Abstract
Cellular membranes can assume a number of highly dynamic shapes. Many cellular processes also require transient membrane deformations. Membrane shape is determined by the complex interactions of proteins and lipids. A number of families of proteins that directly bend membranes have been identified. Most associate transiently with membranes and deform them. These proteins work by one or more of three types of mechanisms. First, some bend membranes by inserting amphipathic domains into one of the leaflets of the bilayer; increasing the area of only one leaflet causes the membrane to bend. Second, some proteins form a rigid scaffold that deforms the underlying membrane or stabilizes an already bent membrane. Third, some proteins may deform membranes by clustering lipids or by affecting lipid ordering in membranes. Still other proteins may use novel but poorly understood mechanisms. In this review, we summarize what is known about how different families of proteins bend membranes.
Keywords: Membrane, bending, curvature, bilayer, binding
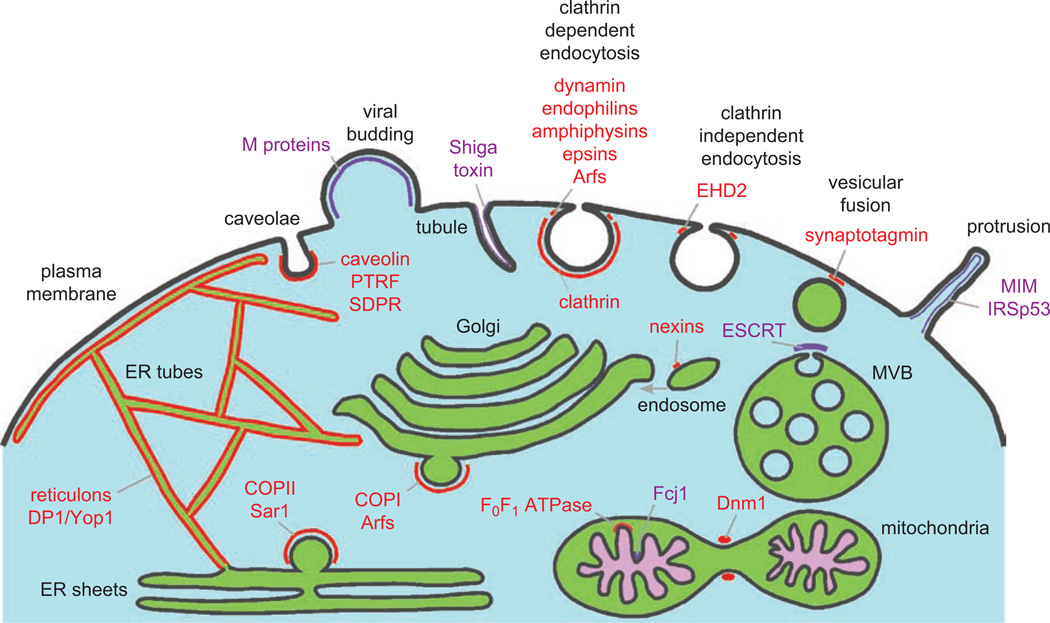
All eukaryotic cells contain internal membrane-bound organelles. Most have elaborate yet highly dynamic shapes that are essential for optimal organelle function. These shapes can include tubules, sheets, vesicles, fenestrated sheets, and cisternae, often in the same membrane. In addition, transient membrane deformations are required for many essential cellular processes, including vesicular trafficking, cell movement, and cell division. Establishing and altering organelle morphology requires numerous proteins and lipids. Figure 1 shows some of the regions and processes in cells that necessitate membrane bending and locations of some of the proteins discussed in this review.
Figure 1.
Membrane bending caused by proteins that directly deform cellular membranes. Many processes in the cell require proteins that shape membranes, and some are depicted in this figure. Regions where membrane-deforming proteins bind membranes are shown in either red (proteins that cause positive curvature) or purple (proteins that cause negative curvature). Note that some proteins work at more than one location in cells and not all are shown. A color version of this figure is available online.
Membrane shape is determined by the complex interactions of proteins and lipids. The last few years have seen a dramatic increase in our understanding of how cellular membranes are shaped. Though many membrane-deforming processes remain poorly understood, at least three types of mechanism are used. First, membranes can be pulled, pushed, or held in shape by interactions with the cytoskeleton, particularly actin filaments and microtubules. Both the assembly and disassembly of the cytoskeleton as well as motors operating on the cytoskeleton can shape membranes. Second, membrane morphology and dynamics can also be affected by the heterogeneous distribution of some lipids within a bilayer. The ability of a lipid to promote positive or negative curvature in membranes is determined by the relative cross-sectional area of its head-group and acyl chains. For some lipids, such as phosophatidyl choline (PC), the cross-sectional areas are similar and they behave essentially as cylinders in the membrane bilayer and do not promote positive or negative curvature. Other lipids, however, are thought to be wedge-shaped, because of differences in the relative size of their head-groups and acyl chains. For example, phosphoinositides (PIPs) have larger head-groups than PC and accumulation of these lipids in a domain of one leaflet of a bilayer can cause positive curvature. Therefore, proteins that affect the distribution of lipids in membranes can bend membranes. These could include proteins that alter the distribution of lipids between the two leaflets of a bilayer (flippases), degrade or modify lipids, or cause lipids to cluster. Third, a growing number of proteins have been shown to directly bend membranes. This review will focus on these proteins and how they promote membrane curvature.
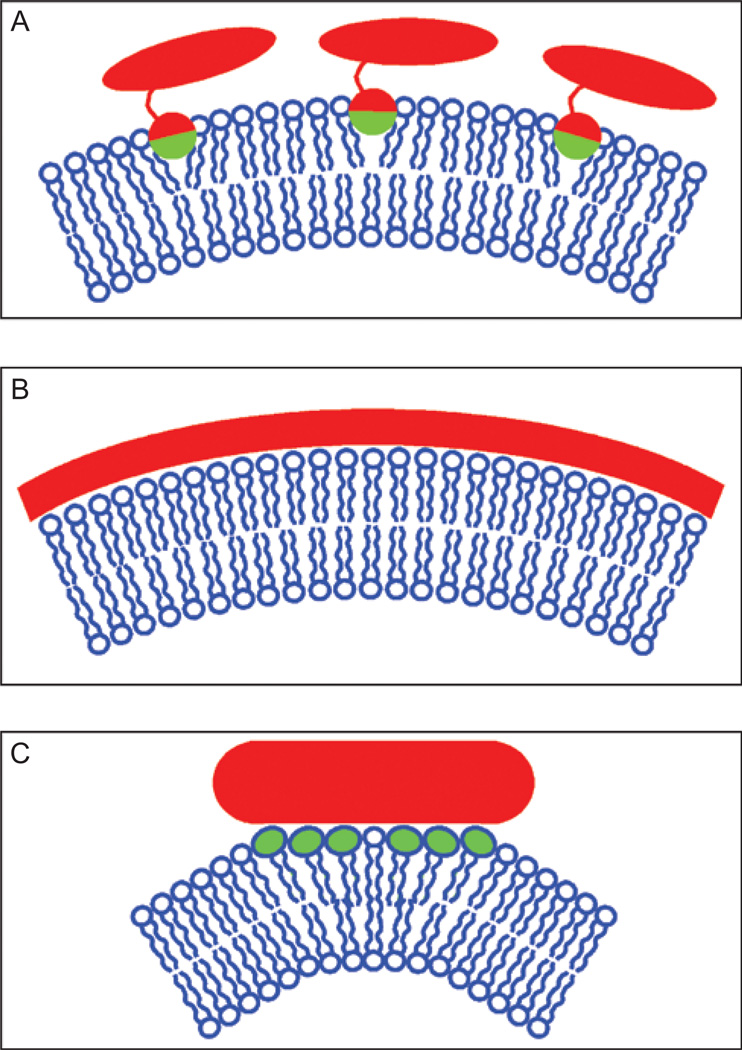
Most membrane-bending proteins transiently associate with membranes and deform them. In general, most have a proposed mechanism to generate membrane curvature and oligomerize to stabilize it. At least three types of mechanism are used. First, some proteins bend membranes by inserting amphipathic domains partially into the bilayer (Figure 2A). This causes bending because of the bilayer-couple mechanism (Sheetz and Singer, 1974); since strong hydrophobic forces hold the two leaflets of a bilayer together, increasing the area of one leaflet while the other remains unchanged will cause the bilayer to bend. Modeling suggests that shallow insertions that do not penetrate deeply into a bilayer are most effective at membrane bending (Campelo et al., 2008). In theory, any protein that inserts partially into a membrane could deform it. However, while a number of protein domains have been found to transiently insert into bilayers (Lemmon, 2008), in many cases it is not known if this insertion also causes membrane deformation. A second widely used mechanism of membrane bending is the formation of a rigid scaffold that deforms the underlying membrane or stabilizes membranes that have been bent by other mechanisms (Figure 2B). Many scaffold-forming proteins oligomerize to large rigid structures. Finally, there is growing evidence suggesting that some proteins deform membranes by causing lipid clustering. Most membrane-deforming proteins seem to use a combination of these mechanisms. Others, however, may use novel but poorly understood mechanisms. In the following sections, we will summarize what is known about how different families of proteins deform membranes.
