Abstract
Spinocerebellar ataxia type 2 (SCA2) is an autosomal dominant disorder caused by the expansion of a CAG tract in the ATXN2 gene. The SCA2 phenotype is characterized by cerebellar ataxia, neuropathy and slow saccades. SCA2 foreshortens life span and is currently without symptomatic or disease-modifying treatments. Identifying function-specific therapeutics for SCA2 is problematic due to the limited knowledge of ATXN2 function. As SCA2 is likely caused by a gain-of-toxic or gain-of-normal function like other polyglutamine disorders, targeting ATXN2 expression may represent a valid therapeutic approach. This study characterized aspects of ATXN2 expression control using an ATXN2 promoter-luciferase (luc) reporter construct. We verified the fidelity of construct expression by generating transgenic mice expressing the reporter construct. High reporter expression was seen in the cerebellum and olfactory bulb in vivo but there was relatively low expression in other tissues, similar to the expression of endogenous ataxin-2. We verified the second of two possible start codons as the functional start codon in ATXN2. By evaluating deletions in the ATXN2 promoter, we identified an E-twenty six (ETS)-binding site required for ATXN2 expression. We verified that endogenous ETS1 interacted with the ATXN2 promoter by an electromobility supershift assay and chromatin immunoprecipitation polymerase chain reaction. ETS1 overexpression increased ATXN2-luc (ATXN2-luciferase) as well as endogenous ATXN2 expression. Deletion of the putative ETS1-binding site abrogated the effects on the expression of ATXN2-luc. A dominant negative ETS1 and an ETS1 short-hairpin RNA both reduced ATXN2-luc expression. Our study broadens the understanding on the transcriptional control of ATXN2 and reveals specific regulatory features of the ATXN2 promoter that can be exploited therapeutically.
INTRODUCTION
Spinocerebellar ataxia type 2 (SCA2) is an autosomal dominant neurodegenerative disease characterized by progressive functional and cell loss of neurons in the cerebellum, brain stem and spinal cord. The cause of SCA2 is CAG expansion in the ATXN2 gene resulting in polyglutamine (polyQ) expansion in the ataxin-2 protein. Patients with SCA2 are characterized by progressive cerebellar ataxia, slow saccadic eye movements and other neurologic features such as neuropathy (1). Moderate CAG expansion in the ATXN2 gene is also associated with parkinsonism or amyotrophic lateral sclerosis (ALS) indistinguishable from the idiopathic forms of these diseases (2–6).
The pathogenic functions of polyQ disease proteins that occur with polyQ expansion may be attributed to the gain of toxicity associated with the development of intranuclear inclusion bodies or with soluble toxic oligomers (7). While SCA2 patient brains are characterized by loss of Purkinje cells, SCA2 Purkinje cells lack inclusion bodies indicating polyQ-expanded ataxin-2 may cause toxicity that is unrelated to inclusion body formation (8). Functions gained in polyQ-expanded ataxin-2 may include anomalous accumulation in Golgi bodies (9), gain-of-normal functions (10) and sequestering of transcription factors (TFs) and glyceraldehyde-3-phosphate dehydrogenase like for other polyQ proteins (11–13). Some normal functions of ataxin-2 have been characterized. Ataxin-2 is present in stress granules and P-bodies suggesting functions in sequestering mRNAs and protein translation regulation during stress (14). Ataxin-2 overexpression interfered with the P-body assembly, while underexpression interfered with stress granule assembly (14). Interactions with polyA-binding protein 1, the RNA splicing factor A2BP1/Fox1 and polyribosomes further support roles for ataxin-2 in RNA metabolism (15–17). Ataxin-2 is a regulator of EGF receptor internalization and signaling by the way of its interactions with SRC kinase and the endocytic protein CIN85 (18). Ataxin-2 also interacts with the ALS-related protein TDP-43 in an RNA-dependent manner and familial and sporadic ALS associates with the occurrence of long normal CAG repeat expansion ATXN2 (5,6).
Mutant ataxin-2 is also an effector of Ca2+ efflux from the endoplasmic reticulum by binding inositol 1,4,5-triphosphate receptor (InsP3R). Greater Ca2+ efflux was observed in cultures of mouse primary neurons possessing ataxin-2 Q58 than Q22 and the SCA2-related phenotype of ataxin-2 Q58 transgenic mice was resolved with dantrolene, an inhibitor of the ryanodine receptor (19). Previous work by our group also showed that long normal CAG repeats in the CACNA1A calcium channel gene lowered SCA2 age of onset (20). Collectively, these findings suggest a functional connection between ataxin-2 and calcium metabolism that with further characterization might be exploited therapeutically for SCA2.
The concept of developing a treatment for SCA2 by reducing total ATXN2 expression was the primary motivation for this study. Studies in rodents on SCA2 and other polyQ disease genes provided support for this approach. For Huntington's disease, SCA1 and SCA3, reductions in the mutant proteins for these diseases (huntingtin, ataxin-1 and ataxin-3, respectively) resulted in phenotype reversals after phenotype onset in inducible mice (21–23). In a rat model of Machado-Joseph disease, silencing of both the wild-type and mutant ataxin-3 genes improved neuropathology, while allele-specific silencing of just the wild-type ataxin-3 gene did not worsen pathology (24). We also noted no neuropathology in ATXN2 knockout mice (25). The regulatory features of the ATXN2 gene remain poorly described. In one study of the ATXN2 promoter, various regulatory elements including predictions for Sp1-binding sites were identified in the exon 1 region as well as a promoter-localized CpG island (26). Another study demonstrated that the KRAB-containing zinc-finger transcriptional regulator ZBRK1 forms a complex with ataxin-2 that transactivates ATXN2 (27).
In the present study, we further characterize regulatory features of the ATXN2 gene by introducing mutational changes in various ATXN2 promoter-luciferase (luc)-ATXN2-3′-UTR plasmid constructs, with the purpose to identify regulatory features that might be exploited therapeutically. Deletions in the upstream region and 5′-UTR resulted in the identification of specific regions in ATXN2 for expression control. Within the 5′-UTR, we identified a 14 bp region required for ATXN2 expression that spans an 8 bp sequence that has 100% consensus with the E-twenty six (ETS) family TF-binding site. We found that this region directed the regulation of ATXN2 expression by the TF ETS1. We also characterized the effects of CAG repeat expansion on ATXN2 expression and the ataxin-2 expression in ATXN2-luc (ATXN2-luciferase) transgenic mice.
