Abstract
Although some animals are capable of regenerating organs, the mechanisms by which this is achieved are poorly understood. In planarians, pluripotent somatic stem cells called neoblasts supply new cells for growth, replenish tissues in response to cellular turnover, and regenerate tissues after injury. For most tissues and organs, however, the spatiotemporal dynamics of stem cell differentiation and the fate of tissue that existed prior to injury have not been characterized systematically. Utilizing in vivo imaging and bromodeoxyuridine pulse-chase experiments, we have analyzed growth and regeneration of the planarian intestine, the organ responsible for digestion and nutrient distribution. During growth, we observe that new gut branches are added along the entire anteroposterior axis. We find that new enterocytes differentiate throughout the intestine rather than in specific growth zones, suggesting that branching morphogenesis is achieved primarily by remodeling of differentiated intestinal tissues. During regeneration, we also demonstrate a previously unappreciated degree of intestinal remodeling, in which pre-existing posterior gut tissue contributes extensively to the newly formed anterior gut, and vice versa. By contrast to growing animals, differentiation of new intestinal cells occurs at preferential locations, including within newly generated tissue (the blastema), and along pre-existing intestinal branches undergoing remodeling. Our results indicate that growth and regeneration of the planarian intestine are achieved by coordinated differentiation of stem cells and the remodeling of pre-existing tissues. Elucidation of the mechanisms by which these processes are integrated will be critical for understanding organogenesis in a post-embryonic context.
Keywords: planarian, regeneration, remodeling, neoblast, intestine, organogenesis
Introduction
How regenerating animals rebuild organs in a post-embryonic context is not well understood. Although many organisms can renew tissues homeostatically in response to normal cellular turnover or minor damage, only some animals are capable of repairing or even completely replacing organs after more serious injury (Poss, 2010). Gastrointestinal organs exemplify this range of regenerative abilities. In many animals, epithelial cells lining the digestive tract turn over rapidly, and are replaced by resident somatic stem cells (Casali and Batlle, 2009; Faro et al., 2009; Illa-Bochaca and Montuenga, 2006; Ishizuya-Oka, 2007; van der Flier and Clevers, 2009). In response to insults such as irradiation or cytotoxic damage, these stem cells can increase their rates of proliferation and differentiation in order to regenerate the digestive epithelium, for example, in the small intestine of mouse (Potten, 1992; Potten et al., 1997) and the midgut of Drosophila melanogaster (Amcheslavsky et al., 2009; Chatterjee and Ip, 2009; Jiang et al., 2009).
Many animals are capable of much more extensive repair or even replacement of their gastrointestinal tracts. For example, some amphibians can recover from complete transection of the intestine, restoring the integrity of the intestinal tract and full functionality within 1-2 months (Goodchild, 1956; O'Steen, 1958; O'Steen, 1959; O'Steen and Walker, 1962). Even more impressively, other organisms can regenerate part or all of their digestive systems after spontaneous evisceration (sea cucumbers), amputation (the ascidian Polyandrocarpa misakiensis), or during asexual reproduction (the annelids Enchytraeus japonensis and Pristina leidyi) (García-Arrarás et al., 1998; Kaneko et al., 2010; Mashanov and Dolmatov, 2001; Shukalyuk and Dolmatov, 2001; Takeo et al., 2008; Zattara and Bely, 2011). Despite the obvious biomedical implications of understanding such regenerative feats, the cellular and molecular mechanisms underlying animals' abilities to achieve such dramatic de novo organogenesis are almost completely uncharacterized.
Freshwater planarians can also regenerate tissues in response to nearly any type of amputation (Reddien and Sánchez Alvarado, 2004), and are thus ideally suited for investigating organ regeneration. Planarians' regenerative prowess is conferred in part by a population of pluripotent somatic stem cells called neoblasts that give rise to missing tissues and organs after injury (Newmark and Sánchez Alvarado, 2002). In the last decade, planarians have become more tractable experimental models owing to the introduction of cellular, molecular, and genomic technologies (Forsthoefel and Newmark, 2009; Newmark and Sánchez Alvarado, 2002; Robb et al., 2008). In planarians, as in many regenerating animals, two distinct events occur after amputation (Brockes and Kumar, 2008; Gurley and Sánchez Alvarado, 2008; Newmark and Sánchez Alvarado, 2002; Poss, 2010). First, new tissue (called a regeneration blastema) is generated by the proliferation and differentiation of neoblasts. Second, old tissue remodels and integrates with newly produced cells to complete the restoration of morphology and function.
Both processes occur in planarians (Reddien and Sánchez Alvarado, 2004), but have not been rigorously analyzed at the level of individual organs. For example, although the dynamics of neoblast proliferation in response to feeding and injury have been documented (Baguñà, 1974; Baguñà, 1976a; Baguñà, 1976b; Baguñá and Romero, 1981; Newmark and Sánchez Alvarado, 2000; Salò and Baguñà, 1984; Wenemoser and Reddien, 2010), spatiotemporal analyses of neoblast differentiation have only been conducted for a limited number of cell types (Eisenhoffer et al., 2008; Newmark and Sánchez Alvarado, 2000; Reddien et al., 2005; Sakai et al., 2002). Similarly, remodeling has not been studied extensively. After amputation, apoptosis occurs to reduce overall cell numbers as polarity and symmetry of small tissue fragments are restored (Pellettieri et al., 2009). Furthermore, there is some evidence that organs such as the intestine can reorganize after amputation (Gurley et al., 2010; Morgan, 1902). However, systematic experiments examining the contribution of pre-injury tissue to regenerating organs have been lacking, due in part to a lack of techniques for labeling and monitoring differentiated cells over extended time periods after injury.
In this study, we examine the spatiotemporal dynamics of differentiation and remodeling during growth and regeneration of the planarian intestine. The intestine is responsible for digestion of ingested food; its highly branched morphology is thought to facilitate body-wide distribution of metabolites, serving part of the role that vasculature serves in higher organisms (Brøndsted, 1969). Cells of the intestinal epithelium, or gastrodermis, are organized into a single columnar layer surrounded by a basal lamina and enteric muscles (Bueno et al., 1997; Garcia-Corrales and Gamo, 1986; Garcia-Corrales and Gamo, 1988; Kobayashi et al., 1998; Orii et al., 2002). Histological analyses suggest the existence of only two intestinal cell types – absorptive phagocytes that engulf food particles for intracellular digestion, and secretory goblet cells that release digestive enzymes into the lumen (Bowen, 1980; Bowen et al., 1974; Garcia-Corrales and Gamo, 1986; Garcia-Corrales and Gamo, 1988; Ishii, 1965). Decades of ultrastructural and physiological studies have characterized the role of these cells during digestion and nutrient storage (Bowen, 1980; Bowen et al., 1974; Garcia-Corrales and Gamo, 1986; Garcia-Corrales and Gamo, 1988; Ishii, 1965). However, in contrast to other organs such as the central nervous system (Cebrià, 2007), and aside from several studies of axial polarity (Adell et al., 2009; Cebrià et al., 2007; Gurley et al., 2010; Gurley et al., 2008; Iglesias et al., 2008; Nogi and Levin, 2005; Oviedo et al., 2010), the intestine has received limited attention during developmental studies of growth and regeneration. Here, we characterize these processes in the planarian Schmidtea mediterranea, utilizing in vivo imaging and bromodeoxyuridine pulse-chase experiments. Our work suggests that differentiation and remodeling are coordinated during intestinal morphogenesis, and that future studies of the intestine will be broadly informative in unraveling the mechanisms by which non-resident stem cells can be recruited and integrated with pre-existing structures to achieve organogenesis in a post-embryonic context.
Materials and Methods
Animal maintenance and care
Asexual Schmidtea mediterranea (clonal line ClW4) were maintained as described (Cebrià and Newmark, 2005). Unless otherwise noted, animals 3-6 mm in length were starved for at least seven days prior to amputation and/or fixation in all experiments.
In situ hybridization
Animals were fixed and whole-mount in situ hybridizations on Carnoy's-fixed samples were conducted as described (Umesono et al., 1997), with the exception that signal:noise was improved with ethanol treatment (Pearson et al., 2009). Antisense riboprobe was synthesized using a cofilin-like EST (PL030005A10G01, GenBank ID: DN290811) as the template (Zayas et al., 2005).
Dextran feeding
A 1:3 (liver:water) homogenate (83 μL) and 2% ultra-low melting point agarose (15 μL, Sigma) were mixed with 2 μL (1 mg/mL) 10,000 MW dextrans conjugated to Alexa 546 (Molecular Probes), spotted onto Petri dishes and allowed to solidify on ice. For growth experiments, animals were fed dextrans 1-2 days prior to imaging, and otherwise regular pureed calf liver every 2-3 days. Analysis of branching morphogenesis was limited to animals that grew during the course of the experiment. For regeneration time courses, animals were fed 2-3 times over 5-6 days, amputated 2 days after the last feeding and imaged periodically. Regenerates were fed Alexa 488-conjugated dextrans 10 days after amputation and imaged again two days later. For irradiation experiments, animals were fed dextrans 2-3 times, gamma irradiated 24 h later with 30 Gy as described (Guo et al., 2006), and amputated one day later.
Phosphotyrosine/muscle double immunofluorescence
Intact and regenerating samples were killed for 3 min in ice cold 2% HCl, fixed for 1-2 hr in methacarn (6:3:1 methanol:chloroform:acetic acid), rinsed for 15 minutes in – 20°C methanol, and bleached O/N in 6% H2O2 in methanol. After rehydration to PBT× (1× PBS + 0.3% Triton X-100), samples were blocked O/N in blocking buffer (0.6% BSA/0.45% fish gelatin/PBT×), then incubated sequentially in a fortuitously discovered rabbit preimmune serum that labels planarian enteric and body wall muscles (“rabbit anti-muscle,” 1:500, F. Cebrià and T. Guo, unpublished), goat anti-rabbit 568 (1:1000, Molecular Probes), mouse anti-phosphotyrosine (1:500, Cell Signaling), and goat anti-mouse 488 (1:400, Molecular Probes) with 6-8 PBT× washes over 8 hr followed by O/N reblocking between antibodies.