Figure 2.
Mechanisms of membrane bending. (A) Some proteins deform membranes by inserting amphiphatic domains, often α-helices, into membranes. The hydrophobic part of the protein (shown in green) inserts partially into the membrane. (B) Proteins can form rigid scaffolds that either deform the underlying membrane or stabilize curvature in membranes that are already bent. Many scaffold-forming proteins oligomerize to form large rigid structures. (C) Proteins can also cause bending by affecting the distribution of lipids in the bilayer. Clustering lipids with large head-groups, for example, could cause membrane bending. Some proteins may also order lipids in the membrane and cluster to bend membranes. A color version of this figure is available online.
Coat forming proteins
Most proteins known to deform membranes are involved in vesicular trafficking. Generation of transport vesicles requires extensive membrane remodeling. First, membrane deformation is usually initiated by a protein that inserts an amphipathic domain into the bilayer or a concave protein that binds the membrane, stabilizing a bent membrane. Next, a self-assembling protein coat forms a cage-like structure that scaffolds the membrane, increasing the volume of the vesicle and determining its size. Finally, other proteins must further deform the membrane to complete vesicle fission.
The three major coat-forming protein complexes in cells are coat protein complex I (COPI), coat protein complex II (COPII), and clathrin. All three have been extensively studied and reviewed (Antonny, 2006; Edeling et al., 2006; Fromme et al., 2008; Hughes and Stephens, 2008; Spang, 2008; Doherty and McMahon, 2009); here we focus on how they bend membranes to form vesicles with a defined size. COPII is the simplest of the three types of coat-forming complexes and has three elements: the small GTPase Sar1p, the Sec23/24 complex, and the Sec13/31 complex. Sar1p initiates assembly of the coat. It is a member of the Arf family of small GTPases, some of which also initiate the assembly of other protein coats (Pasqualato et al., 2002). In the GTP state, Sar1p binds membranes and exposes an amphipathic helix that inserts into the bilayer (Goldberg, 1998; Antonny et al., 2001; Huang et al., 2001) and causes it to bend (Bielli et al., 2005; Lee et al., 2005). That Sar1p bends membranes was demonstrated with a widely used assay; addition of a membrane-deforming protein to liposomes increases their positive curvature, causing them to tubulate (or even vesiculate into smaller liposomes). Thus, tubulation of liposomes by a protein indicated that it bends membranes. It was found that addition of Sar1-GTP but not Sar1-GDP causes liposomes to tubulate. This tubulation requires the N-terminal amphipathic helix of Sar1p, suggesting that membrane insertion by this domain causes bending. After Sar1p initiates membrane bending, it subsequently recruits the Sec23/24 complex to membranes. This complex has a concave surface containing positively charged basic resides that can interact with a negatively charged membrane (Bi et al., 2002). It therefore acts as a scaffold that stabilizes curvature in the underlying membrane. By themselves, Sar1p and Sec23/24 promote tubule formation rather than spherical membrane buds; spherical coat-bearing vesicles are only seen when Sec13/31 is also present (Lee et al., 2005). Cryo-electron microscopy (cryo-EM) has revealed that this complex assembles into cage-like structures (Stagg et al., 2006). The Sec13/31 complex forms a tetramer that is about 30 nm long. These can be joined to make triangles and squares that are combined to make the closed cage around the vesicle, shaping it. Sar1p is also required to drive the final membrane bending needed for vesicle fission once the COPII coat has been formed (Bielli et al., 2005; Lee et al., 2005).
Membrane deformation by COPI and clathrin coats is probably similar to that of COPII. The assembly of COPI and clathrin coats often requires a small, Sar1-like GTPase called Arf1 (D’Souza-Schorey and Chavrier, 2006). Like Sar1p, Arf1 associates with membranes when it binds GTP and inserts an amphipathic helix, which, in contrast to Sar1p, is myristoylated (Antonny et al., 1997). More recently, it has been shown that Arf1 directly tubulates liposomes in vitro, indicating that it can deform membranes (Beck et al. 2008). It is not yet known if the amphipathic helix and myristoylation are needed for membrane bending by Arf1. Numerous other proteins regulate the assembly of Arf1 and various adaptor proteins that recruit coats (Edeling et al., 2006; Anders and Jurgens, 2008; Doherty and McMahon, 2009). Clathrin can self-assemble into a cage-like lattice that is similar to the one formed by Sec13/31 and scaffolds the underlying membrane, maintaining the size and shape of vesicles (Fotin et al., 2004; 2006).
BAR domain proteins
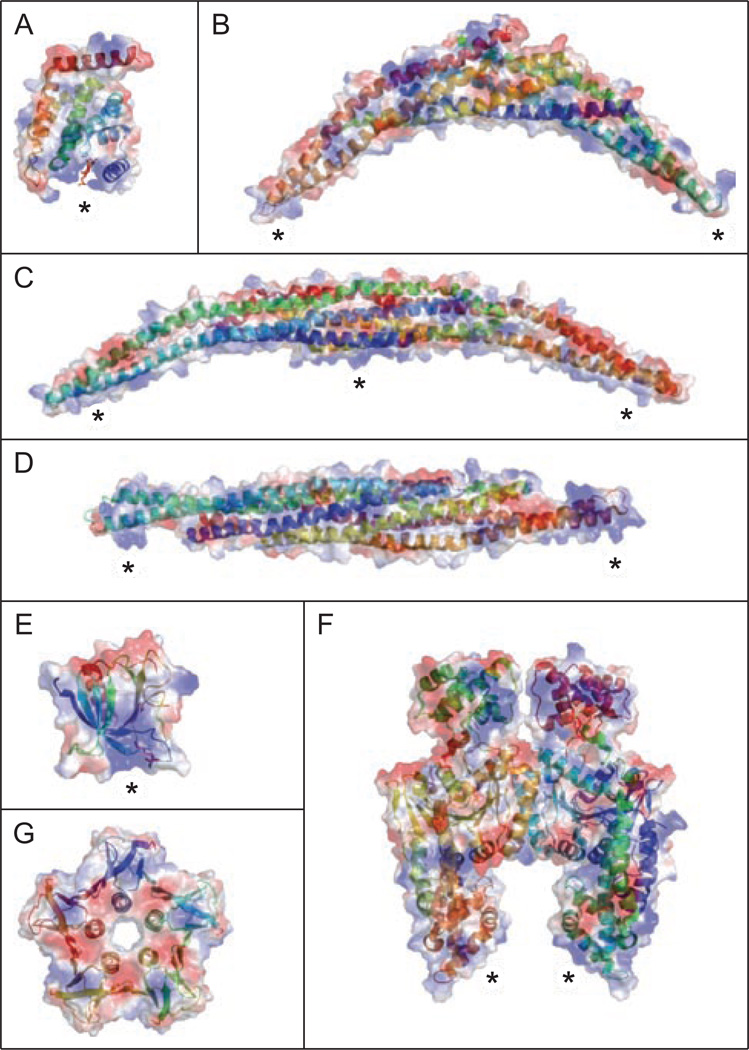
BAR (Bin/amphiphysin/Rvs) domain proteins are found associated with numerous membrane surfaces throughout the cell and confer membrane curvature in a range of diameters (Frost et al., 2009). Based on the degree of curvature, BAR domains are subdivided into three classes, BAR, F-BAR (Fes/CIP4 homology BAR) and I-BAR (Inverse-BAR). Crystal structures from all classes reveal a common structural motif: a three-helix coiled-coiled stretch that dimerize forming a 6-helix bundle with a positively charged surface (Figures 3B–3D). The dimer is banana shaped, with a positively charged surface on the concave (BAR and F-BAR) or slightly convex (I-BAR) surface that mediates binding to negatively charged lipid head-groups in the membrane. The degree of curvature for each BAR domain is believed to dictate the radius of membrane bending. In support of this model, BAR domain proteins form tubules in vitro with varying diameters that are consistent with the shape of the BAR domain (Farsad et al., 2001; Peter et al., 2004; Itoh et al., 2005; Henne et al., 2007; Mattila et al., 2007; Shimada et al., 2007).