RESULTS
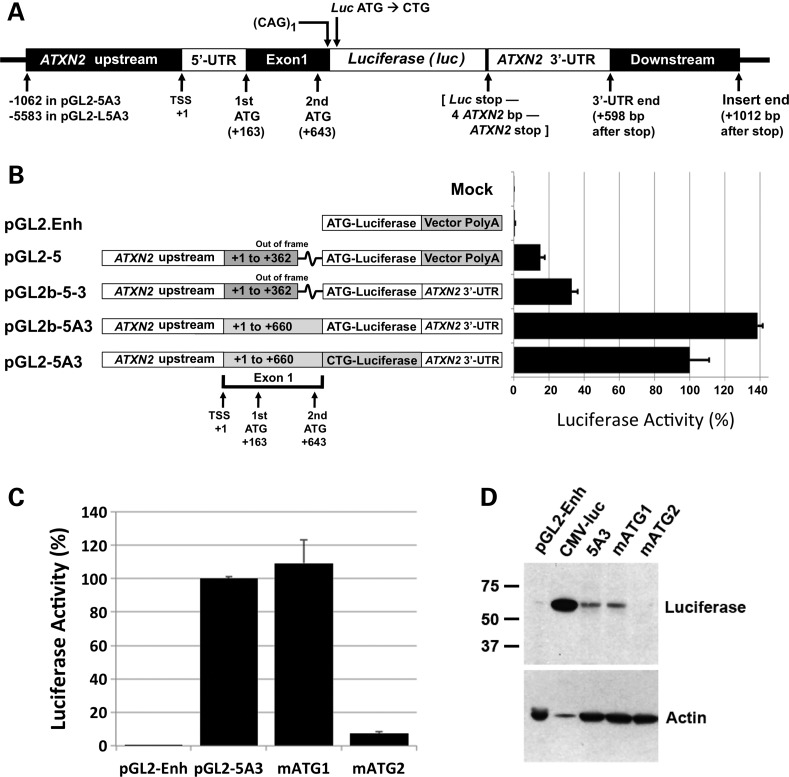
Luciferase expression from pGL2-5A3
Our approach to cloning pGL2-5A3 (Fig. 1) involved six steps resulting in various clones with different promoters and polyA features. We conducted luciferase assays comparing among the different plasmids made in this cloning process and pGL2-5A3, providing insight on the behavior of ataxin-2-luciferase expression from this plasmid (Fig. 1B). Three primary observations were made: first, luciferase expression increased by 2-fold when the pGL2-Enhancer vector provided polyA and the SV40 enhancer sequence was replaced by the ATXN2-3′-UTR. This observation indicated that the ATXN2-3′-UTR imparts considerable RNA stability, since the vector-encoded SV40 polyA enhancer was designed to increase expression. Second, adding an additional 298 bp of ATXN2 exon 1 including the second ATG while simultaneously placing the ATXN2 exon 1 fragment in frame with luciferase, markedly improved expression increasing luciferase activity by 4-fold compared with lack of this fragment. This indicated that the ATXN2-luc transcript preferentially utilizes its own ATG over the luciferase ATG. Third, luciferase expression was reduced by 25% when the vector-encoded ATG was mutated to CTG creating a fusion of the ataxin-2 exon 1-encoded protein fragment with luciferase.
Figure 1.
Cloning of pGL2-5A3 and evaluation of start codons. (A) Features of pGL2-5A3, the principal clone from which most other constructs in the study were made. pGL2-5A3 includes −1062 to +660 of the ATXN2 gene ending on the first CAG of the CAG tract followed by an XhoI restriction site. The luciferase start codon was mutated to CTG, encoding leucine, so to fuse the ataxin-2 fragment with luciferase. Downstream of the luciferase stop codon is an AgeI restriction site followed by 1019 bp of ATXN2 3′-UTR and downstream sequence (+4098 to +5116). ATXN2 bp positions are relative to the TSS except as indicated in the figure. (B) Representation of the cloning process from top to bottom, comparing pGL2-5A3 to three of its ‘ancestral’ plasmids and the vector from which they were made, pGL2.Enhancer (pGL2.Enh). Colored boxes indicate differences among the cloned regions. Note that addition of the ‘A’ fragment of ATXN2 exon 1 in pGL2b-5A3 placed the longer exon 1 fragment in frame with luciferase. On the right are the results of assays showing how expression of luciferase differed among these plasmids. Shown are the mean ± SD of RLUfirefly/RLURenilla values from three independent transfections each read in triplicate, expressed as a percentage. (C) Relative use of the two start codons in ATXN2. Luciferase assays were conducted to compare expression from pGL2-5A3 with two plasmids where either of the two start codons were mutated to CTG, pGL2-5A3(mATG1) and pGL2-5A3(mATG2). Mutation of the second start codon resulted in nearly complete loss of ATXN2-luc expression, while mutation of the first start codon resulted in no expression change. The data shown are for HEK293 cells. The experiment conducted in SH-SY5Y cells showed a nearly identical result. (D) Western blot analysis with anti-luciferase antibody of proteins expressed from pGL2-5A3, pGL2-5A3(mATG1) and pGL2-5A3(mATG2) in HEK293 revealed ATXN2-luc proteins of the size predicted for the usage of the second start codon. Note that lane 2 was purposefully underloaded.
The second ATG in ATXN2 is the ATXN2 start codon
In order to understand the relative usage of the two in-frame start codons located in ATXN2 exon 1, we generated two additional clones of pGL2-5A3, each with one of the codons changed to CTG. Luciferase assays and immunoblotting analysis showed that expression was abolished upon the mutation of the second start codon but not the first (Fig. 1C and D). Thus, the second ATG in the reading frame represents the ATXN2 start codon. We have never observed by western blotting evidence of two ataxin-2 bands or two ataxin-2-luciferase bands indicative of the utilization of two start codons.
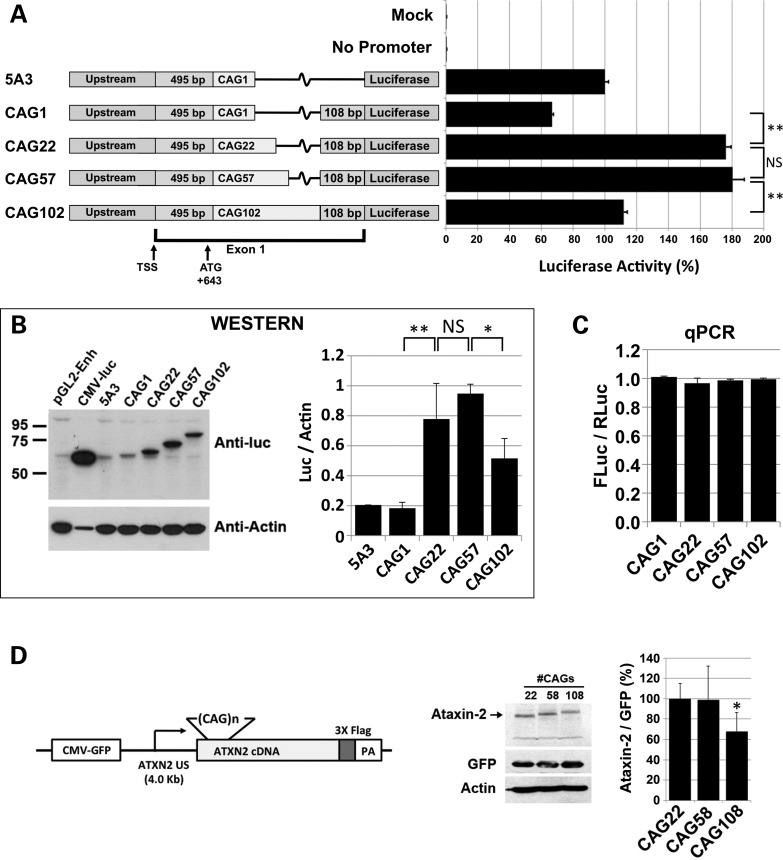
Effect of CAG expansion on ATXN2-luc expression
We evaluated the effect of CAG expansion on ATXN2 expression, with an experimental design to distinguish between CAG length effects on the ATXN2 message and the ataxin-2 protein. We did this because we predicted that the CAG repeat length controls ATXN2 expression, since the CAG repeat is located within a predicted CpG island (26) and because we now localized the start codon to just 15 bp upstream of the repeat (Fig. 1C and D). We observed that addition of a 108 bp fragment just downstream of the single CAG in our plasmid pGL2-5A3 caused a slight decrease in ATXN2-luc expression in luciferase assays that, however, was not evident by western blotting (Fig. 2A and B). The 108 bp fragment was added because it harbors CpG island characteristics and therefore may support transcription (26). Increasing the CAG repeat from 1 to 22 or 57 CAGs significantly increased the expression of ATXN2-luc in luciferase assays (2.7-fold) and as detected by immunoblotting (4.1-fold; Fig. 2A and B). Further increase in the CAG tract to 102 CAGs resulted in a significant reduction in ATXN2-luc expression in luciferase assays and immunoblots (Fig. 2A and B). The length of the CAG tract caused no significant differences in ATXN2-luc expression as determined by quantitative polymerase chain reaction (qPCR; Fig. 2C). To support these observations, we conducted two additional western blot experiments with an entirely different set of plasmids with full-length ATXN2 and varying CAG repeat lengths. The additional set of plasmids included CAG lengths 22, 58 and 108, had 4.0 kb of ATXN2 upstream sequence/native promoter and included a control cytomegalovirus-green fluorescent protein (CMV-GFP) gene encoded on the same plasmids (Fig. 2D). Western blotting demonstrated significantly reduced ataxin-2 expression for CAG108 only (Fig. 2D) and the result was nearly identical to that shown in Fig. 2B.
