Abstract
Two distinct cDNA clones encoding for the glutamate decarboxylase (GAD) isoenzymes GAD1 and GAD2 from Arabidopsis (L.) Heynh. were characterized. The open reading frames for GAD1 and GAD2 were expressed in Escherichia coli and the recombinant proteins were purified by affinity chromatography. Analysis of the recombinant proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis suggest that GAD1 and GAD2 encode for 58- and 56-kD peptides, respectively. The enzymatic activities of the pure recombinant GAD1 and GAD2 proteins were stimulated 35- and 13-fold, respectively, by Ca2+/calmodulin but not by Ca2+ or calmodulin alone. Southern-blot analysis of genomic DNA suggests that there is only one copy of each gene in Arabidopsis. The GAD1 transcript and a corresponding 58-kD peptide were detected in roots only. Conversely, the GAD2 transcript and a corresponding 56-kD peptide were detected in all organs tested. The specific activity, GAD2 transcript, and 56-kD peptide increased in leaves of plants treated with 10 mm NH4Cl, 5 mm NH4NO3, 5 mm glutamic acid, or 5 mm glutamine as the sole nitrogen source compared with samples from plants treated with 10 mm KNO3. The results from these experiments suggest that in leaves GAD activity is partially controlled by gene expression or RNA stability. Results from preliminary analyses of different tissues imply that these tendencies were not the same in flower stalks and flowers, suggesting that other factors may control GAD activity in these organs. The results from this investigation demonstrate that GAD activity in leaves is altered by different nitrogen treatments, suggesting that GAD2 may play a unique role in nitrogen metabolism.
GAD (EC 4.1.1.15) catalyzes the conversion of Glu to GABA in the presence of the cofactor PLP. GAD is present in Escherichia coli (Smith et al., 1992), mammals (Erlander and Tobin, 1991), and plants (Satyanarayan and Nair, 1990). In plants the enzyme has a unique feature, a CaM-binding domain at the carboxy terminus (Baum et al., 1993; Arazi et al., 1995; Gallego et al., 1995). CaM binding has been demonstrated in GAD isolated from petunia (Baum et al., 1993) and fava bean (Ling et al., 1994). In addition, the last 30 amino acids of the GAD1 gene product from Arabidopsis has been shown to bind CaM (Arazi et al., 1995). In vitro analyses have shown that Ca2+ and CaM stimulate GAD activity 1- to 9-fold (Ling et al., 1994; Snedden et al., 1995; Cholewa et al., 1997; Johnson et al., 1997) in partially purified protein preparations, and nearly 20-fold in purified preparations (Snedden et al., 1996). These findings suggest that GAD may be stimulated in vivo by Ca2+ signal pathways. This hypothesis is consistent with data collected from studies demonstrating the rapid increase in cytoplasmic Ca2+ concentrations (Knight et al., 1991, 1992; Price et al., 1994; Cholewa et al., 1997) and GABA titers (Wallace et al., 1984; Mayer et al., 1990; Cholewa et al., 1997) in plant cells upon exposure to various environmental stimuli.
Despite a better understanding of the cellular factors that may stimulate GAD activity, the physiological roles of the enzyme or the product, GABA, have not been clearly established in plants. Since elevated GAD activity is usually seen in tissues with low cytoplasmic pH (Satyanarayan and Nair, 1990), and the synthesis of GABA consumes a proton, GABA metabolism has been proposed to regulate cytoplasmic pH in plant tissues subjected to various stress conditions (Streeter and Thompson, 1972; Davies, 1980). However, Cholewa et al. (1997) demonstrated that GABA accumulation may be stimulated by Ca2+ and not by decreased cytoplasmic pH when plants are subjected to an abrupt cold-shock treatment. But several other physiological roles for GABA have been proposed. Selman and Cooper (1978) suggested that GABA may provide a direct temporary reserve of carbon and nitrogen for Glu or an indirect reserve for protein synthesis. Since GABA is an inhibitor of neuron transmission in animals, Wallace et al. (1984) suggested that increased levels of GABA could alter the eating habits of insects. Recently, Ramputh and Bown (1996) demonstrated that elevated levels of GABA in the diet of oblique-banded leaf-roller larvae decreased their growth, development, and survival. In addition, Chen et al. (1994) questioned whether GABA in plants was involved in the control of ion channels, as in animal neurons.
Baum et al. (1996) overexpressed a truncated version of a petunia GAD gene, which lacked the CaM-binding site, in transgenic tobacco plants and demonstrated that the CaM-binding domain was required for normal plant development and for the maintenance of GABA and Glu levels. These results provide some evidence that GAD is involved in nitrogen metabolism. Other investigators demonstrated that GAD may not be solely involved in the maintenance of cytoplasmic pH. Robinson et al. (1991) and Carroll et al. (1994) showed that GABA was a major nitrogen source for freshly assimilated ammonium in nonstressed cell suspensions. Tuin and Shelp (1994) demonstrated that the in situ synthesis of GABA in developing soybean cotyledons was via GAD, but that GABA was rapidly metabolized to provide tricarboxylic acid cycle intermediates for amino acid metabolism, possibly via the GABA shunt. In addition, Cholewa et al. (1997) reported a Ca2+/CaM-independent increase in GABA when cells were treated with Glu or butyrate. Together, these results suggest not only that GABA synthesis may be a response to stress but that GAD may perform a unique physiological role in nitrogen and/or carbon metabolism via the GABA shunt.
Although several of the factors that stimulate GAD activity have been identified, there is still a gap in the knowledge of the role(s) of transcriptional and/or posttranslational events in the control of GAD activity. Chen et al. (1994) demonstrated that a 58-kD CaM-binding GAD was expressed in various floral parts, seeds, stems, roots, and leaves of petunia. In addition, their data suggested that there was a correlation between in vitro GAD activity, abundance of the 58-kD peptide, and levels of GAD transcript in developing leaves and germinating seeds. However, this phenomenon was not observed in developing flowers. In open flowers the 58-kD peptide was abundant and in vitro GABA synthesis was high, but the level of GAD transcript was almost undetectable. The authors suggested that posttranslational regulation in leaves and seeds may differ from that of flowers, and the process may play a role in controlling GAD activity in flowers.
Chen et al. (1994) used immunoblot analysis to monitor the accumulation of GAD in germinating petunia seeds. They identified three peptides of 48, 58, and 66 kD that cross-reacted with the antiserum to a recombinant petunia GAD (58 kD). There appeared to be a synchronous increase and decrease of the 48- and 66-kD proteins, which was inversely proportional to levels of the 58-kD GAD throughout germination. These data suggest that there may be several GAD-like peptides or isoenzymes in plants. Molecular mass determinations of GAD from potato (45.5 kD) (Satyanarayan and Nair, 1985), and fava bean (62 kD) (Ling et al., 1994) also suggest that there may be distinct GAD isoenzymes. The presence of several GAD isoenzymes could explain the discrepancies in apparent molecular masses of GAD from different plant species and could explain the apparent contradictions concerning physiological role(s) of GAD in plants. The existence of several isoenzymes could be due to posttranslational modifications of a single peptide or to the existence of multiple loci or genes encoding GAD. To gain a greater understanding of the function and role of the GAD isoenzymes in plant nitrogen metabolism, we initiated a study of the isoenzymes and genes in Arabidopsis. In this study we combined molecular biological approaches with biochemical and immunological techniques to identify the cDNA clones and the protein/peptide components that they encode, and to demonstrate that the gene products bind CaM. In addition, we determined the effects of different nitrogen sources on GAD activity in Arabidopsis.