Figure 3.
Crystal structures of membrane bending proteins. Electrostatic surface of each domain with blue and red regions indicating positive and negative patches, respectively. Within each surface structure is the sequence in ribbon format, color coded based on distance from the N-terminus. Each domain contains positive patches that are potential lipid binding sites (marked with *). (A) ENTH domain from epsin with Ins(1,4,5)P3 bound to a positively charged surface (Ford et al., 2002). (B) Traditional BAR domain from Arfactin (Tarricone et al., 2001). (C) F-BAR from Cdc42-interacting protein 4 (Shimada et al., 2007). (D) I-BAR from IRSp53 (Millard et al., 2005). (E) PH domain from dynamin (Ferguson et al., 1994). (F) EHD2 (Daumke et al., 2007). (G) Shiga-like toxin I B subunit (pdb ID:1czg). A color version of this figure is available online.
Traditional BAR domain proteins can be further subdivided according to flanking modules that modify their function. For example, several BAR domain proteins contain an N-terminal amphiphathic helix next to the BAR domain that is believed to insert into the lipid bilayer (N-BAR). Both amphiphysins and endophilins contain N-BAR domains and have been associated with several membrane remodeling events. Amphiphysin-1 and endophilin-1 are involved in clathrin-mediated endocytosis and bind to dynamin through SH3 domains. These proteins bind to lipid and induce tubulation with a diameter potentially dictated by their N-BAR structure, 20–30 nm (Farsad et al., 2001; Gallop et al., 2006; Masuda et al., 2006). Both proteins have been localized to the long necks of budding vesicles in GTPγS treated synaptosomes and have been suggested to promote neck formation (Bauerfeind et al., 1997; Ringstad et al., 1999). Amphiphysin-2 is associated with T-tubules in skeletal muscle (Lee et al., 2002) and endophilin-B1 is important for mitochondrial morphology and the formation of autophagosomes (Takahashi et al., 2009). Other proteins have lipid binding motifs adjacent to their BAR domain. These motifs include pleckstrin homology (PH) and phox (PX) domains, which preferentially bind to phosphoinositides. Sorting nexins contain PX-BAR domains and sorting nexin 9 (SNX9) has been shown to also play a direct role in clathrin-mediated endocytosis (Lundmark and Carlsson, 2003; Soulet et al., 2005). The tubular diameter of this protein is ~20–40 nm and may play a role in early neck formation of budding vesicles (Shin et al., 2008; Yarar et al., 2008). Several other sorting nexins (1, 2, 5, and 6) are involved in returning cargo from the endosome to the Golgi possibly through stabilizing a tubular network (Bonifacino and Hurley, 2008). Several of the PH-BAR domain proteins bind to small GTPases and may help regulate either nucleotide exchange or hydrolysis (Frost et al., 2009). These BAR domain proteins may help locate the small GTPases to their sites of action on the membrane as well as sense membrane curvature.
The F-BAR domain is more extended (Figure 3C), forming a shallow arc that potentially leads to a large range of tube sizes, ~25–200 nm in diameter (Itoh et al., 2005; Henne et al., 2007; Shimada et al., 2007), and during endocytosis may also induce membrane curvature early in neck formation (Frost et al., 2009). Structural analysis by cryo-EM of the Toca F-BAR domain supports the model that it binds to the membrane through its concave surface and packs tightly to induce membrane curvature and tubulation (Frost et al., 2008). The F-BAR domain containing protein, Cdc15, is localized to contractile rings during cell division, another region in the cell with extreme membrane curvature (Wu et al., 2006). The I-BAR (Figure 3D) domain proteins induce the opposite curvature of the BAR and F-BAR proteins and potentially play a role in membrane protrusions (Mattila et al., 2007). Both the I-BAR proteins IRSp53 and MIM (missing in metastasis) are associated with the cellular protrusions of filopodia (Yamagishi et al., 2004; Bompard et al., 2005). The I-BAR domain proteins also interact with actin filaments, a property shared with other BAR domain proteins, and suggests a direct link between membrane bending and the actin cytoskeleton (discussed in detail in the following reviews: Peter et al., 2004; Dawson et al., 2006; Frost et al., 2009).
Numerous BAR domain proteins partner with dynamin during the vesicle formation stage of endocytosis (clathrin-mediated and clathrin-independent) (amphiphysin, endophilin, SNX9, CIP4, Tuba, syndapin, GRAF1) (Farsad et al., 2003; Cestra et al., 2005; Gallop et al., 2006; Yarar et al., 2007; Lundmark et al., 2008; Shin et al., 2008). The F-BAR proteins may initiate and stabilize the bud neck by assembling into large helical arrays while the BAR or N-BAR proteins could act in concert with the F-BAR proteins to further constrict the bud neck to a diameter amenable for dynamin assembly.
The dynamin family of proteins
Dynamin, a large GTPase, is involved in numerous vesiculation events throughout the cell, including clathrin-mediated endocytosis, caveolae internalization, and protein trafficking from the endosome and Golgi (Hinshaw, 2000). Dynamin is believed to wrap around and constrict the necks of budding vesicles, which leads to membrane fission and vesicle formation. The driving force for changing the shape of the membrane is the ability of dynamin to self-assemble into spirals and constrict the underlying membrane upon GTP hydrolysis. In support of this model, purified recombinant dynamin self-assembles into helical arrays (50 nm diameter) in the presence of GDP/BeF (Carr and Hinshaw 1997). Dynamin also assembles onto lipid forming dynamin-lipid tubes that constrict (to 40 nm) and twist upon GTP addition (Roux et al., 2006; Sweitzer and Hinshaw, 1998). More recently, fission has been observed when dynamin is added together with GTP to lipid templates (Bashkirov et al., 2008; Pucadyil and Schmid, 2008).
Targeting dynamin to the necks of budding vesicles is most likely enhanced by a preformed neck with a suitable diameter and the PH domain of dynamin (Figure 3E). Dynamin’s PH domain preferentially binds to phosphoinositides (Klein et al., 1998; Lee and Lemmon, 2001), and embeds into the outer leaflet of the lipid bilayer (Burger et al., 2000; Mears et al., 2007). The initial neck formation of the budding vesicle may be driven by several of dynamin partners that contain BAR domains (amphiphysin, endophilin, SNX9, discussed above). These partners bind to dynamin through SH3 motifs that interact with the proline-rich C-terminus of dynamin.
Other dynamin family members are involved in membrane fission and fusion events, plant cell wall formation and cytokinesis (Praefcke and McMahon, 2004). All dynamin family members studied to date have the propensity to self-assemble and the majority have been shown to self-assemble onto lipid to form decorated tubes. The dynamin-related proteins (Drp1/DRPs) are involved in mitochondrial, peroxisome and chloroplast fission while Opa1 and mitofusions are involved in mitochondrial fusion. How dynamin family members are targeted to their specific membrane sites remains unknown for the majority of these proteins. Only one other dynamin family member, DRP3A, contains a PH domain and none contain a proline-rich domain. Interestingly, the proteins involved in membrane fusion contain transmembrane domains, anchoring these proteins to their site of action.