Figure 2.
Effect of CAG length on ATXN2 expression. (A) Luciferase assays using ATXN2-luc constructs with increasing CAG repeat lengths revealed highest expression for CAG22 and CAG57 and lower expression for longer CAG lengths. All constructs were based on pGL2-5A3. Identical results were obtained when the experiment was repeated using constructs modified to include TK-Renilla cassettes on the same plasmids. (B) Western blot analysis of the constructs in (A) revealed a similar pattern of protein expression to that determined by luciferase assays in (A). Note that lane 2 was purposefully underloaded. (C) qPCR analysis using constructs shown in (A) that were modified to include TK-Renilla cassettes on the same plasmids, showed no changes in the abundances of the ATXN2-luc transcript depending upon the CAG length. (D) Constructs expressing full-length ATXN2 with repeat lengths CAG22, CAG58 and CAG108 including 4 kb ATXN2 upstream sequence, analyzed by western blotting with anti-FLAG antibody. Like in (A) and (B), the longest repeat (CAG108) reduced ATXN2 expression. The asterisk indicates a significant difference for CAG108 versus either CAG22 or CAG58 (P < 0.05, Bonferroni post-test, n = 7 transfections). Expression was relative to GFP encoded on the same plasmid as indicated. Actin is also shown. HEK293 cells (A–C) and SH-SY5Y cells (D).
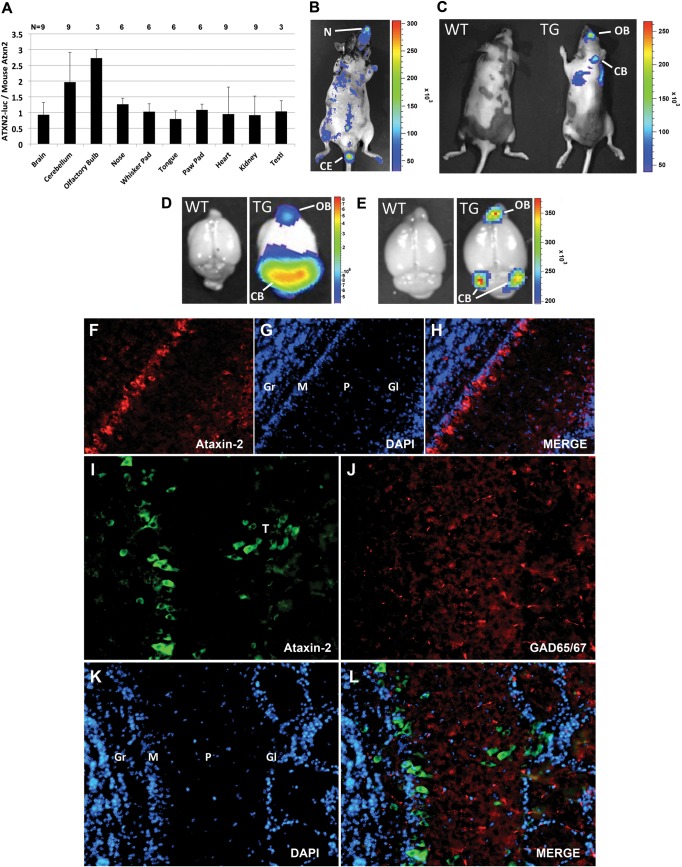
Exogenous ATXN2-luc parallels endogenous ATXN2 expression in vivo
We established two lines of transgenic mice carrying the ATXN2-luc transgene, designated L74 and L75. We initially used luciferase assays of excised brain tissues to verify that the ATXN2-luc transgene was expressed (data not shown). Both L74 and L75 had the highest expression of ATXN2-luc in the cerebellum while low expression was observed in other tissues, including the heart and total brain. Subsequently, we evaluated only line L75 with the highest ATXN2-luc expression. qPCR analysis of multiple mice showed uniform ATXN2-luc expression relative to mouse Atxn2 in several tissues, including the total brain, nose, whisker pad, tongue, paw pad, heart, kidney and testis, but high relative expression in the cerebellum and olfactory bulb (OB; Fig. 3A). in vivo imaging system (IVIS) imaging of luciferase in whole animals revealed high ATXN2-luc expression in the nose (Fig. 3B) which was regularly observed among mice, although by qPCR ATXN2-luc expression in the nose was not elevated (Fig. 3A). We also observed high expression in the cauda epididymis (Fig. 3B and Supplementary Material, Fig. S1). Additionally, we observed luciferase signals in whole animals consistent with expression in the cerebellum and OB that were absent in wild-type controls (Fig. 3C). Excised whole brains showed elevated expression in the OB and cerebellum that was more widespread in 2-month-old animals (Fig. 3D) compared with 7-month-old animals (Fig. 3E). We characterized the distribution of ATXN2 in the OB of wild-type C57Bl/6 mice by indirect immunofluorescent (IF) labeling to verify that ATXN2-luc expression in OB was consistent with endogenous ATXN2 expression. ATXN2 was strongly expressed in mitral cells and in tufted cells of the glomerular layer (Fig. 3F–L).
Figure 3.
ATXN2-luc expression in vivo was elevated in the cerebellum and OB compared with endogenous ATXN2. (A) For most tissues, the average expression of ATXN2-luc to endogenous mouse Atxn2 was 1:1 by qPCR. ATXN2-luc expression was high in some mice in the cerebellum and OB relative to mouse Atxn2 but the average ratio in the cerebellum was not significantly different from that for other tissues. qPCRs were run in triplicate and the reported values represent the means among mice ± SD. (B–E) Evaluation of ATXN2-luc expression on the IVIS. (B) ATXN2-luc expression in the nose (N) and cauda epithelium (CE) of a ATXN2-luc transgenic mouse. (C) Detection of ATXN2-luc in the OB and cerebellum (CB) in a 7-month-old ATXN2-luc transgenic mouse (TG) and lack of signal in a wild-type littermate (WT). (D) ATXN2-luc detected in the OB and CB of a 2-month-old ATXN2-luc transgenic mouse and lack of signal in a wild-type littermate. (E) ATXN2-luc detected in the OB and CB of a 7-month-old ATXN2-luc transgenic mouse and lack of signal in a wild-type littermate. (F–L) IF detection of endogenous Atxn2 in the OB of wild-type C57Bl/6 mice. (F) Ataxin-2 detected with ataxin-2 mAb in mitral cells and tufted cells in the glomerular layer (×10 magnification). (G) Same section as (F) stained with DAPI. (H) Merge of (F) and (G). (I) Ataxin-2 detected with polyclonal antibody 1080 in mitral cells and tufted cells in the glomerular layer (×20 magnification). (J) Same section as (I) stained with anti-GAD65/67. (K) Same section as (I) stained with DAPI. (L) Merge of (I)–(K). Note that no ataxin-2 was detected in GAD65/67-positive interneurons. Labels: T, tufted cells; Gr, granule layer; M, mitral layer; P, external plexiform layer; Gl, glomerular layer. Bars = 100 μm.
