 and α = 0.05 experiment-wise Type I error rates) were used as the threshold for declaring whether the observed F statistic indicated a suggestive or significant chromosome-phenotype association. We did separate analyses for the data from 2001 and 2007 because the variances differed between years.
To determine whether the effect of individual H. subflexa-origin chromosomes varied among environments or genetic backgrounds, we conducted a mixed-model analysis of variance for each chromosome and trait. We used PROC GLIMMIX (SAS 9.2) to evaluate the effect of chromosome (fixed), sex (fixed), family (random), and possible interactions on the observed phenotype. Appropriate distributions were specified where the data were non-normally distributed (e.g. Poisson distribution for count data). Effects that were not significant were dropped from the model, and a reduced model used to estimate the effect of chromosome on phenotype. Our sample sizes were unequal, so we used least squares means to examine differences between means. We corrected for multiple comparisons within each dependent variable by using the SIMULATE option in the LSMEANS statement. SIMULATE is a simulation-based method for controlling the family-wise error rate by estimating the precise value of the adjusted p-values given the number of tests performed, and is both more precise and more liberal than Bonferroni correction (Edwards and Berry 1987).
Phenotypic differences associated with chromosome state within each level of sex, lineage, and family were examined using the LSMEANS SLICE option in GLIMMIX. SLICE performs a partitioned analysis of a given factor at different levels of the other factors (i.e., simple main effects Winer 1971), allowing us to evaluate the statistical significance of a chromosome’s effect at each level. We used the SIMULATE option to obtain p-values corrected for the number of tests performed.
We evaluated chromosome × chromosome interactions among chromosomes that were significant in either permutation or mixed model analysis. We used ANOVA to test for pairwise interactions between significant chromosome. For each trait, all significant chromosomes were included in a single model.
Because stringent significance tests can eliminate causal loci when the amount of variance they explain is small (Yang et al. 2010), we evaluated the effects of both significant (p < 0.05) and suggestive (0.05 < p < 0.1) chromosomes (Kruglyak and Lander 1995). Although suggestive chromosomes are at an increased risk of being false positives, we feel their inclusion is important if we wish to understand the overall genetic architecture of host use.
ii. Chromosome effects
The effect of each H. subflexa-origin chromosome on the associated trait(s) was measured in two ways. First, we estimated the percent of variation explained (PVE) in a backcross population by comparing the phenotypes of H. subflexa chromosome-present (Hs+) individuals to those of H. subflexa chromosome-absent (Hs−) individuals, using regression analysis to estimate PVE (expressed as r2) for each chromosome. A chromosome’s effect was calculated as the average difference between Hs+ and Hs− individuals.
Second, because the aim of these experiments was to understand the genetic basis of differences between species (rather than variation within the backcross population), we calculated the percent of the phenotypic gap between H. subflexa and H. virescens accounted for by a given chromosome (Fishman et al. 2002; Lexer et al. 2005). The equation used was: percentage species difference = (average effect of chromosome ÷ average difference between H. subflexa and H. virescens) × 100.
Results
Linkage Mapping and Determination of Homology
Independent linkage maps were generated for each of the seven backcross families. This was necessary because not all AFLP markers were informative in all families, and some gene-based markers were only used in one or two families. The distribution of autosomes among the backcross progeny did not differ from expected Mendelian ratios. See Supplementary Tables 2 and 3 for details.
After the seven independent maps with all 30 autosomes were complete, a single homologous map was derived by comparison of linkage groups among families. The resulting map includes 29 autosomes that can be identified in all seven families (Supplementary Table 3). For the one autosome where homology could not be determined across families, a conservative analysis within each family showed no association between this autosome and any of the traits examined. Thus, we are confident that conclusions about the relationship between chromosomes and phenotypes can safely be drawn from our homologous map.
We were unable to map the female sex chromosome because all backcross females had both W and Z chromosomes from H. virescens, but we could map the male sex chromosome because all backcross males had one copy of Z from each species (Figure 1). Because of this inheritance pattern, any phenotypic effects of sex chromosomes on host use traits could not be mapped, but differences between the sexes could result from the presence of the H. subflexa Z chromosome in backcross males.
Phenotypic Analysis of Quantitative Traits
