Abstract
A key interest in clinical diagnosis and pharmaceutical industry is to have a repertoire of noninvasive biomarkers to—individually or in combination—be able to infer or predict the degree of liver injury caused by pathological conditions or drugs. Metabolomics—a comprehensive study of global metabolites—has become a highly sensitive and powerful tool for biomarker discovery thanks to recent technological advances. An ultra-performance liquid chromatography/time-of-flight tandem mass spectrometry (UPLC/TOF MS/MS)-based metabolomics approach was employed to investigate sera from galactosamine-treated rats to find potential biomarkers for acute liver injury. Hepatic damage was quantified by determining serum transaminase activity and in situ liver histological lesions. Principal component analysis in combination with coefficient of correlation analysis was used for biomarker selection and identification. According to the data, serum levels of several metabolites including glucose, amino acids, and membrane lipids were significantly modified, some of them showing a high correlation with the degree of liver damage determined by histological examination of the livers. In conclusion, this study supports that UPLC-MS/MS based serum metabolomics in experimental animal models could be a powerful approach to search for biomarkers for drug- or disease-induced liver injury.
Keywords: Galactosamine, Hepatotoxicity, Acute-liver injury, DILI, Biomarkers, Serum metabolic profiling, UPLC-MS/MS metabolomics
1 Introduction
Drug-induced liver injury (DILI) remains a major cause of worldwide mortality (Boelsterli and Lim 2007; Ulrich 2007) and represents a serious clinical and financial problem because it is the single greatest cause of attrition in drug development and withdrawal of approved drugs from the market (Kaplowitz 2001). The cost of introducing a new drug to the market (estimated at hundred of millions of dollars) generates a high socio-economical pressure to generate new tools and approaches for the identification of highly sensitive biomarkers of liver toxicity. Currently, besides liver biopsy the activity of a series of hepatic enzymes and proteins released into the blood by damaged liver cells are the most useful tools available in detecting liver damage in a non-invasive manner. Hepatic screening test measuring bilirubin (total and unconjugated), alkaline phosphatase, aspartate aminotransferase (AST), alanine aminotransferase (ALT), albumin, gamma-glutamyl transferase (GGT), prothrombin time and glutathione-S-transferase are routinely used in both preclinical and human clinical drug studies (Ozer et al. 2008). The inherent deficiencies and limitations in sensitivity or specificity of each of these existing markers preclude the use of any one marker as the “standard” for detecting liver damage or DILI (Ozer et al. 2008; Muller and Dieterle 2009). The development of novel diagnostic biomarkers with greater sensitivity and specificity can enhance the efficiency in the detection of low-degree liver damage, increasing the confidence of drug safety for release into the market.
Metabolomics refers to the comprehensive analysis of endogenous small molecules present in a biological system. The recent technological development of analytical instruments combined with rapid progress in bioinformatics has led to new opportunities to quickly and simultaneously measure and model huge numbers of metabolites in biological samples (Nicholson and Wilson 2003; Fernie et al. 2004; Chen et al. 2007; Clarke and Haselden 2008). In consequence, metabolomics has found broad application in the identification of biomarkers, and in the unravelling of pathophysiological mechanism in many scientific fields, such as plant biology (Schauer and Fernie 2006), toxicology (Coen et al. 2007; Clarke and Haselden 2008) and disease diagnosis (Brindle et al. 2002). Currently, metabolomic profiling has been performed using a number of techniques, including high-field nuclear magnetic resonance (NMR) (Nicholls et al. 2003; Duarte et al. 2005; Coen et al. 2007), and gas or liquid chromatography/mass spectrometry (GC–MS, LC-MC)(Lin et al. 2010; Lutz et al. 2006; Major et al. 2006; Zhang et al. 2007; Kushnir et al. 2009). While NMR demonstrates its advantages in highly selective and non-destructive analysis, LC–MS possesses much higher sensitivity and resolution. The development of ultra-performance liquid chromatography (UPLC) has made it possible to achieve even higher resolutions, higher sensitivities, and rapid separations when compared to conventional LC methods (Nordstrom et al. 2006; Novakova et al. 2006). UPLC combined with orthogonal acceleration time-of-flight mass spectrometry (oaTOF–MS), which enables the exact mass measurements, is undoubtedly a suitable system for metabolomics (Chan et al. 2007). However, because of this high sensitivity and capacity to detect subtle changes, the use of well-characterized in vitro (Dan and Yeoh 2008; Gomez-Lechon et al. 2008) and in vivo (Newsome et al. 2000; Tunon et al. 2009) experimental models are advisable in order to minimize the impact of variables such as age, gender, genetic and lifestyle heterogeneity.
For the identification of non-invasive biomarkers enabling direct inference of the extent of liver-injury, an UPLC/TOF-MS-based metabolomics approach was applied to characterize the serum metabolic profile associated with acute liver injury. D-galactosamine (galN)-treated rats were chosen as the experimental model for two reasons. First, it is a classical “model” hepatotoxin resembling human viral hepatitis (Keppler et al. 1968) that has been widely used in studies on different aspects such as mechanism of damage induction, biomarker discovery and therapy of acute liver injury (El-Mofty et al. 1975; Stramentinoli et al. 1978; Maezono et al. 1996; Coen et al. 2007; Feng et al. 2007; Lv et al. 2007; Komano et al. 2008; Miller et al. 2008; Yovchev et al. 2008; Kitazawa et al. 2009). Second, studies in rats revealed an extreme inter-animal variability in the extent of the induced liver injury (Coen et al. 2007; Miller et al. 2008), which is convenient since it gives the opportunity to correlate different changes in metabolite levels to different stages of liver injury. Our results indicate that serum levels of many membrane lipids including mono and diacylglycerolphospholipids, and sphingomyelins were significantly affected by the treatment with the hepatotoxin. Remarkably, a number of them were identified by using exact mass measurements, and found to be in high correlation with the degree of cell death in the liver as assessed by histological examination. These results constitute a clear support to use UPLC-MS-based metabolomics to identify non-invasive biomarkers for liver damage.
2 Materials and methods
2.1 Animal model and samples collection
All the animal experimentation was conducted in accordance with the Spanish Guide for the Care and Use of Laboratory Animals (RD 1201/2005—BOE 21/10/05). Twenty-three male Wistar rats (14 weeks old, body weight 300–400 g), were maintained in an environmentally controlled room at 22°C on a 12 h light/dark cycle and provided with standard diet (Rodent Maintenance Diet, Harlan Teklad Global Diet 2014) and water ad libitum. The rats were randomly allocated in two groups. The test group (n = 11) received an intraperitoneal injection of 1 g/kg/5 ml of D(+)-galactosamine (2-amino-2-deoxy-D-galactose) hydrochloride (Sigma-Aldrich Chemical Co., Steinheim Germany) while animals in the control group (n = 12) were injected with the same volume of saline solution (5 ml/Kg of 0.9% NaCl sterile). Four days before (untreated samples) and 18-h after the injection, individualized blood samples of each animal were drawn under anesthesia (4% isoflurane), after a 12-h food fasting period and, the samples were quickly processed to obtain the sera by using gel serum separator tubes (BD Microtainer SST Tubes). The obtained sera were transferred to a fresh Eppendorf tubes and stored at −80°C. Immediately after blood extraction, the liver of each animal was excised and stored in 10% formalin for histopathological analysis.
2.2 Clinical serum biochemistry and histopathological analysis of liver tissue
Aliquots of 10 or 1 μl serum from saline- or galN-treated animals, respectively, were analyzed for alanine aminotransferase (ALT) activity using Infinity ALT (GPT) reagent (Thermo Electron, Waltham, MA). Formalin-fixed liver sections were embedded into paraffin, sectioned to 5 μm slices, mounted on slides and stained with hematoxylin-eosin (American HistoLabs Inc., Gaithersburg, MD, USA). The hematoxilin-eosin-stained tissue sections were examined by light microscopy for morphological changes following treatment with galN. Colour images were captured using a 40× objective on a Zeiss Axioplan light microscope equipped with an AxioCam HR CCD camera and Axiovision 3.1 software (Carl Zeiss, Inc., Thornwood, NY, USA). DNA fragmentation, characteristic of apoptosis, was assessed in formalin-fixed liver sections by direct terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling (TUNEL assay) by using ApoTag® Peroxidase in situ Apoptosis Detection Kit (Chemicon Int., Temecula, CA, USA) performed in accordance with manufacturer's instructions. A quantitative value referred as cell death score (CDS) was determined for each animal by counting the number of apoptotic cells that were detected in 1 square millimetre of liver corresponding to the area covered by 10 frames acquired using a 40× objective. Each of the 10 frames were physically separated by at least 1 mm in order to cover different regions of the liver, and they were captured by using a “blind test” in which the examiner was not aware of the identity of the sample present in each slide.
