Abstract
The naturally occurring polyamines putrescine, spermidine or spermine are ubiquitous in all cells. Although polyamines have prominent regulatory roles in cell division and growth, precise molecular and cellular functions are not well established in vivo. In this work we have performed microarray experiments with a spermidine synthase, spermine oxidase mutant (Δspe3 Δfms1) strain to investigate the responsiveness of yeast genes to supplementation with spermidine or spermine. Expression analysis identified genes responsive to the addition of either excess spermidine (10−5 M) or spermine (10−5 M) compared to a control culture containing 10−8 M spermidine. 247 genes were up-regulated >2-fold, and 11 genes were up-regulated more than 10-fold after spermidine addition. Functional categorization of the genes showed induction of transport related genes, and genes involved in methionine, arginine, lysine, NAD and biotin biosynthesis. 268 genes were down-regulated more than 2-fold, and 6 genes were down-regulated more than 8-fold after spermidine addition. A majority of the down-regulated genes are involved in nucleic acid metabolism and various stress responses. In contrast, only few genes (18) were significantly responsive to spermine. Thus, results from global gene expression profiling demonstrate a more major role for spermidine in modulating gene expression in yeast than spermine.
Keywords: Microarray, polyamines, spermidine, spermine, yeast
Introduction
Polyamines are found in essentially all cellular organisms and intracellular concentrations are controlled at various steps (Tabor and Tabor, 1984; Pegg, 1986; Cohen, 1998; Igarashi and Kashiwagi, 2000). The essential role of polyamines has been shown by using various inhibitors of polyamine biosynthesis or by using mutants in the biosynthetic pathways and they have been shown to have a variety of phenotypic effects (Mamont et al., 1981; Diala et al., 1980; Tabor and Tabor, 1984; Pitkin and Davis, 1990; Cohen, 1998). In addition to the ionic interaction of polyamines with negatively charged macromolecules (such as nucleic acids, membranes and ion channels), polyamines can be covalently incorporated into proteins. One of the important functions of spermidine is its role as a substrate for the hypusine modification of a 17–18,000 Da protein, known as eukaryotic initiation factor 5A (eIF5A), which is a ubiquitous initiation factor in eukaryotic cells (Park et al., 1981; Park, 2006). In our previous studies we have constructed mutants of S. cerevisiae where different steps in the biosynthetic pathway for polyamines have been deleted, and have shown that spermidine is essential for the growth of these auxotrophs (Balasundaram et al., 1991; Balasundaram et al., 1993; Balasundaram et al., 1999; Hamasaki-Katagiri et al., 1997; Chattopadhyay et al., 2002; Chattopadhyay et al., 2006). More recently we have found that very low concentrations of spermidine (10−8 M) are sufficient to support near-normal growth and hypusine formation of these auxotrophs of S. cerevisiae, even though under these conditions the cells contain only about 1/1000 of the internal spermidine concentration found in a wild type strain (Chattopadhyay et al., 2008). In the current study we have taken advantage of global gene expression analysis using microarrays to address the question of why wild type cells normally contain concentrations of spermidine that are so much higher than is required for the growth of a spermidine auxotroph and what are the molecular targets or metabolic pathways affected by excess polyamines. We have compared the changes in gene expressions when a higher concentration of either spermidine or spermine was added to cultures growing at a near normal growth rate with 10−8 M spermidine. We have purposely not made this comparison between cells that were not growing because they were completely deprived of spermidine and cells growing normally after spermidine supplementation, since such a comparison would obviously be complicated by the profound changes in gene expression expected in growing versus non-growing cells.
Materials and methods
Growth condition
For all the experiments in this paper Y549 (Matα met15 leu2 ura3 spe3Δ::URA3 fms1Δ::KANMX, (Chattopadhyay et al., 2003) strain was used and all cultures were grown at 30° C with shaking in air. As described earlier, this strain cannot synthesize spermidine due to lack of spermidine synthase (SPE3, Hamasaki-Katagiri et al., 1997) and cannot oxidize spermine to spermidine due to deletion of FMS1, the gene encoding yeast spermine oxidase (White et al., 2001; Chattopadhyay et al., 2003). Yeast cultures were grown routinely in plates containing yeast extract, peptone, dextrose and agar (YPAD). To deplete intracellular amines carried over from YPAD plates, the cultures were inoculated into SD medium (0.67% yeast nitrogen base, required amino acid supplements, 2% dextrose) supplemented with 10−8 M spermidine and grown for more than 24 h with several dilutions. The culture was then diluted 1:2000 fold into SD medium containing 10−8 M spermidine, divided into three parts and incubated overnight. When the cultures reached to an OD600 of 0.1, in one part 10−5 M spermidine was added; in another part 10−5 M purified spermine (purified by chromatography on a Dowex 50 column, (Chattopadhyay et al., 2003) was added; the third part was kept as a control without any addition. These three cultures were then grown for another 6 h at 30° C and harvested for RNA isolation. Intracellular concentrations of polyamines after incubation were determined as previously described (Chattopadhyay et al., 2008).
Isolation of RNA
Total RNA was isolated according to the protocol described in the RNeasy mini kit (Qiagen, Germantown, MD). 107–108 cells from the yeast cultures, grown and harvested as described above, were washed with 1.2 M sorbitol solution (1.2 M sorbitol, 0.1% β-mercaptoethanol, 0.1M EDTA) and resuspended in the same solution containing 50U of lyticase (Sigma) per 107 cells. The cell suspensions were incubated for 10 min. at 30° C and the resulting spheroplasts were immediately processed for RNA isolation. The quantity and quality of RNA were evaluated by OD260/OD280 assays and by RNA capillary electrophoresis (Agilent Biotechnologies).