Because we are interested in discovering the genetic basis of variation in the traits that distinguish H. subflexa from H. virescens, we first examined interspecific differences in the measured traits. We evaluated these differences for each year, family, and sex. The backcross progeny were also evaluated for each year, sex, and family. For traits that did not differ significantly between H. subflexa and H. virescens, we made no attempt to discover the genetic basis of backcross variation.
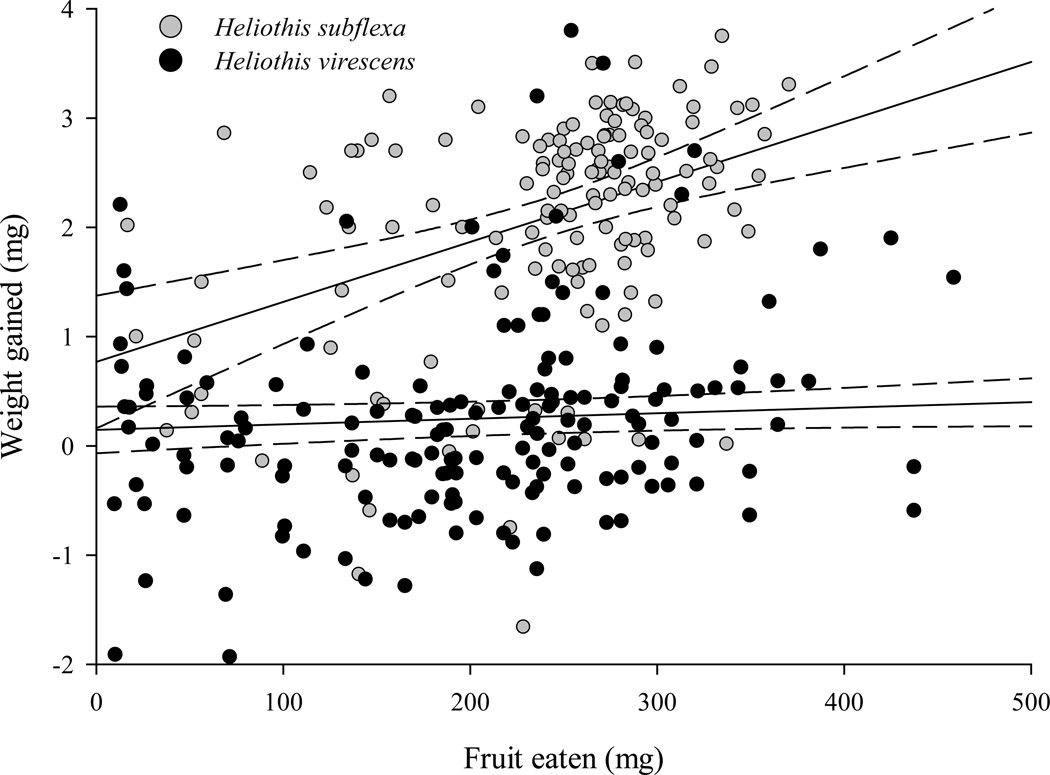
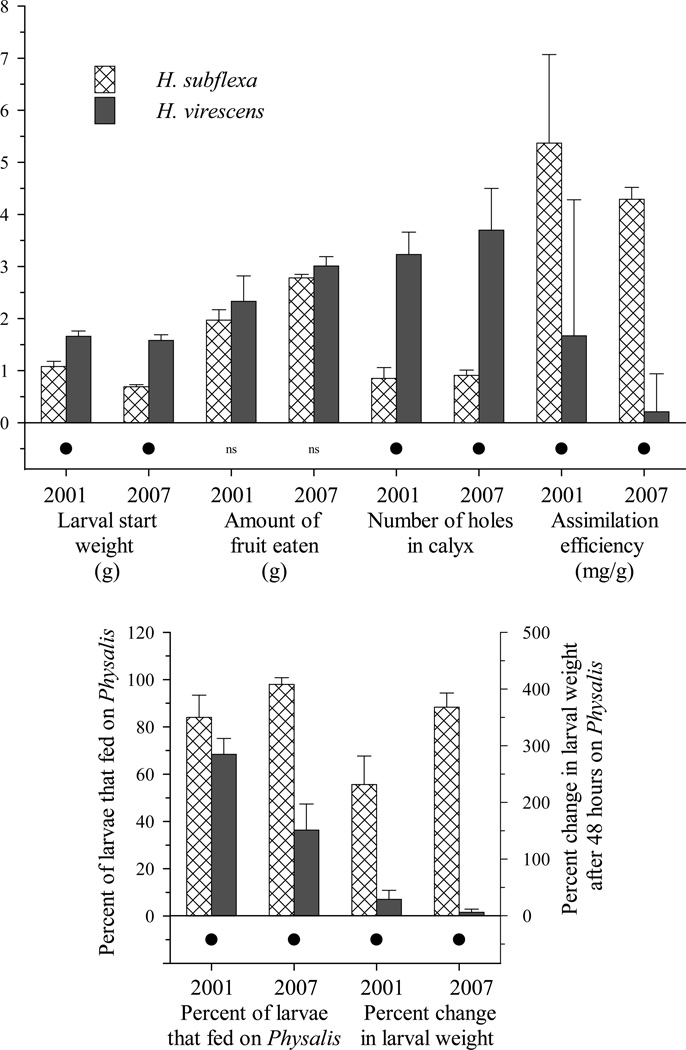
Among control insects, most variation was explained by species. Heliothis subflexa were more willing than H. virescens to feed on Physalis, but the two species did not differ in the amount of fruit they consumed. The percent change in larval weight during feeding assays was greater for H. subflexa than H. virescens. Interestingly, percent change in larval weight increased linearly with amount of fruit eaten for H. subflexa, but not for H. virescens (Figure 2). For H. subflexa, the amount of fruit eaten explained 13 percent of the variance in larval weight change; for H. virescens, the effect of amount of fruit eaten on larval weight change was not significant. H. subflexa had higher assimilation efficiency and bored fewer holes in the calyx surrounding Physalis fruit than H. virescens. See Figure 3 for the phenotypic differences between species.
Figure 2.
Relationship between the amount of fruit eaten and the amount of weight gained in H. subflexa and H. virescens. Regression line and 95% confidence interval are shown for each species. For H. subflexa (grey dots) larval weight change = 0.8 + 5.5 × (amount of fruit eaten); p < 0.0001. For H. virescens (black dots) larval weight change = 0.15 + 0.5 × (amount of fruit eaten); p = 0.1.
Figure 3.
Phenotypic means for H. subflexa and H.virescens for the traits measured in this study. Error bars represent the 95% confidence interval. Significance levels are indicated by the symbol below each comparison: black dot = interspecific difference significant (p < 0.05); ns = interspecific difference not significant.
Among backcross progeny, most traits varied with year, family, larval start weight, and sex. The willingness of larvae to feed, the amount of fruit consumed, and the proportion change in larval weight differed between years, families, and larval start weights, but not between cold treatments or fruit sources. The number of holes in the calyx varied with family and with larval start weight (heavier larvae bored more holes), and larvae that did not feed bored more holes than did feeders (non-feeders = 3.0 holes, feeders = 1.6 holes, p < 0.0001). The number of holes varied weakly with fruit source (p = 0.01), but none of the pairwise comparisons between individual plants were significant. For all VS07 (but not for VS01) backcross progeny, assimilation efficiency varied with family and with larval start weight (larger insects had higher assimilation efficiency, p < 0.0001). Additional phenotypic results are described in Supplementary Materials. See Supplementary Table 4 for phenotypic means in the VS families.
We examined the correlations among traits in the H. subflexa, H. virescens, and backcross insects. A number of correlations were found in each group (Table 2); of particular interest, the number of holes bored in the calyx of Physalis by backcross larvae was negatively correlated with willingness to feed, percent change in larval weight, and assimilation efficiency (p ≤ 0.0001). Within H. virescens, there was no correlation between these traits; within H. subflexa, the correlations, where significant, were in the opposite direction: increase in the number of holes was correlated with an increased willingness to feed and higher assimilation efficiency.
Table 2.
Kendall correlation coefficients between phenotypic traits measured in Heliothis subflexa, Heliothis virescens, and VS backcross insects.
| Larval Start Weight |
Larval Feeding |
Percent Change in Larval Weight |
Change in Fruit Weight |
Assimilation Efficiency |
||
|---|---|---|---|---|---|---|
| VS backcross insects | Larval Start Weight | — | ||||
| Willingness to feed on Physalis | 0.18*** | — | ||||
| Percent Change in Larval Weight | −0.12*** | —1 | — | |||
| Change in Fruit Weight | −0.07** | —1 | 0.25*** | — | ||
| Assimilation Efficiency | 0.13*** | —1 | 0.70*** | 0.11*** | — | |
| Number of Holes in Calyx | 0.06** | −0.20*** | −0.18*** | −0.07* | −0.14*** | |
| H. subflexa control insects | Larval Start Weight | — | ||||
| Willingness to feed on Physalis | −0.10† | — | ||||
| Percent Change in Larval Weight | −0.53*** | —1 | — | |||
| Change in Fruit Weight | −0.06† | —1 | 0.20** | — | ||
| Assimilation Efficiency | −0.12* | —1 | 0.41*** | −0.15* | — | |
| Number of Holes in Calyx | −0.08† | 0.28*** | 0.12† | −0.02† | 0.18* | |
| H. virescens control insects | Larval Start Weight | — | ||||
| Willingness to feed on Physalis | 0.10† | — | ||||
| Percent Change in Larval Weight | −0.06† | —1 | — | |||
| Change in Fruit Weight | 0.22*** | —1 | 0.15* | — | ||
| Assimilation Efficiency | −0.06† | —1 | 0.82*** | 0.10† | — | |
| Number of Holes in Calyx | −0.11† | 0.002† | −0.07† | −0.006† | −0.09† | |
p ≤ 0.0001;
p ≤ 0.001;
p ≤ 0.05
Correlation not significant
Only measured when larval feeding occurred
Chromosome Analysis
For each trait, 8–11 H. subflexa-origin chromosomes were associated with phenotypic variation, and several had different effects or significance levels in different years, sexes, or families. Supplementary Tables 5–8 give statistical results for all chromosomes affecting the traits below.