The structural and biochemical properties of other dynamin family members are beginning to be unraveled. For example, the dynamin-related protein in yeast (Dnm1) involved in mitochondrial fission assembles into large spirals, ~110 nm, in the presence of non-hydrolysable GTP analogs and also associates with negatively charged lipids, even though it does not contain a PH domain, to form large protein decorated tubes (Ingerman et al., 2005). Evidence suggests it is targeted to the outer mitochondria membrane through linker proteins, Mdv1 or Caf4, which then associate with a transmembrane protein, Fis1 (Hoppins et al., 2007). Upon initiation of mitochondrial fission, Dnm1 encircles the constricting outer mitochondrial membrane, where it is believed to play a direct role in membrane fission in a similar mechanism as described for dynamin. Future studies on additional dynamin family members will determine if a common mechanism of action is conserved and which features diverge for specific cellular functions. It is intriguing that dynamins are involved in both fission and fusion events in the cell. However, both processes may involve common properties of dynamin family members; self-assembly and successive disassembly upon GTP hydrolysis. Both fusion and fission requires membrane bilayers to come close together. In fission, constriction of the bud necks by dynamin and subsequent disassembly may lead to fission, while in fusion, the helical assembly of a dynamin may span the space between two opposing membranes and bring them together, and again the subsequent GTP induced release may lead to fusion.
ENTH domains
Epsin1 participates in clathrin-mediated endocytosis and was initially discovered because it interacts with Eps15, another protein involved in this process (Chen et al., 1998). Subsequently, a number of other isoforms and epsin-like proteins have been identified in mammals and other eukaryotes including Xenopus, Drosophila and yeast (Horvath et al., 2007). All these proteins share an epsin N-terminal homology (ENTH) domain, which is also found in other proteins including some clathrin adaptor proteins (Itoh and De Camilli, 2006). The ENTH domain in epsin1 binds PI(4,5)P2, a lipid that is enriched in the plasma membrane, while ENTH domains in other proteins, such as epsinR, bind monophosphorylated PIPs (Itoh et al., 2001; Ford et al., 2001; 2002; Kalthoff et al., 2002; Mills et al., 2003). The PI(4,5)P2 head-group is bound in a pocket on the surface of epsin1 (Figure 3A). Upon binding, an N-terminal amphipathic helix becomes ordered and probably inserts into the membrane, causing it to deform (Ford et al., 2002). Like other proteins that deform membranes, the ENTH domain of epsin1 causes vesicles to tubulate but, interestingly, the protein probably does not oligomerize (Ford et al., 2002). In addition to the membrane-bending ENTH domain, epsin contains other regions that interact with clathrin coat forming proteins. It is possible that after the ENTH domain initiates membrane bending, other regions of epsin1 subsequently recruit clathrin and other proteins that stabilize and further shape the membrane during endocytosis.
EHD domains
Eps15 homology (EH)-domain containing proteins (EHDs, also known as receptor-mediated endocytosis RME-1 proteins) are ATPases that play roles in endosomal recycling pathways and clathrin-independent endocytosis (Grant and Caplan, 2008). Mammals have four of these proteins and homologs have been identified in many other eukaryotes. The structure of one of these proteins, mouse EHD2, has been solved and revealed how they might bend membranes (Daumke et al., 2007). The nucleotide-binding domain of this protein dimerizes and forms a highly curved putative membrane-binding surface lined with a number of basic residues that could interact with charged lipid head-groups as well as two hydrophobic residues that may insert into the interior of the membrane (Figure 3F). Both the curvature of the binding surface and membrane insertion could cause positive membrane bending. Consistent with this, the protein was also found to tubulate vesicles.
Remarkably, the nucleotide-binding domain of EHD2 was found to be similar to that of GTPase domain of dynamin. Although there is no sequence homology, both domains share a similar three-dimensional structure and low basal rate of nucleotide hydrolysis that is simulated by membrane binding (Daumke et al., 2007). Based on this similarity and the structure of the EHD2 dimer, it was proposed that the dimer oligomerizes into larger rings. Consistent with this, ring-like structures can been by EM when liposomes in the presence of EHD2. Thus, despite lacking sequence similarity, EHD proteins may bind and bend membranes in a way similar to dynamins.
C2 domains
The C2 domain was originally identified as one of two conserved regulatory regions of proteins kinase C (Nishizuka, 1988). It was subsequently shown to be a Ca2+-dependent membrane-binding domain (Cho and Stahelin, 2006). A large number of proteins have been found to contain C2 domains and, while the roles of many have not been identified, most function in signal transduction and membrane trafficking. There are a number of subgroups of C2 domains, not all bind membranes and some do not bind Ca2+ (Cho and Stahelin, 2006). Most C2 domains probably have more than one membrane binding site and, interestingly, Ca2+ concentration can affect the specificity of the lipid binding surfaces (Bai et al., 2004; Schiavo et al., 1996). The C2 domains from three proteins have been shown by electron spin resonance and X-ray reflectivity measurements to insert into membranes to a range of depths (Ball et al., 1999; Frazier et al., 2002; Frazier et al., 2003; Kohout et al., 2003; Malmberg et al., 2003; Malkova et al., 2005; Rufener et al., 2005).
One of these, synaptotagmin, was recently shown to tubulate vesicles in vitro, suggesting that it bends them by causing positive curvature (Martens et al., 2007). This protein plays a role in synaptic vesicle exocytosis (Fernandez et al., 2001), though its function remains controversial. It contains a transmembrane domain and two C2 domains. Interestingly, both C2 domains are necessary for tubulation and they must be linked together, indicating that the spacing of the two domains is important. Consistent with the idea that membrane insertion is important for bending, altering the residues in the C2 domains that enter the membrane affected tubulation; when four of these were changed into smaller alanines tubulation was ablated, while altering them to more bulky tryptophans caused increased tubulation and fragmentation (Martens et al., 2007). In addition, three other synaptotagmin-like proteins, that contain two C2 domains, were also found to tubulate vesicles, suggesting that membrane bending may be a common feature of these proteins. However, another study failed to find any significant membrane tubulation by synaptotagmin (Arac et al., 2006) and there are other models of synaptotagmin function unrelated to its ability to induce membrane curvature (Sudhof and Rothman, 2009).
ESCRT-III
The endosomal sorting complex required for transport (ESCRT) is essential for a number of processes in the cell including multivesicular body (MVB) biogenesis, the budding of some enveloped viruses, and daughter cell scission in cytokinesis (Piper and Katzmann, 2007; Williams and Urbe, 2007; Hurley, 2008; Raiborg and Stenmark, 2009; Carlton and Martin-Serrano, 2009). MVBs are part of the endosomal trafficking pathway and contain intraluminal vesicles (ILVs). Thus MVB formation, like viral budding, requires membrane budding away from the cytosol. This is the opposite direction from vesicle budding from organelles in the secretory pathway. A number of heteromeric protein complexes are required for protein trafficking in the MVB pathway. One of these, called ESCRT-III, is responsible for invaginating the outer membrane of MVBs, leading to the formation of ILVs. In this way it is similar to Shiga toxin (see below) and I-BAR-containing proteins that cause tubular invaginations or protrusion of membranes away from the surface they interact with. However, unlike these proteins, ESCRT-III deforms membranes but does not enter the invaginations it forms (Babst et al., 1998; 2002). This allows it to catalyze multiple rounds of ILV formation. How ESCRT-III performs this remarkable feat is not yet understood but a number of recent papers have begun to reveal how it might work.
ECSRT-III in yeast consists of four subunits: Vps20, Vps24, Vps2, and Snf7 (also called Vps32). Homologs of these proteins are found in all eukaryotes. All of the subunits of this complex are highly charged and assemble on endosome membranes in an ordered manner (Babst et al., 2002; Zamborlini et al., 2006; Shim et al., 2007; Lata et al., 2008; Teis et al., 2008). The order of assembly on membranes is probably Vps20, Snf7, Vps24, and Vps2 (Teis et al., 2008).