Microarray, Statistics and Bioinformatics
Total RNAs were reverse transcribed by Superscript II (Invitrogen). Oligo (dT) linked to T7 RNA polymerase promoter sequence was used to prime cDNA synthesis. After 2nd strand synthesis, biotinylated cRNA was made by in vitro transcription using biotinylated UTP and CTP (Bioarray high-yield RNA transcript labeling kit, ENZO Diagnostics) and purified with RNeasy columns (Qiagen). The biotinylated cRNA was heated in a buffer containing100 mM potassium acetate, 30 mM magnesium acetate, 40 mM Tris-acetate, pH 8.0 to produce 35–200 base fragments. Affymetrix (Santa Clara, CA) GeneChip S-98 arrays (~7000 genes) (n= 5 for control and spermidine treated samples and 3 for spermine treated samples) were hybridized for 16 h at 45° C with 15 μg of fragmented cRNA. Arrays were stained with streptavidin-phycoerythrin conjugate (Molecular Probes) and visualized with Gene array scanner (Agilent, Palo Alto, CA). Probe profiler software version 3.4 was used to convert hybridization intensity data into quantitative estimates of gene expression Gene probes that were not expressed in any of the samples were considered absent and were not included for further analyses The microarray signals were analyzed using the Affymetrix RMA algorithm. Comparisons of expression analyses were performed using Affimetrix MAS version 5.0 software according to the manufacturer’s method. Up- and down-regulated genes were selected based on P values of <0.05 and fold change >+2 or −2 as assessed by two-way ANOVA using Partek Pro software (Partek, St. Charles, MO, USA). Hypothetical genes and unknown ORFs were not considered in the current analyses. The complete microarray data can be obtained from (GEO accession number GSE15269). Enrichment in specific pathways were determined by combined annotations taken from the Saccharomyces genome (http://db.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl), Affymetrix NetAffx and gene ontology consortium (http://www.geneontology.org) databases. Significance of enrichment was assessed using hypergeometric distribution.
Real time PCR analysis
For validation of the microarray results five genes from the up-regulated gene list were selected for real time PCR analyses. Cultures were grown as above with 10−8 M spermidine (control) or with 10−5 M spermidine, and cells were harvested after 1, 2, 4 and 6h. Each RT-PCR assay was repeated using three biological replicates and each analysis consisted of three technical replicates. Before PCR, each total RNA was processed with RNase free DNase (Qiagen). RNA was reverse transcribed by superscript II (Invitrogen). The primers were designed using Applied Biosystems (Foster City, CA, USA) Primer Express™ design software. Primers and fluorescence resonance energy transfer probes were purchased from Applied Biosystems. The RT-PCR reagents, including the 18S rRNA, assay plates and 7900 HT Fast Real Time PCR system were obtained from Applied Biosystems. Relative quantitation of transcript signals was performed by using 5-log10 standard curves with 18 S rRNA as the normalizer for each assay. Fold change was calculated by the delta Ct method (Livak and Schmittgen, 2001). Statistical significance for the calculated fold changes were set with an alpha value of 0.05 and the P-values were then calculated for the real time PCR results.
Results
Global gene expression in response to spermidine addition
We have applied microarray analysis to study the global gene expression profile of logarithmically growing yeast polyamine auxotroph (spe3Δ fms1Δ) supplemented with spermidine. Our results showed that a considerable number of genes were responsive to the addition of 10−5 M spermidine to a spermidine auxotroph growing in 10−8 M spermidine. The changes in gene expression were measured 6 hours after the higher concentration of spermidine was added to the cultures. As shown in Figure 1, there was little difference in growth rate resulting from these differences in spermidine concentration, and thus the effects found were not due to marked growth changes.
Fig. 1. Effect of 10−5 M spermidine or 10−5 M spermine on growth of a yeast auxotroph growing in minimum spermidine (10−8 M).
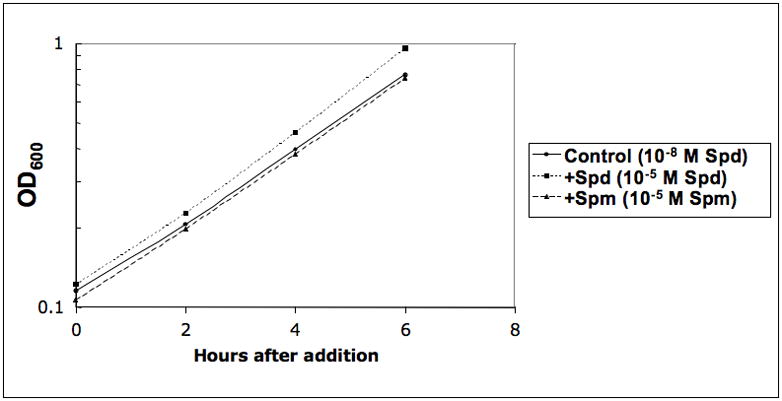
Y549 culture was grown in 10−8 M spermidine as mentioned in the text and the culture was diluted into three parts and grown overnight to an OD 600 of 0.1; to one part spermidine or spermine at 10−5 M concentration was added and the third culture was kept as control without further addition and incubated for 6h. Optical density at 600 nm was noted at different time points as mentioned in Materials and Methods.
A comparison of the effects of spermidine and spermine treatment is shown in Fig. 2A. It is evident from the volcanic graph, that spermidine treatment has a more pronounced effect than spermine treatment. After the addition of 10−5 M spermidine, transcripts of 247 genes (3.5%1) were up-regulated more than 2-fold. 92 genes (1.3%) were up-regulated >3-fold, 33 genes were up-regulated >5-fold and 11 genes were up-regulated more than 10-fold. 268 genes (4.3 %) were down-regulated more than 2-fold, 50 genes (0.7%) were down-regulated more than 4-fold, and 6 genes were down-regulated more than 8-fold. To facilitate data analysis, the genes were grouped into functional categories based on Saccharomyces genome database, Affymetrix gene ID and gene ontology (GO). Top GO categories of up-regulated genes are shown in Fig. 2B. Most significant categories include sulfur amino acid metabolism (including methionine and siroheme biosynthesis), arginine biosynthesis, biotin biosynthesis, lysine and NAD biosynthesis.
Fig. 2. Changes in gene expression after addition of spermidine or spermine.
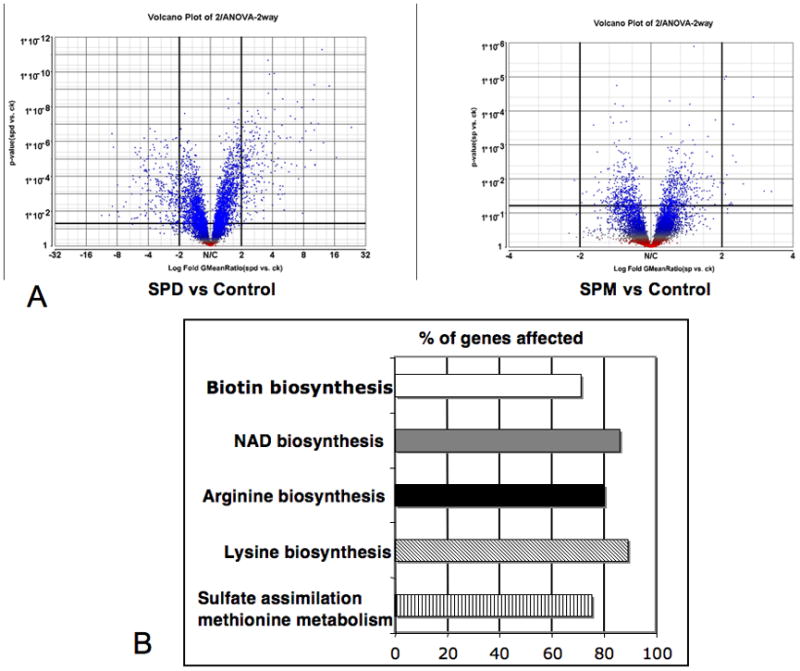
A. Volcanic plots depicting p-values obtained from a two-way ANOVA on the y-axis and fold change on the x-axis of either spermidine (SPD, left panel) or spermine (SPM, right panel) treated cells compared to control. B. Top 5 enriched gene ontology categories (p<0.001) showing the major metabolic pathways induced by spermidine. The graph shows the percentage of genes affected out of a total within each functional category.