i. Willingness to feed on Physalis
In the VS07 insects, eight chromosomes were associated with variation in willingness to feed on Physalis (no significant or suggestive chromosomes were found in the VS01 population) (Supplementary Figure 1). Individual chromosomes accounted for 38 to 165 percent of the difference between H. subflexa and H. virescens; PVE ranged from 0.2 to 5 percent. Most chromosomes had effects in the H. subflexa direction (i.e. their presence increased willingness to feed on Physalis), but for one chromosome (H26), all significant effects were H. virescens-like; another chromosome (H01) had H. subflexa-like effects in one family and H. virescens-like effects in another.
ii. Percent change in larval weight
Ten chromosomes were associated with variation in the percent change in larval weight after 48h on Physalis (Supplementary Figures 2a & b). Each chromosome accounted for 1 to 5 percent of the interspecific difference; PVE ranged from 0.3 to 3.6 percent. Two chromosomes (H19 and H29) had significant effects in both the VS01 and VS07 populations, and one (H05) had significant effects in VS01 but not in VS07. The remaining seven chromosomes had effects only in the VS07 population. Two chromosomes (H06 and H13) had exclusively H. virescens-like effects (their presence was associated with a decrease in percent change in larval weight).
iii. Number of holes in the calyx of Physalis
Eleven chromosomes were associated with variation in the number of holes bored (Supplementary Figures 3a & b). Individual chromosomes accounted for 4 to 14 percent of the interspecific difference; PVE ranged from 0.3 to 3.7 percent. Three chromosomes (H05, H17 and H19) had effects in both VS01 and VS07, and two (H01 and H18) had effects only in VS01; the remaining six chromosomes had effects in VS07 only. Six chromosomes had effects only in the H. virescens direction (their presence was associated with an increase in the number of holes), and most of the remaining chromosomes had mixed effects (H. subflexa-like in some populations, H. virescens-like in others).
iv. Assimilation efficiency
Eleven chromosomes were associated with variation in assimilation efficiency (Supplementary Figures 4a & b). Each chromosomes accounted for 6 to 20 percent of the interspecific difference; PVE ranged from 0.4 to 4.6 percent. Only one chromosome (H03) had effects in both the VS01 and VS07 populations, and two (H19 and H20) had effects in VS01 but not in VS07. The eight remaining chromosomes had effects in VS07 only. One chromosome (H13) had exclusively H. virescens-like effects (its presence was associated with a decrease in assimilation efficiency), and two others had different effects in different populations: H03 had H. subflexa-like effects in VS07 but H. virescens-like effects in VS01, and H16 had H. subflexa-like effects in females but H. virescens-like effects in males. The remaing chromosomes all had H. subflexa-like effects.
Discussion
Twenty chromosomes were associated with variation in the traits we measured, and forty chromosome-phenotype associations of at least suggestive significance were detected. Several major patterns were apparent in our results. First, our ability to detect the chromosomes affecting each trait depended on sample size; second, most chromosomes had small effects; third, most chromosomes affected more than one trait; fourth, some chromosomes had effects in the “wrong” direction; finally, many chromosomes had context-dependent effects. We discuss these patterns below.
The effect of sample size on chromosome detection
In 2007, we analyzed 1,147 insects, an almost four-fold increase compared to our 2001 study. The increase in sample size was nearly matched in scale by the increase in the number of chromosomes detected (11 in VS01, 35 in VS07), a striking result that has been observed in at least one other study (Turri et al. 2001a; Turri et al. 2001b).
Chromosome effect sizes
The effects of suggestive (0.05 < p < 0.1) and significant (p < 0.05) chromosomes were quite similar, although the chromosomes of largest effect tended to be significant rather than suggestive. Suggestive chromosomes explained an average of 21 percent of the interspecific difference, with values ranging from 1.5 to 86 percent, while significant chromosomes explained an average of 32 percent and ranged from 1.7 to 165 percent.
Chromosome effects were small when measured as PVE, but larger in relation to the interspecific difference explained. The two measures of effect size, though of different magnitudes, were correlated (Supplementary Table 9). This tendency for loci to explain more of the phenotypic difference between species than within the mapping population is common in studies of non-domesticated animals (Morjan and Rieseberg 2004).
Chromosome effect sizes were larger in VS01 than in VS07, consistent with the “Beavis effect,” in which QTL effects are overestimated, and the number of QTL underestimated, when sample sizes are small (Beavis 1998; Xu 2003). This pattern held true for all of traits for which chromosomes were found in both years, and across the suggestive and significant chromosome groups: in VS01, the average chromosome explained 37 percent of the interspecific difference, while in VS07, the average was 29 percent. We compared the effect sizes of the six chromosomes which affected the same trait in both years, and found the same pattern.