BrdU labeling and detection
Animals were fed 5-Bromo-2′-deoxyuridine (Sigma) blended with a 1:3 (liver:water) homogenate at a final concentration of 2-2.5 mg/mL as described (Newmark and Sánchez Alvarado, 2000), with the exception that the final concentration of ultra-low melting point agarose (Sigma) was reduced to 0.3%. For whole-mount labeling, animals were killed in ice-cold 2% HCl for 30 sec to 1 min, fixed in methacarn for 2 hr, rinsed for 15 minutes in –20°C methanol, then bleached overnight (12-15 hours) in 6% H2O2 in methanol. 3% H2O2 was used for 24 hr & 48 hr chases, since overbleaching at these time points negatively affected BrdU signal detection and animal integrity. Following rehydration, animals were treated with 2N HCl for 5-15 minutes, quenched in 0.1 M borax for 2 minutes, rinsed in PBT×, then incubated in blocking buffer (above) for 2-4 hr. The following antibodies were used: mouse anti-BrdU (1:25, BD Biosciences); rabbit anti-muscle (above, 1:500); goat anti-mouse HRP (1:100, Invitrogen); and goat anti-rabbit Alexa 546 (1:1000, Molecular Probes). Antibody incubations were conducted overnight, followed by 6-8 PBT× washes over 8 hr the following day. Tyramide signal amplification (1:1500 FITC-tyramide plus 0.006% H2O2 for 10 min in PBS+.01% Tween-20) to detect BrdU was conducted as described (Pearson et al., 2009).
For cryosections, animals were fixed in methacarn as above, but then rehydrated without bleaching and transferred to 30% sucrose in PBT×, embedded in Tissue Freezing Medium (TBS), and cryosectioned at 20 μm. When not processed immediately, animals were stored in sucrose at 4°C or slides were stored at –80°C; storage by either method for up to 3 months did not affect BrdU detection efficiency (not shown). After cryosectioning, embedding medium was removed and slides were rehydrated with three 10 min incubations in deionized water. In order to detect muscles on unbleached specimens, antigen retrieval (AR) was performed by treating slides in 20 mM sodium citrate (pH 6.0) at 100°C for 10 min in a microwave, and allowed to cool to room temperature. We found that BrdU detection was possible after AR alone, but was improved by brief denaturation with 2 N HCl for 2 min, followed by a 2 min neutralization in 0.1 M borax. After equilibration in PBT×, slides were blocked for 30 min, incubated in primary antibodies diluted as above in blocking buffer for 2 hr at room temperature, washed 4 times for 10 min each in PBT×, incubated in secondary antibodies and DAPI (1 μg/μL) in blocking buffer for 1 hr at room temperature, and washed again. TSA was conducted on slides, as above, with 10 min development. Specimens were mounted in Vectashield (Vector Labs) and imaged.
Imaging and image processing
BrdU-labeled samples, cryosections, and phosphotyrosine/muscle-labeled regenerates were imaged using a Zeiss LSM 710 confocal microscope running ZEN 2009, or on a Nikon Eclipse TE200 inverted epifluorescent scope with a MicroFIRE camera (Optronics) and Picture Frame v2.3. Whole-mount ISH samples were imaged with a Leica M205A running LAS 3.6.0. Living, dextran-fed animals and regenerates were imaged using a Zeiss SteREO Lumar.V12. Short (30 second) movies of animals were captured using the multi-dimensional time lapse module in AxioVision 4.6.3; the best individual frames (in which animals were well extended to see gut branches) were used in figures. Images were processed using ImageJ 1.40g and Adobe Photoshop CS4.
Results
Live imaging reveals intestinal branching morphogenesis in growing planarians
Planarians grow by the addition of cells, rather than an increase in cell size (Baguñá and Romero, 1981; Newmark and Sánchez Alvarado, 2002). However, while the modulation of cell number and proportion has been documented for dissociated planarian tissues, whole organism studies of alterations in organ morphology and complexity as cell number increases have been rare (Oviedo et al., 2003; Takeda et al., 2009). Accordingly, we initiated our studies of the intestine in growing animals.
The planarian intestine is comprised of a single anterior and two posterior primary branches, from which secondary, tertiary, and quaternary branches ramify toward the lateral margins of the animal (Figs. 1A-C). The digestive systems of large planarians possess more intestinal branches and occupy greater area (proportional to animal size) than small animals (von Bertalanffy, 1940). However, detailed characterization of the addition of intestinal branches has not been conducted. One reason is that immunohistochemical methods (Figs. 1B, C) and other approaches (Pearson et al., 2009; von Bertalanffy, 1940) require fixation, and do not allow linear studies of morphological changes in living, individual animals over time.
Figure 1. Anatomy of the planarian intestine.
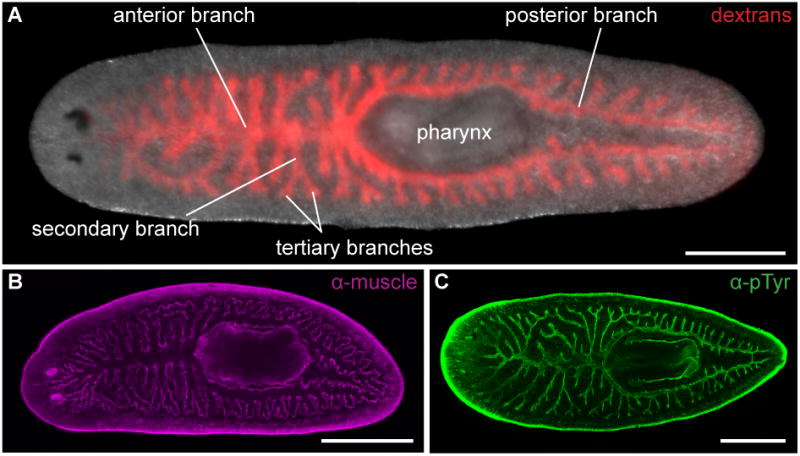
(A) Animals were fed Alexa 546-conjugated dextrans (red) to visualize all intestinal branches. The intestine possesses three primary branches – one anterior and two posterior – from which secondary and tertiary branches project laterally. These branches meet at a junction just anterior to the pharynx, the muscular feeding organ through which food enters the intestine and waste is excreted. (B) Enteric muscles (magenta) surround all intestinal branches. The anti-muscle antibody also labels pharyngeal and outer body wall muscles. (C) Anti-phosphotyrosine (green) labels the lumenal surface (specifically apical cell-cell junctions) of intestinal epithelial cells. B and C are confocal projections. All panels are dorsal views, with anterior to the left. Scale bars, 500 μm.
Accordingly, we took advantage of the fact that intestinal phagocytes (the planarian absorptive intestinal epithelial cell) will retain pigments, dyes, and other molecules for extended periods of time after feeding, allowing the visualization of the digestive system in vivo (Hyman, 1925; Lin, 1931; Morgan, 1900; Salò and Baguñà, 1985). In our experiments, we fed animals periodically with fluorescently conjugated dextrans, enabling straightforward imaging of intestinal branches in living planarians (Figs. 1A and 2A). Between dextran feedings, we fed animals every 2-3 days to promote growth over a month-long time course.
Figure 2. New intestinal branches are added in growing planarians.
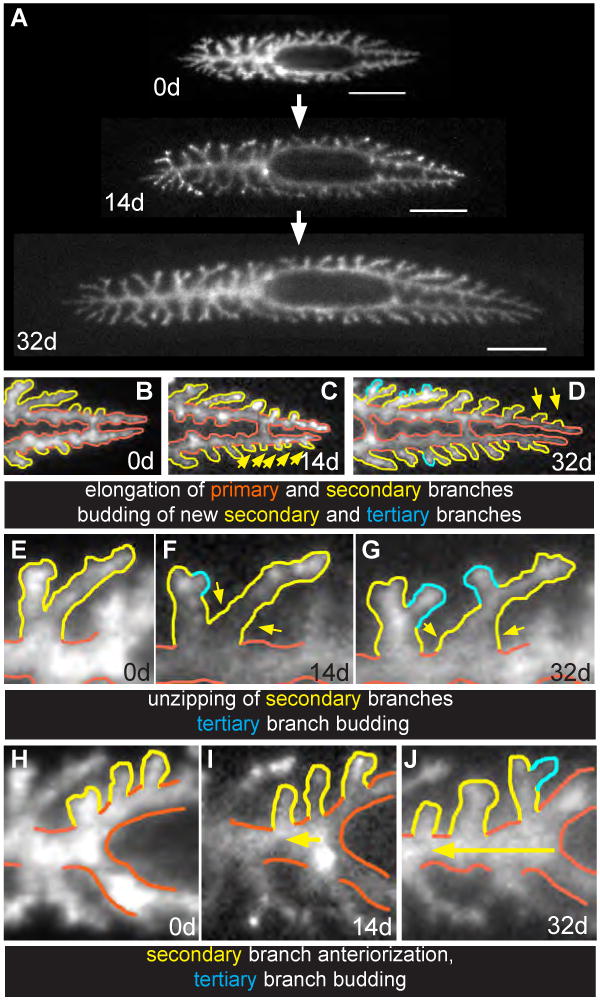
Animals were fed fluorescent dextrans 1-2 days prior to imaging at 0, 14, and 32 days, and fed calf liver every 2-3 days routinely between dextran feedings. One of the 11 animals analyzed is shown. (A) Over the month-long time course, intestinal branches elongated and animals elaborated new secondary and tertiary branches as they grew. (B-D) In posterior regions, primary branches (orange) elongate, new secondary branches (yellow, arrows) bud from the lateral surfaces of primary branches, and tertiary branches (cyan) bud from the sides of secondary branches. (E-G) In anterior regions, new secondary branches are generated by an “unzipping” mechanism, during which tertiary branches (long yellow branch, right, arrows) migrate medially to eventually become new secondary branches anchored on the primary branch (orange). In addition, new tertiary branches (cyan) appear by budding from secondary branches. (H-J) New anterior secondary branches are also generated by the anteriorization of secondary branches from posterior regions near the pharynx to positions along the primary anterior branch. Scale bars in A, 500 μm. Orange, primary branches; yellow, secondary branches; cyan, tertiary branches. Anterior is to the left in all panels.
As expected, primary and secondary branches elongated in growing animals, proportional to overall animal size (Figs. 2A-D, Supplementary Table 1). In addition, new intestinal branches were elaborated by a number of distinct processes. The type of branching morphogenesis observed varied depending on anteroposterior location. For example, in the tail, new secondary branches emerged de novo from the lateral surfaces of posterior primary branches (Figs. 2B-D), in a process similar to “budding” or “side branching” observed during branching morphogenesis in other organisms (Andrew and Ewald, 2010). In the anterior of the animal, however, secondary branches often appeared by the “splitting” or “unzipping” of tertiary branches, which gradually migrated more medially to the primary anterior branch, becoming new secondary branches (Figs. 2E-G). Alternatively, new secondary branches also arose by “anteriorization,” in which branches from peripharyngeal regions of posterior branches were displaced over time to more anterior positions (Figs. 2H-J). Tertiary branches were generated throughout the animal by either branching from the sides of secondary branches, or by bifurcation at the tips of secondary branches (Figs. 2B-G).