2.3 UPLC® -MS analysis
Proteins were precipitated from the defrosted serum samples (50 μl) by adding four volumes of methanol in 1.5 ml microtubes at room temperature. After brief vortex mixing the samples were kept overnight at −20°C. Supernatants were collected after centrifugation at 13,000 rpm for 10 min, and transferred to vials for UPLC®-MS analysis. Chromatography was performed on a 1 mm i.d. × 100 mm ACQUITY 1.7 μm C8 BEH column (Waters Corp., Milford, USA) using an ACQUITY UPLC system (Waters Corp., Milford, USA). The column was maintained at 40°C and eluted with a 10 min linear gradient. The mobile phase, at a flow rate of 140 μl/min, consisted of 100% solvent A (0.05% formic acid) for 1 min followed by an incremental increase of solvent B (acetonitrile containing 0.05% formic acid) up to 50% over a further minute, increasing to 100% B over the next 6 min before returning to the initial composition in readiness for the subsequent injection which proceeded a 45 s system re-cycle time. The volume of sample injected onto the column was 1 μl. The eluent was introduced into the mass spectrometer (LCT Premier™, Waters Corp., Milford, USA) by electrospray ionization, with capillary and cone voltages set in the positive and negative ion modes to 3,200 and 30 V, and 2,800 and 50 V respectively. The nebuliser gas was set to 600 l/h at a temperature of 350°C. The cone gas was set to 50 l/h and the source temperature was set to 150°C. Centroid data were acquired from m/z 50–1,000 using an accumulation time of 0.2 s per spectrum. All spectra were mass corrected in real time by reference to leucine enkephalin, infused at 50 μl/min through an independent reference electrospray, sampled every 10 s. A test mixture of standard compounds (acetaminophen, sulfaguanidine, sulfadimethoxine, Val–Tyr–Val, terfenadine, leucine-enkephaline, reserpine and erythromycin—all 5 nM in water) was analyzed before and after the entire set of randomized, duplicated sample injections in order to examine the retention time stability (generally <6 s variation, injection-to-injection), mass accuracy (generally <3 ppm for m/z 400–1,000, and <1.2 mDa for m/z 50–400) and sensitivity of the system throughout the course of the run which lasted a maximum of 28 h per batch of samples injected. For each injection batch, the overall quality of the analysis procedure was monitored using ten repeat extracts of a pooled serum sample. Corresponding root mean square coefficients of variation were 15.3% over all variables detected in the positive ion mode, and 11.7% in the negative ion mode. Online tandem mass spectrometry (MS/MS) experiments for metabolite identification were performed on a Waters QTOF Premier™ (Waters Corp., Milford, USA) instrument operating in both the positive and negative ion electrospray modes; source parameters were identical to those employed in the profiling experiments, except for the cone voltage which was increased (30–70 V) when pseudo MS/MS/MS data were required. During retention time windows corresponding to the elution of the compounds under investigation the quadrupole was set to resolve and transmit ions with appropriate mass-to-charge values. The selected ions then traversed an argon-pressurized cell, with a collision energy voltage (typically between 5 and 50 V) applied in accordance with the extent of ion fragmentation required. Subsequent TOF analysis of the fragment ions generated accurate mass (generally <3 ppm for m/z 400–1,000, and <1.2 mDa for m/z 50–400) MS/MS or pseudo MS/MS/MS spectra corrected in real time by reference to leucine-enkephalin, infused at 50 μl/min through an independent reference electrospray, sampled every 10 s. Centroid data were acquired between m/z 50–1,000 using an accumulation time of 0.2 s per spectrum.
2.4 Data processing
All data were processed using the MarkerLynx application manager for MassLynx 4.1 software (Waters Corp., Milford, USA). The LC/MS data were peak-detected and noise-reduced in both the LC and MS domains such that only true analytical peaks were further processed by the software (e.g., noise spikes are rejected). A list of intensities (chromatographic peak areas) of the peaks detected was then generated for the first chromatogram, using the Rt-m/z data pairs as identifiers. This process was repeated for each LC-MS analysis and the data sorted such that the correct peak intensity data for each Rt-m/z pair were aligned in a table. Detected variables that were found to be zero in both injections of any serum extract corresponding to untreated animals were then removed from the dataset. Although this process would be expected to remove variables associated with xenobiotic metabolites, the influence of these species still has a strong impact on the analysis by way of ion suppression effects. In order to remove this interference, all variables co-eluting with xenobiotic metabolites, found in the retention time region 2.75–3.55 min, were eliminated. In the case of the positive ion mode, the resulting variable ion intensities were then normalized within each injection, to the sum of peak intensities in that injection. In the negative ion mode the normalization factor for each injection was calculated from the ratio of the median intensity to the median intensity of a reference serum injection, as obtained from all variables contained in the retention time bin 6–8 min. All zeros contained within the normalized dataset were substituted with missing values to form a single matrix with Rt-m/z pairs for each injection. The final dataset was mean centered and pareto scaled during multivariate data analysis.
2.5 Multivariate statistical analysis
The first objective in the data analysis process is to reduce the dimensionality of the complex data set to enable easy visualization of any metabolic clustering of the different groups of samples. This has been achieved by principal components analysis (PCA) where the data matrix is reduced to a series of principal components (PCs), each a linear combination of the original Rt-m/z pair peak areas. Each successive PC explains the maximum amount of variance possible, not accounted for by the previous PCs. Hence the scores plots shown in the PCA figures—where the first two principal components, t[1] and t[2], are plotted—represent the most important metabolic variation in the samples captured by the analysis. The corresponding loadings (p[1], p[2] etc.) for each model are the projections of the model variables onto each of the principal components; i.e., variables with positive loadings, p[A], correlate with positively scoring, t[A], samples (injections) on the same component, A. The performance levels of the PCA models were evaluated using the R2X (variance modeled) and Q2 (cross-validated R2X) parameters. Robust trends captured by PCA models are reflected by similar values of R2X and Q2.
2.6 Metabolite identification
Exact molecular mass data from redundant m/z peaks corresponding to the formation of different parent (e.g., cations in the positive ion mode, anions in the negative ion mode, adducts, multiple charges) and product (formed by spontaneous “in-source” CID) ions were first used to help confirm the metabolite molecular mass. This information was then submitted for database searching, either in-house or using the online ChemSpider database where the Kegg, Human Metabolome Database and Lipid Maps data source options were selected. MS/MS data analysis highlights neutral losses or product ions, which are characteristic of metabolite groups and can serve to discriminate between database hits. Specific metabolite group MS/MS characterization methods have been described previously (Barr et al. 2010). Regiochemistry [e.g., PC(18:0/0:0), PC(0:0/18:0)] was only specified if it could be corroborated by mass spectral evidence. However, in many instances these differences can be inferred by relative abundance and retention time information, e.g., in general sn-1 monoacylglycerophosphocholine [PC(X:X/0:0)] is usually much more abundant, and chromatographically retained, than sn-2 monoacylglycerophosphocholine [PC(0:0/X:X)].
2.7 Statistics
The strength of the relationship between the different parameters for liver damage quantization was estimated by a Pearson correlation coefficient. The correlation between each of the serum metabolites detected and the degree of liver damage was assessed by calculation of Spearman rank correlation coefficients. Value of the correlation coefficients being very near to zero, shows that no significant correlation exists between the compared parameters. Fisher's exact test was used to assess the significance of the association between metabolite and degree of liver damage. A statistical significant difference was considered when P value was less than 0.01.
A regression model based on the logistic function f(z) was created to predict the risk of suffering acute-liver injury.
where:
The logit (z) in this model was calculated using the intensity (Imetabolite) of a set of metabolites significantly affected by galN treatment (Table 1, in bold). The arbitrarily chosen metabolites have in common a small relative standard deviation between animals (<2.5%) before treatment with saline solution or galN, and after treatment with the saline solution. The calculated means for the intensity of these metabolites differ significantly (>1.9 times) between animals treated with galN and those treated with the saline solution in all cases.
Table 1. Endogenous serum metabolites significantly affected by galN-treatment.