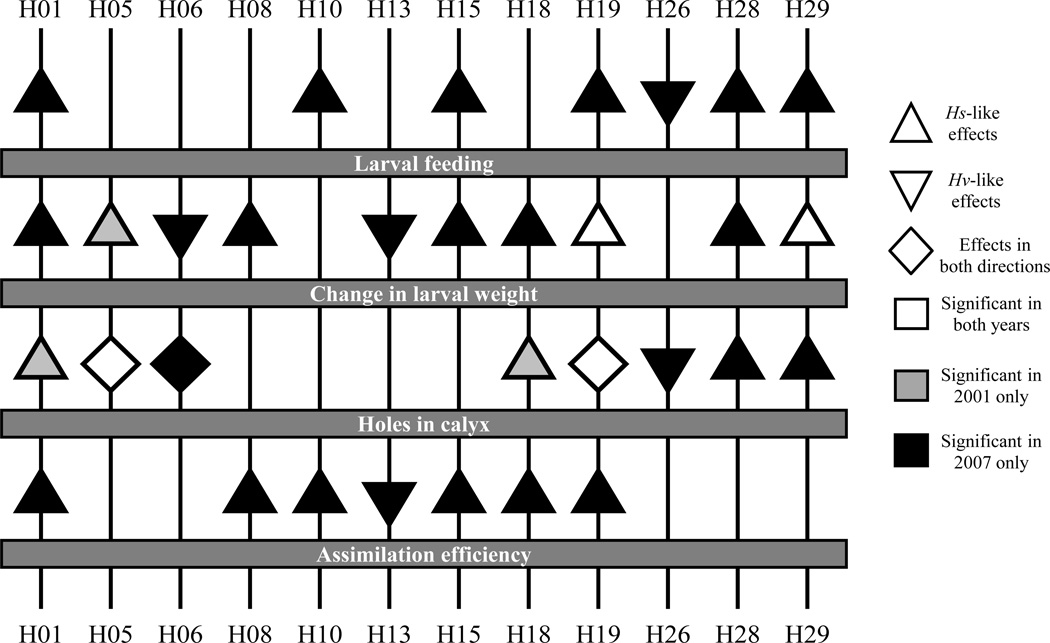
Most chromosomes affect multiple traits
Of the twenty chromosomes of at least suggestive significance, twelve were associated with variation in two or more traits (Figure 4). Given that each chromosome is likely to contain on the order of 500 transcribed genes, it is not surprising that individual chromosomes affect multiple traits, and finding that a single chromosome affects multiple traits does not mean that a single gene affects multiple traits. Indeed, even small genetic regions often harbor more than one causal locus (Kroymann and Mitchell-Olds 2005; Studer and Doebley 2011), so it is quite possible that the “pleiotropy” we observed results from multiple coding or regulatory sequences on a chromosome.
Figure 4.
Twelve chromosomes with effects on two or more of the phenotypic traits measured. Symbol shapes indicate the direction of the effect; symbol fill colors indicate the year(s) in which the effect was significant.
Nonetheless, the strong phenotypic correlation between traits in the VS insects (Table 2) suggests that some loci affecting different aspects of host plant use are, if not pleiotropic, at least genetically correllated, such that single chromosomes contain genes for several different traits. Work in other systems has demonstrated that loci affecting related traits do sometimes cluster together (Shaw and Lesnick 2009; Ferguson et al. 2010; Gould et al. 2010).
Antagonistic chromosomes
Twenty percent of the effects of the introgressed H. subflexa chromosomes were in the H. virescens direction. Effect sizes were similar between chromosomes associated with H. subflexa-like and H. virescens-like phenotypes. The surprising finding that chromosomes from H. subflexa can have H. virescens-like effects suggests that there may be standing variation within H. subflexa that encompasses H. virescens phenotypes. Such “antagonistic” loci (those in the opposite direction of the difference between parental species) have been found in other organisms (Gardner and Latta 2007; Albert et al. 2008), and are expected when variation in adaptive traits is controlled by genes with pleiotropic effects, because selection on a given trait may lead to the incidental fixation of antagonistic effects (Griswold and Whitlock 2003). Alternatively, some H. subflexa chromosomes may have allelic or epistatic interactions with the H. virescens background into which they were introgressed.
Context-dependent chromosome effects
The effects of H. subflexa-origin chromosomes varied between sexes, years, and genetic backgrounds (Figure 5), suggesting that similar phenotypes were not necessarily produced by the same chromosomes.
Figure 5.
The effects and significance of some chromosomes differed between years, sexes, and families. Shown here are examples of chromosomes with similar and opposite effects in different populations. Bars are the additive effect of chromosome presence on larval phenotype (i.e. the mean phenotypic difference between chromosome-present and chromosome-absent larvae). Significance levels are indicated by the symbol above each bar: Green dot = Significant (p < 0.05); Red dot = Suggestive (0.05 < p < 0.1); ns = not significant.
i. Different chromosomes in males and females
More than three-quarters of the chromosomes had different effects in males versus females. Research on model organisms has shown that the same alleles can have opposite effects in males and females (Nuzhdin et al. 1997; Ober et al. 2008). While we found three chromosomes that fit this pattern, the other fourteen that differed between the sexes had effects in the same direction in both sexes but were significant in only one sex. In these cases, it is unclear whether our results are an effect of sample size (such that a larger experiment would find statistical significance in both sexes) or reflect a true difference between the sexes.
ii. Different chromosomes in different years
Thirty-five chromosome-phenotype associations were detected in 2007, eleven in 2001. Six of these involved chromosomes that were significant in both years and for the same traits, though only three had effects in the same direction in both years. One chromosome (H20) was significant only in 2001. The five remaining chromosomes detected in VS01 also had effects in VS07, but affected different traits in each year.
The difference in sample size probably accounts for much of the difference in the number of associations detected, but an alternative explanation is that the effect of H. subflexa-origin chromosomes depends upon the genetic background into which they are introgressed. Because we did not map H. virescens-origin chromosomes, we cannot test the effects of variation within H. virescens on the phenotypes observed. We do, however, know from other experiments (Oppenheim, in prep.) that H. virescens harbors significant intraspecific variation for use of Physalis.
A third possibility is that there is intraspecific variation in H. subflexa for use of Physalis. If multiple alleles are associated with the loci on a chromosome, the effect of the chromosome will differ depending on which allele is present. Such allelic series have been described in maize, where the presence of multiple variants at a shared QTL results in very different effects of the same QTL in different populations (Buckler et al. 2009).
iii. Different chromosomes in different families
Twenty-nine chromosome-phenotype associations had effects that differed among the five VS07 families (Supplementary Figures 1–4). In many cases, the effects were in the same direction in all families, even where their effects were not significant. In other cases, however, a chromosome had strong effects in one family and no effect in others, possibly reflecting truly different effects of the same chromosome in different populations.
Even within the VS07C lineage, we found 20 associations whose effects or significance levels varied among families. The three VS07C families represent a limited set of haplotypes, because they are descendants of a single grandparental cross and because all F1 dams were sisters and all sires were brothers. Thus, we were interested to find variation not just in the significance of the chromosomes associated with phenotypic variation, but also in which chromosomes were associated with a trait and in the direction of phenotypic effects. In several cases, a chromosome’s effects were in opposite directions in different families.
We are unsure how to interpret such diversity of chromosome effects among closely related families. In other systems, intraspecific variation in complementary QTL (different regions within a chromosome that contain QTL acting in opposite directions) results in QTL with individual effects that are far larger than the effect of the chromosome as a whole (Lexer et al. 2005). In our system, the individual effects of complementary QTL would be hidden by the lack of recombination, and only their cumulative phenotypic effect would be seen. For different sets of complementary QTL to explain why the effects of a single chromosome vary within a lineage, these sets would have to be segregating in the F1 population so that a female might inherit either +/+ or +/− QTL on a chromosome.