Real time PCR analysis using specific probes for five of the induced genes (MET2, STR3, BIO5, SEO1, PHO84) are shown in Fig. 3. A comparison of real time PCR and microarray data is shown in Fig. 3A. Both measurements were consistent, and showed induction of gene expression after spermidine treatment, although the fold increase differed in these two different measurements. A time course after the addition of 10−5 M spermidine showed that induction of gene expression occurred only after two hours of spermidine treatment (Fig. 3B, 3C), even though the internal concentration of spermidine was elevated in less than 30 minutes (data not shown).
Fig. 3. Real-time PCR analyses of the several spermidine induced genes.
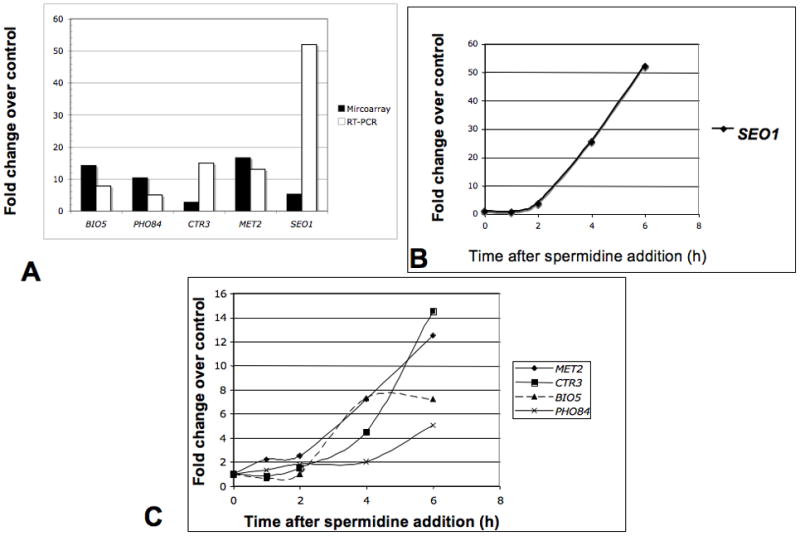
A. Comparison of microarray data and real-time PCR analyses of five spermidine induced genes 6h after spermidine treatment. B and C. Time courses of gene expression after spermidine treatment. Genes selected for validation were BIO5 (7-keto 8-aminopelargonic acid transporter), PHO84 (inorganic phosphate transporter), CTR3 (high affinity copper transporter), MET2 (L-homoserine-O-acetyl transferase), SEO1 (allantoate transporter subfamily).
Upregulation of genes involved in methionine and sulfur amino acid metabolism
As shown in Fig. 4 and Table 1 most of the genes involved in sulfur metabolism and all of the genes in the pathway from the uptake of sulfate to homocysteine and methionine synthesis (Thomas and Surdin-Kerjan, 1997) were induced by spermidine treatment. Several genes involved in the synthesis in siroheme (a molecule that is required for a functional sulfite reductase encoded by MET10), such as MET1, MET8 and MET10 were induced. In addition, genes encoding transporters of methionine (MUP1, MUP3), S-adenosylmethionine (SAM3), and methylmethionine (MMP1) (Isnard et al., 1996) showed enhanced expression.
Fig. 4. Effect of spermidine addition on induction of genes involved in yeast sulfate and sulfur containing amino acid metabolism.
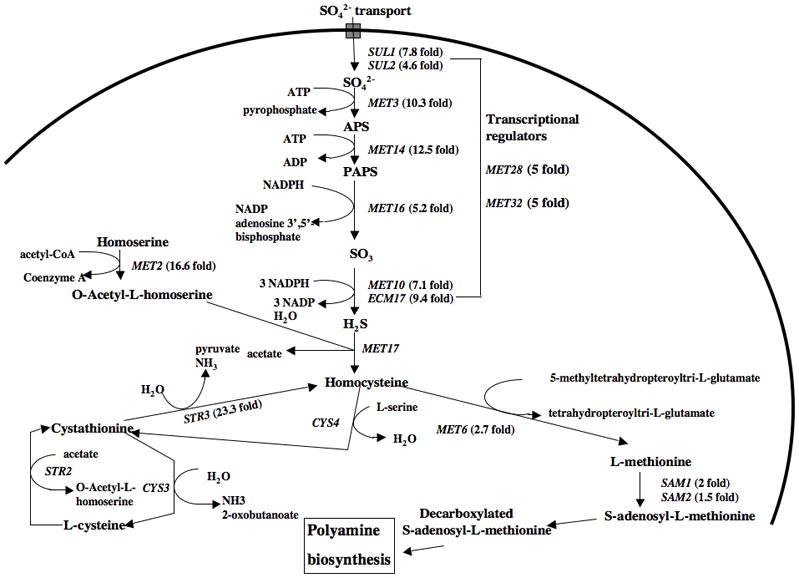
Genes upregulated in the biosynthesis, assimilation and transport of sulfur and sulfur containing amino acids; fold inductions are shown in parenthesis.
Table 1.