Evidence that Snf7 assembles into filaments on membranes and that these filaments likely drive membrane deformation has recently been obtained both in cells and in vitro. When the human homologs of Snf7 are overexpressed in mammalian cells, they form spiral filaments on the plasma membrane, causing the membrane to protrude from the cells and form tubular protrusions similar to those see in MVBs (Hanson et al., 2008). Two groups also reconstituted membrane deformation by Snf7 and the other ESCRT-III components in vitro. One showed that addition of ESCRT-III to liposomes causes them to form inward invaginations (Saksena et al., 2009). Importantly, mutants of Snf7 that fail to oligomerize also failed to invaginate the lipsomes, consistent with the idea that Snf7 oligomerization is required for membrane invagination. A second group demonstrated that ESCRT-III mediates membrane deformation and ILV formations using giant unilamellar vesicles (GUVs), which allow the visualization of changes in membrane shape as they occur (Wollert et al., 2009). The four components of ESCRT-III were added GUVs in the order they are probably assembled on endosomal membranes. After Vps20 was added to GUVs, the subsequent addition of Snf7 led to the rapid formation of internal vesicles. These vesicles were continuous with the outer GUV membrane since an exogenous fluorescent marker could defuse into them. When Vps24 and Vps2 were then added, membrane scission occurred and the vesicles were no longer continuous with the outer membrane of the GUVs. ThusVps20 initiates Snf7 filament assembly on membranes, causing invagination, and Vps24 and Vps2 terminate Snf7 filament assembly, leading to membrane scission.
How ESCRT-III assembly causes membrane deformation remains an intriguing mystery. It may be that Vps20 and Snf7 form a concave surface when they assemble on membranes or perhaps insert slightly into the membrane, leading to membrane deformation. However, this would not explain why these proteins do not enter the invaginations they form. Perhaps assemblies of Vps20 and Snf7 on membrane are rigid and cannot diffuse into the highly curved membrane invaginations they form. How formation of Snf7 filaments cause vesiculation remains an fascinating unanswered question.
Shiga toxin
Shiga toxin is produced by Shigella dysenteriae and can cause a range of enteric illnesses (Zheng and Sadler, 2008). It contains an enzymatic 33 kDa A subunit and a noncovalently attached heptopentomeric 7.7 kDa B subunit (STxB) that carries it into the cell. STxB binds glycosphingolipid globotriaosylceramide (Gb3) on the cell surface and is internalized by a clathrin-independent endocytic pathway. Recent work has demonstrated that STxB itself, without the need for cytosolic proteins, induces the formation of tubular invaginations of the plasma membrane (Romer et al., 2007). Formation of these tubules was stimulated by ATP depletion. They also did not require actin or dynamin, which both play roles in clathrin-dependent endocytosis, or caveolin 1, which has been implicated in some clathrin-independent endocytosis. To test if STxB by itself causes membrane invaginations, it was added to GUVs. Internal tubules rapidly formed only in GUVs containing Gb3, indicating that the STxB must bind to this lipid to form tubules. The tubules were not simply formed by binding Gb3, since an antibody against this lipid did not cause the GUVs to tubulate. These experiments demonstrate that STxB binding to Gb3 was necessary and sufficient to produce the negative membrane curvature required to cause the formation of invaginated tubules. It likely does so by clustering Gb3, which seems to be critical for membrane invagination (Romer et al., 2007). STxB also clusters, perhaps because curvature-inducing proteins can spontaneously aggregate (Reynwar et al., 2007). Interestingly, efficient tubule formation also required that the membrane tension of the GUVs not be high.
STxB bends membranes by a mechanism that probably differs from those employed by proteins discussed in the previous sections. It does not appear to have a curved surface that could form a membrane scaffold and there is no evidence that it inserts a domain into the membrane (Figure 3G) (Stein et al., 1992). In addition, since the head-group of Gb3 is large, clustering of this lipid by STxB on a region of one face of a bilayer might be expected to cause positive membrane curvature rather than the negative curvature needed to form tubular invaginations of the membrane. Instead, STxB probably promotes membrane invagination by ordering the lipids in the membrane it binds to, changing the phase of the underlying lipids and perhaps also increasing membrane thickness since Gb3 has long acyl chains (Romer et al., 2007). The authors speculate that line tension between the lipid domain created by STxB and the surrounding membrane could be used to cluster STxB and drive membrane bending. In support of this model, they found that STxB localizes to more ordered domains of the plasma membrane (i.e., domains in which the lipids are in a different phase from the surrounding lipid). They used the fluorescent dye Laurdan, which can indicate the phase of membranes because it is sensitive to polarity in a bilayer. This showed that STxB on cellular membranes colocalizes with more ordered regions of the membrane. Surprisingly, formation of these ordered regions did not need cholesterol. It also required that the Gb3 have an unsaturated C22:1 acyl chain rather than the fully saturated C22:0, but it is not yet clear why. The nature of the ordered domains caused by STxB membrane binding and how they induce membrane invagination remain to be determined.
Matrix proteins of enveloped viruses
Budding of enveloped viruses from cells requires proteins that deform cellular membranes (Welsch et al., 2007). Although the viral proteins needed for budding have been identified for many viruses, in most cases the mechanisms they use to deform membranes are not well understood. There does not seem to be common budding mechanism employed by all enveloped viruses; in some viruses glycoproteins on the surface drive budding, other viruses use inner core proteins, and still others use both types of proteins (Welsch et al., 2007). For some viruses, cellular proteins also play a role. For example, it has recently been found that retroviruses such as HIV use the ESCRT machinery to facilitate membrane budding and scission at the plasma membrane (Raiborg and Stenmark, 2009). In the past few years a few viral proteins required for budding have been shown to directly deform membranes. Interestingly, they may use a mechanism similar to the one used by Shiga toxin to invaginate the plasma membrane and enter cells.
The matrix (M) protein of rhabdoviruses such as vesicular stomatitis virus (VSV) is required for viral budding from the cell (Mebatsion et al., 1999). M proteins from different viruses do not share sequence similarity or structures but they all bind membranes, particularly those containing negatively charged lipids, and oligomerize (Welsch et al., 2007). Solon et al. (2005) demonstrated that addition of VSV M protein directly deforms membranes by adding it to GUVs. They found that 2–3 µm invaginations were formed by the protein. These are similar to the invaginations the protein would need to form at the plasma membrane to allow viral budding. The GUVs contained the zwitterionic lipid phosphatidylcholine and the negatively charged lipid phosphatidylserine. Remarkably, the M-protein clustered on the GUVs together with the phosphatidylserine at the invaginations on the GUVs. It is possible that, using a mechanism similar to that of Shiga toxin (see the previous section), lipid clustering by M protein alters the order of the membrane and drives membrane invagination. However, in contrast to Shiga toxin, M protein produces large invaginations rather than narrow membrane tubules.
Strong evidence that M proteins can in fact impose constraints on lipid mobility and change the order of membranes were obtained by Shnyrova et al. (2007) working with the M protein of Newcastle disease virus (NDV). Using fluorescence microscopy and measurements of electrical admittance, they showed that addition of the NDV M protein to GUVs caused invagination and vesicle scission. The addition of M protein to liposomes also caused dequenching of a number of fluorescent lipids, which the authors suggest indicates the formation of membrane domains when the M protein binds to membranes. Thus, by decreasing the mobility of lipids they bind to and by causing lipid clustering, M proteins likely promote domain formation in membranes. This process might drive invagination, perhaps by a mechanism similar to that used by Shiga toxin.
Reticulons and DP1/Yop1
Most of the membrane-deforming proteins discussed in the previous sections of this review are peripheral membrane proteins that transiently associate with membranes. Many are involved in vesicular trafficking, which requires temporary changes in membrane shape. However, many organelles have complex shapes that persist for most or all of the cell cycle. Maintaining complex organelle shapes likely requires proteins that reside in the membrane (Voeltz and Prinz, 2007). In the final three sections of this review, we discuss how several families of integral membrane proteins directly shape organelle membranes.