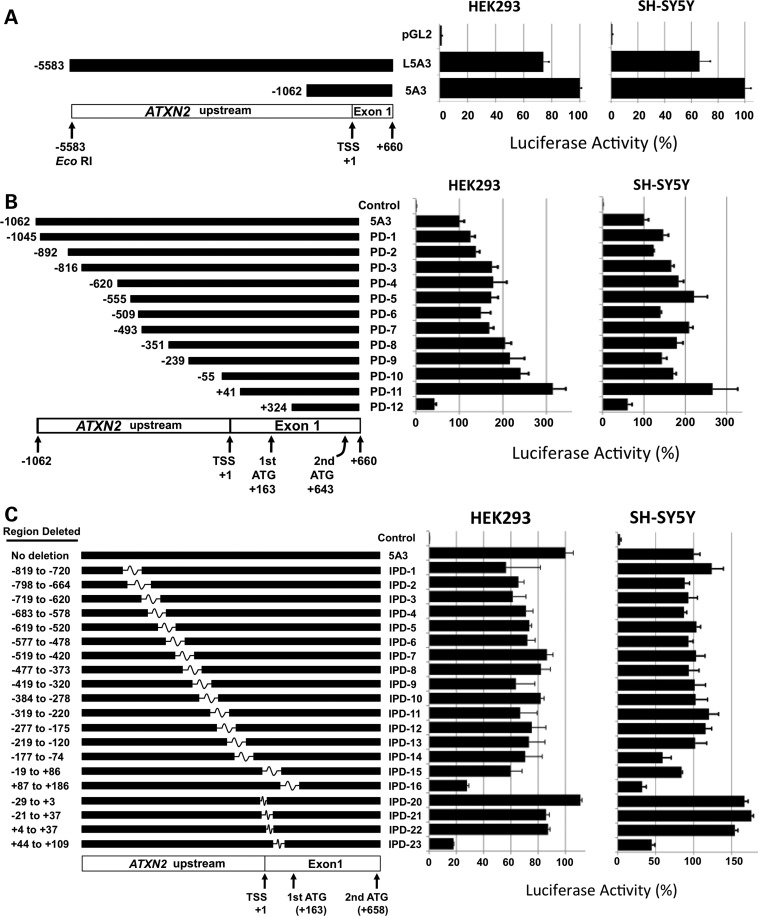
ATXN2 promoter deletion and ATXN2-luc expression
We performed luciferase assays to compare expression between the pGL2-5A3 plasmid containing ATXN2 upstream sequence including −1062 bp from the transcription start site (TSS, as defined for ENST00000377617 in the Ensembl Genome Browser, www.ensembl.org) with the longer pGL2-L5A3 plasmid containing ATXN2 upstream sequence including −5583 bp from the TSS. We observed that the average expression was reduced by 25 and 33% when the longer upstream sequence was included compared with the shorter one, in both HEK293 and SH-SY5Y cells, respectively (t-test 5A3 versus L5A3: P < 0.001 in HEK293 and P < 0.01 in SH-SY5Y; Fig. 4A). We then determined the expression of ATXN2-luc from 12 different pGL2-5A3 plasmids each with different deletions from the upstream end of the cloned ATXN2 promoter fragment [promoter deletions 1–12 (PD1–PD12)]. For deletions PD1–PD11, increasing deletion of the upstream end of the ATXN2 promoter was associated with increased ATXN2-luc expression in HEK293 and SH-SY5Y cells (Fig. 4B). Expression from the non-mutated pGL2-5A3 plasmid compared with that of any of PD1–PD11 was significantly different (t-test: 5A3 versus PD1, P < 0.001, HEK293 and SH-SY5Y; 5A3 versus PD2, P < 0.05, SH-SY5Y). The data suggested the existence of inhibitory promoter elements between −1062 and −816 and −55 and +41 that were progressively removed by increasing PD, whose regulatory proteins are expressed in both HEK293 and SH-SY5Y cells. The deletion of some regions resulted in no change in expression (between −816 and −493 in HEK293 cells and −1045 and −892 and −493 and −55 in SH-SY5Y cells), and one region resulted in significantly reduced expression when deleted (−555 and −493 in SH-SY5Y cells, P < 0.05). For the final deletion PD12, ATXN2-luc average expression was reduced to 57 and 39% of that for the non-deleted pGL2-5A3 control in HEK293 and SH-SY5Y cells, respectively.
Figure 4.
Effects of ATXN2 PDs on ATXN2-luc expression and identification of an ATXN2 5′-UTR region (+87 to +109) accounting for the majority of ATXN2 expression. (A and B) Effect of increasing ATXN2 PD on ATXN2-luc expression. (A) The long ATXN2 upstream sequence contained in plasmid pGL2-L5A3 reduced ATXN2-luc expression similarly in HEK293 and SH-SY5Y cells compared with the shorter upstream sequence in pGL2-5A3. (B) Further decreases in the ATXN2 promoter length in pGL2-5A3 correlated with increases in ATXN2-luc expression. A plateau where no successive increased expression was observed between −816 and −351 in HEK293 cells and −555 and −55 in SH-SY5Y cells. PD12 spans the smaller +87 to +109 5′-UTR region and resulted in expression reduction in both cell lines. (C) Effect of ATXN2 IPDs in pGL2-5A3 on ATXN2-luc expression. Only deletions IPD16 and IPD23 including the +87 to +109 5′-UTR region caused reduced expression in both HEK293 and SH-SY5Y cells compared with the pGL2-5A3 plasmid that contains no interstitial deletion. For each of (A)–(C), a graphic representation is provided (left) where the upstream/promoter sequence present is represented by a black bar and for (C) where the sequence that has been interstitially deleted is represented by a white box. Relative expression in HEK293 and SH-SY5Y cells for each deletion (right), controlled with SV40-Renilla luciferase. Nucleotides are numbered such that +1 corresponds to the transcriptional start site or first base in the 5′-UTR. Values are mean ± SD from three independent transfections each read in triplicate.
ATXN2 interstitial PD and ATXN2-luc expression
We determined the expression of ATXN2-luc from 16 different pGL2-5A3 plasmids each with different overlapping interstitial deletions in the ATXN2 promoter of ∼100 bp in length [interstitial PDs 1–16 (IPD1–IPD16)] and four smaller deletions (IPD20–IPD23; Fig. 4C). In HEK293 cells, all of the IPD1–IPD15 similarly reduced the average ATXN2-luc expression compared with the non-mutated 5A3 control plasmid in multiple independent experiments. We also observed similar expression among IPD1–IPD15 in SH-SY5Y cells except that unlike in HEK293 cells, the non-mutant 5A3 control plasmid expressed similar to constructs with IPD1–IPD15. The IPD16 and IPD20–IPD24 resulted in similar expression changes between HEK293 and SH-SY5Y where, in general, the IPD20–IPD22 resulted in no reduced (HEK293) or slightly elevated (SH-SY5Y) expression, and the IPD16 and IPD23 caused the lowest expression observed among all IPDs tested.