Spermidine induced sulfur or methionine metabolism related genes*
| Gene Names | Annoted functions | Fold Induced |
|---|---|---|
| MET2 | L-homoserine-O-acetyl transferase, methionine biosynthesis | 16.6 |
| STR3 | Sulfur transfer, cystathione beta-lyase activity, methionine biosynthesis | 23.3 |
| MET 14 | Adenyl sulfate kinase, sulfate assimilation pathway; cell | 12.5 |
| MET3 | ATP-sulfurylase, sulfate activation, sulfate to sulfide, methionine metabolism | 10.3 |
| MET10 | Sulfite reductase, sulfite to sulfide | 7.1 |
| MHT1 | S-methylmethionine homocysteine methyltransferase, sulfur amino acid metabolism | 7.3 |
| MXR1 | Protein-methionine-sulfoxide-reductase activity, response to oxidative stress | 8.3 |
| MET22 | Bisphosphate-3-nucleotidase, salt tolerance, methionine biosynthesis | 8.3 |
| MET 28 | Sulfur metabolism, transcriptional activator, | 5 |
| MET8 | Bifunctional dehydrogenase and ferrochelatase, siroheme biosynthesis, sulfate assimilation | 5.8 |
| MET32 | Zinc finger DNA binding transcriptional regulator protein, sulfur amino acid metabolism | 5 |
| MET16 | Phosphoadenylyl-sulfate reductase (thioredoxin) activity, methionine metabolism | 5.2 |
| MET1 | S-adenosyl-L-methionine uroporphyrinogen transmethylase, sulfate assimilation: siroheme biosynthesis | 4.7 |
| MET13 | Methyl tetrahydro folate reductase, sulfur amino acid biosynthesis | 3.2 |
| MET6 | Cobalamine independent methionine synthase, methionine biosynthesis | 2.7 |
| SUL1 | Sulfate transporter activity, control the endogenous concentration of activated sulfur intermediates | 7.8 |
| SUL2 | Sulfate transporter activity, control the endogenous concentration of activated sulfur intermediates | 4.6 |
| SAM3 | S-adenosylmethionine permease, required for utilization of S-adenosylmethionine as a sulfur source | 4 |
| BDS1 | Alkyl sulfatase activity, bacterially-derived sulfatase | 2.8 |
| ECM17 | Sulfite reductase beta subunit, involved in amino acid biosynthesis, repressed by methionine | 9.4 |
| MMP1 | High affinity S-methylmethionine permease | 6.9 |
Induction of genes involved in arginine biosynthesis
The synthesis of arginine in fungi has three main components, the synthesis of ornithine, the synthesis of carbamoyl phosphate and the conversion of the two compounds to arginine (Davis, 1986); (Jauniaux et al., 1978). As shown in Table 2, 8 out of the 10 genes involved in arginine biosynthesis were up-regulated more than 2-fold after spermidine addition. In addition, ORT1, the gene involved in ornithine export from mitochondria was also induced.
Table 2.
Genes induced by spermidine in the arginine, citrulline and ornithine metabolism+
| Gene names | Annoted functions | Fold increase |
|---|---|---|
| ARG1 | Arginosuccinate synthetase, catalyzes formation of L-argininosuccinate from citrulline and L-aspartate | 4.7 |
| ARG2 | Acetylglutamate synthase, first step in the biosynthesis of the arginine precursor ornithine | 2 |
| ARG3 | Ornithine carbamoyltransferase, catalyzes the sixth step in ornithine biosynthesis | 3 |
| ARG4 | Argininisuccinate lyase, this is the final step in arginine biosynthesis | 4.6 |
| ARG5.6 | Processed in the mitochondrion to form acetylglutamate kinase and N-acetyl-gamma- glutamyl-phosphate reductase, catalyzes 2nd and 3rd steps in arginine biosynthesis | 2.8 |
| CPA2 | Large subunit of carbamoyl phosphate synthetase, catalyzes the synthesis of citrulline, an arginine precursor | 2.8 |
| ECM40 | Mitochondrial ornithine acetyltransferase, catalyzes the fifth step in arginine biosynthesis | 2.5 |
| ORT1 | Ornithine transporter to the mitochondria, exports ornithine from mitochondria for arginine biosynthesis | 2 |
Induction of genes involved in biotin, lysine and tryptophan metabolism
Spermidine addition caused the up-regulation of several genes involved in biotin, lysine and tryptophan metabolism (Table 3). All the yeast genes encoding biotin synthesizing enzymes (BIO2, BIO3, BIO4), transporter of either biotin (VHT1) or precursor of biotin (BIO5), were up-regulated by 10−5 M spermidine addition (Table 3). A study of the time course of the induction of the BIO5 gene by Real-time PCR showed that induction of this gene occurred as early as 2 h after the addition of the higher concentration of spermidine. (Fig. 3C).
Table 3.
Genes induced by spermidine in the biotin, lysine and NAD (tryptophan) metabolism⊥
| Gene Names | Annoted functions | Fold induced |
|---|---|---|
| BIO5 | Putative transmembrane protein, responsible for uptake of 7-keto 8-aminopelargonic acid | 14.2 |
| BIO3 | Adenosylmethionine-8-amino-7-oxononanoate terminase, biotin biosynthesis | 10.2 |
| BIO4 | Dithiobiotin synthase activity, biotin biosynthesis | 9.9 |
| BIO2 | Biotin synthase activity, biotin biosynthesis | 4.9 |
| LYS9 | Saccharopine dehydrogenase (L-glutamate forming), lysine biosynthesis, aminoadipic pathway | 12 |
| LYS1 | Saccharopine dehydrogenase (L-lysine forming), lysine biosynthesis, aminoadipic pathway | 10.1 |
| LYS4 | Homoaconitase, catalyzes the conversion of homocitrate homoisocitrate | 4.4 |
| LYS2 | L-aminoadipate-semialdehyde dehydrogenase, lysine biosynthesis, aminoadipic pathway | 3.7 |
| LYS20 | Homocitrate synthase, lysine biosynthesis, aminoadipic pathway | 4.3 |
| LYS12 | Homo-isocitrate dehydrogenase, lysine biosynthesis | 3 |
| LYS21 | Homocitrate synthase isozyme, catalyzes condensation of acetyl CoA and alpha-ketoglutarate to form homocitrate | 3.4 |
| BNA2 | Tryptophan 2,3-dioxygenase, biosynthesis of nicotinic acid from tryptophan via kynurenine pathway | 11.1 |
| BNA1 | 3-hydroxyanthranilic acid dioxygenase, biosynthesis of nicotinic acid from tryptophan via kynurenine pathway, NAD biosynthesis | 3.4 |
| BNA3 | Arylformamidase, biosynthesis of nicotinic acid from tryptophan via kynurenine pathway, NAD biosynthesis; | 3.7 |
| BNA4 | Kynurenine 3-mono oxygenase, biosynthesis of nicotinic acid from tryptophan via kynurenine pathway | 3 |
| BNA5 | Kynureninase, required for de novo biosynthesis of NAD from tryptophan. | 2.7 |
Eight genes involved in lysine biosynthesis from aminoadipic acid and homoisocitrate showed 3-fold to 12-fold increased expression after the addition of spermidine. Several genes involved in tryptophan degradation were also induced by the addition of spermidine.