The endoplasmic reticulum (ER) forms a dynamic network of tubules that extends throughout the cytosol. In the past few years, it was found that two families of conserved proteins, the reticulons and DP1/Yop1, are needed to maintain the tubular ER in a variety of organisms (De Craene et al., 2006; Voeltz et al., 2006; Audhya et al., 2007; Tolley et al., 2008). The reticulons and DP1/Yop1 are highly abundant integral membrane proteins in the ER. These two families do not share any sequence similarity but are likely to be structurally similar. Either alone is sufficient to maintain ER tubular structure in yeast (Voeltz et al., 2006). Reconstitution of these proteins with phospholipids revealed that they are not only required but also sufficient to shape membranes into tubules (Hu et al., 2008).
How the reticulons and DP1/Yop1 shape the ER membrane is not yet known but they probably promote membrane bending in two ways. First, they have an unusual membrane topology. All family members share a conserved reticulon homology domain that has two hydrophobic regions that anchor the proteins in the membrane. Unlike many integral membrane proteins they do not completely span the membrane but probably form a wedge-shape in the membrane (Voeltz et al., 2006). Thus, like soluble proteins that bend membrane by inserting amphipathic helices into membranes, the reticulons and DP1/Yop1 probably increase the area of one of the two leaflets of a bilayer and therefore cause bending. A second feature of the reticulons and DP1/Yop1 is that they form oligomers, and oligomerization is required for membrane tubulation (Hu et al., 2008; Shibata et al., 2008). These oligomers could be rigid and curve membranes by a scaffolding mechanism. However, the tubules formed in vitro are smaller in diameter (about 17 nm) than those in cells (about 35 nm) (Hu et al., 2008). Therefore, the scaffold must have some flexibility. Interestingly, rather than forming very large oligomers that might completely encircle ER tubules, current evidence suggests that the reticulon and DP1/Yop1form oligomers with 5–8 monomers (Hu et al., 2008; Shibata et al., 2008). It has been proposed that these oligomers form arcs perpendicular to the length of the ER tubule. If these arcs are evenly spaced on ER tubules, they could maintain the tubules while covering only 10% of the surface (Hu et al., 2008).
Caveolin and other caveolae-associated proteins
Caveolae are sac-like invaginations of the plasma membrane that are found in many mammalian cells types. They have been implicated in a number of cellular functions including signaling, endocytosis, and lipid regulation (Parat, 2009). Formation of caveolae requires caveolin. There are three caveolins in mammals. One of these, caveolin-1, is expressed in most cell types and cells lacking this protein also lack caveolae (Drab et al., 2001). Caveolin-3 is expressed only in skeletal and cardiac muscle and is needed for caveolae formation in these cells (Hagiwara et al., 2000; Galbiati et al., 2001). In contrast, caveolin-2 is not needed for caveolae formation and, though it does associate with the other caveolins, its role in caveolae formation is less clear (Parat, 2009).
Caveolins almost certainly play a direct role in generating the highly curved membranes of caveolae, but this has not yet been conclusively demonstrated. A number of structure studies indicate that caveolae contain a protein coat (Rothberg et al., 1992; Parton et al., 2006; Richter et al., 2008) and it has been suggested that caveolin could constitute this coat, by itself or together with other proteins (Monier et al., 1995). Caveolins may bend membranes in a manner similar to the reticulons and DP1/Yop1p. Like these proteins, the caveolins have an unusual membrane topology; they form a single hairpin in the membrane that does not completely span the bilayer and have both their N- and C-termini in the cytosol. They could therefore cause membrane bending by occupying more space in one of the two leaflets of a membrane. A second property caveolins share with reticulons and DP1/Yop1 is that they oligomerize and oligomerization is necessary for caveolae formation. Caveolins assemble into detergent-resistant oligomers (Monier et al., 1995; Fernandez et al., 2002) and it has been calculated that each caveolae contains a fixed number of caveolins, roughly 150 molecules (Pelkmans and Zerial, 2005). These oligomers probably form a scaffold that helps generate or stabilizes the high curvature of the caveolae membrane.
It seems possible that caveolins could also bend the caveolae membranes by altering the clustering or ordering of lipids. Caveolin-1 binds cholesterol (Murata et al., 1995; Thiele et al., 2000) and caveolae are known to be enriched in this and other lipids (Parat, 2009). There is also evidence that a region of caveolin-1 can induce formation of membrane domains enriched in phosphatidylserine, PI(4,5)P2, and cholesterol (Wanaski et al., 2003). Whether lipid clustering by caveolins alters membrane curvature is not yet known.
Other proteins may work together with caveolins to bend membranes. Two groups have recently found a critical role for cavin, also known as polymerase I and transcript release factor (PTRF), in caveole function and biogenesis (Hill et al., 2008; Liu and Pilch, 2008; Liu et al., 2008). PTRF-cavin is a soluble protein and is recruited to caveolae. Interestingly, it is a phosphatidylserine-binding protein and it may have affinity for the lipid environment of caveolae (Hill et al., 2008). It is possible that it plays a role in generating or stabilizing the highly curved caveolae. A homolog of PTFR-cavin has been suggested to have a role in generating membrane curvature and forming caveolae (Hansen et al., 2009). Overexpression of this protein, called serum deprivation response (SDPR), causes tubulation of caveolae in vivo. It will be interesting to determine how SDPR, PTFR-cavin, and caveolins work together to bend membranes and generate caveolae.
The F1F0-ATP synthase and cristae morphology
Mitochondria have an outer and an inner membrane. The inner membrane has an elaborate structure; portions are close apposed to the outer membrane while other regions form cristae that extend into the mitochondrial matrix. These cristae can have complex tubular or lamellar shapes that vary in different cell types (Zick et al., 2009). How they are formed is not understood but a number of proteins have been implicated in maintaining cristae morphology (Zick et al., 2009). One of these is the F1F0-ATP synthase complex, which is a highly abundant protein complex in the mitochondrial inner membrane. Yeast lacking subunits of this complex or with reduced amounts of the complex have aberrant cristae (Giraud et al., 2002; Paumard et al., 2002; Arselin et al., 2004; Bornhovd et al., 2006). There is considerable evidence that the F1F0-ATP synthase dimerizes with the monomers tilted at a angle relative to one another, though the angle may vary in different organisms (Minauro-Sanmiguel et al., 2005; Dudkina et al., 2005; 2006; Buzhynskyy et al., 2007). Because the F1F0-ATP synthase is highly abundant in the membrane, tilted dimers could bend the bilayer and affect cristae shape. The F1F0-ATP synthase dimers probably also oligomerize into higher order structures that could further stabilize positive curvature of the cristae membrane.
Recently, the protein Fcj1 was found to regulate the oligomerization of the F1F0-ATP synthase and to be required for proper cristae structure (Rabl et al., 2009). The authors propose that Fcj1 promotes negative membrane curvature while the F1F0-ATP synthase promotes positive curvature. Using immuno-EM they show that Fcj1 and the F1F0-ATP synthase are enriched in different regions of cisternae and could therefore work together to control cisternae shape.
Conclusions
Membrane bending is required for many processes in the cell (Figure 1) and the past few years have seen remarkable increases in the number of proteins that have been shown to directly deform membranes. There are probably many more yet to be identified. In particular, there must be more proteins required to shape organelles. It seems likely that all organelles that have defined shapes will require a number of membrane-bending proteins to keep these shapes.
We are still just beginning to understand how membrane-bending proteins function. Many seem to use one or more of at least three basic mechanisms: inserting amphipathic domains, forming rigid scaffolds, and affecting lipid distribution in the membrane (Figure 2). Much remains to be learned. In particular, very little is known about how proteins affect lipid distribution and order in membranes and how this can cause membrane bending. Membrane bending by integral membrane proteins, such as the reticulons, is also poorly understood. Finally, some membrane-bending proteins, such as Snf7, seem to use novel, poorly understood mechanisms. Understanding how the various ways that proteins can deform membranes and how these processes are regulated will be a major challenge for the future.
Acknowledgment
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content of this paper.