MATERIALS AND METHODS
Plant Material
Arabidopsis (L.) Heynh. ecotype Columbia (Col-0) seeds were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). Plants were grown in (20 × 10 × 6 cm [length × width × depth]) plastic pots, either in a peat-vermiculite mixture (Jiffy Mix, Jiffy Products of America, Batavia, IL)1, or in vermiculite. Plants grown in soil were watered as needed by subirrigation throughout the experiment. Plants grown in vermiculite were subirrigated with a complete mineral nutrient solution (Cammaerts and Jacob, 1985) with nitrate (10 mm KNO3) as a sole nitrogen source. After 15 d, the pots containing vermiculite were flushed daily for 5 d with 25 mL of sterile distilled water, and the plants were subirrigated for 4 d with different nitrogen treatments (10 mm KNO3 [control], 10 mm NH4Cl, 5 mm NH4NO3, 5 mm Glu, or 5 mm Gln), as described by Turano et al. (1997). Plants were maintained at 20°C to 21°C and 60% to 70% RH under cool-white lights (120–140 μmol PPFD m−2 s−1) in a 16-h light/8-h dark cycle.
Crude Protein Extractions
Proteins were extracted from frozen or fresh samples. Frozen samples (approximately 500 mg) were ground to a fine powder with a mortar and pestle. The powder was transferred to 1 mL of extraction buffer (50 mm Tes, pH 5.8, 5 mm EDTA, 1 mm MgCl2, 0.1 mm PLP, 1 mm DDT, 0.5% [w/v] PVP-40, and 4 mm Cys). Fresh PMSF was added to a final concentration of 1 mm in all extracts. The fresh samples (200 mg) were ground in 400 μL of extraction buffer as described above. The samples were incubated on ice for 15 to 30 min. Debris were removed from the sample by centrifugation at 13,000g for 10 min. The supernatant containing the crude protein extract was used to determine the protein concentration and GAD activity.
Protein Determinations and Enzymatic Activity Assays
Protein concentrations were determined using a modification of the Lowry method as described by Markwell et al. (1978) or by NanoOrange as described by the manufacturer (Molecular Probes, Inc., Eugene, OR). GAD activity was determined using a method similar to that of Snedden et al. (1992). Assays were conducted in 17- × 52-mm glass vials plugged with a soft rubber stopper. An 18-gauge syringe needle was used to pierce the stopper and a folded piece of Whatman paper no. 3 (7 × 75 mm) held in place by a small piece of rubber tubing. The Whatman paper was saturated with 180 μL of 1 n KOH and suspended in the vial when the rubber stopper was put in place to trap the CO2 that evolved from the GAD reaction. A small stopper was placed in the end of the syringe needle to prevent the escape of CO2 from the reaction. The vials were maintained on ice prior to the initiation of the reaction. Reactions from crude protein extracts from plant tissues were conducted in 500 μL with 100 mm pyridine-HCl, pH 5.8, 10 mm NaCl, 0.1 mm PLP, and 20 mm Glu containing 0.02 μCi/μmol l-(1-14C)Glu with 25 to 50 μL of enzyme extract. Crude protein extracts were assayed at pH 5.8 and not 7.3 because there were problems associated with CaM binding and the effects of proteases on the CaM-binding domain of GAD in these extracts. Pure rGAD1 and rGAD2 were assayed in 500 μL with 100 mm 1,3-bis-Tris-propane-HCl, pH 7.3, 10 mm NaCl, 0.1 mm PLP, and 20 mm Glu containing 0.02 μCi/μmol l-(1-14C)Glu with 25 to 50 μL of enzyme extract.
To test Ca2+ and/or CaM stimulation, CaCl2 and CaM were added alone or in combination to a final concentration of 1 mm and 0.4 μm, respectively. The CaM antagonist TFP was dissolved in water and added to a final concentration of 100 μm. Assays were initiated with the addition of the substrate. The vials were incubated in a gently shaking water bath at 30°C for 1 h. Vials were immediately placed on ice and 100 μL of 2 n H2SO4 was added to the vessels through the 18-gauge syringe to terminate the reaction. The reactions were incubated on ice for 30 to 45 min to allow complete absorption of the CO2. The filter paper for each vial was placed into a scintillation vial containing 4.5 mL of Formula A-989 high-flash-point cocktail (Packard Instrument Co., Inc., Meriden, CT) and counted in a scintillation counter (model LS 2800, Beckman). In each experiment, enzyme determinations were performed in triplicate.
Gel Electrophoresis and Immunoblot Analysis
Proteins were separated by SDS-PAGE (8.0% or 9.0% polyacrylamide) using a Mini-Protean II (Bio-Rad) system. Silver staining and immunoblot analysis were conducted as described by Turano et al. (1990). Rabbit serum raised against petunia rGAD was kindly provided by Dr. Hillel Fromm (Baum et al., 1993). For the identification of GAD isoenzymes in different organs, an equal amount of protein (150 μg) from roots, leaves, flower stalks, flowers, and siliques of 42-d-old plants was added per lane. Similarly, 150 μg of protein was added per lane to determine the effects of different nitrogen sources on GAD isoenzymes in leaves of 24-d-old plants. To determine the effects of different nitrogen sources on GAD isoenzymes in roots of 24-d-old plants, 75 μg of protein was added per lane.
GAD Clones
Two expressed sequence tag clones encoding GAD1 (35B3T7) and GAD2 (5B2T7P) were obtained from the Arabidopsis Biological Resource Center. The clones were sequenced by the dideoxy chain-termination method using modified T7 DNA polymerase (Sequenase 2.0, United States Biochemical), as described by the manufacturer. The data were analyzed with the IntelliGenetics Suite (IntelliGentics, Inc., Mountain View, CA) on a Sun system.
DNA and RNA Isolation and Gel Electrophoresis
Total DNA was isolated, digested with restriction enzymes, and blotted to nitrocellulose as described by Turano et al. (1992). For the determination of organ-specific expression, total RNA was isolated from roots, leaves, flower stalks, flowers, and siliques of 42-d-old Arabidopsis plants. To determine the effects of nitrogen on GAD, total RNA was isolated from roots and leaves of 24-d-old plants. Total RNA was isolated from each organ, separated by gel electrophoresis using formaldehyde-formamide gels, blotted to nitrocellulose, and hybridized as described by Turano et al. (1997).
Construction of rGAD1 and rGAD2 for Expression in Escherichia coli
The open reading frames for GAD1 and GAD2 were amplified and cloned in the E. coli expression vector pKK223-3 (Pharmacia). Oligonucleotide primers were synthesized with HindIII sites at the 5′ (5′-GCCCAAGCTTAGGAAACAGAAATGGTGCTCTCCCACGCCG-3′) and 3′ (5′-GCCCAAGCTTAGCAGATACCACTCG-3′) for rGAD1 and the 5′ (5′-GCCCAAGCTTAGGAAACAGAAATG-GTTTTGACAAAAACCG-3′) and 3′ (5′-GCCCAAGC-TTTAGCACACACCATTCA-3′) for rGAD2. Separate amplification reactions were conducted with 5′- and 3′-specific primers with the appropriate cDNA clone as a template using a gene-amplification kit (PanVera Corp., Madison, WI). Amplification reactions were conducted as follows: 94°C for 30 s, 55°C for 30 s, and 72°C for 4 min, for 25 cycles. The amplified fragments were digested with HindIII, ligated into the vector, and transformed into XL1-Blue MRF′ (Stratagene) competent cells. The correct orientation was verified by restriction endonuclease analysis.