| Metabolite ID | m/z | RT | ID methoda | Salineb log(NS) | galNb log(NS) | log(FC)c | −log(p)d |
|---|---|---|---|---|---|---|---|
| AA Methionine | 150.06 | 0.87 | Standard | −3.3 | −2.8 | 0.58 | 3.21 |
| AA Proline | 116.07 | 0.61 | Standard | −0.7 | −0.2 | 0.56 | 2.75 |
| AA Threonine | 120.06 | 0.59 | Standard | −1.1 | −0.7 | 0.42 | 2.50 |
| AA Tyrosine | 182.08 | 0.98 | Standard | 0.0 | 1.0 | 1.00 | 4.25 |
| FFA Linoleic acid | 281.25 | 5.52 | Standard | −0.8 | −0.7 | 0.11 | 1.92 |
| Glucose | 383.12 | 0.59 | Standard | −0.1 | −0.3 | −0.16 | 2.74 |
| LysoPC(0:0/18:0) | 524.37 | 4.54 | MS/MS | 0.3 | −0.3 | −0.64 | 13.45 |
| LysoPC(14:0/0:0)) | 468.31 | 3.60 | Standard | −0.4 | −0.7 | −0.30 | 7.91 |
| LysoPC(15:0/0:0) | 482.32 | 3.83 | Standard | −0.3 | −0.7 | −0.33 | 7.31 |
| LysoPC(16:0/0:0)) | 496.34 | 4.09 | Standard | 1.7 | 1.3 | −0.42 | 11.14 |
| LysoPC(16:1/0:0) | 494.32 | 3.77 | MS/MS | 0.0 | −0.2 | −0.15 | 2.86 |
| LysoPC(17:0/0:0) | 510.36 | 4.37 | Standard | −0.1 | −0.6 | −0.47 | 7.14 |
| LysoPC(18:1/0:0) | 522.36 | 4.28 | Standard | 0.9 | 0.5 | −0.34 | 8.75 |
| LysoPC(18:3) | 518.32 | 3.98 | Database | −1.5 | −2.1 | −0.54 | 7.40 |
| LysoPC(19:0/0:0) | 538.38 | 4.96 | Standard | −0.5 | −1.0 | −0.51 | 6.39 |
| LysoPC(20:0/0:0) | 552.40 | 5.26 | Standard | −0.8 | −1.2 | −0.41 | 7.31 |
| LysoPC(20:1/0:0) | 550.39 | 4.81 | MS/MS | −0.2 | −1.0 | −0.75 | 10.16 |
| LysoPC(20:2/0:0) | 548.37 | 4.44 | MS/MS | −0.7 | −1.1 | −0.40 | 6.73 |
| LysoPC(20:3) | 546.36 | 4.54 | Database | −1.2 | −1.5 | −0.31 | 6.50 |
| LysoPC(20:4) | 544.34 | 3.99 | MS/MS | 1.2 | 1.0 | −0.22 | 4.34 |
| LysoPC(22:0) | 580.43 | 5.86 | Database | −1.1 | −1.3 | −0.23 | 6.83 |
| LysoPC(22:1/0:0) | 578.42 | 5.38 | MS/MS | −1.3 | −1.7 | −0.38 | 8.85 |
| LysoPC(22:5/0:0) | 570.36 | 4.12 | MS/MS | −1.1 | −0.9 | 0.21 | 2.71 |
| LysoPC(24:0) | 608.46 | 6.46 | Database | −0.9 | −1.0 | −0.09 | 1.71 |
| LysoPC(24:1/0:0) | 606.45 | 5.96 | MS/MS | −1.0 | −1.3 | −0.35 | 8.93 |
| LysoPC(O-16:0/0:0) | 482.36 | 4.21 | MS/MS | −0.6 | −1.1 | −0.57 | 12.59 |
| LysoPC(O-18:0/0:0) | 510.39 | 4.80 | MS/MS | −1.1 | −1.6 | −0.48 | 11.61 |
| LysoPC(O-20:0/0:0) | 538.42 | 5.41 | MS/MS | −1.5 | −1.8 | −0.37 | 9.32 |
| LysoPC(O-22:0/0:0) | 566.46 | 6.02 | MS/MS | −1.9 | −2.0 | −0.19 | 3.62 |
| LysoPC(P-18:0/0:0) | 508.37 | 4.39 | MS/MS | −0.7 | −1.4 | −0.69 | 13.95 |
| LysoPE(16:0) | 454.29 | 4.07 | MS/MS | −0.3 | 0.3 | 0.53 | 3.72 |
| LysoPE(18:0) | 482.32 | 4.64 | MS/MS | −0.1 | 0.2 | 0.37 | 3.44 |
| LysoPE(18:2) | 478.29 | 3.93 | MS/MS | −0.7 | 0.0 | 0.75 | 6.92 |
| LysoPE(20:4) | 502.29 | 3.97 | MS/MS | −0.6 | 0.2 | 0.74 | 4.52 |
| LysoPE(22:6) | 526.29 | 3.97 | Database | −1.2 | −0.4 | 0.83 | 4.15 |
| LysoPE(P-16:0/0:0) | 438.30 | 4.25 | MS/MS | −0.9 | −1.4 | −0.54 | 9.19 |
| PC(14:0/16:0) | 706.54 | 7.39 | Standard | −0.4 | −0.1 | 0.31 | 9.80 |
| PC(14:0/18:1) | 754.55 | 7.50 | MS/MS | −0.9 | −0.5 | 0.38 | 6.50 |
| PC(14:0/20:4) | 754.54 | 7.15 | MS/MS | −0.2 | 0.0 | 0.21 | 6.48 |
| PC(15:0/18:2) | 744.56 | 7.38 | MS/MS | 0.2 | 0.5 | 0.37 | 3.64 |
| PC(16:0/16:0) | 734.57 | 7.80 | MS/MS | 0.5 | 0.8 | 0.29 | 5.83 |
| PC(16:0/18:1) | 760.59 | 7.87 | Standard | 1.3 | 1.6 | 0.31 | 8.68 |
| PC(16:0/18:2) | 758.57 | 7.58 | Standard | 1.9 | 2.1 | 0.20 | 9.44 |
| PC(16:0/20:5) | 780.56 | 7.25 | MS/MS | 0.3 | 0.5 | 0.20 | 4.58 |
| PC(17:0/18:2) | 772.59 | 7.77 | MS/MS | 0.3 | 0.7 | 0.44 | 4.76 |
| PC(18:0/18:2) | 786.61 | 7.97 | Standard | 1.5 | 1.8 | 0.24 | 10.96 |
| PC(18:0/20:4) | 810.60 | 7.92 | Standard | 2.0 | 1.8 | −0.15 | 7.61 |
| PC(18:0/22:6) | 834.60 | 7.68 | Standard | −0.3 | −0.9 | −0.60 | 1.85 |
| PC(18:1/20:4) | 808.59 | 7.63 | MS/MS | 1.4 | 1.2 | −0.20 | 4.83 |
| PC(18:2/18:2) | 782.57 | 7.56 | Standard | 2.0 | 2.0 | −0.03 | 3.02 |
| PC(P-16:0/20:4) | 766.58 | 7.71 | MS/MS | 0.3 | 0.1 | −0.20 | 7.64 |
| PC(P-18:0/20:4) | 794.61 | 7.79 | MS/MS | 0.4 | 0.2 | −0.25 | 8.72 |
| PC(P-18:1/20:2) | 796.62 | 7.98 | Database | −0.6 | −1.0 | −0.41 | 5.72 |
| PE(16:0/18:2) | 716.52 | 7.61 | MS/MS | −0.8 | 0.3 | 1.14 | 3.51 |
| SM (d18:1/16:0) | 703.58 | 6.99 | MS/MS | 1.2 | 0.9 | −0.26 | 9.07 |
| SM (d18:1/18:0) | 731.61 | 7.49 | MS/MS | 0.2 | 0.0 | −0.15 | 3.35 |
| SM(d18:1/14:0) | 675.54 | 6.50 | MS/MS | −0.8 | −1.0 | −0.20 | 5.88 |
| SM(d18:1/15:0) | 689.56 | 6.75 | MS/MS | −0.5 | −0.8 | −0.28 | 7.11 |
Metabolite identification method is indicated
Log of the normalized signal (NS)
Log of the average fold change (FC) between galN vs saline-treated rats (negative value = down regulation, positive value = up regulation)
−log(p) value of Welch's t-test between galN and saline-treated rats
The regression coefficients (βi) were defined as follows:
where Īuntreated samples represents the mean intensity of the chosen metabolite (i) in all samples before treatment. The values in the quotient (ΣIn) represent the sum of intensities observed for a set of stable metabolites, and it is used as an internal normalizing step to facilitate inter-individual evaluation. The set includes six metabolites that showed a minimal relative standard deviation (<2.5%) between all samples before and after saline or galN treatments, and it is formed by those with m/z 328,318; 358,367; 372,381; 450,750; 516,788 and 696,649. These metabolites were detected in all injected samples but discarded in the identification process of PCA analysis, a tool used to identify significant altered metabolites. This normalization step in the algorithm used allows the grouping of information and precludes the possible variations due to different instrumental setup. It is a normalization of signals within an experiment, so it is done for every animal sample independently.
3 Results
3.1 Serum metabolomics profiling of galN-treated rat model for acute liver injury
Acute liver injury in a group of male rats was induced by one-single intraperitoneal injection of galN. The control group received an injection of saline solution only. After 18 h, one of the galN-treated rats died and was not further considered in the subsequent study. Individualized blood samples from the remaining of the animals were drawn, analysed by UPLC-MS/MS and subjected of multivariate data analysis. Due to the larger number of mass spectrometric peaks (1,351) detected in the mass spectrometer working in the positive mode compared to the negative one (143), only data obtained in the former mode was further considered. Principal component analysis (PCA) of these data clearly discriminated between sera of galN-treated rats (Fig. 1, open triangles) and sera of saline-treated rats (open squares). In addition, PCA analysis showed that saline solution injections did not significantly modify the metabolic profile of rat sera as judged by the clustering together with the serum samples of untreated rats (Fig. 1, filled triangles and squares). This supports the idea that modifications in the endogenous serum metabolome of the different groups were mainly due to the treatment with the hepatotoxin. The loading plot of the PCA analysis (Fig. 2) reveals more than 100 serum metabolites grouped in different chemical families with a significant weight in the separation of the galN-treated from saline-treated rats. They included mainly membrane lipids such as phosphatidylcholine, phosphatidylethanolamine, lysophosphatidylcholine or lysophosphatidylethanolamine (Table 1). The serum levels of sphingomyelins, amino acids (methionine, threonine or tyrosine) and free fatty linoleic (18:2) acid were also significantly altered by the treatment with galN (Table 1). Amongst the metabolites that were identified by MS/MS or analytical standards, a number of clear tendencies were observed; in general sn-1 monoacylglycerophosphocholine, sn-2 arachidonyl diacylglycerophosphocholine and sphingolipids were less abundant in animals treated with galN, whilst diacylglycerophosphocholines, monoacylglycerophosphoethanolamines and amino acids showed the opposite trend.
Fig. 1.
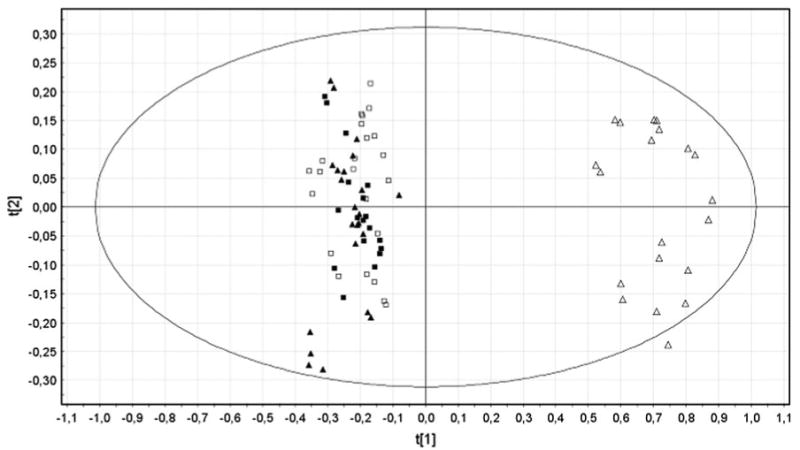
PCA scores plots discriminating galN-treated rats from saline-treated or untreated rats from positive ion mode UPLC™-MS data (t[1]: R2X = 0.609 and Q2 = 0.585, t[2]: R2X = 0.057 and Q2 = 0.042): untreated rats (n = 23), filled squares and triangles; saline-treated rats (n = 12), open squares; and galN-treated rats (n = 10), open triangles
Fig. 2.