Induction of transport related genes after spermidine addition
Spermidine treatment for 6 h enhanced the expression of genes for several transporters, such as SUL1, SUL2 (involved in sulfate transport), BIO5 (a transporter of 7,8 aminopelargonic acid, a substrate of biotin biosynthesis), SEO1 (a putative permease member of the allantoate transporter family), CTR3 (which encodes the high-affinity copper transporter of the plasma membrane) and PHO84 (a gene encoding a high affinity inorganic phosphate transporter) (Table 4). Real time PCR analysis of several of these genes (BIO5, SEO1 and PHO84) showed that increased expression occurred 2 h after spermidine treatment (Fig. 3). The genes for oligo-peptide transporter OPT2 increased 3-fold and gene expression of small peptide transporter PTR2 increased 2-fold after spermidine treatment.
Table 4.
Spermidine induced genes involved in various transport functionsΠ
| Gene Names | Annoted functions | Fold Induced |
|---|---|---|
| SUL1 | Sulfate transporter activity, control the endogenous concentration of activated sulfur intermediates; PM | 7.8 |
| BIO5 | Transmembrane regulator of KAPA/DAPA transport, permease activity, vitamin/cofactor transport: PM | 14.2 |
| CTR3 | High affinity copper transporter: PM | 2.7 |
| SEO1 | Permease, suppressor of Sulfoxyde Ethionine resistance; PM | 5.2 |
| GIT1 | Member of yeast sugar permease, phospholipids transporter activity: PM | 3.3 |
| PHO84 | Inorganic phosphate transporter, Phosphate metabolism: PM | 10.3 |
| MMP1 | S-methylmethionine Permease, S-methylmethionine transport: PM | 6.9 |
| SUL2 | Sulfate transporter activity, control the endogenous concentration of activated sulfur intermediates; PM | 4.6 |
| OPT2 | Oligo peptide transporter: PM | 3 |
| VHT1 | Vitamin H (biotin) transporter; PM | 4.1 |
| MCH5 | Monocarboxylate permease, transporter activity; Membrane | 4 |
| FET4 | Low affinity iron transporter: PM | 3.2 |
| PHO89 | Na/Pi symporter, phosphate transport; PM | 2.4 |
| PTR2 | Small peptide transport into the cell, peptide transporter | 2.1 |
| MEP3 | Ammonia permease, ammonia transporter activity: PM | 2.2 |
| TPO5 | Protein involved in polyamine transport | 2.1 |
Effect on transcription factors
Increased transcription of more than 240 genes may not reflect a direct effect of spermidine addition; spermidine may indirectly regulate altered expression of some of the transcription factors, which in turn control the expression of various genes induced by spermidine. Indeed, spermidine treatment resulted in increased expression of 16 transcription factors as shown in Table 5. These include factors involved in amino acid metabolism (Met28p, Met32p, Gat1p), inositol metabolism as well as Zap1P, Hal9p, Tea1p that are involved in zinc regulation, increased salt tolerance and Ty1 enhancer activation, respectively.
Table 5.
Spermidine induced genes involved in encoding transcription factorsτ
| Gene names | Annoted function of encoded protein | Fold change |
|---|---|---|
| MET28 | Transcriptional activator in the Cbf1p-Met4p-Met28p complex, regulator of sulfur metabolism | 5 |
| MET32 | Zinc-finger DNA-binding protein, transcriptional regulation of methionine biosynthetic genes | 5 |
| HIR1 | Transcriptional co-repressor, transcription of histone H2A, H2B, H3 and H4 genes | 2 |
| ZAP1 | Zinc-regulated transcription factor, binds to zinc responsive element, zinc ion homeostasis | 2.4 |
| PHO4 | Basic helix-loop-helix transcription factor, regulated by phosphorylation, cellular response to phosphate starvation | 2.2 |
| GAT1 | Regulation of nitrogen utilization, contains a GATA-1 type DNA-binding motif | 2.1 |
| NRG2 | Negative regulator of glucose-controlled genes | 2.3 |
| INO2 | Component of Ino2p/Ino4p basic helix-loop-helix transcription activator, required for derepression of phospholipid biosynthetic genes | 1.9 |
| HAL9 | Zinc-finger transcription factor, over expression increases salt tolerance | 1.5 |
| WTM2 | WD repeat containing transcriptional modulator, involved in regulation of meiosis and silencing | 1.9 |
| TEA1 | Ty1 enhancer activator, required for full levels of Ty enhancer-mediated transcription, zinc cluster DNA-binding protein | 1.5 |
| TFC4 | One of six subunits of RNA polymerase III transcription initiation factor complex, part of TauA domain | 1.6 |
Gene expressions that are repressed after spermidine treatment
The repressed genes can be grouped into a few major categories; DNA, RNA binding, transcription (Table 6A) and stress related genes (Table 6B). Several genes involved in encoding transcription factors, such as CIN5, MIG2, SUT1, GAT2, STP4, are down-regulated more than 2-fold. A few genes (such as HTL1, GIS1, ARP8, SNF11) that wereinvolved in chromatin remodeling complex were down-regulated. Also repressed were genes encoding pre-mRNA splicing (SNT309, ECM2), telomerase functions (HLT1), and genes involved in frameshift suppression (MBF1). Another major category of spermidine-repressed genes included various stress responsive genes, such as HSP12, HSP104, GPD1, CYC7, GPX2.
Table 6A.
Spermidine repressed genes involved in DNA, RNA binding and regulation of gene expression, splicing±
| Gene names | Annoted functions | Fold change |
|---|---|---|
| MBF1 | Transcriptional coactivator that bridges the DNA-binding region of Gcn4p and TATA-binding protein Spt15p, frameshift suppressor | −3.1 |
| GIS1 | JmjC domain-containing histone demethylase, transcription factor involved in the expression of genes during nutrient limitation | −2.1 |
| HTL1 | Component of the RSC chromatin remodeling complex, may function in chromosome stability, telomerase maintenance | −1.9 |
| RAD51 | Strand exchange protein, forms a helical filament with DNA that search for homology, involved in double-strand DNA break repair | −1.8 |
| MIG2 | Protein with zinc fingers, involved in repression of SUC2, multicopy inhibition of GAL gene expression | −1.8 |
| RPA135 | RNA polymerase I subunit A135, DNA directed RNA polymerase act | −1.6 |
| ARP8 | Nuclear actin-related protein involved in chromatin-remodeling enzyme complexes | −1.9 |
| SNF11 | Subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation | −1.7 |
| RSA1 | Protein involved in the assembly of 60S ribosomal subunits | −1.7 |
| GAT2 | Protein containing GATA family zinc-finger motifs, expression repressed by leucine | −1.9 |
| SNT309 | Component of NineTeen complex, involved in RNA splicing | −2 |
| STP4 | Protein containg a Kruppel-type zinc-finger domain | −2.4 |
| MED7 | Subunit of the RNA polymerase II mediator complex, essential for transcriptional regulation | −2.1 |
| CIN5 | Basic leucine zipper transcriptional factor of yAP-1 family, involed in drug resistance and salt tolerance | −2.4 |
| TOF2 | Topoisomerase I-interacting factor, may be involved in DNA topological changes | −1.8 |
| SUT1 | Transcription factor of the zinc family, involved in induction of hypoxic gene expression | −2 |
| ECM2 | Pre-mRNA splicing factor | −1.8 |
| NPL3 | RNA-binding protein, carries poly(A)+ mRNA from nucleus into the cytoplasm, phosphorylated by Sky1p | −1.8 |
Table 6B.