E. coli cultures containing rGAD1 or rGAD2 were grown overnight, approximately 16 h, at 37°C in Luria-Bertani medium with 100 μg/mL ampicillin. Overnight cultures were diluted 1:100 in fresh Luria-Bertani medium with 100 μg/mL ampicillin and incubated at 25°C with shaking at 200 rpm. After 4 h, isopropylthio-β-galactoside was added to a final concentration of 100 μm to induce expression of the recombinant protein, and incubation was continued for 3 to 4 h for rGAD1 and 20 h for rGAD2. Bacterial cells were collected by centrifugation at 6000g for 10 min. Pellets were immediately frozen in liquid nitrogen and stored at −80°C. The E. coli pellets could be stored at −80°C for 2 months without a significant decrease in GAD activity. Pellets containing cells were resuspended in 5 to 7 mL of extraction buffer (50 mm Tris-HCl, pH 7.5, 0.2 mm EDTA, 2.5% [v/v] glycerol, 2 mm DTT, 0.05 mm PLP, 0.05% [v/v] Triton, 10 μm leupeptin, 20 μg/mL lysozyme, and 1 mm PMSF). The cells were lysed by sonication with 12 2-s bursts at 150 W. Cellular debris were removed by centrifugation at 12,000 rpm for 20 min. The supernatant was filtered through a 0.45-μm filter. CaCl2, DTT, and PMSF were added to final concentrations of 1, 2, and 1 mm, respectively. The supernatant was loaded onto an approximately 1-mL bed volume CaM-Sepharose column (Pharmacia) and preequilibrated in binding buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2.5% [v/v] glycerol, 1 mm CaCl2, 2 mm DTT, 10 μm leupeptin, and 1 mm PMSF). The column was washed with 20 bed volumes of wash buffer minus calcium (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2.5% [v/v] glycerol, 2 mm DTT, 10 μm leupeptin, and 1 mm PMSF). CaM-binding proteins were eluted with 50 mm Tris-HCl, pH 7.5, 2 mm EDTA, 150 mm NaCl, 2.5% [v/v] glycerol, and 1 mm PMSF. Samples were immediately assayed as described above.
Amplification and Radioactive Labeling of GAD Probes and Hybridization
One oligoprimer (17-mer) was synthesized (Bio-Synthesis, Inc., Lewisville, TX) to the 3′ regions of each of the GAD genes. The primers, designated 3primGAD1 (5′-GATCGATATAGAGAAAG-3′) and 3primGAD2 (5′-TAGATACCTTGCCTTCC-3′), were used in separate PCRs with the M13-forward primer and their corresponding cDNA clones as the templates for use in a gene-amplification kit (Perkin-Elmer). Reactions were conducted as follows: 94°C for 30 s, 45°C for 30 s, and 72°C for 2 min, for 25 cycles. Each of the amplified DNA fragments, 404 and 332 bp, unique to the 3′ regions of GAD1 and GAD2 (Fig. 1), respectively, were gel purified, amplified a second time, and used as gene-specific probes in genomic Southern and RNA-blot analyses. Amplification of the DNA probes, total RNA isolation, blotting, prehybridizations, and hybridizations were performed as described by Turano et al. (1997). A probe encoding for a 26S rRNA gene (F.J. Turano, unpublished data) was used as an internal control on RNA blots to ensure equal loading of RNA per lane. Data were quantified on a beta scanner (Betagen, IntelliGentics, Inc.).
Figure 1.
Nucleotide sequence of Arabidopsis (At) GAD cDNA clones. The nucleotide sequence of the 3′ noncoding region of the cDNA clone corresponding to GAD1 (A) and the full-length cDNA clone corresponding to GAD2 (B) are shown in the sense strand. A, Nucleotides are numbered from the putative stop codon of GAD1. B, The amino acids are numbered from the putative start codon of GAD2. C, Schematic representation of the 3′ gene-specific probes. Thin lines represent either the 5′ or 3′ noncoding regions, the open boxes represent the coding regions, and the thick bars represent the gene-specific probe for either GAD1 or GAD2.
RESULTS
Sequence Analysis of GAD Clones
The nucleotide sequence (1509 bp) for the coding region of 35B3T7 (GAD1) was available in GenBank (accession no. U10034), and an additional 250 bp of sequence was determined for the 5′ (data not shown) and 3′ noncoding (Fig. 1A) regions. The full-length GAD1 clone was 1760 bp. A partial nucleotide sequence (465 bp) for 5B2T7P (GAD2) was available in GenBank (accession no. T04804). The remainder of the cDNA was sequenced; the full-length clone was 1676 bp long and encoded a 494-amino acid peptide (Fig. 1B). The nucleotide identity between GAD1 and GAD2 was 74% and 28% in the coding and 3′ noncoding regions, respectively. Oligoprimers (17-mer) specific for GAD1 and GAD2, designated 3primGAD1 and 3primGAD2 (see Methods), were synthesized to the 3′ regions of each of the cDNA clones. The primers were used to make 404- and 332-bp DNA probes unique to the 3′ regions of GAD1 and GAD2 (Fig. 1C), respectively.
GAD1 and GAD2 have open reading frames that encode for proteins containing 502- and 494-amino acid residues, respectively. Theoretically (estimated using PROSITE [version 14.0, Medical Biochemistry Department, Geneva, Switzerland]), GAD1 encodes for a 57.1-kD peptide and GAD2 encodes for a 56.1-kD peptide. The deduced amino acid sequences of the two Arabidopsis GAD cDNA clones were aligned with the deduced amino acid sequences of petunia (Baum et al., 1993) and tomato (Gallego et al., 1995) GAD cDNA clones (Fig. 2). The GAD1 and GAD2 gene products had 82% amino acid identity. The gene product for GAD1 had 85% and 76% amino acid identity with the petunia and tomato GAD sequences, respectively. The gene product for GAD2 had 79% and 74% amino acid identity with the petunia and tomato GAD sequences, respectively. Neither of the Arabidopsis peptides contained putative organellar target sequences.
Figure 2.
Comparison of the deduced amino acid sequences for Arabidopsis (At) GAD1 and GAD2 with GAD from tomato (tomGAD; Gallego et al., 1995; accession no. X80840) and petunia (petGAD; Baum et al., 1993; accession no. L16797). The Lys associated with the putative PLP-binding motif and the Trp associated with the putative CaM-binding domains are indicated in bold type.
The amino acid residues Ser-X-X-Lys are conserved at amino acid residues 274 to 277 and 273 to 276 in the GAD1 and GAD2 peptide sequences, respectively. This motif is common among PLP-requiring enzymes (Tanase et al., 1979). The identity and position of the Ser-X-X-Lys motif in the Arabidopsis GAD peptides are conserved with the motif in the products of the gadA and gadB genes from E. coli (Smith et al., 1992), which have been shown to require PLP for GAD activity, and the putative PLP sites of petunia GAD (Baum et al., 1993) and tomato GAD (Gallego et al., 1995). A putative CaM-binding region, located at the carboxy termini of the two peptides, was variable among the sequences. Both peptides contained Trp residues in this region, Trp-488 (GAD1) and Trp-480 (GAD2), which has been shown to be essential for CaM binding in petunia GAD (Arazi et al., 1995). Arazi et al. (1995) demonstrated that the last 30 amino acids of GAD1 were sufficient for CaM binding (Arazi et al., 1995).