Luciferase assays of ATXN2-luc with the ETS1 site mutated or with ETS1 overexpressed demonstrated ETS1 supports ATXN2 expression
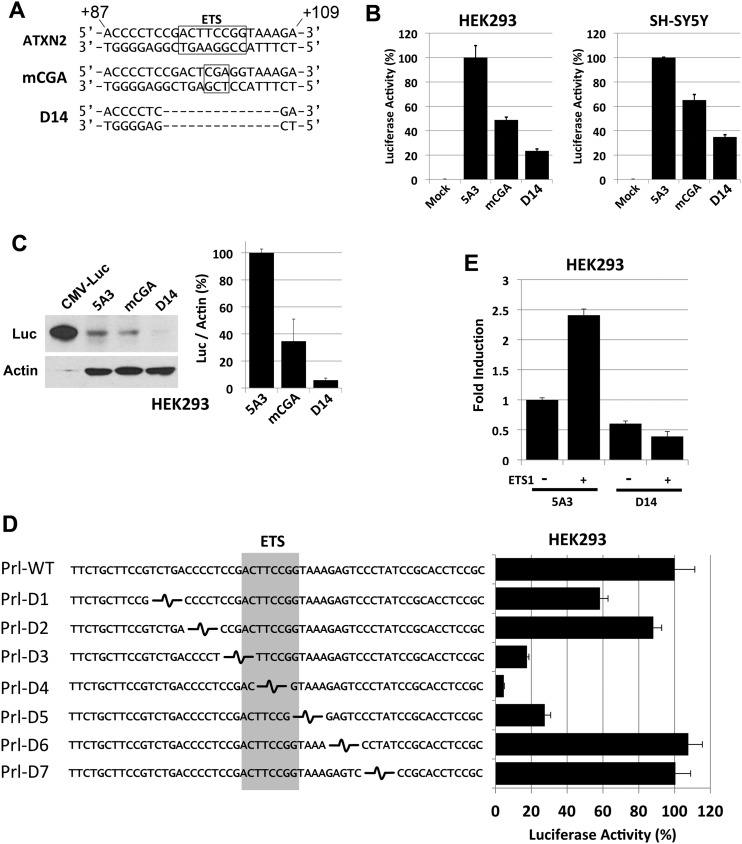
The similarly reduced ATXN2-luc expression observed when IPD16 or IPD23 were present (Fig. 4C) led us to closely evaluate the 23 bp region (+87 to +109) that is shared between these two deletions for TF-binding sites. TF prediction software revealed a consensus sequence for ETS TF binding on the ATXN2 negative strand between +96 and +103 of the ATXN2 gene relative to the TSS. The ETS element sequence in ATXN2 has 100% homology to the ETS element consensus sequence described in the majority of promoters, CCGGAAGT (28). We created two additional pGL2-5A3 construct modifications including a 14 bp deletion encompassing the ETS-binding site (including +94 to +107 sequence CGACTTCCGGTAAA in ATXN2) designated D14, and a 3 bp TCC→CGA mutation (+99 to +101 on the positive strand) in the core GGAA of the ETS element, designated mCGA, corresponding to GGA→TCG on the negative strand (Fig. 5A). The resulting mutated ETS element sequence of CCTCGAGT is predicted to have no affinity for binding by ETS TFs. The mCGA mutation resulted in 51 and 35% reduction in average ATXN2-luc expression compared with the non-mutated 5A3 control construct, while the D14 deletion reduced expression by 76 and 65% in HEK293 and SH-SY5Y cells, respectively (Fig. 5B). The expression of ATXN2-luc with either the mCGA or the D14 mutations were detectable by immunoblotting with anti-luciferase (Fig. 5C). We also made seven additional ATXN2 promoter-luciferase constructs with 5 bp tandem deletions to confirm that the deletion of the ETS element reduced expression greater than any same-sized deletions in the region upstream and downstream of the ETS element. A 5 bp deletion including the ETS element GGAA core (Prl-D4) reduced expression the most (by 95%), while neighboring 5 bp deletions compromising the ends of the ETS element (Prl-D3 and Prl-D5) reduced expression by ∼80%, and other deletions farther away from the ETS element had minimal effect (Fig. 5D). This demonstrated that the ETS element is required for ATXN2 expression. To support this conclusion, we determined the effect of ETS1 overexpression on ATXN2-luc in luciferase assays. Overexpression of ETS1 resulted in a statistically significant 2.4-fold increase in ATXN2-luc expression in luciferase assays, but overexpression of ETS1 had no effect on inducing the expression of ATXN2-luc when the D14 deletion was included (Fig. 5E).
Figure 5.
A 14 bp region within the +87 to +109 ATXN2 5′-UTR region contains an ETS TF-binding site and accounts for the majority of ATXN2 expression. (A) Both strands of the ATXN2 sequence from +87 to +109 are shown and the position of an ETS TF-binding site oriented from +103→+96 on the ATXN2 minus strand is indicated (box). mCGA represents a TCC→CGA mutation with the ETS TF-binding site made in the ATXN2-luc expression plasmid pGL2-5A3. This mutation alters the ETS TF consensus sequence core on the ATXN2 minus strand from GGAA→TCGA to abrogate binding by ETS TFs. D14 represents a 14 bp deletion spanning the ETS TF-binding site in pGL2-5A3. (B) Luciferase assays comparing no mutation to the mCGA and D14 mutations in HEK293 and SH-SY5Y cells. The mCGA mutation resulted in 51 and 35% reduction in the average ATXN2-luc expression in HEK293 and SH-SY5Y cells, respectively, relative to no mutation. The D14 mutation resulted in 76 and 65% reduction in the average ATXN2-luc expression in HEK293 and SH-SY5Y cells, respectively, relative to no mutation. Values are means and SD from three independent transfections each read in triplicate and expression was controlled with SV40-Renilla luciferase. (C) Western blot showing that the mCGA and D14 mutations in pGL2-5A3 reduced ATXN2-luc expression in HEK293 cells compared with no mutation. The D14 mutation reduced ATXN2-luc to background. (D) Effect of seven tandem 5 bp deletions in the ATXN2 promoter on ATXN2-luc expression. Deletions were made by ligating annealed oligonucleotides spanning 58 bp of the ATXN2 promoter into a rat prolactin-luc minimal promoter construct. Deletions overlapping the ETS element reduced expression while those flanking the ETS element had no effect or minimal effect. (E) ETS1 activation of ATXN2 depends on the ETS-binding site. Overexpression of ETS1 increased ATXN2-luc. Inclusion of the D14 deletion in ATXN2-luc blocked the ability for ETS1 to increase ATXN2-luc expression. Values are means and SD from three independent transfections each read in triplicate and expression was controlled with TK-Renilla luciferase.