Spermidine repressed genes involved in various stress responseτ
| Gene names | Annoted functions | Fold change |
|---|---|---|
| HSP12 | Plasma membrane localized protein, induced by heat shock, oxidative stress, osmotic stress, glucose depletion | −2.8 |
| HSP104 | Responsive to stress (heat, ethanol, and sodium arsenite), involved in [PSI+] propagation | −3.4 |
| HAP4 | Subunit of the heme-activated, glucose-repressed Hap-protein complex, a transcriptional activator of respiratory gene expression | −3 |
| GPD1 | NAD-dependent glycerol-3-phosphate dehydrogenase, essential for growth under osmotic stress | −2.8 |
| ROX1 | Heme-dependent repressor of hypoxic genes, responsible for DNA bending activity | −3.4 |
| GAD1 | Glutamate decarboxylase, converts glutamate into gamma-aminobutyric acid, involved in response to oxidative stress | −3.4 |
| SDP1 | Stress-inducible dual-specific MAP kinase phosphatase | −2.7 |
| CYC7 | Cytochrome c isoform 2, expressed under hypoxic condition | −5.3 |
| CTT1 | Cytosolic catalase T, has a role in protection from oxidative damage by hydrogen peroxide | −6.6 |
| GPX2 | Glutathione peroxidase, induced by glucose starvation, protects cells from phospholipids hydroperoxides and non-phospholipid peroxides during oxidative stress | −1.8 |
Effect of spermine addition on global gene expression
An important part of this study concerns the differential effects of spermidine and spermine on gene expression. In all of the above experiments, part of the administered spermidine was converted by the cell to spermine by spermine synthase (SPE4) (data not shown). To differentiate the effects due to spermidine from those due to spermine, we also carried out a parallel study on the effects of added spermine on gene expression. In the double mutant that we used (spe3Δ fms1Δ), the added spermine could not be converted to spermidine as this strain lacked spermine oxidase. (As in the other experiments all of these cultures also contained 10−8 M spermidine, since, as we have already shown, spermine does not permit growth of this spermidine auxotroph (Chattopadhyay et al., 2003).
As shown in Table 7, as opposed to the results found after spermidine addition, spermine addition at 10−5 M concentration resulted in a change in the expression of very few genes. Some of the spermine-induced genes were also induced by spermidine as indicated in the Table 7; however, their level of induction was far below than that found after spermidine addition. Only 15 genes were induced 2-fold, and 3-genes were repressed 2-fold by spermine.
Table 7.
Spermine controlled genes⇑ (*also changed by spermidine addition)
| Up-regulated genes | ||
|---|---|---|
| Gene symbol | Fold-change | Annoted Function |
| *CPA2 | 2.7 | Large subunit of carbamoyl phosphate synthetase, synthesis of citrulline, the arginine precursor |
| *ARG4 | 2.3 | Argininosuccinate lyase, arginine biosynthesis |
| YKL218C (SRY1) | 2.2 | 3-hydroxyaspartate dehydrogenase, serine racemase |
| RPL7B | 2.5 | Protein component of the 60S ribosomal subunit |
| *ECM40 | 2 | Mitochondrial ornithine acetyltransferase, ornithine biosynthesis |
| HIS5 | 1.7 | Histidinol-phosphate aminotransferase, histidine biosynthesis |
| *BNA1 | 2.2 | 3-hydroxyanthranilic acid dioxygenase, biosynthesis of nicotinic acid from tryptophan |
| *MEP3 | 2 | Ammonium permease, expresss under nitrogen catabolite repression |
| *MET13 | 1.6 | Isozyme of methylenetetrahydrofolate reductase, methionine biosynthesis |
| *LYS1 | 1.9 | Saccharopine dehydrogenase, lysine biosynthesis |
| SSU1 | 1.7 | Plasmamembrane sulfite pump, efficient sulfite efflux |
| MCH4 | 1.6 | Monocarboxylate transporter homolog, transport of monocarboxylic acid across the plasma membrane |
| YEL073C | 2 | Uncharacterized protein, regulated by inositol/choline |
| Down-regulated genes | ||
| Gene symbol | Fold-change | Annoted Function |
| HHO1 | −1.7 | Histone H1, suppresses DNA repair involving homologous recombination |
| PCL1 | −1.6 | Ph085 cyclin, involved in entry into the mitotic cell cycle |
| CLN2 | −1.7 | G1 cyclin involved in regulation of cell cycle, promotes G1to S phase transition |
| *SNF11 | −1.4 | Subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation |
| BBP1 | −1.4 | Spindle pole body duplication, required for mitotic function of Cdc5p |
| SRL1 | −1.5 | Mannoprotein exhibiting tight association with cell wall, suppressor of Rad53p null lethality |
| CWC25 | −1.5 | Component of a pre-mRNA splicing complex containing Cef1p |
Discussion
In this paper we have reported the first global microarray analyses of the effect of spermidine and spermine in a eukaryotic system. In particular, we have studied these effects in a system designed to minimize any effects of these additions on the growth rate of the cultures; namely, by comparing cultures grown in 10−5 M of spermidine and spermine with cultures grown in 10−8 M spermidine. As seen in Figure 1, the growth rate was only slightly faster in the cultures grown with 10−5 M spermidine, and was not affected at all by the addition of 10−5 M spermine.
Recently, we have reported that yeast cells grown at a nearly optimum growth rate in the presence of 10−8 M spermidine and >50% of the spermidine was used for hypusine modification of eIF5A, so it was clear that one of the major functions of spermidine is the modification of eIF5A (Chattopadhyay et al., 2008). A major stimulus for the current studies was the question of why wild type S. cerevisiae cells normally contain a much higher internal concentration of spermidine than needed for optimum growth (1000-fold). Hence, it was interesting to note that in the current study so many genes were up regulated or down regulated after spermidine addition even though there was little effect on the growth rate. A number of different systems were affected as shown by the data in Table 1–6 and Figure 2, and some of the changes were very large. Particularly striking were the effects on most of the genes involved in sulfur metabolism and on methionine transport and biosynthesis, as well as arginine and lysine and biotin biosynthesis. In these microarray studies, we have found increased expression of Met32p and Met28p by spermidine, which constitute the main transcription activators of the sulfate assimilation pathway, including Cbf1p, Met4p, and Met31p (Kuras et al., 1996; Blaiseau et al., 1997).