Intestinal cells do not divide
Given that intestinal branches elongate and new branches are formed in growing animals, we next sought to identify the source of this new tissue. In vertebrates and some invertebrates, the intestinal epithelium is continuously renewed by resident, lineage-restricted stem cells (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006; Radtke and Clevers, 2005). In planarians, however, all existing data suggest that proliferating somatic cells do not reside within differentiated tissues (Baguñá and Romero, 1981; Eisenhoffer et al., 2008; Guo et al., 2006; Newmark and Sánchez Alvarado, 2000; Orii et al., 2005; Palakodeti et al., 2008; Reddien et al., 2005; Rossi et al., 2006; Rossi et al., 2007; Salvetti et al., 2000; Salvetti et al., 2005).
We formally verified that this was the case for the intestine. The layer of enteric muscles that surrounds the intestine serves to delineate the boundary between the intestinal epithelium and adjacent mesenchymal tissue (Figs. 1 and 3). In intact, uninjured planarians, mitotic neoblasts can be labeled with a marker that recognizes phosphorylation of histone H3 on serine 10 (anti-phospho-histone-H3-Ser10) (Newmark and Sánchez Alvarado, 2000). Using this marker, we find that mitotic cells are found in the mesenchyme and are often closely associated with the enteric muscles (Figs. 3A, D). However, cycling cells are never found on the lumenal side of the muscle boundary (Figs. 3A, D). We further assessed proliferation after feeding, when mitoses increase several-fold over a 24- to 48-hour period (Figs. 3A-C) (Baguñà, 1974; Baguñà, 1976a; Baguñá and Romero, 1981). Again, dividing cells were not observed within the intestine proper, suggesting that there are no quiescent enterocytes that divide in response to nutritional stimuli (Figs. 3E, F).
Figure 3. Intestinal cells do not divide.
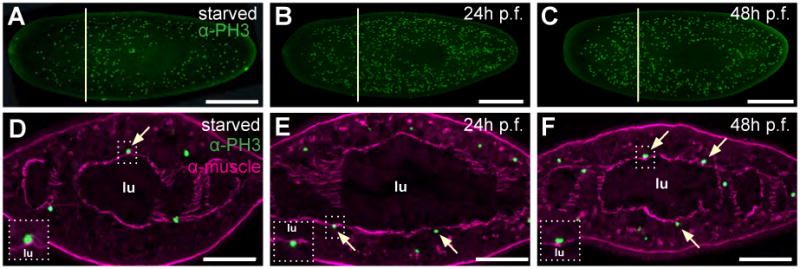
(A-C) Confocal projections of whole-mount planarians labeled with anti-phospho-histone-H3-Ser10 (green). In recently fed animals (i.e. 24h or 48h post-feeding (“p.f.”), B and C), proliferation increases 4-5 fold compared to starved animals (A). (D-F) Epifluorescent images of representative cross sections, taken from approximate regions indicated by yellow lines in A-C. Mitotic cells (green) are often closely associated with enteric muscles, but are never found on the lumenal side of the enteric muscle boundary, even in recently fed animals (E and F). Anterior is to the left (A-C). Dorsal is to the top (D-F). Scale bars: 500 μm, A-C; 100 μm, D-F.
Neoblasts differentiate into intestinal epithelial cells
To establish that intestinal epithelial cells (IECs) are the descendants of planarian stem cells, we labeled S-phase neoblasts by feeding planarians the thymidine analog bromodeoxyuridine (BrdU) (Newmark and Sánchez Alvarado, 2000). BrdU-positive cells are not observed on the lumenal side of the enteric muscle boundary up to 24 hours after the initial pulse (Figs. 4A-B, E-F), further demonstrating that intestinal cells do not actively cycle. Over the course of three to five days, however, BrdU-positive IECs appear in all intestinal branches, and along all polar axes (Figs. 4C-D, G-J, and Supplemental Fig. 1). These data are consistent with previous studies in which BrdU initially labels only neoblasts, based on morphological and molecular characterization (Eisenhoffer et al., 2008; Newmark and Sánchez Alvarado, 2000). Additionally, our observations reinforce the idea that neoblasts are the sole source of differentiated somatic cells. Interestingly, IECs differentiate uniformly along mediolateral, dorsoventral, and antero-posterior axes (Figs. 4G, H, and Supplemental Figs. 1C, D). We do not, for example, observe elevated numbers of BrdU-positive cells at the tips of primary, secondary, or tertiary branches (Supplemental Figs. 1C-F). Thus, new cells integrate uniformly throughout the intestine, suggesting that gut branches increase in length and that branching morphogenesis occurs by orchestrated remodeling of differentiated tissue, not by the birth of new cells at specific growth zones.
Figure 4. Neoblasts differentiate into intestinal cells in uninjured planarians.
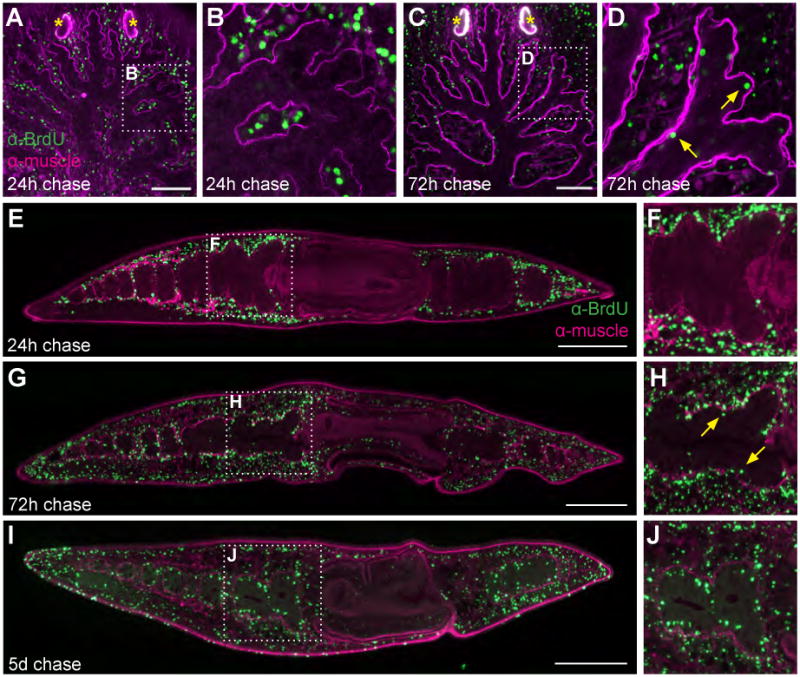
Animals were fed BrdU, and fixed after 1, 3, or 5 days. (A-D) Whole-mount planarians imaged confocally; projections of a subset of optical sections are shown. (A-B) At 24h, BrdU-positive nuclei (green) are found exclusively outside the enteric muscle layer (magenta) (A-B). (C-D) At 72h, BrdU-positive intestinal cells (neoblast progeny) begin to differentiate (C-D, yellow arrows). (E-J) Epifluorescent images of sagittal sections taken near the animal midline. (E-F) At 24h, BrdU-positive nuclei (green) are not found on the luminal side of the enteric muscle boundary. (G-J) Increasing numbers of intestinal cells differentiate over the course of 3-5 days. F, H, and J are insets of boxed regions in E, G, and I, respectively. Anterior is to the top in A-D. Anterior is to the left, dorsal to the top in E-J. Photoreceptor pigment cups (asterisks) label nonspecifically in A and C. BrdU-positive cells have also differentiated in the epidermis and pharynx at 3 and 5 days (G and I). Scale bars, 100 μm (A-D), 250 μm (E-J).
Planarians regenerate intestinal polarity 5-7 days after amputation
In addition to adding new intestinal branches during growth, planarians must also regenerate their digestive system after injury. To characterize the spatial and temporal dynamics of intestinal regeneration we analyzed the re-establishment of intestinal morphology in head, trunk, and tail fragments. Using whole-mount in situ hybridization (WISH) (Umesono et al., 1997), we visualized intestinal morphology with an intestine-specific riboprobe, Smed-cofilin (Fig. 5) (Zayas et al., 2005).
Figure 5. Intestinal regeneration.
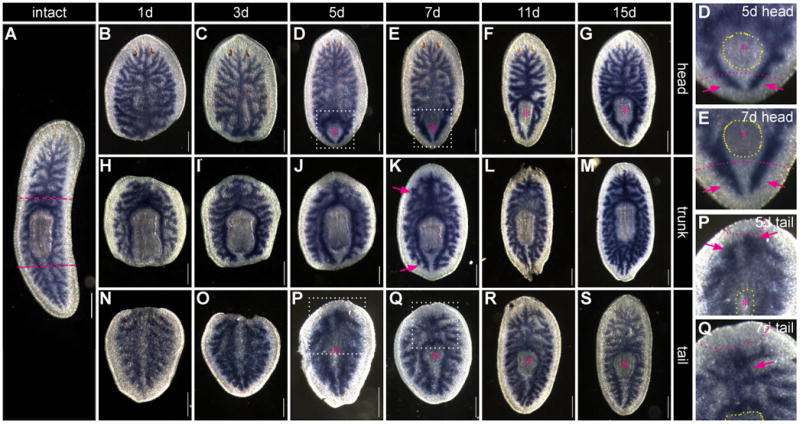
Animals were amputated (A, pink dashed lines), allowed to regenerate, and fixed at the times indicated. The intestine was visualized with a Smed-cofilin riboprobe. (B-G) Head fragments. (H-M) Trunk fragments. (N-S) Tail fragments. Boxed regions in D, E, P, and Q are magnified in insets on the right. Pink asterisks and yellow dashed lines indicate regenerating pharynges in D-G and P-S. Elongating anterior and posterior branches are indicated in D and E insets and panel K (pink arrows). Formerly posterior branches fusing to become the new primary anterior branch are shown in P and Q insets (pink arrows). All scale bars = 250 μm.
In anterior (head) fragments regenerating a new tail (Figs. 5B-G), within five days after injury new posterior branches have begun to project around the regenerating pharynx (Figs. 5B-D). Initially, these posterior branches are quite short, relative to total animal length, but over time, they lengthen as proper symmetry and proportion are restored (Figs. 5E-G). Additionally, at later time points (11-15 days), secondary branches begin to extend laterally from new posterior primary branches (Figs. 5F-G). Accompanying the elaboration of new posterior branches, the pharynx elongates and becomes located more centrally as regeneration proceeds (Figs. 5D-F).
In trunk (pharyngeal) fragments (Figs. 5H-M), anterior and posterior intestinal branches are present after injury, but their length is disproportionately short (Fig. 5H). Beginning around seven days after amputation, these branches grow and extend distally, continuing to lengthen through 15 days of regeneration (Figs. 5K-M). Growth of anterior and posterior branches is accompanied by a gradual reduction in proportional size of the pharynx, a morphological change that is achieved in part by apoptosis (Pellettieri et al., 2009).