Whatever the explanation, the same H. subflexa-like phenotype could be produced by introgressing different H. subflexa chromosomes into the H. virescens background. It is possible that there are many undetected loci contributing to these phenotypes, such that the loci underlying the phenotype are actually the same in all populations, but different subsets happen to be identified in each population. On the other hand, while identical phenotypes may sometimes be controlled by the same genes in different populations (Piertney and Webster 2010), there is accumulating evidence that “genotypic equivalence” (Weiss 2008)—in which a variety of genotypes can confer the same phenotype—is widespread (Arbuthnott 2009; DeFaveri et al. 2011; Elmer and Meyer 2011). Thus it may well be that host use traits in the different families we studied are indeed controlled by different chromosomes.
The evolution and genetic architecture of host use
Host use differences between the specialist H. subflexa and the generalist H. virescens involve variation in neurosensory, physiological, and behavioral traits. Given that we investigated traits in each of these categories, we were curious as to whether different categories would show evidence of differing genetic architectures.
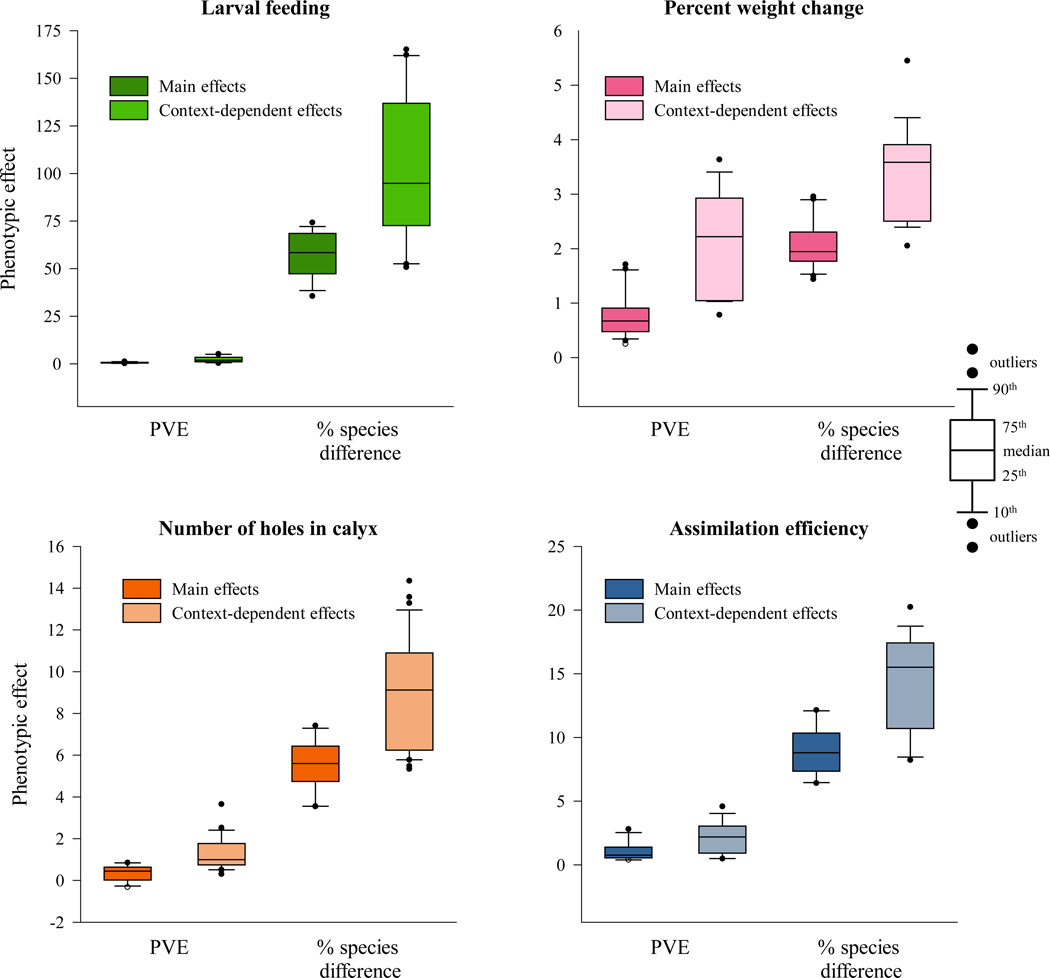
The number of chromosomes affecting each trait was similar, as was the fraction of chromosomes with H. subflexa-like versus H. virescens-like effects. We also found similar levels of context dependence. Effect sizes were strikingly larger within particular populations (e.g. in one sex or one family) than in the population as a whole, and average effect sizes were higher for context-dependent chromosomes than for main effect chromosomes (Figure 6).
Figure 6.
Chromosome effects compared across four phenotypic traits. Results are shown separately for main effect and context-dependent chromosome effects.
Chromosome effect sizes did differ between traits, but not in a manner that suggested an overall distinction between neurosensory/behavioral and physiological traits. For one behavioral trait, larval willingness to feed on Physalis, each chromosome accounted for an average of 91 percent of the interspecific difference, but effect sizes for the second behavioral trait, the number of holes bored in the calyx of Physalis, were more modest (8 percent, on average). Similarly, chromosome effect sizes for one physiological trait, the percent change in larval weight, were noticeably lower than for the other traits examined (accounting for an average of 3 percent of the interspecific difference), but effect sizes were somewhat higher for larval assimilation efficiency, the second physiological trait (13 percent, on average).
Such relatively small effects are hard to reconcile with a simple explanation for physiological adaptation to Physalis, such as tolerance of a host plant defense compound. Instead, it seems that physiological performance on Physalis improves by incremental steps, allowing natural selection to tinker with the component traits. While this pattern reduces the likelihood that a candidate gene approach will succeed in identifying the genetic basis of interspecific variation in performance on Physalis, it is consistent with previous findings that Physalis-specific compounds have anti-feedant rather than acutely toxic effects on insect herbivores (Ascher et al. 1987; Mareggiani et al. 2000; Mareggiani et al. 2001; Bado et al. 2004).
While differences between H. subflexa and H. virescens in willingness to feed on Physalis were much less extreme than those witnessed in the D. sechellia / D. simulans system (in which the generalist is actively repelled by the specialist’s host plant), the cumulative effects of the main effect chromosomes we found completely account for the interspecific difference in this trait.