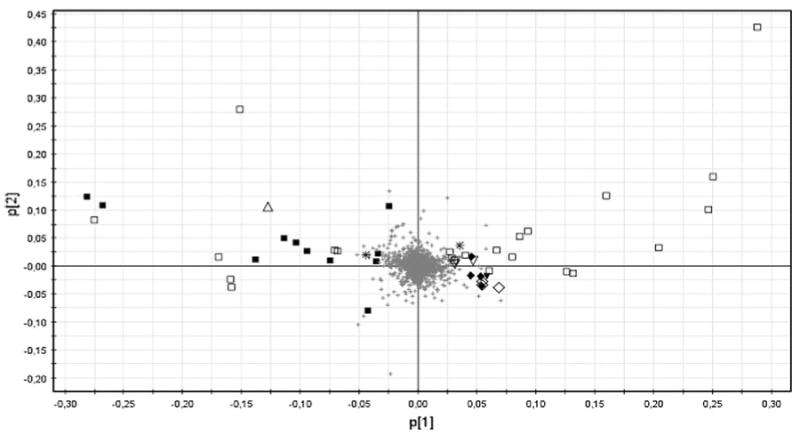
Loading plot for PCA mode positive. The ions with highest contribution to the variance in the scores plot are indicated on the loadings plot. The peaks are labelled according to their identifications; open square PC, filled square LysoPC, open diamond PE, filled diamond LysoPE, filled triangle FA, open triangle SM, open inverted triangle Bile Acid, inverted filled triangle AA, asterisk No determinated
A prediction model was created based on the intensity (I) of a set of metabolites (Table 1, in bold) to predict the risk of suffering acute-liver injury. A logit (Table 2) calculated by the equation described in Sect. 2 was able to differentiate both groups, and the probability of suffering acute-liver injury changes dramatically only for those animals treated with galN, all of them showing liver injury although with different severity.
Table 2. Liver damage assessment. Serum ALT activity and cellular death score (CDS) in liver sections from saline- or galN-treated rats.
| Animal ID | Injection | Predictiona | Visual scoreb,d H&E | CDSb,d (cells/mm2) | ALTc,d (U/L) | |
|---|---|---|---|---|---|---|
|
|
||||||
| Logit | Model | |||||
| 1 | Saline | −2.36 | 0.08 | 1 | 6 | 26.61 |
| 2 | Saline | −2.38 | 0.10 | 1 | 6 | 29.55 |
| 3 | Saline | −2.06 | 0.10 | 1 | 5 | 26.08 |
| 4 | Saline | −1.71 | 0.16 | 1 | 3 | 25.15 |
| 5 | Saline | −1.69 | 0.18 | 1 | 3 | 18.35 |
| 6 | Saline | −2.60 | 0.07 | 1 | 2 | 20.01 |
| 7 | Saline | −2.76 | 0.06 | 1 | 6 | 25.15 |
| 8 | Saline | −1.68 | 0.16 | 1 | 10 | 14.68 |
| 9 | Saline | −2.03 | 0.11 | 1 | 6 | 15.97 |
| 10 | Saline | −1.83 | 0.15 | 1 | 8 | 17.80 |
| 11 | Saline | −1.80 | 0.11 | 1 | 3 | 18.72 |
| 12 | Saline | −2.07 | 0.11 | 1 | 1 | 18.90 |
| 13 | GalN | 8.86 | 1.00 | 6 | 254 | 5807.64 |
| 14 | GalN | 5.05 | 0.99 | 5 | 187 | 1321.58 |
| 15 | GalN | 7.63 | 1.00 | 6 | 149 | 3186.47 |
| 16 | GalN | 6.13 | 1.00 | 4 | 197 | 4790.72 |
| 17 | GalN | 4.21 | 0.98 | 3 | 160 | 1600.58 |
| 18 | GalN | 4.13 | 0.99 | 2 | 87 | 2654.17 |
| 19 | GalN | 4.11 | 0.98 | 2 | 103 | 2848.73 |
| 20 | GalN | 4.76 | 0.99 | 2 | 104 | 3682.06 |
| 21 | GalN | 6.77 | 1.00 | 3 | 142 | 3843.59 |
| 22 | GalN | 3.91 | 0.98 | 2 | 112 | 3601.30 |
Application of a binomial prediction model based on six metabolites (bold, Table 1) to predict acute liver-injury. “Logit” value for each animal is calculated as indicated in Sect. 2 considering the intensity of selected metabolites, and “Model” indicates the probability that the animal suffers acute-liver injury (A value equal to or close to 1 is indicative of high probability of acute-liver injury while a value of 0 or close means low probability to have acute-liver injury)
18-h after intraperitoneal injection of saline solution or galN animals were sacrificed and livers were extracted, formalin-fixed and processed for histology. Liver sections were stained with Hematosylin-eosin (H&E), and analyzed for the presence of apoptotic cells by using ApopTag® Peroxidase In Situ Apoptosis Detection Kit. Liver injury was assessed by visual examinations of H&E-stained sections and scored 1–6 based on hemorragie, necrosis appearance. Liver injury was further quantified by counting apoptotic cells in 10 different randomly chosen fields covering a total area of 1 mm2 ; this value is referred as cellular death score (CDS). Both analyses were done in a blind test in which the analyzer was not aware on the sample identity
Serum alanine aminotransferase (ALT) levels were assayed with the Infinity ALT liquid stable reagent
Correlation analysis (Pearson r coefficient): H&E with CDS (0,91), H&E with ALT (0,75), CDS with ALT (0,88)
3.2 Assessment of liver damage in saline- or galN-treated rats
We next investigated whether there are endogenous serum metabolites that correlate with the degree of liver damage. To quantify this damage biochemical and histological parameters were measured (Table 2). Serum ALT activity was determined for each of the animals and was within the normal range in the saline-treated control group, while ALT activity in the galN-treated group was significantly increased (60–250 fold, Table 2). Histological manifestations such as inflammation, congestion, haemorrhage and hepatocellular degeneration along with necrotic areas were observed in all livers from the galN-treated group, while the control animals showed normal liver morphologies (Fig. 3; Table 2). However, the severity of liver damage, based on histological characteristics showed significant inter-animal variability. Histological quantization of liver damage was undertaken using two approaches. First, a semi-quantitative score from 1 to 6 was assigned based on the severity of the above-mentioned manifestations in the hematoxilin-eosin (H&E) staining (Table 3). Second, TUNEL staining analysis of the characteristic DNA fragmentation of apoptotic cells in liver sections was performed, and a quantitative value referred to as cell death score (CDS) was determined for all animals by counting the number of apoptotic cells in a liver section of 1 mm2 (Fig. 3; Table 3). To avoid possible bias, both analyses were performed in a blind manner in which the examiner was not aware of the sample identity. The data obtained from these quantitative analyses clearly highlight the indicated dissimilar inter-animal response to galN, which is in agreement with previous works that have used this experimental model (Coen et al. 2007, 2009; Miller et al. 2008). The Pearson correlation coefficients between visual H&E score of liver sections and CDS (r = 0.9123) or between serum ALT activity and CDS (r = 0.8863) indicated that CDS is a bona fide quantitative value to define in situ liver injury.
Fig. 3.
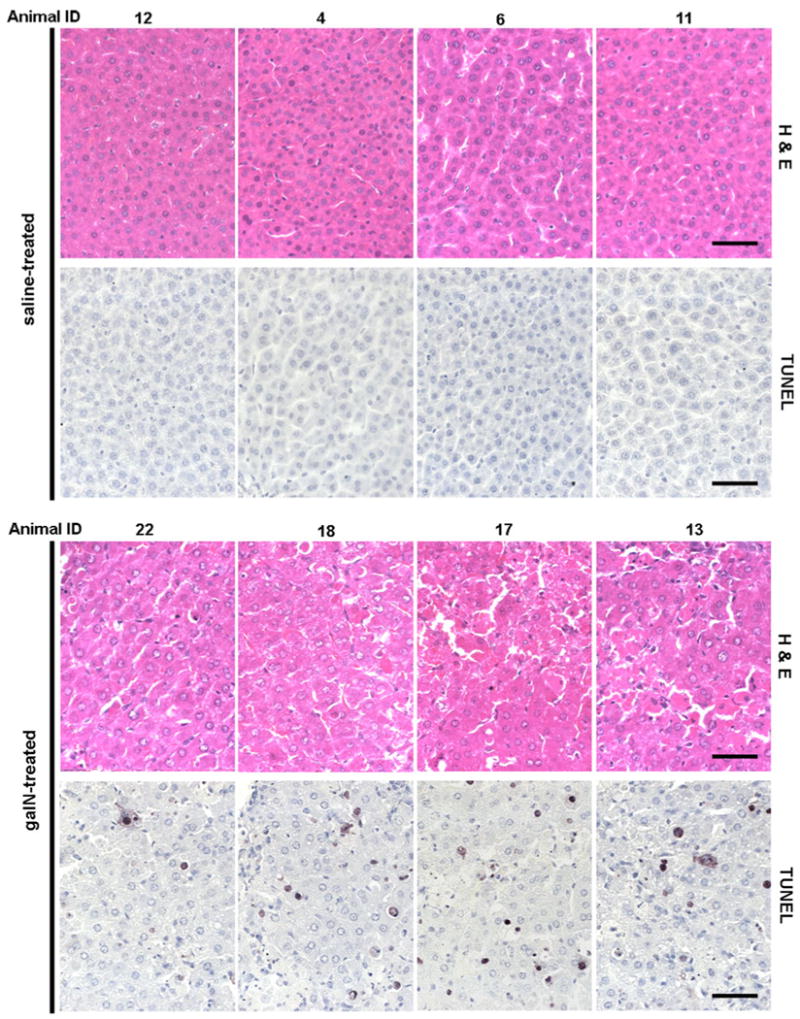
Histopathology and TUNEL immunohistochemistry in liver collected from adult male Wistar rats treated i.p. with saline solution or D-galatosamine (1.0 g/kg). Tissues were taken on 18 h after treatment. Representative photomicrographs of hematoxilin-eosin-stained formalin fixed liver sections from saline- or galN-treated animals. In situ detection of apoptotic cells by direct terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) of fragmented DNA following treatment with saline or galN. Note that all livers from galN-treated rats showed evident signs of lesions, such as loss of liver architecture, severe liver congestion, haemorrhage and necrotic areas. TUNEL analysis shows clearly a higher number of apoptotic cells in liver sections from galN-treated rats compared to saline-treated animals. The darker stains fragmented DNA commonly observed in apoptotic cells
Table 3. Endogenous serum metabolites correlate with liver damage indicators.