In posterior (tail) fragments regenerating a new head (Figs. 5N-S), by five days after amputation, anterior regions of formerly posterior branches begin to converge towards the midline in front of the regenerating pharynx, which develops more centrally in tail fragments than it does in head fragments (Fig. 5P). In addition, midway along the AP axis, primary intestinal branches shift laterally to accommodate the new pharyngeal cavity (Figs. 5P-Q). By seven days of regeneration, the re-establishment of anterior intestinal morphology has progressed considerably, although gaps are clearly visible in the primary branch (Fig. 5Q). Often, two weeks or more are required for the resolution of a single anterior primary branch (Fig. 5S).
We characterized intestinal regeneration further using immunofluorescence to detect two additional markers (Supplemental Fig. 2): phosphotyrosine, which localizes at the apical surface of IECs, effectively marking the intestinal lumen (Fig. 1C) (Cebrià et al., 2007; Guo et al., 2006); and the layer of enteric muscles surrounding all intestinal branches (Fig. 1B). Anti-phosphotyrosine labels the lumen of regenerating branches at the earliest stages of their morphogenesis and throughout regeneration (e.g. in early tail fragments, Supplemental Fig. 2J), while re-establishment of the enteric muscle layer occurs at later time points. As in our initial analysis (Fig. 5), restoration of intestinal polarity visualized by these markers occurred by approximately five days in head and tail fragments (Supplemental Figs. 2B, K). In trunk fragments, gradual elongation of anterior and posterior branches occurred through at least seven days (Supplemental Figs. 2D-I).
Thus, in anterior and posterior fragments, gross anteroposterior intestinal morphology (i.e., the presence of both a single anterior primary branch and two posterior primary branches) is restored five to seven days after injury. In all three fragments, gradual remodeling and growth of these branches occurs over time, completing the restoration of normal proportion and symmetry.
Differentiated intestinal tissues remodel during regeneration
After amputation, small planarian fragments reorganize their bodies considerably to re-establish proportion and symmetry (Fig. 5) (Morgan, 1898; Reddien and Sánchez Alvarado, 2004). T. H. Morgan introduced the term “morphallaxis” to describe this process of transformation of “old” tissue that occurs at a distance from the plane of amputation (Morgan, 1898; Morgan, 1901). However, the degree to which this phenomenon involves the reorganization of old tissue as opposed to cell loss coordinated with differentiation of new tissue is largely unknown. This lack of understanding at the level of specific organs and tissues is due in part to a paucity of methods for clearly monitoring the position of cells that had already differentiated prior to injury. To address this issue, we again took advantage of the fact that intestinal phagocytes retain ingested fluorescent dextrans (Fig. 1A). By feeding dextrans prior to amputation, we were able to observe morphological changes to intestinal tissue in living regenerates over a time course (Fig. 6).
Figure 6. Intestinal tissue remodels during regeneration.
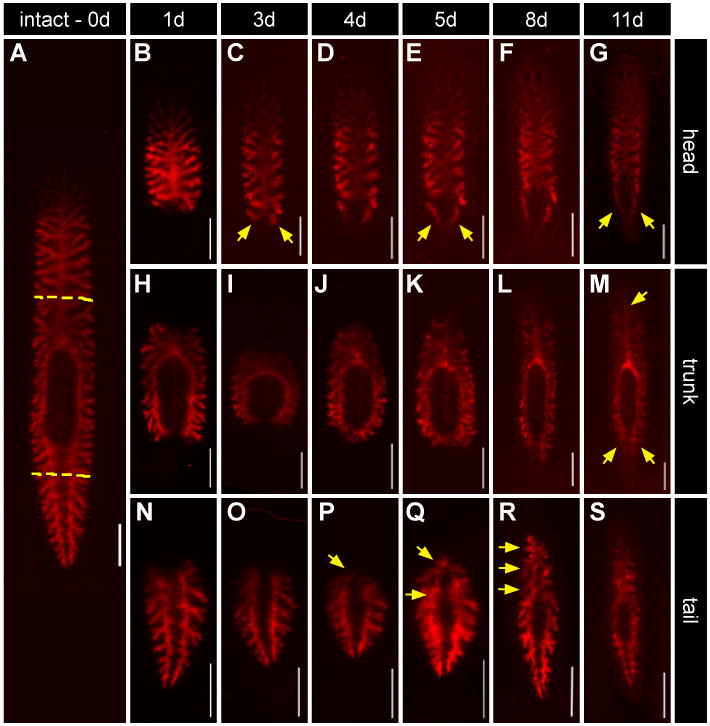
(A) A single animal was fed Alexa 546-conjugated dextrans (red) three times to label differentiated phagocytes. Animals were then amputated one day after the last feeding (yellow dashed lines). Living tissue fragments were imaged on subsequent days after cutting (top panel indicates times). (B-G) Head fragments. Labeled intestinal cells begin to project posteriorly as early as 3 days post amputation (C), contributing significantly to peripharyngeal and posterior branches by 11 days (yellow arrows, C, E, and G). (H-M) Trunk fragments. Anterior and posterior branches gradually elongate over the regeneration time course (yellow arrows, M). (N-S) Tail fragments. Labeled branches project across the midline initially at 4-5 days (yellow arrows, P and Q). Anterior branches eventually (R) coalesce at the midline to reconstitute the primary anterior branch. All scale bars = 500 μm.
Dextran-labeled intestinal branches reorganize extensively along mediolateral and anteroposterior axes, contributing significantly to the regenerating intestine. In general, the timing of morphological changes is similar, or slightly earlier than what was observed during in situ and immunofluorescent analysis. For example, in anterior (head) fragments regenerating their tails (Figs. 6B-G), we found that three to five days after amputation, labeled intestinal branches begin to project posteriorly around the regenerating pharynx (Figs. 6C-E). Eight to 11 days after injury, branches projecting around the pharynx and the anterior portions of posterior branches still fluoresce, suggesting a significant contribution of labeled, pre-existing cells to the restored peripharyngeal and posterior intestine (Figs. 6F, G). In trunk (pharyngeal) regenerates that must develop both a new head and tail, anterior and posterior intestinal branches undergo gradual elongation (Figs. 6H-M). In posterior (tail) fragments regenerating a new head (Figs. 6N-S), the anterior regions of formerly posterior branches coalesce at the midline, eventually merging to reconstitute the primary anterior branch, along with secondary and tertiary branches that project laterally (Figs. 6R-S).
In analysis of multiple specimens, we noticed there was animal-to-animal variability in the remodeling of pre-existing tissue. For example, in some head fragments, red fluorescence extended farther towards the tail in either the left or right posterior branch (Supplementary Figs. 3A-b'), and the posterior extent of the contribution of pre-injury IECs varied as well (Supplementary Figs. 3C-d'). As early as day three in some tail fragments, branches could be observed extending across the midline (Supplementary Figs. 3E-e'); by day five, most fragments had initiated midline fusion of anterior branches (Supplementary Figs. 3F-I). Additionally, at five days the number of these projections across the midline varied from zero to three (Supplementary Figs. 2F-I). Thus, although differentiated tissue always remodels and contributes to regenerating intestinal branches, the timing and precise dynamics of these morphological changes vary from animal to animal.
At later regeneration time points, fluorescence was barely detectable in branches closest to and within the blastema. To define more precisely the location of old (preamputation) and new intestinal cells, we fed animals two different dextran conjugates. Alexa 546-conjugated dextrans (red) were fed to animals prior to amputation, followed by Alexa 488-conjugated dextrans (green) after 10 days, when the pharynx and nervous system had completely regenerated, allowing animals once again to detect and ingest food. This two-colored labeling strategy allowed visualization of intestinal cells that existed prior to injury (red), as well as all functional intestinal branches, including newly differentiated intestinal cells (green) that had not taken up red dextrans (Figs. 7A-F). While most regions of the intestine were labeled by both dextrans, in regions close to the original plane of injury and within the blastema, intestinal branches fluoresced only in the green channel (Figs. 7a'-f'). We conclude that these branches within the blastema (labeled in green only) are comprised of recently differentiated phagocytes, while branches in pre-existing tissue more distant from the plane of amputation (those labeled in red and green) contain IECs that had differentiated and were labeled prior to amputation.
Figure 7. Differentiated intestinal tissue labeled prior to injury contributes minimally to regenerated branches within the blastema.
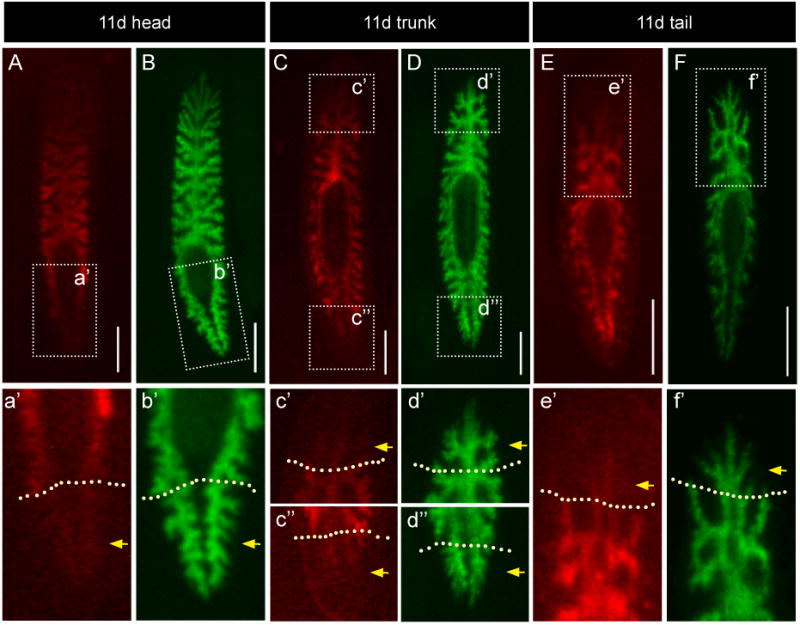
As in Figure 6, single animals were fed Alexa 546-conjugated dextrans (red, A, C, and E, insets in a', c'-c″, and e') to label differentiated phagocytes, and then amputated. At 10 days of regeneration, animals were fed Alexa 488-conjugated dextrans (green, B, D, and F, insets in b', d'-d″, and f') to visualize the entire regenerated intestine. All regenerates were imaged again at 11 days, one day after being fed green dextrans. (A-B) Head fragments. The most posterior regions of tail branches within the blastema (a', yellow arrows, compare to b') fluoresce only weakly in the red channel. (C-D) Pharyngeal fragments. Red fluorescence is reduced in both anterior and posterior branch tips (c'-c″, yellow arrows, compare to d'-d″). (E-F) Tail fragments. Red fluorescence intensity is lowest in the anterior-most branches (e', yellow arrow, compare to f'). Dashed yellow lines in a'-f' delineate the plane of amputation. All scale bars, 500 μm.