The second behavioral trait, the number of holes in the calyx of Physalis, was designed as a laboratory-based measure of a behavioral adaptation we identified in a previous study. The significant chromosomes we found explain an average of eight percent of the interspecific difference in the number of holes, and the main effect chromosomes cumulatively explain thirty-four percent. Interestingly, the value of this behavior may be largely independent of physiological performance on Physalis. While larval feeding and the number of holes were phenotypically correlated (increases in hole number were associated with decreases in the likelihood of feeding), we did not find evidence that the two traits are under shared genetic control. In the proper ecological circumstances (i.e., when selection pressure from natural enemies is high) these two traits might evolve independently: larvae that have poor physiological performance on Physalis but are effective at using it as an escape from natural enemies would be favored over those who feed more efficiently but leave themselves vulnerable to parasitoids.
What implications do our results have for the evolution of host use? When one considers the overall architecture of Physalis use it appears that adaptation to this host may be a mosaic of many minor adaptive loci. Recent work in other insect herbivores (Forister et al. 2007) suggests that host plant use consists of a large number of genetically uncorrelated traits, and Nosil has hypothesized that even relatively weak ecological selection can promote divergence and speciation when many genetically independent traits are subject to selection (Nosil et al. 2009). In the H. subflexa / H. virescens system, it does not appear that genetic changes of large effect (as might be involved in cases where the novel host has novel toxins) are required for the use of Physalis by a non-adapted species like H. virescens. Instead, we suggest that behavioral changes probably paved the way for the generalist ancestor of H. subflexa and H. virescens to adapt to Physalis. Willingness to feed on Physalis is an obvious prerequisite to larval adaptation to Physalis, because any variation in other host use traits will be functionally silent in the absence of feeding. An environment rich in natural enemies, or one in which Physalis species were more abundant (or reliable) than other potential hosts, would exert pressure for even physiologically unadapted larvae to use Physalis. Since Physalis’s defense compounds do not appear to be toxic to unadapted larvae, a genetic change that increased behavioral preference for Physalis could open up a niche with fewer natural enemies (or competitors) and result in selection for improved physiological performance. Whether such selection on a generalist like H. virescens could lead to H. subflexa-like phenotypes, and whether the genetic architecture of these phenotypes would resemble that seen in the present study, is the subject of ongoing research in our lab.
Supplementary Material
Acknowledgements
We thank M. Estock, J. L. Emerson, and M. Cortes-Cruz for developing primers and for their assistance in genotyping many insects, and are grateful to Kathryn Lanier for help in rearing and maintaining them. We thank L. Delph, C. Peichl, and two anonymous reviewers for their thoughtful comments and suggestions, and appreciate the help of J. Mahar in navigating the submission process. SJO thanks George Kennedy, Trudy Mackay, and Coby Schal in the warmest possible terms for service beyond the call of duty as her Ph.D. committee members. Funding was provided by National Science Foundation grant DEB9981671 to FG, and by National Institutes of Health grant GM068991, an Eloise Gerry Fellowship from Sigma Delta Epsilon Graduate Women in Science, and a Professional Research Enhancement Award from the W. M. Keck Center for Behavioral Biology, North Carolina State University, to SJO.
Literature cited
- Albert AYK, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Balabhadra S, Kingsley DM, Schluter D. The genetics of adaptive shape shift in stickleback: Pleiotropy and effect size. Evolution. 2008;62:76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- Althoff DM, Gitzendanner MA, Segraves KA. The Utility of Amplified Fragment Length Polymorphisms in Phylogenetics: A Comparison of Homology within and between Genomes. Systematic Biology. 2007;56:477–484. doi: 10.1080/10635150701427077. [DOI] [PubMed] [Google Scholar]
- Arbuthnott D. The genetic architecture of insect courtship behavior and premating isolation. Heredity. 2009;103:15–22. doi: 10.1038/hdy.2009.22. [DOI] [PubMed] [Google Scholar]
- Ascher KRS, Eliyahu M, Glotter E, Goldman A, Kirson I, Abraham A, Jacobson M, Schmutterer H. The antifeedant effect of some new withanolides on 3 insect species, Spodoptera littoralis, Epilachna varivestis, and Tribolium castaneum. Phytoparasitica. 1987;15:15–29. [Google Scholar]
- Bado S, Mareggiani G, Amiano N, Burton G, Veleiro AS. Lethal and sublethal effects of withanolides from Salpichroa origanifolia and analogues on Ceratitis capitata. Journal of Agricultural and Food Chemistry. 2004;52:2875–2878. doi: 10.1021/jf035508a. [DOI] [PubMed] [Google Scholar]
- Bateman ML. Entomology. Raleigh: North Carolina State University; 2006. Impact of Plant Suitability, Biogeography, and Ecological Factors on Associations between the Specialist Herbivore Heliothis subflexa G. (Lepidoptera: Noctuidae) and the Species in its Host Genus, Physalis L. (Solanaceae), in West-Central Mexico. [Google Scholar]
- Beavis WD. QTL analyses: Power, precision, and accuracy. Molecular dissection of complex traits. 1998:145–162. [Google Scholar]
- Berenbaum MR, Feeny PP. Chemical mediation of host-plant specialization-the papillionid paradigm. In: Tilmon KJ, editor. Specialization, Speciation and Radiation: the Evolutionary Biology of Herbivorous Insects. Berkeley: University of California Press; 2008. [Google Scholar]
- Bernays E, Graham M. ON THE EVOLUTION OF HOST SPECIFICITY IN PHYTOPHAGOUS ARTHROPODS. Ecology. 1988;69:886–892. [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, Goodman MM, Harjes C, Guill K, Kroon DE, Larsson S, Lepak NK, Li HH, Mitchell SE, Pressoir G, Peiffer JA, Rosas MO, Rocheford TR, Romay MC, Romero S, Salvo S, Villeda HS, da Silva HS, Sun Q, Tian F, Upadyayula N, Ware D, Yates H, Yu JM, Zhang ZW, Kresovich S, McMullen MD. The Genetic Architecture of Maize Flowering Time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- Burton RL. A Low-Cost Artificial Diet for the Corn Earworm. Journal of Economic Entomology. 1970;63:1969–1970. [Google Scholar]
- Caillaud MC, Via S. Quantitative genetics of feeding behavior in two ecological races of the pea aphid, Acyrthosiphon pisum. Heredity. 2012;108:211–218. doi: 10.1038/hdy.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LX, He H, Qiu F. Natural withanolides: an overview. Natural Product Reports. 2011;28:705–740. doi: 10.1039/c0np00045k. [DOI] [PubMed] [Google Scholar]