| RT | m/z | ION | ID metabolite | Confidencea | Fold change | SD | CDSd | ALTd | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| rhoe | P valuef | rhoe | P valuef | ||||||||
| 7.39 | 706.5384 | [M+H]+ | PC(14:0/16:0) | Standard | 2.0 | 0.16 | 0.71 | 2.2E-04 | 0.74 | 7.4E-05 | |
| 7.80 | 734.5724 | [M+H]+ | PC(16:0/16:0) | MS/MS | 1.9 | 0.06 | 0.79 | 1.5E-05 | 0.70 | 3.0E-04 | |
| 7.50 | 754.5466 | [M+Na]+ | PC(14:0/18:1) | MS/MS | 2.4 | 0.44 | 0.72 | 1.8E-04 | 0.77 | 2.5E-05 | |
| 7.87 | 760.5910 | [M+H] | PC(16:0/18:1) | Standard | 2.1 | 0.23 | 0.67 | 6.5E-04 | 0.73 | 1.3E-04 | |
| 7.38 | 744.5561 | [M+H]+ | PC(15:0/18:2) | MS/MS | 2.4 | 0.82 | 0.70 | 3.0E-04 | 0.72 | 1.6E-04 | |
| 7.58 | 758.5671 | [M+H] | PC(16:0/18:2) | Standard | 1.6 | 0.14 | 0.70 | 2.9E-04 | 0.73 | 1.3E-04 | |
| 7.77 | 772.5880 | [M+H]+ | PC(17:0/18:2) | MS/MS | 2.8 | 0.78 | 0.67 | 6.5E-04 | 0.76 | 4.1E-05 | |
| 7.97 | 786.6082 | [M+H] | PC(18:0/18:2) | Standard | 1.7 | 0.24 | 0.71 | 1.9E-04 | 0.81 | 4.7E-06 | |
| 7.15 | 754.5386 | [M+H] | PC(14:0/20:4) | MS/MS | 1.6 | 0.32 | 0.75 | 5.0E-05 | 0.70 | 3.0E-04 | |
| 7.71 | 766.5758 | [M+H]+ | PC(P-16:0/20:4) | MS/MS | 0.6 | 0.09 | −0.86 | 2.2E-07 | −0.70 | 3.2E-04 | |
| 7.79 | 794.6074 | [M+H] | PC(P-18:0/20:4) | MS/MS | 0.6 | 0.10 | −0.96 | 1.2E-12 | −0.80 | 6.5E-06 | |
| 7.92 | 810.6025 | [M+H] | PC(18:0/20:4) | Standard | 0.7 | 0.09 | −0.90 | 1.1E-08 | −0.74 | 7.4E-05 | |
| 7.25 | 780.5568 | [M+H] | PC(16:0/20:5) | MS/MS | 1.6 | 0.35 | 0.77 | 2.4E-05 | 0.85 | 4.7E-07 | |
| 7.98 | 796.6198 | [M+H]+ | PC(P-18:1/20:2) | Database | 0.4 | 0.19 | −0.78 | 1.7E-05 | −0.83 | 1.5E-06 | |
| 3.60 | 468.3075 | [M+H] | LysoPC(14:0/0:0) | Standard | 0.5 | 0.14 | −0.85 | 6.2E-07 | −0.71 | 2.4E-04 | |
| 3.83 | 482.3231 | [M+H] | LysoPC(15:0/0:0) | Standard | 0.5 | 0.16 | −0.87 | 1.3E-07 | −0.70 | 3.2E-04 | |
| 4.21 | 482.3602 | [M+H] | LysoPC(O-16:0/0:0) | MS/MS | 0.3 | 0.10 | −0.86 | 3.5E-07 | −0.74 | 9.0E-05 | |
| 4.09 | 496.3378 | [M+H] | LysoPC(16:0/0:0) | Standard | 0.4 | 0.10 | −0.88 | 5.8E-08 | −0.70 | 3.1E-04 | |
| 3.98 | 496.3406 | [M+H]+ | LysoPC(0:0/16:0) | MS/MSb | 0.3 | 0.11 | −0.87 | 1.3E-07 | −0.73 | 1.0E-04 | |
| 4.37 | 510.3551 | [M+H] | LysoPC(17:0/0:0) | Standard | 0.3 | 0.18 | −0.90 | 1.3E-08 | −0.78 | 1.5E-05 | |
| 4.80 | 510.3917 | [M+H] | LysoPC(O-18:0/0:0) | MS/MS | 0.3 | 0.11 | −0.96 | 6.9E-13 | −0.75 | 5.9E-05 | |
| 4.39 | 508.3749 | [M+H] | LysoPC(P-18:0/0:0) | MS/MS | 0.2 | 0.09 | −0.87 | 1.4E-07 | −0.74 | 8.7E-05 | |
| 4.66 | 524.3714 | [M+H] | LysoPC(18:0/0:0) | Standard | 0.3 | 0.08 | −0.83 | 1.4E-06 | −0.73 | 1.1E-04 | |
| 4.54 | 524.3718 | [M+H]+ | LysoPC(0:0/18:0) | MS/MS | 0.2 | 0.08 | −0.95 | 1.9E-11 | −0.69 | 4.2E-04 | |
| 4.96 | 538.3838 | [M+H] | LysoPC(19:0/0:0) | Standard | 0.3 | 0.15 | −0.88 | 7.5E-08 | −0.70 | 3.0E-04 | |
| 5.26 | 552.3993 | [M+H]+ | LysoPC(20:0/0:0) | Standard | 0.4 | 0.13 | −0.92 | 1.3E-09 | −0.63 | 1.7E-03 | |
| 5.86 | 580.4316 | [M+H] | LysoPC(22:0) | Database | 0.6 | 0.14 | −0.94 | 1.8E-10 | −0.69 | 3.7E-04 | |
| 6.46 | 608.4629 | [M+H] | LysoPC(24:0) | Database | 0.8 | 0.22 | −0.76 | 4.1E-05 | −0.38 | 8.4E-02 | |
| 4.81 | 550.3853 | [M+H] | LysoPC(20:1/0:0) | MS/MS | 0.2 | 0.07 | −0.96 | 4.7E-12 | −0.69 | 4.3E-04 | |
| 5.38 | 578.4156 | [M+H]+ | LysoPC(22:1/0:0) | MS/MSb | 0.4 | 0.13 | −0.92 | 2.3E-09 | −0.76 | 3.9E-05 | |
| 5.96 | 606.4473 | [M+H]+ | LysoPC(24:1/0:0) | MS/MSb | 0.4 | 0.12 | −0.96 | 1.2E-12 | −0.69 | 3.7E-04 | |
| 4.28 | 522.3562 | [M+H]+ | LysoPC(18:1/0:0) | Standard | 0.5 | 0.13 | −0.87 | 1.4E-07 | −0.67 | 6.3E-04 | |
| 4.44 | 548.3699 | [M+H] | LysoPC(20:2/0:0) | MS/MS | 0.4 | 0.12 | −0.82 | 2.4E-06 | −0.70 | 2.6E-04 | |
| 4.09 | 518.3254 | [M+H] | LysoPC(18:3) | Database | 0.3 | 0.11 | −0.85 | 5.0E-07 | −0.72 | 1.6E-04 | |
| 3.98 | 518.3219 | [M+H] | LysoPC(18:3) | Database | 0.3 | 0.13 | −0.85 | 4.0E-07 | −0.70 | 2.6E-04 | |
| 3.99 | 544.3407 | [M+H]+ | LysoPC(20:4/0:0) | MS/MS | 0.6 | 0.21 | −0.86 | 3.5E-07 | −0.67 | 6.0E-04 | |
| 7.61 | 716.5239 | [M+H]+ | PE(16:0/18:2) | MS/MS | 13.7 | 7.76 | 0.79 | 1.4E-05 | 0.76 | 4.2E-05 | |
| 4.25 | 438.2967 | [M+H] | LysoPE(P-16:0) | MS/MS | 0.3 | 0.15 | −0.82 | 2.4E-06 | −0.88 | 5.8E-08 | |
| 4.07 | 454.2922 | [M+H]+ | LysoPE(16:0) | MS/MS | 3.4 | 1.36 | 0.85 | 7.1E-07 | 0.79 | 1.2E-05 | |
| 4.52 | 482.3227 | [M+H]+ | LysoPE(18:0) | MS/MS | 3.1 | 1.47 | 0.76 | 4.1E-05 | 0.74 | 9.0E-05 | |
| 4.64 | 482.3237 | [M+H]+ | LysoPE(18:0) | MS/MS | 2.4 | 0.84 | 0.75 | 6.4E-05 | 0.82 | 3.8E-06 | |
| 3.93 | 478.2927 | [M+H]+ | LysoPE(18:2) | MS/MS | 5.7 | 1.35 | 0.79 | 1.4E-05 | 0.90 | 1.5E-08 | |
| 3.97 | 502.2924 | [M+H]+ | LysoPE(20:4) | MS/MS | 5.5 | 1.96 | 0.73 | 1.0E-04 | 0.89 | 3.3E-08 | |
| 3.97 | 526.2932 | [M+H] | LysoPE(22:6) | Database | 6.7 | 2.92 | 0.75 | 5.4E-05 | 0.78 | 2.1E-05 | |
| 6.50 | 675.5420 | [M+H] | SM(d18:1/14:0) | MS/MS | 0.6 | 0.14 | −0.86 | 3.8E-07 | −0.81 | 5.9E-06 | |
| 6.75 | 689.5582 | [M+H] | SM(d18:1/15:0) | MS/MS | 0.5 | 0.15 | −0.93 | 4.1E-10 | −0.83 | 1.7E-06 | |
| 6.99 | 703.5782 | [M+H]+ | SM (d18:1/16:0) | MS/MS | 0.5 | 0.11 | −0.92 | 1.3E-09 | −0.80 | 7.2E-06 | |
| 7.49 | 731.6091 | [M+H]+ | SM (d18:1/18:0) | MS/MS | 0.7 | 0.20 | −0.76 | 3.6E-05 | −0.68 | 4.7E-04 | |
| 0.87 | 150.0572 | [M+H]+ | Methionine | Standard | 3.8 | 1.79 | 0.85 | 4.4E-07 | 0.80 | 9.8E-06 | |
| 0.61 | 116.0698 | [M+H]+ | Proline | Standard | 3.6 | 1.99 | 0.83 | 1.9E-06 | 0.79 | 1.5E-05 | |
| 0.98 | 182.0792 | [M+H]+ | Tyrosine | Standard | 10.0 | 4.36 | 0.83 | 1.8E-06 | 0.84 | 8.2E-07 | |
| 0.59 | 120.0647 | [M+H]+ | Threonine | Standard | 2.6 | 1.38 | 0.85 | 7.1E-07 | 0.82 | 3.4E-06 | |
Metabolites confirmed by MS/MS or standard are indicated
Regiochemistry assigned based on the elution order
Average Fold Change and standard deviation, GalN- versus saline- treated rats
Correlation analysis with Liver damage indicators (CDS score and serum ALT activity)
Spearman's rank correlation coefficient GalN- versus saline- treated rats
F-distribution probability
3.3 Serum endogenous metabolites correlate with the degree of liver cell death