We also found that remodeling of pre-existing branches requires neoblasts. Ionizing irradiation selectively ablates the neoblasts, blocking the production of a blastema after amputation due to the absence of proliferating cells (Brøndsted, 1969). In animals that have been irradiated prior to amputation, remodeling of dextran-labeled intestinal tissue is either severely delayed (Supplemental Figs. 4C, c' and Supplemental Figs. 4G, g'), or is completely blocked (Supplemental Figs. S4D, d' and Supplemental Figs. 4H-h') in both anterior and posterior fragments. Our results provide in vivo confirmation of independently conducted experiments in which, despite the appropriate reestablishment of axial polarity cues in irradiated animals, intestinal reorganization does not occur in tail pieces analyzed by FISH (Gurley et al., 2010).
During regeneration neoblasts differentiate into IECs within the blastema and in remodeling branches
To verify that amputation does not stimulate cell division by otherwise quiescent enterocytes, we assessed proliferation in regenerates two and five days after amputation. In these experiments, we did not observe phosphohistone-H3-positive cells within the enteric muscle boundary in regenerates, either at the tips of severed (Supplemental Figs. 5A, B) or elongating (Supplemental Fig. 5C) intestinal branches, or in regions of remodeling (Supplemental Fig. 5D). These data suggest that during regeneration, as in intact and recently fed animals (Fig. 3), new intestinal cells arise as the progeny of neoblasts.
In order to determine the spatial pattern of intestinal cell differentiation during regeneration, we fed BrdU to uninjured planarians, labeling S-phase neoblasts. Animals were then amputated and the location of BrdU-positive intestinal cells (i.e. BrdU-positive neoblast progeny) was assessed in histological sections of regenerating head and tail fragments (Figs. 8 and 9). At three days after amputation, BrdU-positive cells are found primarily at the distal tips of severed branches within the partially restored enteric muscle boundary (Figs. 8A, B and 8E-H). In five day regenerates, IEC differentiation is more widespread, occurring preferentially in new branches within and near the blastema, but also in regions where dextran labeling experiments showed a significant degree of remodeling by IECs that existed prior to amputation (Figs. 6 and 7). For example, in head fragments these sites include new posterior branches (Figs. 8C and I-J), but also more anterior regions where the intestine must re-establish continuity with the regenerating pharynx (Figs. 8C and K-L). In tail fragments, new IECs differentiated at locations where new branches projected across the anterior midline (Figs. 8M, N and Supplemental Fig. 6A), and along the medial wall of branches on either side of the regenerating pharynx (Supplemental Fig. 6B).
Figure 8. Differentiation of intestinal cells at three and five days after amputation.
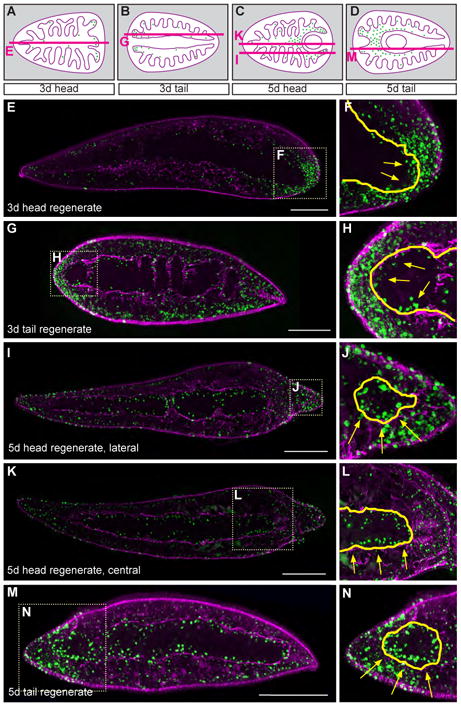
Animals were fed BrdU, amputated 24h later, and allowed to regenerate for the times indicated. They were then fixed, cryosectioned, and labeled with anti-BrdU (green) and anti-muscle (magenta) antibodies. (A-D) Schematics of sagittal planes (red lines) examined for 3- and 5- day head and tail regenerates in E-N. Green dots, BrdU-positive nuclei. (E-H) In 3-day anterior and posterior regenerates, new intestinal cells (arrows, green nuclei within the yellow boundary, F and H) have differentiated primarily at the severed ends of intestinal branches. (I-L) In 5-day head regenerates, intestinal cells have differentiated preferentially within new posterior branches in the blastema (arrows, J) as well as more anteriorly (arrows, L) where the primary anterior branch integrates with the regenerating pharynx. (M-N) In 5-day tail regenerates, intestinal cells have differentiated anteriorly, in new branches that are remodeling and extending across the anterior midline (arrows, N). Anterior is to the left in all panels. Scale bars, 200 μm.
Figure 9. Enterocytes differentiate in both new and remodeling intestinal branches.
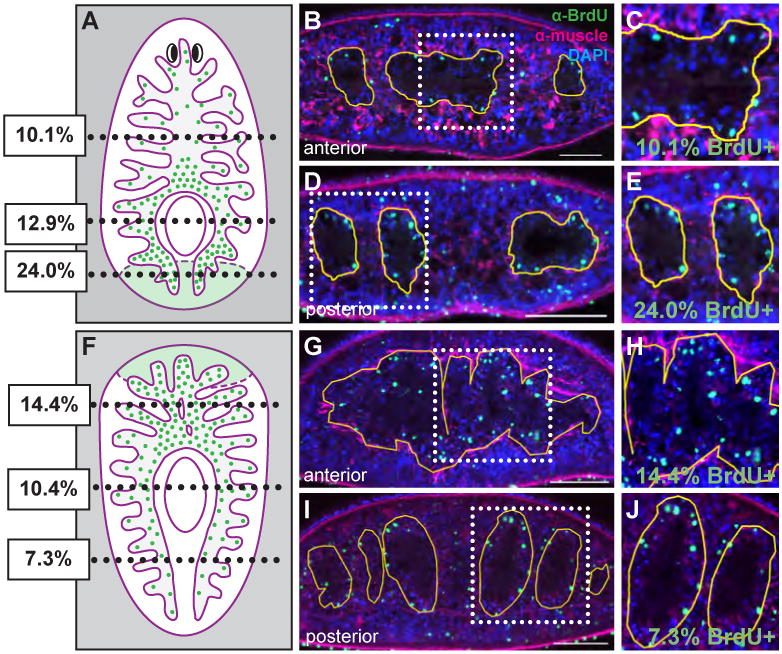
(A and F) Schematics demonstrating the location of cross sections of seven day head (A) and tail (F) regenerates examined quantitatively (see Results). The average percentage of BrdU-positive enterocytes within the muscle boundary are indicated. The blastema is shaded in light green, and locations where differentiation occurs preferentially are represented by increased numbers of BrdU-positive nuclei (green dots). (BE) Representative sections and magnifications of anterior (B and C) and posterior (D and E) regions of head regenerates. Over twice as many intestinal cells have differentiated in posterior branches (E) than in anterior branches (C). (G-J) Representative sections from tail regenerates. Approximately twice as many new intestinal cells have differentiated in anterior regions (G and H) than in posterior branches (I and J). Green, BrdU; magenta, muscle; blue, DAPI. Anterior is to the top in A and F; dorsal is to the top in all other panels. Scale bars in B, D, G, and I, 100 μm.
In order to quantitatively analyze differentiation, we assessed BrdU-labeling indices in head and tail regenerates at seven days after amputation, a time point at which the three primary intestinal branches (i.e., anteroposterior polarity) have been restored (Fig. 5). In head fragments regenerating their tails, recently regenerated posterior intestinal branches contain over twice as many BrdU-positive nuclei (24.0%, n = 6 sections from 2 animals, 716 cells) than in more anterior (i.e. pre-existing) branches (10.1%, n = 8 sections from 2 animals, 2152 cells) (Figs. 9A-E). In tail fragments regenerating their heads, a similar increase in differentiation is observed in regenerating tissue. In anterior branches, twice as many nuclei are BrdU-positive (14.4%, n = 4 sections from 2 animals, 1305 cells) as in posterior (pre-existing) branches (7.3%, n = 5 sections from 2 animals, 1582 cells) that did not undergo significant remodeling during regeneration (Figs. 9F-J). These data correlate with our dextran labeling experiments, in which branches closest to the plane of amputation retain the lowest levels of fluorescence (Fig. 7).
Intriguingly, in both anterior and posterior regenerates, differentiation is also moderately elevated in peripharyngeal branches (i.e., posterior branches that project around both sides of the regenerating pharynx) located at a considerable distance from the plane of amputation. For example, in head fragments, 12.9% (n = 4 sections from 2 animals, 1430 cells) of cells in peripharyngeal branches were BrdU positive. In tail fragments, 10.4% (n = 6 sections from 2 animals, 1158 cells) peripharyngeal IECs were BrdU positive. Thus, although dextran labeling experiments (Figs. 6 and 7) suggest a significant contribution of pre-existing IECs to peripharyngeal branches, differentiation of new intestinal cells occurs at a higher frequency during regeneration of these regions of the intestine as well. Taken together, our data indicate that during regeneration, neoblasts differentiate into new intestinal cells not only within intestinal branches that form entirely within the blastema (e.g. posterior branches in head regenerates), but also in regions of the intestine that remodel within pre-injury tissue at some distance from the blastema.
Discussion
Planarians possess a population of pluripotent somatic stem cells (the neoblasts) that are distributed throughout the body. As such, these animals offer a unique opportunity to understand how non-resident stem cells respond to traumatic injury and rebuild organs postembryonically, a phenomenon that is not well understood in regenerating organisms. However, the dynamics of stem cell differentiation, and the integration of new cells with tissue that existed prior to injury, have not been well characterized at a cellular level.
In this investigation, we have investigated these processes (differentiation and remodeling) in the context of growth and regeneration of the planarian intestine. In growing animals, we have found that intestinal cells differentiate uniformly along all intestinal branches, and that new gut branches are elaborated by the remodeling of differentiated tissue. By contrast, after injury, differentiation of new intestinal cells is spatially restricted, occurring both within the blastema, but also wherever intestinal branches remodel to restore intestinal polarity and integrate with regenerating organs such as the pharynx. Remodeling is similarly restricted, occurring in regions where missing intestinal branches must be re-established; however, differentiated enterocytes that existed prior to injury do not contribute significantly to new branches within the blastema. In regions far from the plane of amputation, for example in the posterior gut branches of tail fragments, both remodeling and differentiation are minimal.