- Cho S, Mitchell A, Mitter C, Regier J, Matthews M, Robertson R. Molecular phylogenetics of heliothine moths (Lepidoptera: Noctuidae: Heliothinae), with comments on the evolution of host range and pest status. Systematic Entomology. 2008;33:581–594. [Google Scholar]
- Cho SW, Mitchell A, Regier JC, Mitter C, Poole RW, Friedlander TP, Zhao SW. A highly conserved nuclear gene for low-level phylogenetics - elongation factor-1-alpha recovers morphology-based tree for heliothine moths. Molecular Biology and Evolution. 1995;12:650–656. doi: 10.1093/oxfordjournals.molbev.a040244. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFaveri J, Shikano T, Shimada Y, Goto A, Merila J. Global analysis of genes involved in freshwater adaptation in threespine sticklebacks (Gasterosteus aculeatus) Evolution. 2011;65:1800–1807. doi: 10.1111/j.1558-5646.2011.01247.x. [DOI] [PubMed] [Google Scholar]
- Dekker T, Ibba I, Siju KP, Stensmyr MC, Hansson BS. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D-sechellia. Current Biology. 2006;16:101–109. doi: 10.1016/j.cub.2005.11.075. [DOI] [PubMed] [Google Scholar]
- Dethier VG. Evolution of feeding preferences in phytophagous insects. Evolution. 1954;8:33–54. [Google Scholar]
- Dworkin I, Jones CD. Genetic Changes Accompanying the Evolution of Host Specialization in Drosophila sechellia. Genetics. 2009;181:721–736. doi: 10.1534/genetics.108.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley EJ, Jones CD. Next-Generation Mapping of Complex Traits with Phenotype-Based Selection and Introgression. Genetics. 2011;189:1203. doi: 10.1534/genetics.111.129445. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Rollmann SM, Morgan TJ, Mackay TFC. Quantitative genomics of aggressive behavior in Drosophila melanogaster. Plos Genetics. 2006;2:1386–1395. doi: 10.1371/journal.pgen.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D, Berry JJ. THE EFFICIENCY OF SIMULATION-BASED MULTIPLE COMPARISONS. Biometrics. 1987;43:913–928. [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, Kruglyak L. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039-U1101. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich PR, Raven PH. Butterflies and plants--a study in coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- Eich E. Solanaceae and convolvulaceae : secondary metabolites : biosynthesis, chemotaxonomy, biological and economic significance : a handbook. Berlin: Springer; 2008. [Google Scholar]
- Elmer KR, Meyer A. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends in Ecology & Evolution. 2011;26:298–306. doi: 10.1016/j.tree.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Etges WJ, de Oliveira CC, Noor MAF, Ritchie MG. GENETICS OF INCIPIENT SPECIATION IN DROSOPHILA MOJAVENSIS. III. LIFE-HISTORY DIVERGENCE IN ALLOPATRY AND REPRODUCTIVE ISOLATION. Evolution. 2010;64:3549–3569. doi: 10.1111/j.1558-5646.2010.01096.x. [DOI] [PubMed] [Google Scholar]
- Fang QQ, Cho S, Regier JC, Mitter C, Matthews M, Poole RW, Friedlander TP, Zhao S. A new nuclear gene for insect phylogenetics: DOPA carboxylase is informative of relationships within Heliothinae (Lepidoptera:Noctuidae) Systematic Biology. 1997;46:269–283. doi: 10.1093/sysbio/46.2.269. [DOI] [PubMed] [Google Scholar]
- Ferguson L, Lee SF, Chamberlain N, Nadeau N, Joron M, Baxter S, Wilkinson P, Papanicolaou A, Kumar S, Kee TJ, Clark R, Davidson C, Glithero R, Beasley H, Vogel H, Ffrench-Constant R, Jiggins C. Characterization of a hotspot for mimicry: assembly of a butterfly wing transcriptome to genomic sequence at the HmYb/Sb locus. Molecular Ecology. 2010;19:240–254. doi: 10.1111/j.1365-294X.2009.04475.x. [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Willis JH. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution. 2002;56:2138–2155. doi: 10.1111/j.0014-3820.2002.tb00139.x. [DOI] [PubMed] [Google Scholar]
- Forister ML, Ehmer AG, Futuyma DJ. The genetic architecture of a niche: variation and covariation in host use traits in the Colorado potato beetle. Journal of Evolutionary Biology. 2007;20:985–996. doi: 10.1111/j.1420-9101.2007.01310.x. [DOI] [PubMed] [Google Scholar]
- Gardner KM, Latta RG. Shared quantitative trait loci underlying the genetic correlation between continuous traits. Molecular Ecology. 2007;16:4195–4209. doi: 10.1111/j.1365-294X.2007.03499.x. [DOI] [PubMed] [Google Scholar]
- Gould F, Estock M, Hillier NK, Powell B, Groot AT, Ward CM, Emerson JL, Schal C, Vickers NJ. Sexual isolation of male moths explained by a single pheromone response QTL containing four receptor genes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8660–8665. doi: 10.1073/pnas.0910945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold CK, Whitlock MC. The genetics of adaptation: The roles of pleiotropy, stabilizing selection and drift in shaping the distribution of bidirectional fixed mutational effects. Genetics. 2003;165:2181–2192. doi: 10.1093/genetics/165.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics. 1922;12:101–109. [Google Scholar]
- Hawthorne DJ, Via S. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature. 2001;412:904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- Heidel-Fischer HM, Vogel H, Heckel DG, Wheat CW. Microevolutionary dynamics of a macroevolutionary key innovation in a Lepidopteran herbivore. Bmc Evolutionary Biology. 2010;10 doi: 10.1186/1471-2148-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henniges-Janssen K, Reineke A, Heckel DG, Groot AT. Complex inheritance of larval adaptation in Plutella xylostella to a novel host plant. Heredity. 2011;107:421–432. doi: 10.1038/hdy.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpenko CP, Proshold FI. Fertility and mating performance of interspecific crosses between Heliothis virescens and H. subflexa (Lepidoptera: Noctuidae) backcrossed for 3 generations to H. subflexa. Annals of the Entomological Society of America. 1977;70:737–740. [Google Scholar]
- Kroymann J, Mitchell-Olds T. Epistasis and balanced polymorphism influencing complex trait variation. Nature. 2005;435:95–98. doi: 10.1038/nature03480. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES. A nonparametric approach for mapping quantitative trait loci. Genetics. 1995;139:1421–1428. doi: 10.1093/genetics/139.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laster ML, Pair SD, Martin DF. Acceptance and development of Heliothis subflexa and Heliothis virescens (Lepidoptera, Noctuidae), and their hybrid and backcross progeny on several plant species. Environmental Entomology. 1982;11:979–980. [Google Scholar]