In order to detect serum metabolites correlating with the degree of liver damage, Spearman's rank correlation coefficients between CDS scores of each animal and each of the serum detected-metabolites were calculated (Table 3). As result 169 endogenous metabolites were found to correlate with CDS score (rho > 0.71, P value < 0.001) and from this group 49 could be identified by using databases, chemical standards or MS/MS data (Table 3). In addition to the significant correlation observed with the degree of liver damage, all of these 49 metabolites showed also a high correlation with serum ALT activity (Table 3) further supporting the direct connection of these metabolites with liver damage. The average fold change between galN- and saline-treated groups for many of the metabolites was by a factor higher than 2 (Table 3), and as it is exemplified by box plots in Fig. 4 the range of the serum levels was different among the metabolites. In this sense, the highest ranges were observed for phosphatidylethanolamine (16:0/18:2) and tyrosine in which serum levels increased between 6 and 17, and 8 and 22 folds after treatment with galN, respectively (Fig. 4). Another interesting observation was that the highest correlation (rho > 0.91) of liver cell death occurs with serum levels of long-chain containing phospholipids such as LysoPC(0:0/18:0), LysoPC(20:0/0:0), LysoPC(22:0), LysoPC(20:1/0:0), LysoPC(22:1), LysoPC(24:1/0:0), LysoPC(O-18:0/0:0), PC(P-18:0/20:4) (Table 3). In this sense, an increase in the degree of liver cell death reduces the level of these metabolites in serum, highlighting that these metabolites could be used to define a serum metabolic signature for liver cell death in acute liver injury.
Fig. 4.
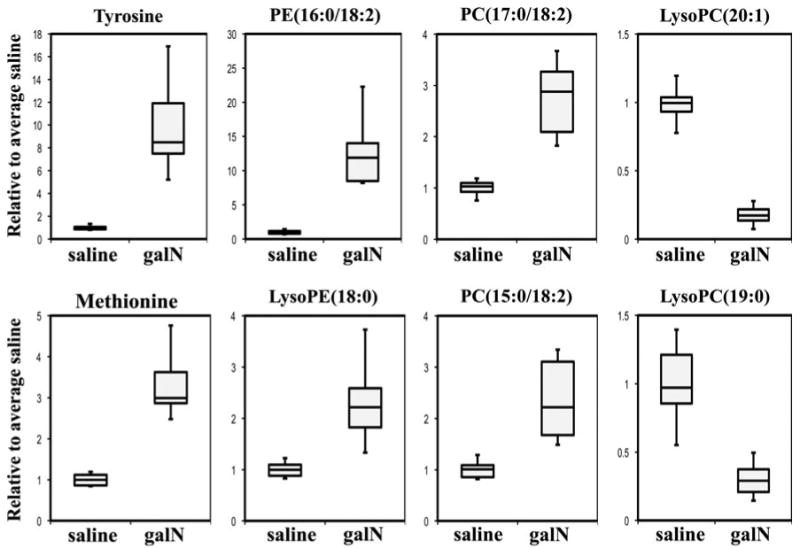
Representative serum metabolites significantly altered by galN-induced acute liver injury. Examples of metabolites showing high correlation with CDS score are represented by box plots. For the box plots the top and bottom of the boxes represent the 75th and 25th percentile, respectively. The top and bottom bars (whiskers) represent the entire spread of the data points for the animals. The hyphen indicates the median value. The y-axis is the level of metabolite relative to the average of the level of that metabolite in serum of saline-treated animals
4 Discussion
In the present study, we have performed an UPLC-based metabolomics analysis to detect candidate endogenous metabolites suitable for the direct assessment of liver damage degree using a minimally invasive technique. We have used a well-characterized experimental model for acute liver injury; i.e., rats treated with a single high dose of galN that has been shown to create a hepatic lesion resembling human viral hepatitis (Keppler et al. 1968). The underlying molecular mechanism involves a severe depletion of hepatic uridine nucleotide pool including UTP, UDP, UMP, UDP-glucose, UDP-galactose, UDP-glucuronate (Decker and Keppler 1972; Coen et al. 2007) which results in inhibition of protein and RNA synthesis. This molecular mechanism causes a compendium of histopathological hepatic lesions described by several groups (Keppler et al. 1968; Medline et al. 1970; Koff et al. 1971; Miyahara et al. 1982), characterized by acidophilic degeneration of the cytoplasm at 4 h, apoptosis and cell death at 6 h and more extensively at 24 h. Also observed is an inflammatory response with neutrophil accumulation and activation of Kuppfer cells starting 6 h after toxin administration. Hepatocytes as evidenced by light microscopy, show extreme variation in nuclear and cytoplasmic size and shape, develop lipid accumulation (steatosis) that is maximal at 24 h (Koff et al. 1971). Electron microscopy additionally showed and increased number of autophagic vacuoles, a reduction of glycogen particles, hypertrophy of the smooth endoplasmic reticulum, and mitochondrial abnormalities in size and shape (Miyahara et al. 1982). In general, maximal response to galN administration is seen 48 h post-administration, with complete recovery within 7–12 days if the regulation of the apoptotic, necrotic and inflammatory processes work properly (Decker and Keppler 1972). Different groups have used galN-based rodent models to characterize the mechanisms causing acute liver injury, and to identify putative diagnostic biomarkers by using metabolomics approaches. The NMR approach was applied in order to investigate how galN is metabolized and what its metabolic effect is in rat livers (Coen 2010). GC–MS/MS analysis of serum from LPS/galN-treated mice detect 267 distinct endogenous compounds and showed that some of them including glucose, β-hydroxybutiric acid (a ketone body), 5-hydroxyindolacetic acid (the main serotonin-derived metabolite) and several amino acids could be used as blood metabolic markers for fulminant hepatic failure (Coen et al. 2007, 2009; Feng et al. 2007).
We have performed an UPLC–MS/MS-based metabolomics analysis of serum samples from galN- and saline-treated animals, and we were able to detect and quantify more than 1,351 mass spectrometric peaks in each of them providing a very comprehensive serum metabolome for acute liver injury. Because of the huge amount of data generated by this UPLC–MS/MS-based approach multivariate statistical analysis was applied in order to determine principal components that clearly discriminate between animals that were treated with galN from the control group. By applying the principal component analysis metabolites that significantly contributed to this discrimination were identified, including the amino acids methionine, threonine and tyrosine, which were enriched in the serum of the injured rats, in accordance with previous studies using other technologies. This increment in the serum levels of amino acids after acute liver injury has been suggested to be caused by an inhibition of the uptake and utilization of amino acids, especially aromatic amino acids and an increment in the degradation of proteins in the liver (Arai et al. 2001). In agreement also with these findings metabolism of methionine has been involved in liver disease (Mato et al. 2008) and rats treated with galN were previously shown to affect these metabolism (Cabrero et al. 1988). In addition, it has been reported that livers from control animals released glucose, whilst the livers from galN-injured animals took up glucose reflecting a switch in the carbohydrate metabolism, changing from gluconeogenesis to glycolysis (Arai et al. 2001). In concordance with that result glucose was one of the identified metabolites in our study that clearly distinguished the control from the test group in that galN treated animals showed reduced glucose levels (Table 1). Thus, the UPLC–MS/MS analysis, as presented here, corroborates well with previous reports regarding amino acids and glucose metabolism.