Electromobility supershift assays demonstrated that ETS1 bound an ATXN2 probe including the ETS1-binding site
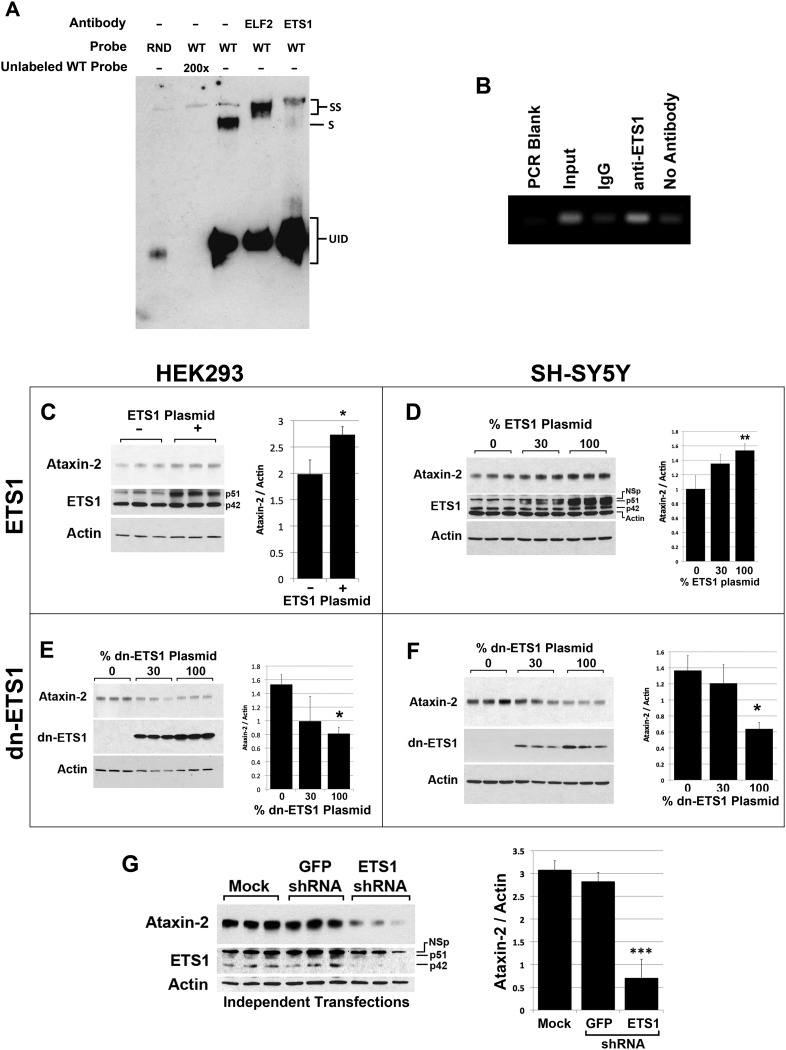
We conducted electromobility shift assays (EMSAs) with antibody supershift to test that the D14 region of the ATXN2 gene interacted with the ETS1 TF and other ETS TFs. We utilized a 21 bp probe containing ATXN2 sequence encompassing the D14 region. Supershifted bands were observed for ETS1 and ELF2 from SH-SY5Y nuclear lysate that were not shifted when antibodies to these TFs were excluded (Fig. 6A). Additional EMSAs are provided as Supplementary Material. These supplemental figures show the following: positive supershift for ETS1, ELF2 and GABPα but not for ELF1 or STAT3 (the latter used as a negative control) in both HEK293 and SH-SY5Y cell nuclear lysates (Supplementary Material, Fig. S2A and B); greater probe shift for the wild-type ATXN2 probe versus mutant mCGA ATXN2 probe and lack of shift bands when SH-SY5Y protein lysate was excluded (Supplementary Material, Fig. S2C).
Figure 6.
ETS1 interacts with the ETS1 site in ATXN2 and regulates ATXN2 expression. (A) EMSA verified ETS1 binds a probe containing the ETS1 as well as ELF2 in SH-SY5Y nuclear lysate included as a positive control. EMSA supershift assays using HEK293 and SH-SY5Y nuclear lysates with various electrophoresis times and additional controls (including ELF2) are shown in Supplementary Material, Fig. S2. (B) ChIP PCR analysis evaluated by agarose gel electrophoresis demonstrated endogenous ETS1 in SH-SY5Y cells bound the ETS1 site in ATXN2, as indicated by pull-down of the complex with anti-ETS1 but not rabbit IgG. When evaluated by qPCR binding ratios ranged from 7 to 30. Western blots showing that the overexpression of the 51 kDa isoform of ETS1 in HEK293 (C) and SH-SY5Y cells (D) increased Ataxin-2 expression. A non-specific band (NSp) was sometimes observed. Experiments were loaded in triplicate. Western blots showing that the overexpression of dn ETS1 in HEK293 (E) and SH-SY5Y cells (F) reduced Ataxin-2 expression. Experiments were loaded in triplicate. (G) Western blot showing that the underexpression of ETS1 by transfection of an ETS1 shRNA resulted in reduced Ataxin-2 expression in SH-SY5Y cells. No change for Ataxin-2 or ETS1 expression was observed when a control GFP shRNA was used. Each lane represents an independent transfection. Significance was determined by ANOVA Bonferroni post-tests; *P < 0.05, **P < 0.01 and ***P < 0.001. Effect of ETS1 overexpression on endogenous ataxin-2 expression in HEK293 cells (H–K) and SH-SY5Y cells (L) determined by IF labeling: cells overexpressing the 51 kDa isoform of ETS1 had increased ataxin-2 expression localized to a perinuclear space (H). Some cells overexpressing ETS1 showed diffusely distributed ataxin-2 expression (I) or aggregations of ataxin-2 (J). HEK293 cells not transfected with ETS1 showed low and diffuse expression of both ETS1 and ataxin-2 (K). ×30 magnification; bar, 100 μm; equal exposure settings were used. (L) Nearly all ETS1-transfected SH-SY5Y cells overexpressing ETS1 had detectable ataxin-2 expression. DAPI was used to identify untransfected cells. ×20 magnification; bar, 100 μm.
Chromatin immunoprecipitation PCR assays demonstrated endogenous ETS1 was bound to the native ATXN2 promoter
To demonstrate that endogenous ETS1 interacted with the native ATXN2 promoter, we conducted chromatin immunoprecipitation (ChIP) PCR analysis. ChIP PCR experiments using SH-SY5Y cell lysates demonstrated increased ATXN2 amplicon abundance for ChIPs conducted with anti-ETS1 versus rabbit IgG control or no antibody control after two or three protein-A-agarose column washes, when either examined by agarose gel electrophoresis (Fig. 6B) or quantitatively by qPCR. qPCR demonstrated binding ratios of 7–30. The ChIP experiment was repeated three times using SH-SY5Y cells with the same result.