Over one hundred years ago, Thomas Hunt Morgan introduced the concepts “epimorphosis” (generation of new tissue by proliferation) and “morphallaxis” (remodeling of pre-existing tissue without proliferation at the plane of amputation) to characterize regeneration in planarians and other organisms (Morgan, 1898; Morgan, 1901; Morgan, 1902). Although the these terms do not precisely describe events at a cellular level (Agata et al., 2007; Reddien and Sánchez Alvarado, 2004; Salò and Baguñà, 2002), our observations are nonetheless broadly consistent with Morgan's descriptions and elucidate these processes further in two ways. First, we do not observe a significant contribution of pre-existing, differentiated intestinal tissue to the blastema, supporting the idea that differentiated tissues produced at the plane of amputation are generated de novo as the progeny of neoblasts (epimorphosis). Second, our demonstration that intestinal tissue remodels indicates that morphallaxis to re-establish proportion and symmetry in small tissue fragments is achieved not only by cell loss (Pellettieri et al., 2009; Reddien and Sánchez Alvarado, 2004) but by orchestrated reorganization of differentiated cells and increased differentiation in remodeling tissues.
The idea that new tissue is produced not only at the site of injury, but also in other regions of the body, is not novel; for example, the pharynx often regenerates at some distance from the blastema (Brøndsted, 1969; Kobayashi et al., 1999; Reddien and Sánchez Alvarado, 2004). We have demonstrated that new intestinal cells are also produced at sites distant from the plane of amputation, and suggest that functional integration of new tissue with old occurs over a wider region of the body than has been previously appreciated, not just at the boundary between the blastema and pre-existing structures. Furthermore, because irradiation to ablate the neoblasts blocks intestinal remodeling, differentiation and remodeling must be coordinated to re-establish organ morphology within pre-existing tissue.
Neoblasts are the source of new intestinal cells
In most model organisms in which the renewal of intestinal cells has been studied, the gut epithelium is replenished by lineage-restricted stem cells that reside within the digestive tract (Crosnier et al., 2006; Faro et al., 2009; Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). By contrast, we have formally demonstrated that enterocytes do not divide in uninjured, recently fed, or injured planarians, but rather, that neoblasts differentiate into intestinal cells under these conditions. Our results are consistent with the fact that mitotic cells have not been reported in ultrastructural studies of the planarian intestine (Bowen et al., 1974b; Garcia-Corrales and Gamo, 1988; Ishii, 1965). Furthermore, with few exceptions, planarian genes expressed by neoblasts are not expressed in the intestine (Eisenhoffer et al., 2008; Guo et al., 2006; Palakodeti et al., 2008; Reddien et al., 2005; Rossi et al., 2006; Rossi et al., 2007; Rouhana et al., 2010; Salvetti et al., 2000; Salvetti et al., 2005; Shibata et al., 1999; Solana et al., 2009).
Thus, in planarians, mesenchymally located neoblasts must commit to intestinal cell fates, cross a layer of enteric muscles and basement membrane, integrate into the gastrodermal epithelium, and elaborate the specialized organelles and cytoplasmic volume required for secretory or absorptive functions (Fig. 10). These morphological differences between planarians and other organisms raise a number of intriguing questions. How are stem cells that reside outside of differentiated tissues recruited and instructed to commit to intestinal cell fates? Does the intestine release cues that promote neoblast lineage restriction and/or stimulate migration into the intestine proper? Do evolutionarily conserved mechanisms that regulate intestinal cell differentiation function in planarians? Do these mechanisms differ depending on whether differentiated cells are produced within the blastema or along pre-existing gut branches? We address several of these issues in greater detail, below.
Figure 10. Model of intestinal differentiation.
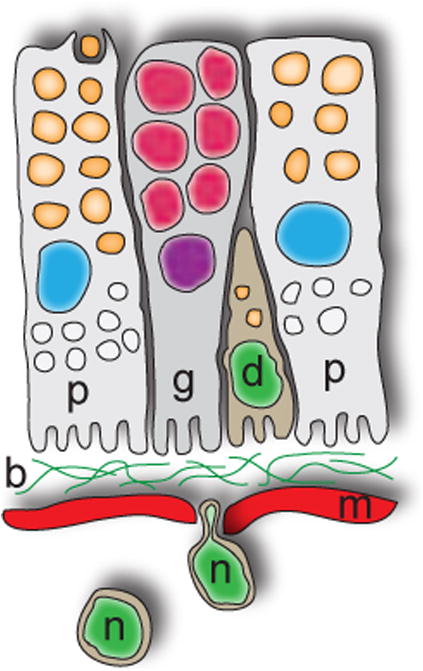
Our data indicate that neoblasts (n) must commit to intestinal cell fates, cross the layer of enteric muscles (red, m) and basal lamina (b), and integrate into the epithelium, eventually differentiating (d) into either absorptive phagocytes (p) or secretory goblet cells (g). The precise location at which neoblasts commit to intestinal lineages awaits identification of early differentiation markers. Orange, phagocytic vacuoles; white, lipid droplets; red, secretory granules. In contrast to enterocytes in vertebrates and other organisms, planarian absorptive cells do not possess a microvillar or ‘brush’ border, but do have an elaborate, infolded basal membrane. Apical is to the top, basal is to the bottom.
First, although lineage-restricted stem cells do not exist in the intestine, do neoblasts whose fates are restricted to intestinal cell types exist? Mitotic cells are often closely associated with the intestine. However, these cells are always found on the mesenchymal side of the enteric muscle boundary, and are not located at stereotypical positions along intestinal branches. Furthermore, in uninjured animals, differentiation of BrdU-positive intestinal cells does not occur at any particular axial location. Thus, if lineage-restricted neoblasts exist, they do not appear to be confined to a stereotypical anatomical region along the digestive tract. Analyses of gene expression both in vivo and of purified neoblasts have revealed considerable heterogeneity of the stem cell population (Eisenhoffer et al., 2008; Hayashi et al., 2006; Hayashi et al., 2010; Rossi et al., 2006; Rossi et al., 2007). In almost all cases, however, data from these investigations suggest that markers of differentiation are expressed by cells that have exited the cell cycle (Eisenhoffer et al., 2008; Hayashi et al., 2010). Ultimately, identification of genes expressed during early phases of intestinal cell differentiation and analysis of co-expression with cell cycle and stem cell markers will be required to formally rule out the existence of a neoblast population committed to intestinal cell fates.
Second, it will be important to determine the molecular details of intestinal cell fate commitment and differentiation. In vertebrates and Drosophila, a variety of evolutionarily conserved signaling pathways regulate the proliferation and differentiation of resident intestinal stem cells (Casali and Batlle, 2009; Crosnier et al., 2006; van der Flier and Clevers, 2009). As one example, in mouse, secreted Wnt proteins play a central role in promoting proliferation of both crypt base columnar cells and transient amplifying cells (van der Flier and Clevers, 2009). Additionally, Wnt signaling is crucial for the development of secretory cell types (Crosnier et al., 2006; van der Flier and Clevers, 2009). In planarians, Smed-wnt11-2, Smed-βcatenin-1, and Smed-APC play crucial roles in the determination of anteroposterior polarity; disruption of expression of these genes by RNAi leads to altered polarity of the intestine, central nervous system, and other tissues (Adell et al., 2009; Gurley et al., 2010; Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008). Other planarian wnt genes are expressed by cells within or near the intestine (Gurley et al., 2010). However, it is currently unknown whether these genes play additional roles in the promotion of neoblast proliferation or differentiation into intestinal cells, or whether the intestine releases other, as-yet unidentified cues to regulate neoblast dynamics.
More generally, it will be important to understand how signaling is regulated in different contexts, for example, after injury or during growth. For example, in regions where IEC differentiation is elevated during regeneration (e.g., the severed tips of intestinal branches and remodeling branches), are factors that influence neoblast cell fate commitment (e.g., secreted regulators) simply expressed at higher levels? Or do planarians possess additional upstream signaling programs that link wound healing and the generation of various cell types, including IECs? Answers to these questions may be key to understanding why some organisms are more adept at repairing damage to their digestive tracts than others.
One potential source of regeneration-specific extracellular cues might be apoptotic cells. Apoptosis is dramatically upregulated near the amputation site in planarians, although whether enterocytes at the severed ends of gut branches are among these dying cells is not yet known (Pellettieri et al., 2009). Intriguingly, in amputated planarian tail fragments, proliferating neoblasts localize antero-dorsally, around the severed tips of intestinal branches, suggesting that the damaged intestine may in fact have a more general role in the regulation of neoblast dynamics during regeneration (Wenemoser and Reddien, 2010). The phenomenon of compensatory proliferation (cell division induced by nearby apoptotic cells) may be a widespread mechanism during the initiation of regeneration in Hydra, Drosophila, and other organisms (Bergmann and Steller, 2010; Galliot and Chera, 2010; Martin et al., 2009). For example, in the midgut of Drosophila, apoptotic cells release cytokines that promote the proliferation of intestinal stem cells (Amcheslavsky et al., 2009; Jiang et al., 2009). Likewise, in the mouse intestine, irradiation and cytotoxic damage cause increased proliferation and replacement of lost cells, although the molecular mechanisms responsible are currently less well understood (Merritt et al., 1994; Potten, 1992; Potten, 1998; Potten, 2004). Whether similar mechanisms function to promote intestinal regeneration in planarians remains to be determined.
Branching morphogenesis of the planarian intestine occurs through remodeling of differentiated cells
We visualized the morphogenesis of new branches in vivo in both growing and regenerating animals by labeling differentiated intestinal phagocytes with fluorescently conjugated dextrans. In growing animals, remodeling of differentiated tissue appears to be the major mechanism by which new branches are generated, and we observed a number of distinct modes of branching morphogenesis along the anteroposterior axis. In regenerating animals, tissue labeled prior to amputation also reorganizes, contributing significantly to new intestinal branches. We did not detect disassembly or reassembly of branches into individual cells during regeneration; thus, remodeling would seem to occur at the level of entire branches.
In organisms across the animal kingdom, a variety of organs depend on the development and maintenance of tubular structures (Affolter et al., 2009; Andrew and Ewald, 2010). Some of the modes of branching morphogenesis we describe here resemble those observed in other model systems. For example, budding morphogenesis, side branching, and elongation have been described in the mouse lung, Drosophila trachea, and the mammalian vasculature, to name just a few (Andrew and Ewald, 2010). Similarly, the medial migration of anterior tertiary branches in growing animals (which we term “unzipping”) bears some resemblance to clefting of salivary glands (Andrew and Ewald, 2010). To our knowledge, however, there is no similar description of anteriorization of peripharyngeal secondary branches (migration of entire branches along the anteroposterior axis) or remodeling of entire, fully developed tubular organ systems after amputation.