Importantly, our results show that serum levels of major and essential structural components of membrane lipid bilayers including phosphatidylcholines, phosphatidylethanolamines and sphingomyelins were significantly increased in acute liver injury. This increase, associated mostly with PCs carrying short-chain unsaturated (14:0, 15:0 and 16:0) acyl groups in the sn-1 position, could be explained by the massive destruction of membranes as judged by the cell death observed in the histological examination of the livers. In contrast, lysophosphatidylcholines were reduced in the serum samples from injured rats (Table 1). This family of lipids are formed from PCs by PLA2 activity that has been reported to be enhanced in galN-treated rats (Petkova et al. 1987). Contrary to what is observed in this study the increase in PLA2 should be reflected in an increase in LysoPCs. Lyso-phospholipids are known to be highly bio-active molecules that can signal through G-protein-coupled receptors (Fox et al. 2009) so a tight regulation of them during a regenerative process will be critical for the final outcome of an injured tissue. In this sense, it has been shown that endothelial cells are able to degrade extracellular lysoPCs (Tosti et al. 1999) reducing plasma and tissue levels of these pro-inflammatory lipid molecules. This activity could provide an explanation for the low levels observed in the serum of acute liver injury, but further experiments are needed to clarify this aspect.
Finally, a logistic regression prediction model based on our metabolomic study was created using a selection of six metabolites from the groups mentioned above, including methionine, three lysophosphocholines and two phosphocholines. The model is able to predict the high probability of acute-liver injury in all galN-treated animals with more than 98% accuracy, with no false positives observed between saline-treated animals. However, a full validation of this prediction model will require a bigger test set. Also it would be preferable to include samples from other experimental models for hepatic and non-hepatic injury in order to establish that these six metabolites are indeed specific biomarkers for acute liver injury.
Interestingly, PE (16:0/18:2) is among the identified metabolites the one that suffer the highest increase in the serum of galN-treated animals. The source of this increase could be also the massive liver cell death induced by galN. Interestingly, PEs are enriched in the inner leaflet of mitochondria and is well-known that galN causes severe damage to the mitochondria (Miyahara et al. 1982), which raises the possibility that the increment in the levels of this metabolite could be more specifically a consequence of a degeneration of the mitochondrial membrane.
Liver damage was observed in 100% of the animals treated with galN as judged by serum ALT activity and liver histology. The quantization of this damage showed inter-individual variability in the response of the animals to the treatment with galN ranging from lethal to low (but visible) damage. Although we found a strong correlation (r > 0.75) between the three parameters (Table 2) that we used to quantify the damage, there were outliers (e.g., animal number 18, Table 2) with H&E and CDS scores not correlating with serum ALT activity. A possible explanation of this could be the fact that high levels of serum ALT activity is not exclusively produced by liver injury, but could also be indicative of muscle damage (Lott and Landesman 1984). Due to this specificity feature of the transaminase activity we have focused our work in the correlation with CDS scores. We took advantage of the inter-individual variability detected in the liver damage from the galN-treated rats to identify a serum metabolic signature to assess noninvasively the degree of cell death that occurred during acute liver injury. The highest correlations (rho > 0.91, P value < 0.001) were observed for long-chain fatty acids containing lipids such as LysoPC(0:0/18:0), LysoPC(20:0/0:0), LysoPC(22:0), LysoPC(20:1/0:0), LysoPC(22:1), LysoPC(24:1/0:0), LysoPC(O-18:0/0:0), PC(P-18:0/20:4), SM (d18:1/15:0) and SM (d18:1/16). Thus, quantization of these 10 metabolites in serum could constitute a very sensitive method to establish the degree of liver cell death, however further investigations in other experimental models are required to truly establish that these metabolites are specific biomarkers of cell death in the liver.
5 Conclusions
Our results highlight the power of UPLC–MS/MS-based metabolomics approach in combination with an experimental animal model for studying metabolic changes of acute liver injury. We were able to identify potential non-invasive biomarkers correlating with a specific lesion. Extending this approach to other hepatic models with different liver injuries would help to provide a repertoire of highly sensitive and specific biomarkers for different liver affections. Although the clinical applicability of these studies on diagnosis and prognosis of liver diseases will still require time-consuming translational research, in a short period of time this set of hepatic biomarkers could assist pharmaceutical research and development, accelerating the launch of safer drugs to the market.
Acknowledgments
We gratefully thank B. Martínez de la Pera, C. Oceja, I. Iturriza and L. María for their technical assistance and FAES FARMA S.A. for its support with rat experimentation procedures and sample collection. This work was supported by grants from the Fondo de Investigaciones Sanitarias (Institute of Health Carlos III, 06/0621 & PS09/00526 to J.M.F.P.); Program “Ramon y Cajal” of Spanish Ministry (to J.M.F.P); Diputación Foral de Bizkaia (612TK20100014); National Institute of Health Grant R01 AT004896 (to S.C.L. and J.M. M.); Centro de Investigación Biomédica en Red en el Área temática de Enfermedades Hepáticas y Digestivas (CIBERehd) is funded by the Institute of Health Carlos III.
Contributor Information
Esperanza Gonzalez, Metabolomics Unit, CIC bioGUNE, CIBERehd, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain.
Sebastiaan van Liempd, Metabolomics Platform, CIC bioGUNE, CIBERehd, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain.
Javier Conde-Vancells, Metabolomics Unit, CIC bioGUNE, CIBERehd, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain.
Virginia Gutierrez-de Juan, Metabolomics Unit, CIC bioGUNE, CIBERehd, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain.
Miriam Perez-Cormenzana, OWL Genomics, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain.
Rebeca Mayo, OWL Genomics, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain.
Agustin Berisa, R&D and Innovation Department, FAES FARMA S.A., 48940 Leioa, Bizkaia, Spain.
Cristina Alonso, OWL Genomics, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain.
Cesar A. Marquez, MD Renal, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain
Jonathan Barr, OWL Genomics, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain.
Shelly C. Lu, Division of Gastrointestinal and Liver Diseases, Keck School of Medicine, University Southern California, Los Angeles, CA 90033, USA
Jose M. Mato, Metabolomics Unit, CIC bioGUNE, CIBERehd, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain
Juan M. Falcon-Perez, Email: jfalcon@cicbiogune.es, Metabolomics Unit, CIC bioGUNE, CIBERehd, Bizkaia Technology Park, 48160 Derio, Bizkaia, Spain, IKERBASQUE, Basque Foundation for Science, 48011 Bilbao, Spain.
References
- Arai K, Lee K, Berthiaume F, Tompkins RG, Yarmush ML. Intrahepatic amino acid and glucose metabolism in a D-galactosamine-induced rat liver failure model. Hepatology. 2001;34:360–371. doi: 10.1053/jhep.2001.26515. [DOI] [PubMed] [Google Scholar]
- Barr J, Vazquez-Chantada M, Alonso C, Perez-Cormenzana M, Mayo R, Galan A, et al. Liquid chromatography-mass spectrometry-based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. Journal of Proteome Research. 2010;9:4501–4512. doi: 10.1021/pr1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelsterli UA, Lim PL. Mitochondrial abnormalities— a link to idiosyncratic drug hepatotoxicity? Toxicology and Applied Pharmacology. 2007;220:92–107. doi: 10.1016/j.taap.2006.12.013. [DOI] [PubMed] [Google Scholar]
- Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HW, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nature Medicine. 2002;8:1439–1444. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- Cabrero C, Duce AM, Ortiz P, Alemany S, Mato JM. Specific loss of the high-molecular-weight form of S-adenosyl-L-methionine synthetase in human liver cirrhosis. Hepatology. 1988;8:1530–1534. doi: 10.1002/hep.1840080610. [DOI] [PubMed] [Google Scholar]
- Chan EC, Yap SL, Lau AJ, Leow PC, Toh DF, Koh HL. Ultra-performance liquid chromatography/time-of-flight mass spectrometry based metabolomics of raw and steamed Panax notoginseng. Rapid Communications in Mass Spectrometry. 2007;21:519–528. doi: 10.1002/rcm.2864. [DOI] [PubMed] [Google Scholar]
- Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metabolism Reviews. 2007;39:581–597. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CJ, Haselden JN. Metabolic profiling as a tool for understanding mechanisms of toxicity. Toxicologic Pathology. 2008;36:140–147. doi: 10.1177/0192623307310947. [DOI] [PubMed] [Google Scholar]
- Coen M. A metabonomic approach for mechanistic exploration of pre-clinical toxicology. Toxicology. 2010;278:326–340. doi: 10.1016/j.tox.2010.07.022. [DOI] [PubMed] [Google Scholar]
- Coen M, Hong YS, Clayton TA, Rohde CM, Pearce JT, Reily MD, et al. The mechanism of galactosamine toxicity revisited; a metabonomic study. Journal of Proteome Research. 2007;6:2711–2719. doi: 10.1021/pr070164f. [DOI] [PubMed] [Google Scholar]