References
- Anders N, Jurgens G. Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell Mol Life Sci. 2008;65:3433–3445. doi: 10.1007/s00018-008-8227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B. Membrane deformation by protein coats. Curr Opin Cell Biol. 2006;18:386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]
- Arac D, Chen X, Khant HA, Ubach J, Ludtke SJ, Kikkawa M, Johnson AE, Chiu W, Sudhof TC, Rizo J. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- Arselin G, Vaillier J, Salin B, Schaeffer J, Giraud MF, Dautant A, Brethes D, Velours J. The modulation in subunits e and g amounts of yeast ATP synthase modifies mitochondrial cristae morphology. J Biol Chem. 2004;279:40392–40399. doi: 10.1074/jbc.M404316200. [DOI] [PubMed] [Google Scholar]
- Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178:43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Bai J, Tucker WC, Chapman ER. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat Struct Mol Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- Ball A, Nielsen R, Gelb MH, Robinson BH. Interfacial membrane docking of cytosolic phospholipase A2 C2 domain using electrostatic potential-modulated spin relaxation magnetic resonance. Proc Natl Acad Sci USA. 1999;96:6637–6642. doi: 10.1073/pnas.96.12.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerfeind R, Takei K, De Camilli P. Amphiphysin I is associated with coated endocytic intermediates and undergoes stimulation-dependent dephosphorylation in nerve terminals. J Biol Chem. 1997;272:30984–30992. doi: 10.1074/jbc.272.49.30984. [DOI] [PubMed] [Google Scholar]
- Beck R, Sun Z, Adolf F, Rutz C, Bassler J, Wild K, Sinning I, Hurt E, Brugger B, Bethune J, Wieland F. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc Natl Acad Sci USA. 2008;105:11731–11736. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Bielli A, Haney CJ, Gabreski G, Watkins SC, Bannykh SI, Aridor M. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J Cell Biol. 2005;171:919–924. doi: 10.1083/jcb.200509095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J Cell Sci. 2005;118:5393–5403. doi: 10.1242/jcs.02640. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhovd C, Vogel F, Neupert W, Reichert AS. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J Biol Chem. 2006;281:13990–13998. doi: 10.1074/jbc.M512334200. [DOI] [PubMed] [Google Scholar]
- Burger KN, Demel RA, Schmid SL, de Kruijff B. Dynamin is membrane-active: lipid insertion is induced by phosphoinositides and phosphatidic acid. Biochemistry. 2000;39:12485–12493. doi: 10.1021/bi000971r. [DOI] [PubMed] [Google Scholar]
- Buzhynskyy N, Sens P, Prima V, Sturgis JN, Scheuring S. Rows of ATP synthase dimers in native mitochondrial inner membranes. Biophys J. 2007;93:2870–2876. doi: 10.1529/biophysj.107.109728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F, McMahon HT, Kozlov MM. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J. 2008;95:2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- Carr JF, Hinshaw JE. Dynamin assembles into spirals under physiological salt conditions upon the addition of GDP and gamma-phosphate analogues. J Biol Chem. 1997;272:28030–28035. doi: 10.1074/jbc.272.44.28030. [DOI] [PubMed] [Google Scholar]
- Cestra G, Kwiatkowski A, Salazar M, Gertler F, De Camilli P. Tuba, a GEF for CDC42, links dynamin to actin regulatory proteins. Methods Enzymol. 2005;404:537–545. doi: 10.1016/S0076-6879(05)04047-4. [DOI] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- Dawson JC, Legg JA, Machesky LM. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–498. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- De Craene JO, Coleman J, Estrada de Martin P, Pypaert M, Anderson S, Yates JRR, Ferro-Novick S, Novick P. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, Schedl A, Haller H, Kurzchalia TV. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun HP. Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett. 2005;579:5769–5772. doi: 10.1016/j.febslet.2005.09.065. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Sunderhaus S, Braun HP, Boekema EJ. Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett. 2006;580:3427–3432. doi: 10.1016/j.febslet.2006.04.097. [DOI] [PubMed] [Google Scholar]
- Edeling MA, Smith C, Owen D. Life of a clathrin coat: insights from clathrin and AP structures. Nat Rev Mol Cell Biol. 2006;7:32–44. doi: 10.1038/nrm1786. [DOI] [PubMed] [Google Scholar]
- Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsad K, Slepnev V, Ochoa G, Daniell L, Haucke V, De Camilli P. A putative role for intramolecular regulatory mechanisms in the adaptor function of amphiphysin in endocytosis. Neuropharmacology. 2003;45:787–796. doi: 10.1016/s0028-3908(03)00306-x. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- Fernandez I, Arac D, Ubach J, Gerber SH, Shin O, Gao Y, Anderson RG, Sudhof TC, Rizo J. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- Fernandez I, Ying Y, Albanesi J, Anderson RG. Mechanism of caveolin filament assembly. Proc Natl Acad Sci USA. 2002;99:11193–11198. doi: 10.1073/pnas.172196599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Fotin A, Kirchhausen T, Grigorieff N, Harrison SC, Walz T, Cheng Y. Structure determination of clathrin coats to sub-nanometer resolution by single particle cryo-electron microscopy. J Struct Biol. 2006;156:453–460. doi: 10.1016/j.jsb.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AA, Wisner MA, Malmberg NJ, Victor KG, Fanucci GE, Nalefski EA, Falke JJ, Cafiso DS. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 2002;41:6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- Frazier AA, Roller CR, Havelka JJ, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry. 2003;42:96–105. doi: 10.1021/bi0268145. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Orci L, Schekman R. Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol. 2008;18:330–336. doi: 10.1016/j.tcb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Engelman JA, Volonte D, Zhang XL, Minetti C, Li M, Hou HJ, Kneitz B, Edelmann W, Lisanti MP. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J Biol Chem. 2001;276:21425–21433. doi: 10.1074/jbc.M100828200. [DOI] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud MF, Paumard P, Soubannier V, Vaillier J, Arselin G, Salin B, Schaeffer J, Brethes D, di Rago JP, Velours J. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim Biophys Acta. 2002;1555:174–180. doi: 10.1016/s0005-2728(02)00274-8. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Grant BD, Caplan S. Mechanisms of EHD/RME-1 protein function in endocytic transport. Traffic. 2008;9:2043–2052. doi: 10.1111/j.1600-0854.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara Y, Sasaoka T, Araishi K, Imamura M, Yorifuji H, Nonaka I, Ozawa E, Kikuchi T. Caveolin-3 deficiency causes muscle degeneration in mice. Hum Mol Genet. 2000;9:3047–3054. doi: 10.1093/hmg/9.20.3047. [DOI] [PubMed] [Google Scholar]
- Hansen CG, Bright NA, Howard G, Nichols BJ. SDPR induces membrane curvature and functions in the formation of caveolae. Nat Cell Biol. 2009;11:807–814. doi: 10.1038/ncb1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Kent HM, Ford MG, Hegde BG, Daumke O, Butler PJ, Mittal R, Langen R, Evans PR, McMahon HT. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, Hancock JF, Parton RG. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132:113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE. Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Horvath CA, Vanden Broeck D, Boulet GA, Bogers J, De Wolf MJ. Epsin: inducing membrane curvature. Int J Biochem Cell Biol. 2007;39:1765–1770. doi: 10.1016/j.biocel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- Huang M, Weissman JT, Beraud-Dufour S, Luan P, Wang C, Chen W, Aridor M, Wilson IA, Balch WE. Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export. J Cell Biol. 2001;155:937–948. doi: 10.1083/jcb.200106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H, Stephens DJ. Assembly, organization, and function of the COPII coat. Histochem Cell Biol. 2008;129:129–151. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kalthoff C, Groos S, Kohl R, Mahrhold S, Ungewickell EJ. Clint: a novel clathrin-binding ENTH-domain protein at the Golgi. Mol Biol Cell. 2002;13:4060–4073. doi: 10.1091/mbc.E02-03-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DE, Lee A, Frank DW, Marks MS, Lemmon MA. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J Biol Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- Kohout SC, Corbalan-Garcia S, Gomez-Fernandez JC, Falke JJ. C2 domain of protein kinase C alpha: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 2003;42:1254–1265. doi: 10.1021/bi026596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Lemmon MA. Analysis of phosphoinositide binding by pleckstrin homology domain from dynamin. Methods Enzymol. 2001;329:457–468. doi: 10.1016/s0076-6879(01)29107-1. [DOI] [PubMed] [Google Scholar]