Another interesting change was in the cluster of genes involved in biotin biosynthesis such as BIO5, BIO3, BIO4 (Phalip et al., 1999) along with BIO2; all these genes were induced by spermidine addition. The genes for biotin biosynthesis in yeast are present as a gene cluster on chromosome XIV and are regulated by environmental stress (Gasch et al., 2000), iron deprivation (Shakoury-Elizeh et al., 2004), glucose limitation (Ferea et al., 1999) or histone modification (Wyrick et al., 1999). Biotin is essential for all living organisms and is a cofactor for several carboxylase family of enzymes. Saccharomyces cerevisiae is auxotrophic to biotin, however, can be complemented by addition of KAPA 7-keto 8-aminopelargonic acid (KAPA), 7,8-diaminopelargonic acid (DAPA) or dithiobiotin to the medium. Biotin biosynthesis is also increased by S-adenosylmethionine, which serves as an amino group donor in the synthesis of KAPA from DAPA (Fontecave et al., 2004). Spermidine addition upregulated genes in the methionine biosynthesis including the synthesis of S-adenosylmethionine; on the other hand, spermidine treatment also repressed S-adenosylmethionine decarboxylase (SPE2) by 1.5-fold (see below). Thus, it is possible that the accumulated S-adenosylmethionine can trigger the increased expression of biotin biosynthesizing genes as an indirect effect of spermidine treatment.
Surprisingly, some of the genes induced by spermidine treatment (especially those involved in methionine metabolism) were found by Aranda and Olmo (Aranda and del Olmo, 2004) to be induced by acetaldehyde treatment. They also found that acetaldehyde treatment resulted in the induction of polyamine transport genes (TPO2, TPO3). Other genes involved in vitamin-B1 biosynthesis and in aryl alcohol metabolism were also induced both by acetaldehyde treatment, and by spermidine addition (Aranda and del Olmo, 2004), (Table 3 and for complete microarray data see GEO accession NO GSE15269). In a different study, Santiago and Mamoun (Santiago and Mamoun, 2003) observed that several genes involved in inositol, methionine, and biotin biosynthesis were down-regulated along with genes implicated in polyamine transport (TPO1, TPO2) by addition of inositol or choline to yeast cultures. Although we have no explanation for this similarity in the effects of these three very different treatments, it seems possible from these global gene-expression studies, and our current study that polyamine levels might play a role in the regulation of these gene expression.
Our microarray data showed 1.5-fold down regulation of S-adenosylmethionine decarboxylase (SPE2) gene expression after spermidine treatment. Expressions of the known polyamine transporters were unchanged except TPO5, which showed a 2.1-fold induction of gene expression after spermidine addition. There was no change in gene expression of ornithine decarboxylase (SPE1) after spermidine or spermine treatment, which suggests a post-translational control of ornithine decarboxylase by antizyme (Palanimurugan et al., 2004).
The large number of very different systems affected by the addition of spermidine emphasizes the importance of the higher internal concentration of spermidine normally present in wild type yeast cells. However, at this time we are unable to define which of the systems represent the primary effect of the addition of spermidine or the results of indirect effects of other gene expression. Real time PCR analyses of five of the induced genes (Fig. 3B, 3C) suggest the notion that spermidine addition may result in an indirect effect on gene expression of various pathways, as most of the gene expressions were changed after 2h of spermidine addition. Of particular interest is the increased expression of a number of transcription factors after spermidine addition, particularly those involved in the expression of several genes in methionine, arginine, and other amino acid metabolizing genes (Table 5). A study by Yoshida et al (Yoshida et al., 2004) in E. coli has postulated the involvement of polyamine modulation in the expression of genes responsible for bacterial growth (Yoshida et al., 2004; Igarashi and Kashiwagi, 2006). In their experiment they have shown that most of the genes enhanced by polyamines were not under the direct control of polyamines, but were due to indirect effect of transcription factors, whose synthesis was enhanced by polyamines. Thus, it seems possible that increased transcription of various metabolic pathways might be regulated indirectly by change in the transcription factors, whose expressions are high due to the higher concentration of spermidine.
Some of the genes repressed by spermidine, also include genes in nucleic acid function (Table 6A), which may be due to binding of excess amount of spermidine to the negatively charged nucleic acids (Cohen SS, 1998, and references there in; Igarashi and Kashiwagi, 2000; Vijayanathan et al., 2001). Spermidine and spermine repressed the SWI-SNF chromatin-remodeling complex of yeast (Table 6A, 7). Polyamine involvement in the regulation of gene expression through the modulation of chromatin remodeling complex has been demonstrated in yeast and other systems (Pollard et al., 1999; Huang et al., 2007).
Yeast polyamine mutants grown in 10−8 M spermidine did not show any obvious defect in growth or any indication of oxidative stress (Chattopadhyay et al., 2006), when the spermidine level was restored to wild type level by adding 10−5 M spermidine to the culture medium, various stress responsive genes were repressed such as HSP12, HSP104, ROX1, CYC7, and others (Table 6B).
Our work provides new insights into the responsiveness of yeast mutant lacking spermidine synthase to millimolar level of polyamines, which are present in wild type yeast cells and may suggests specific molecular targets of the high intracellular concentration of spermidine that is normally present in wild type yeast. More analyses and experimental data, as well as comparable studies with different yeast mutants, are needed to distinguish between direct and indirect effects of the polyamines and to explain the physiological connections between the different pathways affected by spermidine. Moreover, these results are complicated by the fact that when spermidine concentration is in excess (10−5 M), the modified eIF5A level is more than 20-fold than needed for optimum growth (Chattopadhyay et al., 2008). In addition, it is likely that the added spermidine would repress the enzymes involved in polyamine biosynthesis with a resultant decrease in the level of intracellular putrescine and decarboxylated S-adenosylmethionine, and that these changes might affect other systems involved in arginine and methionine metabolism. In conclusion, even though it is not possible to define with certainty all of the direct effects of spermidine, the data in this paper clearly indicate that spermidine has a profound effect on the expression of a large number of genes, either directly or indirectly.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH (National Institute of Diabetes, Digestive and Kidney Diseases).