Changes of ETS1 expression affect endogenous ataxin-2 abundance
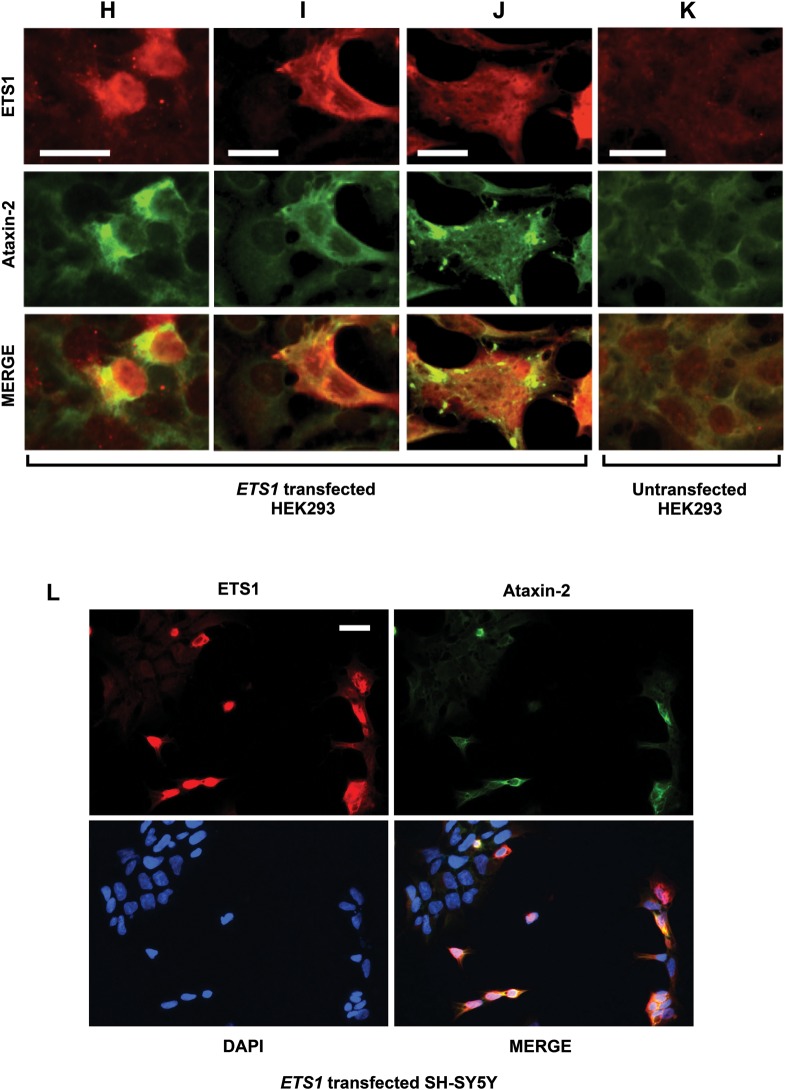
To demonstrate that ETS1 regulates ATXN2 expression, we overexpressed full-length ETS1, a dominant negative (dn)-ETS1 fragment or an ETS1 short-hairpin RNA (shRNA) and assessed ataxin-2 abundance by western blotting. Overexpression of ETS1 resulted in significant increases in ataxin-2 abundance in both HEK293 and SH-SY5Y cells (Fig. 6C and D), while that of dn ETS1 resulted in a significant reduction in ataxin-2 expression in these cell lines (Fig. 6E and F). Additionally, the overexpression of an shRNA plasmid targeting the expression of ETS1 resulted in significantly lower ataxin-2 expression, while an shRNA plasmid against green fluorescent protein (GFP) had no effect (Fig. 6G). IF labeling demonstrated that ETS1 overexpression increased endogenous ataxin-2 expression and localization. We observed a subset of cells overexpressing ETS1 with increased endogenous ataxin-2 expression with ataxin-2 often localizing to a perinuclear region or forming aggregate bodies (Fig. 6H–K). IF labeling also increased ataxin-2 abundance in nearly all SH-SY5Y cells overexpressing ETS1 (Fig. 6L). Note that, we were unable to demonstrate an effect of mutant ATXN2 CAG repeats on ETS1 ability to transactivate ATXN2 (not shown).
DISCUSSION
A promising approach for developing therapies for polyQ disorders is to lower the disease gene expression. The rationale for this is that mutations in polyQ diseases primarily result in a gains of normal or toxic functions, although there can also be partial functional losses (29). Other therapeutic modalities for polyQ diseases are problematic due to lack of knowledge on gene functions that are specifically related to disease onset or progression. To support our efforts to develop therapeutics for SCA2, we have rigorously characterized the control of ATXN2 expression, including validation of our study construct in vivo. We identified features of ATXN2 expression control that might be exploited therapeutically and developed hypotheses on how CAG length controls expression with relevance to other polyQ diseases.
We discovered evidence for negative regulatory elements distributed throughout the ATXN2 gene upstream region that may support expression control and tissue-specific expression. Promoter-luciferase assay studies showed that ATXN2-luc constructs with longer upstream sequences generally had lower ATXN2-luc expression than those with shorter upstream sequences. This is likely due to the presence of multiple inhibitory elements throughout the ATXN2 promoter. This conclusion is consistent with observations made by Aguiar et al. (2,26) who showed that an ATXN2-luc construct possessing ATXN2 upstream sequence −436 to +362 expressed 1.6 ± 0.1 times higher than one possessing a longer ATXN2 upstream sequence of −2912 to +362 (mean and SD for F9 cells and SH-SY5Y cells). These findings suggest the existence of inhibitory elements distributed throughout the ATXN2 promoter that may be critical for fine-tuning expression in a developmental or tissue-specific manner.
We created ATXN2-luc transgenic mice to verify that the region of the ATXN2 promoter and 3′-UTR in our study replicated expression similar to the endogenous Atxn2 gene. Queries of the Allen Brain Atlas (Allen Institute for Brain Science) (30,31) showed that Atxn2 is highly expressed in Purkinje cells, hippocampus (CA1–3 and dentate gyrus) and the mitral layer of the OB compared with other mouse brain tissues. Analysis of ATXN2-luc expression in ATXN2-luc transgenic mice by qPCR and IVIS imaging demonstrated consistencies with the Allen Brain Atlas in situ hybridization data. For most tissues, ATXN2-luc expression was 1:1 versus endogenous Atxn2 by qPCR, and ATXN2-luc was elevated in the cerebellum for a subset of mice and in the OB, but on average ATXN2-luc in the cerebellum was not statistically different from 1:1 versus Atxn2. We next preformed immunohistochemical (IHC) staining of the OB sections from wild-type mice to confirm the Allen Brain Atlas data, showing that ATXN2 is expressed high in mitral cells, and to support biological relevance for ATXN2-luc expression in the OB. IHC confirmed highest expression of ataxin-2 in mitral cells and equally high expression in tufted cells. Collectively, these data suggest that ATXN2-luc in transgenic mice is expressed much like endogenous mouse ATXN2, with the caveat that differences in cerebellar ATXN2-luc expression among mice might reflect reduced stability of expression control when the promoter is truncated. IVIS imaging also identified ATXN2 expression outside the central nervous system including high expression in the nose and cauda epididymis. Previously, ATXN2 knockout mice were characterized as having reduced fertility (32) that may relate to loss of ATXN2 expression in developing and mature sperm of the cauda epididymis.
Strong expression of ATXN2 in the OB and nose are interesting in light of marked obesity in ATXN2 knockout mice (25,32). In unpublished experiments, we have now shown that obesity is the cause of severe hyperphagia that is even present in heterozygote animals. It is possible that changes in olfaction caused by ATXN2 loss in mitral cells of the OB and other cells involved in olfaction may decrease reward signals mediated by smell. This hypothesis is supported by a central role of dAtx2 in olfactory habituation in the fly (33) and the finding of impaired olfaction in SCA2 patients (34).
The impact of CAG repeat expansion on gene expression and protein abundance is not completely understood for other polyQ disease genes. For ATXN2, features that might affect expression include (i) the location of the ATXN2 CAG repeat within a CpG island in exon 1 (26,35) raising the possibility that CAG expansion can influence transcription by altering the proximity of TFs acting on either side of the repeat and (ii) the close proximity of the CAG repeat to the start codon potentially influencing ATXN2 translation. The increase in the CAG repeat length from 1 CAG to 22 CAGs increased the expression of ataxin-2-luciferase 4-fold. We speculate that this may be due to increased protein stability or facilitation of non-ATG initiated translation as was observed for SCA8 (36). The longest CAG repeats in both ATXN2 and ATXN2-luc caused reduced protein abundance when evaluated by luciferase assays or western blotting, but we could find no evidence for long CAG repeats influencing transcription by qPCR. This observation may reflect reduced solubility for the polyQ-expanded ataxin-2 and ataxin-2-luc proteins or may be due to other factors related to the RNA including abnormal translation, pre-mRNA processing caused by thermodynamically stable CAG hairpinning (37) or RNA aggregation as for DMPK (38). Translational regulation by the ATXN2 CAG repeat might be facilitated by its close proximity to the start codon (separated by only four codons) and could be regulated by RNA-binding proteins interacting with the CAG repeat hairpin (39). Both ATXN7 and HTT have CAG repeats near their start codons, at codons 30 and 18, respectively, and we hypothesize that CAG repeat expansion might also regulate the translation of these genes. Our study of CAG repeat length on ATXN2 expression raises new hypotheses that will require evaluation in future work.