CaM Binding and Ca2+/CaM Stimulation of rGAD1 and rGAD2
To demonstrate CaM binding and/or Ca2+/CaM stimulation, the open reading frame of GAD1 or GAD2 was overexpressed in E. coli. The recombinant proteins were purified by affinity chromatography (Fig. 3) and tested for Ca2+/CaM stimulation (Table I). E. coli extracts overexpressing the open reading frames for GAD1 or GAD2 were loaded onto CaM-Sepharose columns. The columns were washed extensively to remove nonspecific proteins (see Methods for details). CaM-binding proteins were eluted with 2 mm EGTA. Purified CaM-binding proteins were analyzed by silver staining SDS-polyacrylamide gels and by immunoblot analysis (Fig. 3). The estimated molecular masses for rGAD1 and rGAD2 were 58 and 56 kD, respectively. These findings are similar to the estimated molecular masses derived from the deduced amino acid sequences of the two cDNA clones described above. In crude and purified extracts, the 58 and 56-kD peptides cross-reacted with rabbit serum raised against petunia rGAD. Furthermore, neither the 58- nor the 56-kD peptides was observed in crude E. coli extracts containing only the vector pKK223-3. These results confirm that the recombinant proteins were expressed in E. coli, were purified to homogeneity, and were CaM-binding proteins. The purified recombinant proteins were assayed for GAD activity in the presence of Ca2+, CaM, Ca2+/CaM, or Ca2+/CaM/TFP (Table I). Neither rGAD1 nor rGAD2 was stimulated by Ca2+ or CaM alone. However, both rGAD1 and rGAD2 were stimulated 35- and 13- fold, respectively, by Ca2+/CaM. In both cases, the Ca2+/CaM stimulation was significantly reduced by the CaM antagonist TFP. These data confirm that both rGAD1 and rGAD2 encode for CaM-binding proteins and that both are enzymatically stimulated by Ca2+/CaM.
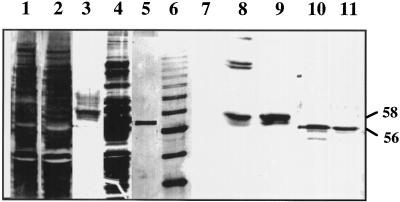
Figure 3.
Characterization of rGAD1 and rGAD2 by SDS-PAGE and immunoblot analysis. Crude protein extracts (20 μg/lane) from E. coli containing the expression vector pKK223-3 (lanes 1 and 7) or the open reading frame for GAD1 (lanes 2 and 8) or GAD2 (lanes 4 and 10) cloned into pKK223-3 were resolved in a SDS 8.0% polyacrylamide gel. Protein extracts from E. coli expressing the open reading frame for GAD1 or GAD2 were purified by affinity chromatography using CaM-Sepharose. The eluted peptides (0.075 μg/lane) rGAD1 (lanes 3 and 9) and rGAD2 (lanes 5 and 11) were resolved by SDS-PAGE. Lane 6 contains a molecular mass marker (10-kD ladder, Life Technologies). The proteins in lanes 1 through 6 were stained with silver; the proteins in lanes 7 through 11 were blotted onto nitrocellulose and incubated with antiserum raised against petunia rGAD. The positions of the rGAD1 (58 kD) and rGAD2 (56 kD) peptides are indicated.
Table I.
Stimulation of pure rGAD1 or rGAD2 at pH 7.3
| Treatment | rGAD1
|
rGAD2
|
||
|---|---|---|---|---|
| GAD activity | Stimulation | GAD activity | Stimulation | |
| μmol CO2 min−1 mg−1 protein | -fold | μmol CO2 min−1 mg−1 protein | -fold | |
| −Ca2+/CaM (control) | 1.95 ± 0.4 | — | 1.9 ± 0.4 | — |
| +Ca2+ | 2.0 ± 0.6 | 0 | 5.6 ± 0.8 | 1.9 |
| +CaM | 5.5 ± 1.6 | 1.8 | 5.5 ± 0.9 | 1.9 |
| +Ca2+/CaM | 71.0 ± 7.1 | 35.4 | 26.9 ± 3.4 | 13.2 |
| +Ca2+/CaM/TFP | 32.0 ± 4.0 | 15.4 | 5.2 ± 1.2 | 1.7 |
rGAD1 and rGAD2 were assayed in 100 mm 1,3-bis-Tris-propane-HCl, pH 7.3, 10 mm NaCl, 100 μm PLP, and 20 mm glutamate ([1-C14]l-glutamate 0.02 μCi/mmol). When indicated, CaCl2 and CaM were added to final concentrations of 1 mm and 0.4 μm, respectively. The values are the averages ± se of two separate experiments.
Southern-Blot Analysis
Southern-blot analysis was used to determine the number of copies of GAD1 and GAD2 genes in Arabidopsis (Fig. 4). Specific 3′ probes for the cDNA corresponding to GAD1 and GAD2 were hybridized under stringent conditions to Southern blots of Arabidopsis genomic DNA digested with BamHI, EcoRI, or HindIII. It was evident that under the hybridization conditions used in these experiments, the probes were gene specific, since there were different hybridization patterns for each probe. The hybridization patterns for both GAD1 and GAD2 were simple; one band was apparent in each digest. Based on the simplicity of the hybridization patterns, comparisons of the intensities of these Southern blots with those of GDH1 (data not shown), a single-copy gene (Melo-Oliveira et al., 1996), and the initial results from the identification of genomic clones (data not shown), it appears that both GAD1 and GAD2 represent single-copy genes.
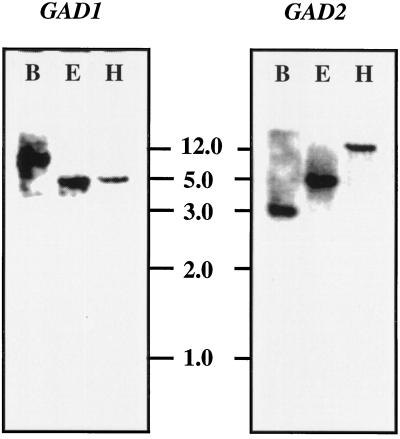
Figure 4.
Southern-blot analysis of Arabidopsis genomic DNA digested with BamHI (B), EcoRI (E), or HindIII (H), separated by electrophoresis, transferred to nitrocellulose membranes, and hybridized under stringent conditions with gene-specific probes for either GAD1 or GAD2 (see Methods for details). Molecular masses are indicated.