- Lee E, Marcucci M, Daniell L, Pypaert M, Weisz OA, Ochoa GC, Farsad K, Wenk MR, De Camilli P. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–1196. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Liu L, Pilch PF. A critical role of cavin (polymerase I and transcript release factor) in caveolae formation and organization. J Biol Chem. 2008;283:4314–4322. doi: 10.1074/jbc.M707890200. [DOI] [PubMed] [Google Scholar]
- Liu L, Brown D, McKee M, Lebrasseur NK, Yang D, Albrecht KH, Ravid K, Pilch PF. Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab. 2008;8:310–317. doi: 10.1016/j.cmet.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR. Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J Biol Chem. 2003;278:46772–46781. doi: 10.1074/jbc.M307334200. [DOI] [PubMed] [Google Scholar]
- Lundmark R, Doherty GJ, Howes MT, Cortese K, Vallis Y, Parton RG, McMahon HT. The GTPase-activating protein GRAF1 regulates the CLIC/GEEC endocytic pathway. Curr Biol. 2008;18:1802–1808. doi: 10.1016/j.cub.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova S, Long F, Stahelin RV, Pingali SV, Murray D, Cho W, Schlossman ML. X-ray reflectivity studies of cPLA2{alpha}-C2 domains adsorbed onto Langmuir monolayers of SOPC. Biophys J. 2005;89:1861–1873. doi: 10.1529/biophysj.105.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg NJ, Van Buskirk DR, Falke JJ. Membrane-docking loops of the cPLA2 C2 domain: detailed structural analysis of the protein-membrane interface via site-directed spin-labeling. Biochemistry. 2003;42:13227–13240. doi: 10.1021/bi035119+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006;25:2889–2897. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T, Weiland F, Conzelmann KK. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol. 1999;73:242–250. doi: 10.1128/jvi.73.1.242-250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Futterer K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills IG, Praefcke GJ, Vallis Y, Peter BJ, Olesen LE, Gallop JL, Butler PJ, Evans PR, McMahon HT. EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J Cell Biol. 2003;160:213–222. doi: 10.1083/jcb.200208023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minauro-Sanmiguel F, Wilkens S, Garcia JJ. Structure of dimeric mitochondrial ATP synthase: novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis. Proc Natl Acad Sci USA. 2005;102:12356–12358. doi: 10.1073/pnas.0503893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S, Parton RG, Vogel F, Behlke J, Henske A, Kurzchalia TV. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Peranen J, Schreiner R, Wieland F, Kurzchalia TV, Simons K. VIP21/caveolin is a cholesterol-binding protein. Proc Natl Acad Sci USA. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature. 1988;334:661–665. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- Parat MO. The biology of caveolae: achievements and perspectives. Int Rev Cell Mol Biol. 2009;273:117–162. doi: 10.1016/S1937-6448(08)01804-2. [DOI] [PubMed] [Google Scholar]
- Parton RG, Hanzal-Bayer M, Hancock JF. Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J Cell Sci. 2006;119:787–796. doi: 10.1242/jcs.02853. [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO Rep. 2002;3:1035–1041. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Zerial M. Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae. Nature. 2005;436:128–133. doi: 10.1038/nature03866. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Korner C, Jagasia R, Keil T, Baumeister W, Cyrklaff M, Neupert W, Reichert AS. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Reynwar BJ, Illya G, Harmandaris VA, Muller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- Richter T, Floetenmeyer M, Ferguson C, Galea J, Goh J, Lindsay MR, Morgan GP, Marsh BJ, Parton RG. High-resolution 3D quantitative analysis of caveolar ultrastructure and caveola-cytoskeleton interactions. Traffic. 2008;9:893–909. doi: 10.1111/j.1600-0854.2008.00733.x. [DOI] [PubMed] [Google Scholar]
- Ringstad N, Gad H, Low P, Di Paolo G, Brodin L, Shupliakov O, De Camilli P. Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron. 1999;24:143–154. doi: 10.1016/s0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Romer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D, Aly MR, Fraisier V, Florent JC, Perrais D, Lamaze C, Raposo G, Steinem C, Sens P, Bassereau P, Johannes L. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–675. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- Rufener E, Frazier AA, Wieser CM, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin C2B domain determined by site-directed spin labeling. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo G, Gu QM, Prestwich GD, Sollner TH, Rothman JE. Calcium-dependent switching of the specificity of phosphoinositide binding to synaptotagmin. Proc Natl Acad Sci USA. 1996;93:13327–13332. doi: 10.1073/pnas.93.23.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug–erythrocyte interactions. Proc Natl Acad Sci USA. 1974;71:4457–4461. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, Liu ZJ, Wang BC, Yamamoto M, Terada T, Miyazawa A, Tanaka A, Sugano S, Shirouzu M, Nagayama K, Takenawa T, Yokoyama S. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Shin N, Ahn N, Chang-Ileto B, Park J, Takei K, Ahn SG, Kim SA, Di Paolo G, Chang S. SNX9 regulates tubular invagination of the plasma membrane through interaction with actin cytoskeleton and dynamin 2. J Cell Sci. 2008;121:1252–1263. doi: 10.1242/jcs.016709. [DOI] [PubMed] [Google Scholar]
- Shnyrova AV, Ayllon J, Mikhalyov II, Villar E, Zimmerberg J, Frolov VA. Vesicle formation by self-assembly of membrane-bound matrix proteins into a fluidlike budding domain. J Cell Biol. 2007;179:627–633. doi: 10.1083/jcb.200705062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon J, Gareil O, Bassereau P, Gaudin Y. Membrane deformations induced by the matrix protein of vesicular stomatitis virus in a minimal system. J Gen Virol. 2005;86:3357–3363. doi: 10.1099/vir.0.81129-0. [DOI] [PubMed] [Google Scholar]
- Soulet F, Yarar D, Leonard M, Schmid SL. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:2058–2067. doi: 10.1091/mbc.E04-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A. The life cycle of a transport vesicle. Cell Mol Life Sci. 2008;65:2781–2789. doi: 10.1007/s00018-008-8349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SM, Gurkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- Stein PE, Boodhoo A, Tyrrell GJ, Brunton JL, Read RJ. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature. 1992;355:748–750. doi: 10.1038/355748a0. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Meyerkord CL, Wang HG. Bif-1/Endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarricone C, Xiao B, Justin N, Walker PA, Rittinger K, Gamblin SJ, Smerdon SJ. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature. 2001;411:215–219. doi: 10.1038/35075620. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Thiele C, Hannah MJ, Fahrenholz F, Huttner WB. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat Cell Biol. 2000;2:42–49. doi: 10.1038/71366. [DOI] [PubMed] [Google Scholar]
- Tolley N, Sparkes IA, Hunter PR, Craddock CP, Nuttall J, Roberts LM, Hawes C, Pedrazzini E, Frigerio L. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic. 2008;9:94–102. doi: 10.1111/j.1600-0854.2007.00670.x. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA. Sheets, ribbons and tubules – how organelles get their shape. Nat Rev Mol Cell Biol. 2007;8:258–264. doi: 10.1038/nrm2119. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Wanaski SP, Ng BK, Glaser M. Caveolin scaffolding region and the membrane binding region of SRC form lateral membrane domains. Biochemistry. 2003;42:42–56. doi: 10.1021/bi012097n. [DOI] [PubMed] [Google Scholar]
- Welsch S, Muller B, Krausslich HG. More than one door – Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N. A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem. 2004;279:14929–14936. doi: 10.1074/jbc.M309408200. [DOI] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL. SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell. 2007;13:43–56. doi: 10.1016/j.devcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9:133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XL, Sadler JE. Pathogenesis of thrombotic microangiopathies. Annu Rev Pathol. 2008;3:249–277. doi: 10.1146/annurev.pathmechdis.3.121806.154311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]