Branching morphogenesis is often achieved by a combination of collective cell migration, cellular rearrangement, changes in cell shape, and modulation of cell:cell junctions (Andrew and Ewald, 2010). Because remodeling can be visualized in vivo, the planarian intestine offers a unique opportunity to identify new molecular mechanisms regulating the morphogenesis of epithelial tubes, and more importantly, to compare these processes in a variety of contexts including homeostatic maintenance, growth, and regeneration. A number of molecules (e.g. secreted Wnt and Slit proteins, as well as Innexin-family gap junction proteins) that regulate anteroposterior and mediolateral polarity in planarians also influence the regeneration of the three-branched morphology of the intestine (Adell et al., 2009; Cebrià et al., 2007; Gurley et al., 2010; Gurley et al., 2008; Iglesias et al., 2008; Nogi and Levin, 2005; Oviedo et al., 2010). However, it is unknown whether these signaling pathways specifically influence remodeling of pre-existing branches, the localized differentiation of new intestinal cells, or both. Identification of the molecules required for intestinal cells (or neoblasts) to respond autonomously to these polarity signals is thus one important avenue for future research.
Intriguingly, we found that irradiation to ablate the neoblasts blocks intestinal remodeling after amputation. Others have observed a similar severe effect on the reorganization of the intestine in regenerating tail fragments, despite the appropriate re-establishment of the expression of anterior-posterior polarity cues such as wnt genes (Gurley et al., 2010). However, that study did not distinguish between the production of new intestinal cells and the remodeling of gut tissue that existed prior to amputation. Our in vivo results explicitly confirm that reorganization of pre-injury tissue is dependent on the neoblasts.
Why would neoblasts be required for remodeling, which should in theory be able to occur via cell-autonomous regulation of intestinal cell shape and/or motility? There are several possibilities, which are not necessarily mutually exclusive. The first is that intestinal regeneration occurs as a highly coordinated process that requires both reorganization of differentiated tissue and the birth of new intestinal cells. Some aspects of regeneration may be primarily dependent on the neoblasts; blocking these events may in turn abolish remodeling that occurs later. As an example, during earlier stages (around 5 days post amputation) of regeneration in tail fragments, new intestinal cells are born preferentially at the midline, where contact between formerly posterior branches must be re-established as a first step in reconstitution of the primary anterior branch. Perhaps the differentiation of new IECs at this specific location somehow stimulates and/or is required for later remodeling events. Second, without the progeny of neoblasts, there may simply not be enough intestinal tissue to re-establish gut morphology. For example, posterior branches in head fragments regenerating their tails may not be able to increase in length without the birth of new intestinal cells. Finally, remodeling of the intestine may be influenced by the neoblast-dependent regeneration of other tissues such as the central nervous system or pharynx. As one possibility, intestinal reorganization in head or tail fragments may not be able to proceed without signals produced by nascent pharynx progenitors. Elucidation of the influence of other tissues on intestinal remodeling will be required to discriminate between these possibilities.
Future perspectives
Recent investigations into the biology of stem cells have informed our understanding of homeostatic tissue maintenance, degenerative diseases, and cancer (Gurley and Sánchez Alvarado, 2008; Poss, 2010). However, in most organisms in which stem cell biology has been studied, regenerative abilities are somewhat limited. An important goal is to understand why some organisms with stem cells can regenerate their organs, while others cannot. Can organs be coaxed to regenerate in vivo? Can stem cells be engineered to produce transplantable tissue ex vivo? As studies of stem cell biology are expanded to organisms with significantly greater regenerative abilities, it will be critical to determine whether animals in different phyla accomplish post-embryonic organogenesis using similar strategies.
In this investigation, we have demonstrated that the planarian intestine will be a useful model for understanding post-embryonic organogenesis. Our work sets the stage for characterization of the molecular mechanisms regulating the differentiation of new cells from a pool of uncommitted, non-resident stem cells, the remodeling of pre-existing tissue, and the integration of old and new tissue during regenerative organogenesis and tissue growth. Work in the planarian intestine will complement investigations in other regenerating organisms (Kaneko et al., 2010; Mashanov et al., 2010; Ortiz-Pineda et al., 2009; Takeo et al., 2008). In the coming years, these efforts are likely to yield important insights into why some animals are capable of restoring functioning organs after severe damage, and how these mechanisms might be harnessed therapeutically.
Supplementary Material
Highlights.
> We examine regeneration and growth of the planarian intestine.
> In vivo imaging reveals morphogenesis and remodeling of intestinal branches.
> Differentiated intestinal tissue remodels during growth and regeneration.
> New intestinal cells are the progeny of neoblasts.
> Differentiation and remodeling are coordinated during growth and regeneration.
Acknowledgments
We would like to thank Dr. Tingxia Guo for initial optimization of labeled dextran feeding to planarians, Drs. Tingxia Guo and Francesc Cebrià for generation and initial characterization of the preimmune serum labeling planarian muscles, and Forrest Waters for technical assistance with analysis of proliferation in regenerating tissue fragments. We are grateful to Drs. Jim Collins, Rachel Roberts-Galbraith, Ryan King, and James Sikes for critical readings of this manuscript, and all members of the Newmark Lab for insightful discussions. This work was supported by a Ruth L. Kirschstein National Research Service Award from the National Institutes of Health (F32-DK077469) to DJF and R01-HD043403 to PAN. PAN is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note added in proof: While this manuscript was under review, Wagner et al. (Science 332 (2011) 811-816) demonstrated that some SMEDWI-1-positive neoblasts located outside of the intestine proper express early endodermal markers. Their data support the idea that neoblasts can commit to intestinal fate before integrating into the gut.
References
- Adell T, Saló E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for β-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–10. doi: 10.1242/dev.033761. [DOI] [PubMed] [Google Scholar]
- Affolter M, Zeller R, Caussinus E. Tissue remodelling through branching morphogenesis. Nat Rev Mol Cell Biol. 2009;10:831–42. doi: 10.1038/nrm2797. [DOI] [PubMed] [Google Scholar]
- Agata K, Saito Y, Nakajima E. Unifying principles of regeneration I: Epimorphosis versus morphallaxis. Dev Growth Differ. 2007;49:73–8. doi: 10.1111/j.1440-169X.2007.00919.x. [DOI] [PubMed] [Google Scholar]
- Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew DJ, Ewald AJ. Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev Biol. 2010;341:34–55. doi: 10.1016/j.ydbio.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñà J. Dramatic mitotic response in planarians after feeding, and a hypothesis for the control mechanism. J Exp Zool. 1974;190:117–22. doi: 10.1002/jez.1401900111. [DOI] [PubMed] [Google Scholar]
- Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. I. Mitotic studies during growth, feeding and starvation. J Exp Zool. 1976a;195:53–64. [Google Scholar]
- Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. II. Mitotic studies during regeneration, and a possible mechanism of blastema formation. J Exp Zool. 1976b;195:65–80. [Google Scholar]
- Baguñá J, Romero R. Quantitative analysis of cell types during growth, de-growth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia. 1981;84:181–194. [Google Scholar]
- Bergmann A, Steller H. Apoptosis, stem cells, and tissue regeneration. Sci Signal. 2010;3:re8. doi: 10.1126/scisignal.3145re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen ID. Phagocytosis in Polycelis tenuis. In: Smith DC, Tiffon Y, editors. Nutrition in the Lower Metazoa. Pergamon Press; Oxford: 1980. pp. 1–14. [Google Scholar]
- Bowen ID, Ryder TA, Thompson JA. The fine structure of the planarian Polycelis tenuis Iijima. II. The intestine and gastrodermal phagocytosis. Protoplasma. 1974;79:1–17. doi: 10.1007/BF02055779. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–49. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- Brøndsted HV. Planarian Regeneration. Pergamon Press; Oxford: 1969. [Google Scholar]
- Bueno D, Baguñà J, Romero R. Cell-, tissue-, and position-specific monoclonal antibodies against the planarian Dugesia (Girardia) tigrina. Histochem Cell Biol. 1997;107:139–49. doi: 10.1007/s004180050098. [DOI] [PubMed] [Google Scholar]
- Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4:124–7. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Cebrià F. Regenerating the central nervous system: how easy for planarians! Dev. Genes Evol. 2007;217:733–48. doi: 10.1007/s00427-007-0188-6. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Guo T, Jopek J, Newmark PA. Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol. 2007;307:394–406. doi: 10.1016/j.ydbio.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Ip YT. Pathogenic stimulation of intestinal stem cell response in Drosophila. J Cell Physiol. 2009;220:664–71. doi: 10.1002/jcp.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Stamataki D, Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet. 2006;7:349–59. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–39. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro A, Boj SF, Clevers H. Fishing for intestinal cancer models: unraveling gastrointestinal homeostasis and tumorigenesis in zebrafish. Zebrafish. 2009;6:361–76. doi: 10.1089/zeb.2009.0617. [DOI] [PubMed] [Google Scholar]
- Forsthoefel DJ, Newmark PA. Emerging patterns in planarian regeneration. Curr Opin Genet Dev. 2009;19:412–20. doi: 10.1016/j.gde.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B, Chera S. The Hydra model: disclosing an apoptosis-driven generator of Wnt-based regeneration. Trends Cell Biol. 2010;20:514–23. doi: 10.1016/j.tcb.2010.05.006. [DOI] [PubMed] [Google Scholar]
- García-Arrarás JE, Estrada-Rodgers L, Santiago R, Torres II, Díaz-Miranda L, Torres-Avillán I. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea:Echinodermata) J Exp Zool. 1998;281:288–304. doi: 10.1002/(sici)1097-010x(19980701)281:4<288::aid-jez5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Garcia-Corrales P, Gamo J. The ultrastructure of the gastrodermal gland cells in the freshwater planarian Dugesia gonocephala s.l. Acta Zool. 1986;67:43–51. [Google Scholar]
- Garcia-Corrales P, Gamo J. Ultrastructural changes in the gastrodermal phagocytic cells of the planarian Dugesia gonocephala s.l. during food digestion (Plathelminthes) Zoomorphology. 1988;108:109–117. [Google Scholar]
- Goodchild CG. Reconstitution of the intestinal tract in the adult leopard frog, Rana pipiens Schreber. J Exp Zool. 1956;131:301–327. [Google Scholar]