Organ Specificity
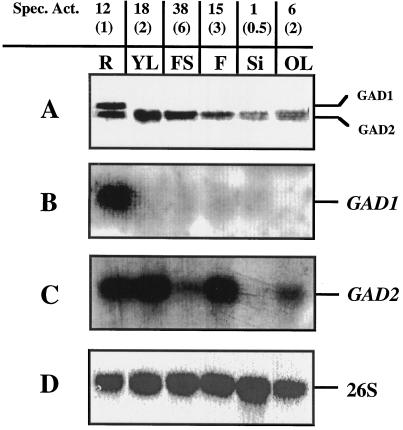
Specific activity, immuno-, and RNA-blot analyses were used to determine the level of activity, peptide, and/or transcript(s) for each GAD isoenzyme in different plant organs (Fig. 5). The specific activity of GAD was highest in flower stalks and lowest in siliques of 42-d-old plants. Protein extracts from different organs were separated by SDS-PAGE (9.0% polyacrylamide), and immunoblot analysis was used to identify the GAD peptides (Fig. 5A). A 58-kD peptide was identified only in root samples, and based on the migration of the peptide in the gel, this subunit was designated GAD1. A 56-kD peptide, designated GAD2, was identified in protein samples from all organs. The transcript corresponding to GAD1 was detected only in root samples. The transcript corresponding to GAD2 was readily detected in all tissues tested, except for siliques, but was visible in samples from siliques upon longer exposure (data not shown). The GAD2 transcript was abundant in samples from roots, young leaves, and flowers, but the level of transcript was relatively lower in flower stalks and old leaves. The results from this experiment suggest that GAD1 encodes for a 58-kD peptide and that GAD2 encodes for a 56-kD peptide. These findings are consistent with the estimated sizes of the peptides deduced from the amino acid sequences and the results obtained from SDS-PAGE and immunoblot analysis of the recombinant proteins described above.
Figure 5.
Specific GAD activity, immunoblot analysis, and expression of GAD1 and GAD2 in different organs of Arabidopsis. Plants were grown in soil and maintained at 20°C to 21°C and 60% to 70% RH under cool-white lights (120–140 μmol PPFD m−2 s−1) in a 16-h light/8-h dark cycle until tissues were harvested. Protein and total RNA extractions were performed on roots (R), flower stalks (FS), flowers (F), siliques (S), leaves (OL) of 42-d old plants, and leaves of 24-d-old plants (YL). The specific activity (Spec. Act.) of each organ is indicated (nanomoles of CO2 per minute per milligram of protein). The values are the averages of three separate experiments (±se). A, For immunoblot analysis, proteins (150 μg/lane) were resolved in an SDS-9.0% polyacrylamide gel, blotted onto nitrocellulose, and incubated with antiserum raised against recombinant petunia GAD. The position of GAD1 (58 kD) and GAD2 (56 kD) peptides are indicated. Total RNA (10 μg) was separated by gel electrophoresis, blotted onto nitrocellulose, and hybridized with DNA probes specific for GAD1 (B) or GAD2 (C). D, A portion of the 26S rRNA probe was used as a control to ensure equal loading of total RNA into the lanes.
Effect of Different Nitrogen Sources on GAD
Plants were maintained in soil or vermiculite with complete nutrient solution and 10 mm KNO3 as a sole nitrogen source. After 20 d the pots containing vermiculite were treated with complete mineral nutrient solution containing 10 mm KNO3, 10 mm NH4Cl, 5 mm NH4NO3, 5 mm Glu, or 5 mm Gln as a sole nitrogen source. Determination of specific GAD activity and immunoblot and northern-blot analyses were performed on protein and total RNA extracts from leaves (Fig. 6) and roots (Fig. 7) 4 d after treatment with different nitrogen sources.
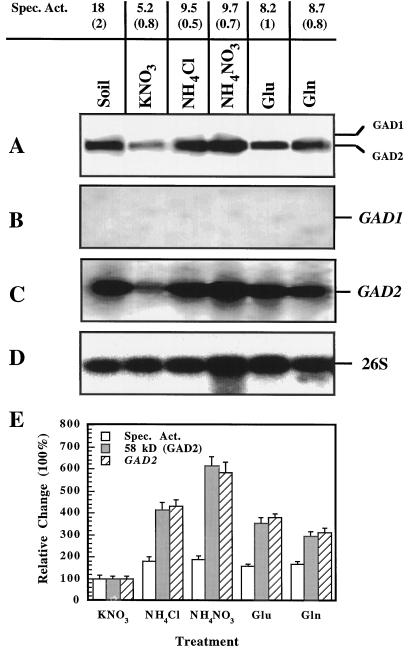
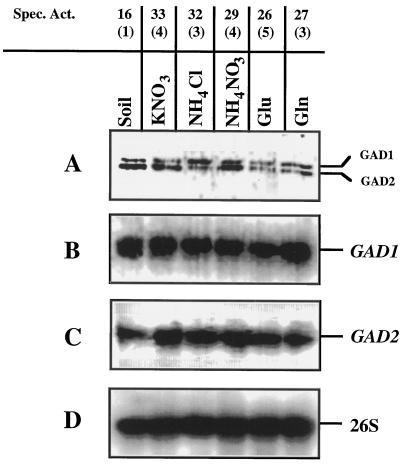
Figure 6.
Effect of various nitrogen treatments on GAD in leaves of Arabidopsis. Plants were grown in vermiculite and maintained in the environmental conditions described in Figure 5. Plants were watered with a mineral nutrient solution containing nitrate (10 mm KNO3) for 20 d. The plants were treated with 10 mm KNO3, 10 mm NH4Cl, 5 mm NH4NO3, 5 mm Glu, or 5 mm Gln for 4 d prior to the harvest of the leaves on d 24. One set of plants was grown in soil and maintained as described in Figure 5. Protein and RNA extractions were collected and data were analyzed as described in Figure 5. The specific activity (Spec. Act.; nanomoles of CO2 per minute per milligram of protein), GAD1 (58 kD) and GAD2 (56 kD) peptides (A), GAD1 (B), GAD2 (C), and 26S (D) transcripts are indicated. The results presented above are representative of three separate experiments. The change in specific activity and the abundance of the 56-kD peptide and GAD2 transcript are presented as a relative change (percentage) in the samples from various nitrogen treatments compared with the levels of GAD activity and abundance of peptide and transcript in controls, which received a 10 mm KNO3 treatment (E). The results are a summary of three separate experiments, and the error bars represent the ses.
Figure 7.
Effect of various nitrogen treatments on GAD in roots of Arabidopsis. Plants were treated as described in Figure 6. Protein and RNA extractions were collected and analyzed as described in Figure 5. The specific activity (Spec. Act.; nanomoles of CO2 per minute per milligram of protein), GAD1 (58 kD) and GAD2 (56 kD) peptides (A), and GAD1 (B), GAD2 (C), and 26S (D) transcripts are indicated. The results are representative of three separate experiments.
The specific activity of GAD in leaves of plants treated with ammonium, either 10 mm NH4Cl or 5 mm NH4NO3, was approximately 85% higher than that of the 10 mm KNO3 (control) treatments (Fig. 6). There were smaller increases, 58% or 67%, respectively, in the specific activity from extracts of plants treated with either Glu or Gln when compared with the control treatment. The 56-kD (GAD2) peptide was observed in all of the treatments (Fig. 6A), but the abundance of the peptide varied among nitrogen treatments. The 56-kD peptide (GAD2) was more abundant in 10 mm NH4Cl- or 5 mm NH4NO3- treated samples than in samples from plants treated with 5 mm Glu or 5 mm Gln, which were more abundant than the control (10 mm KNO3). Neither the 58-kD peptide (GAD1) (Fig. 7A) nor the GAD1 transcript (Fig. 6B) was detected in the leaf samples. The GAD2 transcript was detected in all of the samples (Fig. 6C). The relative change in specific activity and the abundance of 56-kD peptide and GAD2 transcript detected in the various nitrogen treatments were compared with levels in controls, which received a 10 mm KNO3 treatment (Fig. 6E). There was a proportional increase in the amount of 56-kD peptide and GAD2 transcript detected in each treatment compared with controls. However, the relative change in the level of 56-kD peptide and GAD2 transcript were approximately 4 times higher than the increase in the specific activity; i.e. the specific activity increased 83%, whereas the amount of the 56-kD peptide and GAD2 transcript increased 314% and 432%, respectively, in the 5 mm NH4NO3 treatment compared with the control. In summary, these data suggest that GAD activity is altered by different nitrogen sources, and the activity is regulated in part by gene expression or RNA stability.