Footnotes
% expressed per total number of genes in S. cerevisiae
Data are average of five independent experiments (p<0.05); functional annotations are based on Saccharomyces Genome Database (www.yeastgenome.org)
Data are average of five independent experiments (p<0.05); functional annotations are based on Saccharomyces Genome Database (www.yeastgenome.org)
Data are average of five independent experiments (p<0.05); functional annotations are based on Saccharomyces Genome Database (www.yeastgenome.org)
Data are average of five independent experiments (p<0.05); functional annotations are based on Saccharomyces Genome Database (www.yeastgenome.org)
Data are average of five independent experiments (p<0.05); functional annotations are based on Saccharomyces Genome Database (www.yeastgenome.org)
Data are average of five independent experiments (p<0.05); functional annotations are based on Saccharomyces Genome Database (www.yeastgenome.org)
Data are average of five independent experiments (p<0.05); functional annotations are based on Saccharomyces Genome Database (www.yeastgenome.org)
Data are average of three independent experiments (p<0.05); functional annotations are based on Saccharomyces Genome Database (www.yeastgenome.org)
References
- Aranda A, del Olmo ML. Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively. Appl Environ Microbiol. 2004;70:1913–1922. doi: 10.1128/AEM.70.4.1913-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaram D, Tabor CW, Tabor H. Spermidine or spermine is essential for the aerobic growth of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991;88:5872–5876. doi: 10.1073/pnas.88.13.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaram D, Tabor CW, Tabor H. Oxygen toxicity in a polyamine-depleted spe2 delta mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993;90:4693–4697. doi: 10.1073/pnas.90.10.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaram D, Tabor CW, Tabor H. Sensitivity of spermidine-deficient Saccharomyces cerevisiae to paromomycin. Antimicrob Agents Chemother. 1999;43:1314–1316. doi: 10.1128/aac.43.5.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaiseau PL, Isnard AD, Surdin-Kerjan Y, Thomas D. Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol Cell Biol. 1997;17:3640–3648. doi: 10.1128/mcb.17.7.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Park MH, Tabor H. Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc Natl Acad Sci U S A. 2008;105:6554–6559. doi: 10.1073/pnas.0710970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H. Absolute requirement of spermidine for growth and cell cycle progression of fission yeast (Schizosaccharomyces pombe) Proc Natl Acad Sci U S A. 2002;99:10330–10334. doi: 10.1073/pnas.162362899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H. Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermine is converted to spermidine in vivo by the FMS1-amine oxidase. Proc Natl Acad Sci U S A. 2003;100:13869–13874. doi: 10.1073/pnas.1835918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H. Polyamine deficiency leads to accumulation of reactive oxygen species in a spe2Delta mutant of Saccharomyces cerevisiae. Yeast. 2006;23:751–761. doi: 10.1002/yea.1393. [DOI] [PubMed] [Google Scholar]
- Cohen SS. A Guide to the Polyamines. Oxford University Press; New York: 1998. pp. 1–595. [Google Scholar]
- Davis RH. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol Rev. 1986;50:280–313. doi: 10.1128/mr.50.3.280-313.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diala ES, Evans IH, Wilkie D. Primary antimitochondrial activity of the cancer drug methylglyoxal bis(guanylhydrazone) in yeast cells. J Gen Microbiol. 1980;119:35–40. doi: 10.1099/00221287-119-1-35. [DOI] [PubMed] [Google Scholar]
- Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci U S A. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecave M, Atta M, Mulliez E. S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki-Katagiri N, Tabor CW, Tabor H. Spermidine biosynthesis in Saccharomyces cerevisae: polyamine requirement of a null mutant of the SPE3 gene (spermidine synthase) Gene. 1997;187:35–43. doi: 10.1016/s0378-1119(96)00660-9. [DOI] [PubMed] [Google Scholar]
- Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RAJ. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci U S A. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Polyamine Modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J Biochem. 2006;139:11–16. doi: 10.1093/jb/mvj020. [DOI] [PubMed] [Google Scholar]
- Isnard AD, Thomas D, Surdin-Kerjan Y. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J Mol Biol. 1996;262:473–484. doi: 10.1006/jmbi.1996.0529. [DOI] [PubMed] [Google Scholar]
- Jauniaux JC, Urrestarazu LA, Wiame JM. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J Bacteriol. 1978;133:1096–1107. doi: 10.1128/jb.133.3.1096-1107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1996;15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mamont PS, Joder-Ohlenbusch AM, Nussli M, Grove J. Indirect evidence for a strict negative control of S-adenosyl-L-methionine decarboxylase by spermidine in rat hepatoma cells. Biochem J. 1981;196:411–422. doi: 10.1042/bj1960411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanimurugan R, Scheel H, Hofmann K, Dohmen RJ. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 2004;23:4857–4867. doi: 10.1038/sj.emboj.7600473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem (Tokyo) 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci U S A. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalip V, Kuhn I, Lemoine Y, Jeltsch JM. Characterization of the biotin biosynthesis pathway in Saccharomyces cerevisiae and evidence for a cluster containing BIO5, a novel gene involved in vitamer uptake. Gene. 1999;232:43–51. doi: 10.1016/s0378-1119(99)00117-1. [DOI] [PubMed] [Google Scholar]
- Pitkin J, Davis RH. The genetics of polyamine synthesis in Neurospora crassa. Arch Biochem Biophys. 1990;278:386–391. doi: 10.1016/0003-9861(90)90275-4. [DOI] [PubMed] [Google Scholar]
- Pollard KJ, Samuels ML, Crowley KA, Hansen JC, Peterson CL. Functional interaction between GCN5 and polyamines: a new role for core histone acetylation. EMBO J. 1999;18:5622–5633. doi: 10.1093/emboj/18.20.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago TC, Mamoun CB. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J Biol Chem. 2003;278:38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, Rolfes R, Brown PO, Botstein D, Philpott CC. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:1233–1243. doi: 10.1091/mbc.E03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayanathan V, Thomas T, Shirahata A, Thomas TJ. DNA condensation by polyamines: a laser light scattering study of structural effects. Biochemistry. 2001;40:13644–13651. doi: 10.1021/bi010993t. [DOI] [PubMed] [Google Scholar]
- White WH, Gunyuzlu PL, Toyn JH. Saccharomyces cerevisiae is capable of de Novo pantothenic acid biosynthesis involving a novel pathway of beta-alanine production from spermine. J Biol Chem. 2001;276:10794–10800. doi: 10.1074/jbc.M009804200. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kashiwagi K, Shigemasa A, Taniguchi S, Yamamoto K, Makinoshima H, Ishihama A, Igarashi K. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J Biol Chem. 2004;279:46008–46013. doi: 10.1074/jbc.M404393200. [DOI] [PubMed] [Google Scholar]