- Guo T, Peters AHFM, Newmark PA. A bruno-like gene is required for stem cell maintenance in planarians. Dev Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sanchez Alvarado A. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sánchez Alvarado A. β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–7. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Sánchez Alvarado A. Stem cells in animal models of regeneration, StemBook, ed. The Stem Cell Research Community, StemBook. 2008 doi: 10.3824/stembook.1.32.1. http://www.stembook.org. [DOI] [PubMed]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–80. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Shibata N, Okumura R, Kudome T, Nishimura O, Tarui H, Agata K. Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its “index sorting” function for stem cell research. Dev Growth Differ. 2010;52:131–44. doi: 10.1111/j.1440-169X.2009.01157.x. [DOI] [PubMed] [Google Scholar]
- Hyman LH. The reproductive system and other characters of Planaria dorotocephala Woodworth. Trans Am Microsc Soc. 1925;44:51–89. [Google Scholar]
- Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-βcatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–21. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- Illa-Bochaca I, Montuenga LM. The regenerative nidi of the locust midgut as a model to study epithelial cell differentiation from stem cells. J Exp Biol. 2006;209:2215–23. doi: 10.1242/jeb.02249. [DOI] [PubMed] [Google Scholar]
- Ishii S. Electron microscopic observations on the Planarian tissues II. The intestine. Fukushima J Med Sci. 1965;12:67–87. [PubMed] [Google Scholar]
- Ishizuya-Oka A. Regeneration of the amphibian intestinal epithelium under the control of stem cell niche. Dev Growth Differ. 2007;49:99–107. doi: 10.1111/j.1440-169X.2007.00913.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–55. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N, Katsuyama Y, Kawamura K, Fujiwara S. Regeneration of the gut requires retinoic acid in the budding ascidian Polyandrocarpa misakiensis. Dev Growth Differ. 2010;52:457–68. doi: 10.1111/j.1440-169X.2010.01184.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi C, Kobayashi S, Orii H, Watanabe K, Agata K. Identification of two distinct muscles in the planarian Dugesia japonica by their expression of myosin heavy chain genes. Zoolog Sci. 1998;15:861–869. [Google Scholar]
- Kobayashi C, Watanabe K, Agata K. The process of pharynx regeneration in planarians. Dev Biol. 1999;211:27–38. doi: 10.1006/dbio.1999.9291. [DOI] [PubMed] [Google Scholar]
- Lin SW. A new technique for planaria. Bios. 1931;2:115–117. [Google Scholar]
- Martin FA, Perez-Garijo A, Morata G. Apoptosis in Drosophila: compensatory proliferation and undead cells. Int J Dev Biol. 2009;53:1341–7. doi: 10.1387/ijdb.072447fm. [DOI] [PubMed] [Google Scholar]
- Mashanov VS, Dolmatov IY. Ultrastructural Features of Gut Regeneration in Five-Month-Old Pentactulae of the Holothurian Eupentacta fraudatrix. Russ J Mar Biol. 2001;27:376–382. [Google Scholar]
- Mashanov VS, Zueva OR, Rojas-Catagena C, Garcia-Arraras JE. Visceral regeneration in a sea cucumber involves extensive expression of survivin and mortalin homologs in the mesothelium. BMC Dev Biol. 2010;10:117. doi: 10.1186/1471-213X-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt AJ, Potten CS, Kemp CJ, Hickman JA, Balmain A, Lane DP, Hall PA. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res. 1994;54:614–7. [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–9. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Experimental studies of the regeneration of Planaria maculata. Archiv Entwick Mech Org. 1898;7:364–397. [Google Scholar]
- Morgan TH. Regeneration in planarians. Archiv Entwick Mech Org. 1900;10:58–119. [Google Scholar]
- Morgan TH. Regeneration. Macmillan Co.; New York: 1901. [Google Scholar]
- Morgan TH. Growth and regeneration in Planaria lugubris. Arch Entw Mech Org. 1902;13:179–212. [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–53. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Not your father's planarian: a classic model enters the era of functional genomics. Nat Rev Genet. 2002;3:210–9. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- Nogi T, Levin M. Characterization of innexin gene expression and functional roles of gap-junctional communication in planarian regeneration. Dev Biol. 2005;287:314–35. doi: 10.1016/j.ydbio.2005.09.002. [DOI] [PubMed] [Google Scholar]
- O'Steen WK. Regeneration of the intestine in adult urodeles. J Morphol. 1958;103:435–477. [Google Scholar]
- O'Steen WK. Regeneration and repair of the intestine in Rana clamitans larvae. J Exp Zool. 1959;141:449–475. doi: 10.1002/jez.1401410303. [DOI] [PubMed] [Google Scholar]
- O'Steen WK, Walker BE. Radioautographic studies of regeneration in the common newt III. Regeneration and repair of the intestine. Anat Rec. 1962;142:179–187. doi: 10.1002/ar.1091420210. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–4. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Orii H, Ito H, Watanabe K. Anatomy of the planarian Dugesia japonica I. The muscular system revealed by antisera against myosin heavy chains. Zoolog Sci. 2002;19:1123–31. doi: 10.2108/zsj.19.1123. [DOI] [PubMed] [Google Scholar]
- Orii H, Sakurai T, Watanabe K. Distribution of the stem cells (neoblasts) in the planarian Dugesia japonica. Dev Genes Evol. 2005;215:143–57. doi: 10.1007/s00427-004-0460-y. [DOI] [PubMed] [Google Scholar]
- Ortiz-Pineda PA, Ramirez-Gomez F, Perez-Ortiz J, Gonzalez-Diaz S, Santiago-De Jesus F, Hernandez-Pasos J, Del Valle-Avila C, Rojas-Cartagena C, Suarez-Castillo EC, Tossas K, Mendez-Merced AT, Roig-Lopez JL, Ortiz-Zuazaga H, Garcia-Arraras JE. Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genomics. 2009;10:262. doi: 10.1186/1471-2164-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo NJ, Morokuma J, Walentek P, Kema IP, Gu MB, Ahn JM, Hwang JS, Gojobori T, Levin M. Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev Biol. 2010;339:188–99. doi: 10.1016/j.ydbio.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo NJ, Newmark PA, Sánchez Alvarado A. Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev Dyn. 2003;226:326–33. doi: 10.1002/dvdy.10228. [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piR-NA expression in planarians. RNA. 2008;14:1174–86. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–50. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev Biol. 2009;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Smed-βcatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–30. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–22. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS. The significance of spontaneous and induced apoptosis in the gastrointestinal tract of mice. Cancer Metastasis Rev. 1992;11:179–95. doi: 10.1007/BF00048063. [DOI] [PubMed] [Google Scholar]
- Potten CS. Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos Trans R Soc Lond B Biol Sci. 1998;353:821–30. doi: 10.1098/rstb.1998.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS. Radiation, the ideal cytotoxic agent for studying the cell biology of tissues such as the small intestine. Radiat Res. 2004;161:123–36. doi: 10.1667/rr3104. [DOI] [PubMed] [Google Scholar]
- Potten CS, Booth C, Pritchard DM. The intestinal epithelial stem cell: the mucosal governor. Int J Exp Pathol. 1997;78:219–43. doi: 10.1046/j.1365-2613.1997.280362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–9. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–30. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–57. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Robb SM, Ross E, Sánchez Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 2008;36:D599–606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L, Salvetti A, Lena A, Batistoni R, Deri P, Pugliesi C, Loreti E, Gremigni V. DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Dev Genes Evol. 2006;216:335–46. doi: 10.1007/s00427-006-0060-0. [DOI] [PubMed] [Google Scholar]
- Rossi L, Salvetti A, Marincola FM, Lena A, Deri P, Mannini L, Batistoni R, Wang E, Gremigni V. Deciphering the molecular machinery of stem cells: a look at the neoblast gene expression profile. Genome Biol. 2007;8:R62. doi: 10.1186/gb-2007-8-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Shibata N, Nishimura O, Agata K. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev Biol. 341:429–43. doi: 10.1016/j.ydbio.2010.02.037. [DOI] [PubMed] [Google Scholar]
- Sakai T, Kato K, Watanabe K, Orii H. Planarian pharynx regeneration revealed by the expression of myosin heavy chain-A. Int J Dev Biol. 2002;46:329–32. [PubMed] [Google Scholar]
- Salò E, Baguñà J. Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J Embryol Exp Morphol. 1984;83:63–80. [PubMed] [Google Scholar]
- Salò E, Baguñà J. Cell movement in intact and regenerating planarians. Quantitation using chromosomal, nuclear and cytoplasmic markers. J Embryol Exp Morphol. 1985;89:57–70. [PubMed] [Google Scholar]
- Salò E, Baguñà J. Regeneration in planarians and other worms: New findings, new tools, and new perspectives. J Exp Zool. 2002;292:528–39. doi: 10.1002/jez.90001. [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Deri P, Batistoni R. An MCM2-related gene is expressed in proliferating cells of intact and regenerating planarians. Dev Dyn. 2000;218:603–14. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1016>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Lena A, Batistoni R, Deri P, Rainaldi G, Locci MT, Evangelista M, Gremigni V. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132:1863–74. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Shukalyuk AI, Dolmatov IY. Regeneration of the Digestive Tube in the Holothurian Apostichopus japonicus after Evisceration. Russ J Mar Biol. 2001;27:168–173. [Google Scholar]
- Solana J, Lasko P, Romero R. Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev Biol. 2009;328:410–421. doi: 10.1016/j.ydbio.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Nishimura K, Agata K. Planarians maintain a constant ratio of different cell types during changes in body size by using the stem cell system. Zoolog Sci. 2009;26:805–13. doi: 10.2108/zsj.26.805. [DOI] [PubMed] [Google Scholar]
- Takeo M, Yoshida-Noro C, Tochinai S. Morphallactic regeneration as revealed by region-specific gene expression in the digestive tract of Enchytraeus japonensis (Oligochaeta, Annelida). Dev Dyn. 2008;237:1284–94. doi: 10.1002/dvdy.21518. [DOI] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K. A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev Growth Differ. 1997;39:723–7. doi: 10.1046/j.1440-169x.1997.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- von Bertalanffy L. Untersuchungen über die gesetzlichkeit des wachstums. III. Teil Quantitative beziehungen zwischen darmoberfläche und körpergröβe bei Planaria maculata. Wilhelm Roux Arch Entwickl Mech Org. 1940;140:81–89. doi: 10.1007/BF00576248. [DOI] [PubMed] [Google Scholar]
- Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol. 2010;344:979–91. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zattara EE, Bely AE. Evolution of a novel developmental trajectory: fission is distinct from regeneration in the annelid Pristina leidyi. Evol Dev. 2011;13:80–95. doi: 10.1111/j.1525-142X.2010.00458.x. [DOI] [PubMed] [Google Scholar]
- Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci U S A. 2005;102:18491–6. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