The specific activity of GAD in extracts from roots (Fig. 7) of plants treated with different nitrogen sources was 3 to 6 times higher than that of extracts from leaves (Fig. 6) of the same plants. There was one exception: the specific activity in root extracts from plants maintained in soil was similar to that of leaves from the same plants. The specific activity in roots of plants treated with 10 mm NH4Cl, 5 mm NH4NO3, 5 mm Glu, or 5 mm Gln were 3% to 21% lower than that in the controls (10 mm KNO3 treatments). Immunoblot analysis showed (Fig. 7A) that all samples contained both the 58- and 56-kD peptides, GAD1 and GAD2, respectively. In most cases, the level of each peptide detected on immunoblots was approximately the same. However, there was slight variability in the ratio of the level of the 58-kD transcript to that of the 56-kD peptide within each treatment (data not shown). The transcripts for GAD1 and GAD2 were detected on RNA blots (Fig. 7, B and C, respectively) in all of the root samples. After normalization of the data to the 26S probe (data not shown), the amount of GAD1 and GAD2 transcript in each sample remained unchanged. However, there were small decreases (10%–15%) in the level of GAD2 transcript in the roots of Gln-treated plants compared with the control.
DISCUSSION
Two full-length cDNAs encoding GAD in Arabidopsis have been characterized. The two clones appear to encode distinct single-copy genes, GAD1 and GAD2, in Arabidopsis. The small GAD family joins the growing list of small multigene families encoding enzymes involved in Glu metabolism and/or catabolism, i.e. glutamine synthetase (Peterman and Goodman, 1991), glutamate synthase (Coschigano et al., 1998), aspartate aminotransferase (Schultz and Coruzzi, 1995; Wilkie et al., 1995), and glutamate dehydrogenase (Melo-Oliveira et al., 1996; Turano et al., 1997) in Arabidopsis.
The deduced amino acid sequences of both cDNA clones lack target sequences, suggesting that they are located in the cytosol, which is consistent with the cellular location reported for numerous plant GAD isoenzymes (Wallace et al., 1984; Satyanarayan and Nair, 1985; Breitkreuz and Shelp, 1995). The peptides appear to be divided into three distinct regions: (a) a small, variable region at the amino termini (from approximately residues 1–35), (b) a large, highly conserved region (from approximately residues 36–245), and (c) a small, highly variable region at the carboxy termini (from approximately residues 246–493). To date, there does not appear to be a specific function associated with the small, variable region at the amino termini. The large, highly conserved region contains the GAD enzyme domain. In this region, GAD1 and GAD2 contain a putative PLP-binding motif, Ser-X-X-Lys. This motif is conserved in the 58-kD GAD from petunia (Baum et al., 1993) and the putative GAD cDNA from tomato (Gallego et al., 1995), and it aligns with the same motif in the products of the gadA and gadB genes from E. coli (Smith et al., 1992). Furthermore, the sequence analysis is consistent with our biochemical analysis of GAD2, which showed that PLP was required for activity (F.J. Turano and T.K. Fang, unpublished data). The highly variable region at the carboxy termini contains the CaM-binding domain. Both peptides contain a Trp residue in the highly variable carboxy termini, at positions 488 (GAD1) and 480 (GAD2). Similarly, there is a Trp in this region in petunia GAD, which was shown to be essential for CaM binding (Arazi et al., 1995). In addition, Arazi et al. (1995) demonstrated that CaM bound to the carboxy terminus of GAD1. In this report both rGAD1 and rGAD2 were purified by affinity chromatography using CaM-Sepharose. Enzymatic activity for pure rGAD1 and rGAD2 was stimulated 35- and 13-fold, respectively. The stimulation of rGAD1 and rGAD2 by Ca2+/CaM was inhibited 65% and 81%, respectively, by the CaM antagonist TFP. Furthermore, GAD activity was not stimulated by Ca2+ or CaM alone. Together, these data demonstrate that both Arabidopsis isoenzymes, although variable in their carboxy termini, bind Ca2+/CaM. The physiological significance for the variability in the CaM region is not known, but since there are six CaM and CaM-like genes differentially expressed in Arabidopsis (Braam and Davis, 1990; Sistrunk et al., 1994; Ito et al., 1995), it is possible that this region may be involved in specific binding of different CaM and/or CaM-like proteins. It is worth noting that the stimulation of rGAD1 and rGAD by Ca2+/CaM is significantly different, although they were assayed under similar conditions. It is plausible that the distinct CaM-binding domains may play a role in the differential stimulation of the two recombinant proteins. However, it is premature to overlook the possible function of the distinct amino termini and/or variations in the enzymatic domains of the two peptides in the stimulatory process.
Both cDNA clones were expressed in E. coli and the estimated molecular masses for the recombinant proteins were 58 and 56 kD for rGAD1 and rGAD2, respectively (Fig. 3). The migration of the recombinant proteins was similar to those of peptides identified by immunoblot analyses in crude protein extracts from plants. The data suggest that GAD1 encodes a 58-kD peptide (GAD1) and GAD2 encodes a 56-kD peptide (GAD2). This gene/peptide assignment was consistently observed on immunoblots and RNA blots from different organs (Fig. 5) and from similar analyses comparing extracts from leaves (Fig. 6) and roots (Fig. 7) of plants subjected to different nitrogen treatments. Protein and RNA extracts from roots (Figs. 5, lane R, and 7) had detectable levels of both peptides, 58 and 56 kD, and both transcripts, GAD1 and GAD2, whereas all other organs (Figs. 5 and 6) had detectable levels of only the 56-kD peptide (GAD2) and the GAD2 transcript.
In leaves (Figs. 5, lanes YL and OL, and 6) there appeared to be a correlation between the specific activity and the amount of the 56-kD peptide and GAD2 transcript detected on immunoblots and RNA blots, respectively. These data suggest that GAD activity may be controlled by transcriptional events or by RNA stability in leaves. A similar correlation among the accumulation of GAD transcript, GAD peptide, and in vitro GABA synthesis was observed in petunia (Chen et al., 1994). Combined, these data suggest that the transcriptional and posttranscriptional processes that control GAD activity in the leaves of Arabidopsis and petunia may be similar; however, it is too early to state whether these processes occur in the leaves of all plant species. Although there appeared to be a correlation among the in vitro specific activity, the amount of the 56-kD peptide, and the level of GAD2 transcript in leaves, this phenomenon did not appear to be conserved in all plant organs. In flower stalks and flowers (Fig. 5, lanes FS and F), there appeared to be little or no correlation among the level of GAD2 transcript, 56-kD peptide, and specific activity. In flower stalks, the specific activity was higher than that in flowers; likewise, the level of 56-kD peptide was higher in flower stalks than in flowers. However, the level of GAD2 transcript was lower in flower stalks than in flowers. The data suggest that GAD 56-kD protein is more stable in flower stalks than in flowers or leaves of Arabidopsis. These results suggest that other factors such as posttranslational events may regulate GAD activity in these organs. Chen et al. (1994) made similar conclusions when comparing their results concerning developmental studies of petunia flowers with developmental studies of leaves; in their study a 58-kD peptide appeared to be more stable in flowers than in leaves. Also, in the siliques there was little or no GAD activity or transcript detected, but the 56-kD peptide was readily detected, suggesting that there could be other factors, such as posttranslational events, controlling GAD in these organs. However, there could be factors that affect the in vitro determination of GAD activity in samples from siliques. First, the amount of GAD2 detected in samples from siliques by immunoblot analysis was very low compared with other organs and we may be approaching the limits of our assay method. Second, because of the possible release of inhibitory compounds during protein isolations, the level of in vitro GAD activity may not be a true measure of in vivo activity.