- Lexer C, Rosenthal DM, Raymond O, Donovan LA, Rieseberg LH. Genetics of species differences in the wild annual sunflowers, Helianthus annuus and H-petiolaris. Genetics. 2005;169:2225–2239. doi: 10.1534/genetics.104.031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loxdale HD. Rapid genetic changes in natural insect populations. Ecological Entomology. 2010;35:155–164. [Google Scholar]
- Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nature Reviews Genetics. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Marec F, Sahara K, Traut W. Molecular biology and genetics of the Lepidoptera. Boca Raton, FL: CRC Press/Taylor & Francis; 2010. Rise and Fall of the W Chromosome in Lepidoptera; pp. 49–64. [Google Scholar]
- Mareggiani G, Picollo MaI, Veleiro AS, Tettamanzi MaC, Benedetti-Doctorovich MOV, Burton G, Zerba E. Response of Tribolium castaneum (Coleoptera, Tenebrionidae) to Salpichroa origanifolia Withanolides. Journal of Agricultural and Food Chemistry. 2001;50:104–107. doi: 10.1021/jf010766y. [DOI] [PubMed] [Google Scholar]
- Mareggiani G, Picollo MI, Zerba E, Burton G, Tettamanzi MC, Benedetti-Doctorovich MOV, Veleiro AS. Antifeedant activity of withanolides from Salpichroa origanifolia on Musca domestica. Journal of Natural Products. 2000;63:1113–1116. doi: 10.1021/np0001068. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. Plos Biology. 2007;5:985–996. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SG, Huettel MD, Davis M-TB, Weber EH, Weber LA. Male sterility in Heliothis virescens x H. subflexa backcross hybrids. Molecular and General Genetics MGG. 1986;203:451–461. [Google Scholar]
- Mitter C, Farrell B, Futuyma DJ. Phylogenetic studies of insect plant interactions--insights into the genesis of diversity. Trends in Ecology & Evolution. 1991;6:290–293. doi: 10.1016/0169-5347(91)90007-K. [DOI] [PubMed] [Google Scholar]
- Mitter C, Poole RW, Matthews M. Biosystematics of the Heliothinae (Lepidoptera: Noctuidae) Annual Review of Entomology. 1993;38:207–225. [Google Scholar]
- Morjan CL, Rieseberg LH. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Molecular Ecology. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends in Ecology & Evolution. 2009;24:145–156. doi: 10.1016/j.tree.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Pasyukova EG, Dilda CL, Zeng ZB, Mackay TFC. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9734–9739. doi: 10.1073/pnas.94.18.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nature Reviews Genetics. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim SJ, Gould F. Behavioral adaptations increase the value of enemy-free space for Heliothis subflexa, a specialist herbivore. Evolution. 2002a;56:679–689. doi: 10.1111/j.0014-3820.2002.tb01379.x. [DOI] [PubMed] [Google Scholar]
- Oppenheim SJ, Gould F. Is attraction fatal? The effects of herbivore-induced plant volatiles on herbivore parasitism. Ecology. 2002b;83:3416–3425. [Google Scholar]
- Piertney SB, Webster LMI. Characterising functionally important and ecologically meaningful genetic diversity using a candidate gene approach. Genetica. 2010;138:419–432. doi: 10.1007/s10709-008-9322-2. [DOI] [PubMed] [Google Scholar]
- Poole RW, Mitter C, Huettel MD. A revision and cladistic analysis of the Heliothis virescens species group (Lepidoptera: Noctuidae) with a preliminary morphometric analysis of H. virescens. Mississippi Agriculture and Forestry Experiment Station Bulletin. 1993 [Google Scholar]
- Scriber JM, Larsen ML, Allen GR, Walker PW, Zalucki MP. Interactions between Papilionidae and ancient Australian Angiosperms: evolutionary specialization or ecological monophagy? Entomologia Experimentalis Et Applicata. 2008;128:230–239. [Google Scholar]
- Shaw KL, Lesnick SC. Genomic linkage of male song and female acoustic preference QTL underlying a rapid species radiation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9737–9742. doi: 10.1073/pnas.0900229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheck AL, Gould F. The genetic basis of host range in Heliothis virescens - larval survival and growth. Entomologia Experimentalis et Applicata. 1993;69:157–172. [Google Scholar]
- Sheck AL, Groot AT, Ward CM, Gemeno C, Wang J, Brownie C, Schal C, Gould F. Genetics of sex pheromone blend differences between Heliothis virescens and Heliothis subflexa: a chromosome mapping approach. Journal of Evolutionary Biology. 2006;19:600–617. doi: 10.1111/j.1420-9101.2005.00999.x. [DOI] [PubMed] [Google Scholar]
- Studer AJ, Doebley J. Do Large Effect QTLs Fractionate? A Case Study at the Maize Domestication QTL teosinte branched1. Genetics. 2011 doi: 10.1534/genetics.111.126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Zawadzki J, Black B, Kreitman M. Genome Size And Endopolyploidy In Pyrethroid-Resistant And Susceptible Strains Of Heliothis-Virescens (Lepidoptera, Noctuidae) Journal Of Economic Entomology. 1993;86:1030–1034. [Google Scholar]
- Turri MG, Datta SR, DeFries J, Henderson ND, Flint J. QTL analysis identifies multiple behavioral dimensions in ethological tests of anxiety in laboratory mice. Current Biology. 2001a;11:725–734. doi: 10.1016/s0960-9822(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Turri MG, Henderson ND, DeFries JC, Flint J. Quantitative trait locus mapping in laboratory mice derived from a replicated selection experiment for open-field activity. Genetics. 2001b;158:1217–1226. doi: 10.1093/genetics/158.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Hof AE, Marec F, Saccheri IJ, Brakefield PM, Zwaan BJ. Cytogenetic Characterization and AFLP-Based Genetic Linkage Mapping for the Butterfly Bicyclus anynana, Covering All 28 Karyotyped Chromosomes. Plos One. 2008:3. doi: 10.1371/journal.pone.0003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP - A new technique for DNA-fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KM. Tilting at Quixotic Trait Loci (QTL): An Evolutionary Perspective on Genetic Causation. Genetics. 2008;179:1741–1756. doi: 10.1534/genetics.108.094128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat CW, Vogel H, Wittstock U, Braby MF, Underwood D, Mitchell-Olds T. The genetic basis of a plant-insect coevolutionary key innovation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20427–20431. doi: 10.1073/pnas.0706229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer BJ. Statistical Principles in Experimental Design. New York: McGraw-Hill; 1971. [Google Scholar]
- Wink M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry. 2003;64:3–19. doi: 10.1016/s0031-9422(03)00300-5. [DOI] [PubMed] [Google Scholar]
- Wisotsky Z, Medina A, Freeman E, Dahanukar A. Evolutionary differences in food preference rely on Gr64e, a receptor for glycerol. Nat Neurosci. 2011;14:1534–1541. doi: 10.1038/nn.2944. [DOI] [PubMed] [Google Scholar]
- Xu SZ. Theoretical basis of the Beavis effect. Genetics. 2003;165:2259–2268. doi: 10.1093/genetics/165.4.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42:565-U131. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.