- Coen M, Want EJ, Clayton TA, Rhode CM, Hong YS, Keun HC, et al. Mechanistic aspects and novel biomarkers of responder and non-responder phenotypes in galactosamine-induced hepatitis. Journal of Proteome Research. 2009;8:5175–5187. doi: 10.1021/pr9005266. [DOI] [PubMed] [Google Scholar]
- Dan YY, Yeoh GC. Liver stem cells: A scientific and clinical perspective. Journal of Gastroenterology and Hepatology. 2008;23:687–698. doi: 10.1111/j.1440-1746.2008.05383.x. [DOI] [PubMed] [Google Scholar]
- Decker K, Keppler D. Galactosamine induced liver injury. Progress in Liver Diseases. 1972;4:183–199. [PubMed] [Google Scholar]
- Duarte IF, Stanley EG, Holmes E, Lindon JC, Gil AM, Tang H, et al. Metabolic assessment of human liver transplants from biopsy samples at the donor and recipient stages using high-resolution magic angle spinning 1H NMR spectroscopy. Analytical Chemistry. 2005;77:5570–5578. doi: 10.1021/ac050455c. [DOI] [PubMed] [Google Scholar]
- El-Mofty SK, Scrutton MC, Serroni A, Nicolini C, Farber JL. Early, reversible plasma membrane injury in galactosamine-induced liver cell death. The American Journal of Pathology. 1975;79:579–596. [PMC free article] [PubMed] [Google Scholar]
- Feng B, Wu S, Lv S, Liu F, Chen H, Yan X, et al. Metabolic profiling analysis of a D-galactosamine/lipopolysaccharide-induced mouse model of fulminant hepatic failure. Journal of Proteome Research. 2007;6:2161–2167. doi: 10.1021/pr0606326. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: From diagnostics to systems biology. Nature Reviews Molecular Cell Biology. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biology. 2009;7:e1000228. doi: 10.1371/journal.pbio.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lechon MJ, Castell JV, Donato MT. An update on metabolism studies using human hepatocytes in primary culture. Expert Opinion on Drug Metabolism. 2008;4:837–854. doi: 10.1517/17425255.4.7.837. [DOI] [PubMed] [Google Scholar]
- Kaplowitz N. Drug-induced liver disorders: Implications for drug development and regulation. Drug Safety. 2001;24:483–490. doi: 10.2165/00002018-200124070-00001. [DOI] [PubMed] [Google Scholar]
- Keppler D, Lesch R, Reutter W, Decker K. Experimental hepatitis induced by D-galactosamine. Experimental and Molecular Pathology. 1968;9:279–290. doi: 10.1016/0014-4800(68)90042-7. [DOI] [PubMed] [Google Scholar]
- Kitazawa T, Tsujimoto T, Kawaratani H, Fukui H. Therapeutic approach to regulate innate immune response by Toll-like receptor 4 antagonist E5564 in rats with D-galactosamine-induced acute severe liver injury. Journal of Gastroenterology and Hepatology. 2009;24:1089–1094. doi: 10.1111/j.1440-1746.2008.05770.x. [DOI] [PubMed] [Google Scholar]
- Koff RS, Fitts JJ, Sabesin SM, Zimmerman HJ. d-galactosamine hepatotoxicity II. Mechanism of fatty liver production. Proceedings of the Society for Experimental Biology and Medicine. 1971;138:89–92. doi: 10.3181/00379727-138-35837. [DOI] [PubMed] [Google Scholar]
- Komano T, Egashira Y, Sanada H. l-Gln and l-Ser suppress the d-galactosamine-induced IL-18 expression and hepatitis. Biochemical and Biophysical Research Communications. 2008;372:688–690. doi: 10.1016/j.bbrc.2008.05.114. [DOI] [PubMed] [Google Scholar]
- Kushnir MM, Rockwood AL, Bergquist J. Liquid chromatography-tandem mass spectrometry applications in endocrinology. Mass Spectrometry Reviews. 2009;29:480–502. doi: 10.1002/mas.20264. [DOI] [PubMed] [Google Scholar]
- Lin HM, Barnett MP, Roy NC, Joyce NI, Zhu S, Armstrong K, et al. Metabolomic analysis identifies inflammatory and noninflammatory metabolic effects of genetic modification in a mouse model of Crohn's disease. Journal of Proteome Research. 2010;9:1965–1975. doi: 10.1021/pr901130s. [DOI] [PubMed] [Google Scholar]
- Lott JA, Landesman PW. The enzymology of skeletal muscle disorders. Critical Reviews in Clinical Laboratory Sciences. 1984;20:153–190. doi: 10.3109/10408368409165773. [DOI] [PubMed] [Google Scholar]
- Lutz U, Lutz RW, Lutz WK. Metabolic profiling of glucuronides in human urine by LC-MS/MS and partial least-squares discriminant analysis for classification and prediction of gender. Analytical Chemistry. 2006;78:4564–4571. doi: 10.1021/ac0522299. [DOI] [PubMed] [Google Scholar]
- Lv S, Wei L, Wang JH, Wang JY, Liu F. Identification of novel molecular candidates for acute liver failure in plasma of BALB/c murine model. Journal of Proteome Research. 2007;6:2746–2752. doi: 10.1021/pr0701759. [DOI] [PubMed] [Google Scholar]
- Maezono K, Kajiwara K, Mawatari K, Shinkai A, Torii K, Maki T. Alanine protects liver from injury caused by F-galactosamine and CCl4. Hepatology. 1996;24:185–191. doi: 10.1002/hep.510240130. [DOI] [PubMed] [Google Scholar]
- Major HJ, Williams R, Wilson AJ, Wilson ID. A metabonomic analysis of plasma from Zucker rat strains using gas chromatography/mass spectrometry and pattern recognition. Rapid Communications in Mass Spectrometry. 2006;20:3295–3302. doi: 10.1002/rcm.2732. [DOI] [PubMed] [Google Scholar]
- Mato JM, Martinez-Chantar ML, Lu SC. Methionine metabolism and liver disease. Annual Review of Nutrition. 2008;28:273–293. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- Medline A, Schaffner F, Popper H. Ultrastructural features in galactosamine-induced hepatitis. Experimental and Molecular Pathology. 1970;12:201–211. doi: 10.1016/0014-4800(70)90050-x. [DOI] [PubMed] [Google Scholar]
- Miller TJ, Knapton A, Adeyemo O, Noory L, Weaver J, Hanig JP. Cytochrome c: A non-invasive biomarker of drug-induced liver injury. Journal of Applied Toxicology. 2008;28:815–828. doi: 10.1002/jat.1347. [DOI] [PubMed] [Google Scholar]
- Miyahara M, Enzan H, Shiraishi N, Kawase M, Yamamoto M, Hara H, et al. Mitochondrial damage in galactosamine-induced liver intoxication in rats. Biochimica et Biophysica Acta. 1982;714:505–515. doi: 10.1016/0304-4165(82)90161-1. [DOI] [PubMed] [Google Scholar]
- Muller PY, Dieterle F. Tissue-specific, non-invasive toxicity biomarkers: Translation from preclinical safety assessment to clinical safety monitoring. Expert Opinion on Drug Metabolism. 2009;5:1023–1038. doi: 10.1517/17425250903114174. [DOI] [PubMed] [Google Scholar]
- Newsome PN, Plevris JN, Nelson LJ, Hayes PC. Animal models of fulminant hepatic failure: A critical evaluation. Liver Transplantation. 2000;6:21–31. doi: 10.1002/lt.500060110. [DOI] [PubMed] [Google Scholar]
- Nicholls AW, Mortishire-Smith RJ, Nicholson JK. NMR spectroscopic-based metabonomic studies of urinary metabolite variation in acclimatizing germ-free rats. Chemical Research in Toxicology. 2003;16:1395–1404. doi: 10.1021/tx0340293. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Wilson ID. Opinion: Understanding ‘global’ systems biology: Metabonomics and the continuum of metabolism. Nature Reviews Drug Discovery. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- Nordstrom A, O'Maille G, Qin C, Siuzdak G. Nonlinear data alignment for UPLC-MS and HPLC-MS based metabolomics: Quantitative analysis of endogenous and exogenous metabolites in human serum. Analytical Chemistry. 2006;78:3289–3295. doi: 10.1021/ac060245f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova L, Solichova D, Solich P. Advantages of ultra performance liquid chromatography over high-performance liquid chromatography: Comparison of different analytical approaches during analysis of diclofenac gel. Journal of Separation Science. 2006;29:2433–2443. doi: 10.1002/jssc.200600147. [DOI] [PubMed] [Google Scholar]
- Ozer J, Ratner M, Shaw M, Bailey W, Schomaker S. The current state of serum biomarkers of hepatotoxicity. Toxicology. 2008;245:194–205. doi: 10.1016/j.tox.2007.11.021. [DOI] [PubMed] [Google Scholar]
- Petkova DH, Momchilova AB, Markovska TT, Koumanov KS. d-galactosamine induced changes in rat liver plasma membranes lipid composition and some enzyme activities. The International Journal of Biochemistry. 1987;19:289–291. doi: 10.1016/0020-711x(87)90034-6. [DOI] [PubMed] [Google Scholar]
- Schauer N, Fernie AR. Plant metabolomics: Towards biological function and mechanism. Trends in Plant Science. 2006;11:508–516. doi: 10.1016/j.tplants.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Stramentinoli G, Gualano M, Ideo G. Protective role of S-adenosyl-L-methionine on liver injury induced by D-galactosamine in rats. Biochemical Pharmacology. 1978;27:1431–1433. doi: 10.1016/0006-2952(78)90097-7. [DOI] [PubMed] [Google Scholar]
- Tosti E, Dahl L, Endresen MJ, Henriksen T. Endothelial degradation of extracellular lyso-phosphatidylcholine. Scandinavian Journal of Clinical and Laboratory Investigation. 1999;59:249–257. doi: 10.1080/00365519950185616. [DOI] [PubMed] [Google Scholar]
- Tunon MJ, Alvarez M, Culebras JM, Gonzalez-Gallego J. An overview of animal models for investigating the pathogenesis and therapeutic strategies in acute hepatic failure. World Journal of Gastroenterology. 2009;15:3086–3098. doi: 10.3748/wjg.15.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich RG. Idiosyncratic toxicity: A convergence of risk factors. Annual Review of Medicine. 2007;58:17–34. doi: 10.1146/annurev.med.58.072905.160823. [DOI] [PubMed] [Google Scholar]
- Yovchev MI, Grozdanov PN, Zhou H, Racherla H, Guha C, Dabeva MD. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636–647. doi: 10.1002/hep.22047. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang G, Du Y, Zhu L, Jiye A. GC/MS analysis of the rat urine for metabonomic research. Journal of Chromatography B, Analytical Technologies in the Biomedical and Life Sciences. 2007;854:20–25. doi: 10.1016/j.jchromb.2007.03.048. [DOI] [PubMed] [Google Scholar]