In roots (Fig. 7) there was little or no change in the amount of GAD1 or GAD2 transcripts detected in each treatment, but there were small differences in the abundance of the 58- or 56-kD peptides. In addition, there was small variability among the ratio of 58-kD to 56-kD peptides in replica samples from the same treatment. This may be due to differential stability of the two peptides during extraction or the different location of the peptides in the root. To our knowledge, there are no data to support the former hypothesis, but results from Ling et al. (1994) suggest that the GAD isoenzymes may be localized in different regions of the root. They identified a 62-kD peptide in a region 2 to 17 cm from the root tip, but never within it. The small variability in the ratio of 58-kD to 56-kD peptides in our experiments could be attributed to the different proportions of distinct root regions in our samples.
The GAD2 transcript, which encodes a 56-kD peptide, was detected in all tissues tested, suggesting that it is constitutively expressed. Similarly, the transcript corresponding to a 58-kD peptide was identified in all of the tissues of petunia (Chen et al., 1994). Together, these data suggest that these peptides may play a crucial role in plant development. This hypothesis has been supported by results from studies of transgenic plants overexpressing a truncated version of the petunia GAD in tobacco (Baum et al., 1996). Conversely, the GAD1 transcript, which encodes a 58-kD peptide, was detected only in roots. Similarly, a 62-kD peptide was identified in the roots of fava bean (Ling et al., 1994). Together, these data suggest that there are root-specific GAD isoenzymes for which the physiological function is unknown.
One goal of this study was to gain a greater understanding of the function and role of the GAD isoenzymes in nitrogen metabolism. The GAD2 transcript appears to be constitutively expressed; however, the level of transcript detected by RNA-blot analyses was altered by different nitrogen sources. The level of GAD2 transcript increased with 10 mm NH4Cl, 5 mm NH4NO3, 5 mm Glu, or 5 mm Gln treatments. Our data suggest that ammonia, Glu, and Gln affect GAD activity in leaves by increasing gene expression or RNA stability. Other investigators have reported increases in GAD activity or GABA accumulation in plant cells after different nitrogen treatments. Kishinami and Ojima (1980) demonstrated that GABA titers increased significantly in rice cultures after treatment with ammonium or Gln; however, no increase was observed in cell cultures after Glu treatments. They used inhibitors of glutamine synthetase and glutamate synthase to demonstrate that ammonium increased Gln levels, but they did not determine the effects of these compounds on GAD activity. It appeared from their data that GABA biosynthesis was via GAD. Tuin and Shelp (1994) suggested that GAD was involved in nitrogen and carbon metabolism via the GABA shunt. Cholewa et al. (1997) reported a Ca2+/CaM-independent increase in GABA when asparagus mesophyll cells were treated with Glu. Furthermore, Baum et al. (1996) demonstrated that overexpression of a truncated version of a petunia GAD gene lacking the CaM-binding domain in transgenic tobacco had increased titers of free GABA and decreased titers of free Glu. The combined findings from these studies suggest that GAD2 or GAD2 homologs may play a unique physiological role in nitrogen metabolism.
ACKNOWLEDGMENTS
The authors would like to thank Drs. Benjamin F. Matthews, Mark Tucker, Jane Weisemann, and Kenneth G. Wilson for critical review of this manuscript. We appreciate the technical assistance of Ms. Sona S. Thakkar, who performed all of the molecular biology experiments, and Ms. Geraldine G. Glover, who cloned and sequenced rGAD1.
Abbreviations:
- CaM
calmodulin
- GABA
γ-aminobutyric acid
- GAD
glutamate decarboxylase
- PLP
pyridoxal 5-phosphate
- rGAD
recombinant GAD
- TFP
trifluoperazine
Footnotes
The nucleotide sequence data for the 3′ noncoding region of GAD1 and full-length sequence for GAD2 are available in GenBank under accession nos. AF060094 and U46665, respectively.
Mention of trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other products or vendors that may be suitable.
LITERATURE CITED
- Arazi T, Baum G, Snedden WA, Shelp BJ, Fromm H. Molecular and biochemical analysis of calmodulin interactions with the calmodulin-binding domain of plant glutamate decarboxylase. Plant Physiol. 1995;108:551–561. doi: 10.1104/pp.108.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum G, Chen Y, Arazi T, Takatsuji H, Fromm H. A plant glutamate decarboxylase containing a calmodulin binding domain. Cloning, sequence, and functional analysis. J Biol Chem. 1993;268:19610–19617. [PubMed] [Google Scholar]
- Baum G, Lev-Yadun S, Fridmann Y, Arazi T, Katsnelson H, Zik M, Fromm H. Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J. 1996;15:2988–2996. [PMC free article] [PubMed] [Google Scholar]
- Braam J, Davis RW. Rain-, wind-, and touch-induced expression of the calmodulin and calmodulin-related genes in Arabidopsis. Cell. 1990;60:357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- Breitkruz KE, Shelp BJ. Subcellular compartmentation of the 4-aminobutyrate shunt in protoplasts of developing soybean cotyledons. Plant Physiol. 1995;108:99–103. doi: 10.1104/pp.108.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammaerts D, Jacobs M. A study of the role of glutamate dehydrogenase in nitrogen metabolism of Arabidopsis thaliana. Planta. 1985;163:517–526. doi: 10.1007/BF00392709. [DOI] [PubMed] [Google Scholar]
- Carroll AD, Fox GG, Laurie S, Phillips R, Ratcliffe RG, Stewart GR. Ammonium assimilation and the role of γ-aminobutyric acid in pH homeostasis in carrot cell suspensions. Plant Physiol. 1994;106:513–520. doi: 10.1104/pp.106.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Baum G, Fromm H. The 58-kilodalton calmodulin-binding glutamate decarboxylase is a ubiquitous protein in petunia organs and its expression is developmentally regulated. Plant Physiol. 1994;106:1381–1387. doi: 10.1104/pp.106.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholewa E, Cholewinski AJ, Shelp BJ, Snedden WA, Bown AW. Cold stimulated γ-aminobutyric acid synthesis is mediated by an increase in cytosolic Ca2+, not by an increase in cytosolic H+ Can J Bot. 1997;75:375–382. [Google Scholar]
- Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM (1998) Arabidopsis gls mutants and distinct Fd-GOGAT genes; implications for photorespiration and primary nitrogen assimilation. Plant Cell 10: 741–752 [DOI] [PMC free article] [PubMed]
- Davies DD (1980) Anaerobic metabolism and the production of organic acids. In Davies, DD, ed, The Biochemistry of Plants, Vol 2. Academic Press, New York, pp 581–611
- Erlander MG, Tobin AJ. The structural and functional heterogeneity of glutamatic acid decarboxylase: a review. Neurochem Res. 1991;16:215–226. doi: 10.1007/BF00966084. [DOI] [PubMed] [Google Scholar]
- Gallego PP, Whotton L, Picton S, Grierson D, Gray JE. A role for glutamate decarboxylase during tomato ripening: the characterisation of a cDNA encoding a putative glutamate decarboxylase with a calmodulin-binding site. Plant Mol Biol. 1995;27:1143–1151. doi: 10.1007/BF00020887. [DOI] [PubMed] [Google Scholar]
- Ito T, Hirano M, Akama K, Shimura Y, Okada K. Touch-inducible genes for calmodulin and a calmodulin-related protein are located in tandem on a chromosome of Arabidopsis thaliana. Plant Cell Physiol. 1995;36:1369–1373. [PubMed] [Google Scholar]
- Johnson BS, Singh NK, Cherry JH, Locy RD. Purification and characterization of glutamate decarboxylase from cowpea. Phytochemistry. 1997;46:39–44. [Google Scholar]
- Kishinami I, Ojima K. Accumulation of γ-aminobutyric acid due to adding ammonium or glutamine to rice cells. Plant Cell Physiol. 1980;21:581–589. [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature. 1991;352:524–526. doi: 10.1038/352524a0. [DOI] [PubMed] [Google Scholar]
- Knight MR, Smith SM, Trewavas AJ. Wind-induced plant motion immediately increases cytosolic calcium. Proc Natl Acad Sci USA. 1992;89:4967–4971. doi: 10.1073/pnas.89.11.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling V, Snedden WA, Shelp BJ, Assmann SA. Analysis of a soluble calmodulin binding protein from fava bean roots: identification of glutamate decarboxylase as a calmodulin-activated enzyme. Plant Cell. 1994;6:1135–1143. doi: 10.1105/tpc.6.8.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell MAK, Haas SZ, Biebert LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Mayer RR, Cherry JH, Rhodes D. Effects of heat shock on amino acid metabolism of cowpea cells. Plant Physiol. 1990;94:796–810. doi: 10.1104/pp.94.2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Oliveira R, Oliveira IC, Coruzzi GM. Arabidopsis mutant analysis and regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci USA. 1996;93:4718–4723. doi: 10.1073/pnas.93.10.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman TK, Goodman HM. The glutamine synthetase gene family of Arabidopsis thaliana: light-regulation and differential expression in leaves, roots, and seeds. Mol Gen Genet. 1991;230:145–154. doi: 10.1007/BF00290662. [DOI] [PubMed] [Google Scholar]
- Price AH, Taylor A, Ripley SJ, Griffiths A, Trewavas AJ, Knight MR. Oxidative signals in tobacco increase cytosolic calcium. Plant Cell. 1994;6:1301–1310. doi: 10.1105/tpc.6.9.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramputh A-I, Bown AW. Rapid γ-aminobutyric acid synthesis and the inhibition of the growth and development of oblique-banded leaf-roller larvae. Plant Physiol. 1996;111:1349–1354. doi: 10.1104/pp.111.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SA, Slade AP, Fox GG, Phillips R, Ratcliffe G, Stewert GR. The role of glutamate dehydrogenase in plant nitrogen metabolism. Plant Physiol. 1991;95:509–516. doi: 10.1104/pp.95.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayan V, Nair PM. Purification and characterization of glutamate decarboxylase from Solanum tuberosum. Eur J Biochem. 1985;150:53–60. doi: 10.1111/j.1432-1033.1985.tb08987.x. [DOI] [PubMed] [Google Scholar]
- Satyanarayan V, Nair PM. Metabolism, enzymology and possible roles of 4-amino butyrate in higher plants. Phytochemistry. 1990;29:367–375. [Google Scholar]
- Schultz C, Coruzzi GM. The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments. Plant J. 1995;7:61–75. doi: 10.1046/j.1365-313x.1995.07010061.x. [DOI] [PubMed] [Google Scholar]
- Selman IW, Cooper P. Changes in the free amino compounds in young tomato plants in light and darkness with particular reference to γ-aminobutyric acid. Ann Bot. 1978;42:627–636. [Google Scholar]
- Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J. Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell. 1994;6:1553–1565. doi: 10.1105/tpc.6.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DK, Kassam T, Singh B, Elliott S. Escherichia coli has two homologous glutamate deyhdrogenase genes that map to distinct loci. J Bacteriol. 1992;174:5820–5826. doi: 10.1128/jb.174.18.5820-5826.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Arazi T, Fromm H, Shelp BJ. Calcium/calmodulin activation of soybean glutamate decarboxylase. Plant Physiol. 1995;108:543–549. doi: 10.1104/pp.108.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Chung I, Pauls RH, Bown AW. Proton/l-glutamate symport and the regulation of intracellular pH in isolated mesophyll cells. Plant Physiol. 1992;99:665–671. doi: 10.1104/pp.99.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Koutsia N, Baum G, Fromm H. Activation of a petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. J Biol Chem. 1996;271:4148–4153. doi: 10.1074/jbc.271.8.4148. [DOI] [PubMed] [Google Scholar]
- Streeter JG, Thompson JF. In vivo and in vitro studies on γ-aminobutyric acid metabolism with the radish plant (Raphanus sativus) Plant Physiol. 1972;49:572–584. doi: 10.1104/pp.49.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanase S, Kojima H, Morino Y. Pyridoxal 5′-phosphate binding site of pig heart alanine aminotransferase. Biochemistry. 1979;18:3002–3007. doi: 10.1021/bi00581a015. [DOI] [PubMed] [Google Scholar]
- Tuin LG, Shelp AW. In situ [14C] metabolism by developing soybean cotyledons. I. Metabolic routes. J Plant Physiol. 1994;143:1–7. [Google Scholar]
- Turano FJ, Jordan RL, Matthews BF. Immunological characterization of in vitro forms of homoserine dehydrogenase from carrot suspension cultures. Plant Physiol. 1990;92:395–400. doi: 10.1104/pp.92.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano FJ, Thakkar SS, Fang T, Weisemann JM. Characterization and expression of NAD(H) dependent glutamate dehydrogenase genes in Arabidopsis thaliana. Plant Physiol. 1997;113:1329–1341. doi: 10.1104/pp.113.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano FJ, Weisemann JM, Matthews BF. Identification and expression of a cDNA clone encoding aspartate aminotransferase in carrot. Plant Physiol. 1992;100:374–381. doi: 10.1104/pp.100.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace W, Secor J, Schrader E. Rapid accumulation of γ-aminobutyric acid and alanine in soybean leaves in response to an abrupt transfer to lower temperature, darkness or mechanical manipulation. Plant Physiol. 1984;75:170–175. doi: 10.1104/pp.75.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie SE, Roper JM, Smith AG, Warren MJ. Isolation, characterization and expression of a cDNA encoding plastid aspartate aminotransferase from Arabidopsis thaliana. Plant Mol Biol. 1995;27:1227–1233. doi: 10.1007/BF00020897. [DOI] [PubMed] [Google Scholar]