Analysis of the ATXN2 promoter revealed the presence of an ETS element in the ATXN2 5′-UTR identified by detailed deletion analysis. We also noted that none of the predicted sites for ZBRK1 binding localized to this region nor did we identify any other ATXN2 promoter regions that reduced ATXN2 expression upon deletion. As seven ZBRK1 sites were predicted in the ATXN2 promoter (27), our data support that PDs including single ZBRK1 sites are not sufficient to reduce ATXN2 expression. However, our finding is consistent with Aguiar et al. (26) who evaluated two ATXN2-luc constructs lacking the ETS element (−30 and downstream, plasmids C2 and C6 in their study) that had expression close to negative controls. Because the ETS element was 59 bp upstream of the first of two in-frame ATGs that was also within some inhibitory deletions that we made, we mutated the ATGs individually to determine which were functional. Elimination of the upstream ATG had no effect on expression, while that of the downstream ATG reduced expression to a level similar to control assays. This indicated that ATXN2 deletions containing both the ETS element and the first ATG reduced ATXN2-luc expression due to loss of the ETS element and that the second ATG was the functional start codon. This ETS element has 100% match to the consensus sequence for binding by ETS family TFs ETS1, ELF1 and GABPα, defined as CCGGAAGT (28). We demonstrated that ETS1, ELF2 and GABPα interacted with the ETS element by EMSA. Because we could not confirm that the overexpression of ELF1, ELF2 or GABPα could increase ATXN2 expression, we limited ChIP and shRNA validations to the study of ETS1 only. ETS1 is expressed in mouse Purkinje cells higher than in any other cell type in the cerebellum, according to the Allen Brain Atlas (30) (Supplementary Material, Fig. S3) and a study describing ETS1 in developing brain (40). Roles for ETS TFs in ataxias are also supported by the observation that ataxin-1 transcriptionally activates ETV5 (29).
In summary, we verified that the second in-frame ATG in ATXN2 located five codons upstream from the CAG repeat is the functional start codon, resulting in a full-length ataxin-2[Q22] protein of 1152 amino acids, 160 amino acids shorter than assumed previously. We evaluated ATXN2-luc expression in transgenic mice and demonstrated high luciferase expression in the mouse cerebellum, as expected, and high expression in the OB. IHC staining of the mouse OB revealed endogenous ATXN2 expression in mitral cells. We speculate that loss of ATXN2 expression in OB mitral cells in ATXN2 knockout mice may account for obesity due to hyperphagia that we previously observed in these mice. Expression of ATXN2-luc in transgenic mice resembled endogenous ATXN2 supporting that the length of the promoter studied in transgenic mice was sufficient to reflect the true pattern of Atxn2 expression and the potential for using ATXN2-luc mice in the validation of experimental SCA2 therapeutics targeting ATXN2 expression. We demonstrated that an ETS element in ATXN2 is required for ATXN2 expression. Ataxin-2 expression was increased in cells overexpressing ETS1, supporting that ETS1 regulates ATXN2 expression. More work will be required to determine whether other TFs bind this region of the ATXN2 promoter and whether they represent therapeutic targets for SCA2. As the deletion of the ETS element resulted in a major reduction ATXN2 expression, this region may represent an opportune target for decoy oligonucleotide-based therapeutics (41–43).
MATERIALS AND METHODS
Cloning of pGL2-5A3
The preparation of plasmid pGL2-5A3 was a six-step cloning process beginning with our original ATXN2 promoter plasmid pGL2-5. All amplifications for cloning in the study were made using the Expand High FidelityPLUS PCR System (Qiagen) and all clones that were created were verified by sequencing.
To clone plasmid pGL2-5, we amplified (TA = 58°C) a 1431 bp fragment from genomic DNA including ATXN2 upstream-5′-UTR sequence with forward primers ATX2A (5′-GCTAGATTAATTTTTAAAGGGTTTTGCAATG-3′) and ATX2B (5′-TAGCACGCGTAAGAGCTCTG-3′). This fragment was digested with AseI, treated with T4 polymerase to fill the sticky end, redigested with SacI, then ligated to pGL2.Enhancer between the SmaI and SacI sites of the multiple cloning site (MCS). We next created plasmid pGL2a-5 by ligating a linker fragment made by annealing a 48mer (5′-TAAAGGCCAAGAAGGGCGGAAAGTCCAAATTGTA AACCGGTACTGTAG-3′) to a 51mer (5′-TCGACTACAGTACCGGTTTACAATTTGGACTTTCCGCCCTTCTTGGC CTTT-3′) between the EcoNI and SalI sites of pGL2-5. This linker eliminated the vector polyA and incorporated an AgeI site immediately downstream of the vector luc gene stop codon for use in later cloning of the 3′-UTR. In order to remove the sequence downstream of the pGL2 MCS XhoI site, we amplified (TA= 55°C) the luc gene from pGL2 with primers LucXhoIA (5′-GCATCTCGAGATGGAAGACGCCAAAAACATA-3′) and LucAgeIB (5′-GCATACCGGTTTACAATTTGGACTTTCCGCCCTTCT-3′) and ligated this fragment in place of the original luc gene of pGL2a-5 between XhoI and AgeI. We named this clone as pGL2b-5. We then prepared plasmid pGL2b-5-3 by inserting a 1019 bp fragment of ATXN2 3′-UTR and downstream sequence amplified (TA= 53°C) with primers AgeIA (5′-CGCTAACCGGTGTTGTAAGGCTGCCCTGGAGGA-3′) and SalIB (AtxnB2) (5′-GCTAGTCGACAGCCTTCATGCCCTCTGGATTT-3′) into pGL2b-5 between the linker AgeI and the vector SalI site. Because pGL2b-5-3 contained the first ATXN2 start codon but not the second in-frame start codon that we also wanted to study, we next added an additional exon 1 fragment including the second start codon and sequence up to and including the first CAG of the CAG repeat. This fragment was amplified (TA=69°C + 5% dimethyl solfoxide) from genomic DNA with primers SacIv2A (5′-CTCGCCGCTTCGCCGCAGCCAGGT-3′) and XhoIB (5′-GAATGCCCTCGAGAGCCGTGGCCGAGGACGAGGAGAC-3′). This step also removed the vector sequence upstream of the XhoI site which in this plasmid immediately follows the first CAG of the CAG repeat. The resultant plasmid was named pGL2b-5A3. We then changed the start codon of the luciferase gene to CTG (leucine). This was accomplished by amplifying (TA= 55°C) the luc gene from pGL2 with primers LucXhoIA2 (5′-GCATCTCGAGCTGGAAGACGCCAAAAACATA-3′) and LucAgeIB (see above) and ligating it in place of the luc gene of pGL2b-5A3 between XhoI and AgeI. We named the final clone as pGL2-5A3.
Lengthening the ATXN2 promoter in pGL2-5A3 to create pGL2-L5A3
We lengthened the ATXN2 promoter in pGL2-5A3 by replacing an ATXN2 promoter fragment excised from pGL2-5A3 with a longer fragment obtained from a bacterial artificial chromosome (BAC) clone RP11-798L15 (Empire Genomics). A 5656 bp ATXN2 upstream sequence fragment was excised from the BAC plasmid with EcoRI and NotI and ligated into plasmid pCR4-TOPO. The fragment was then excised with PmeI (in the MCS of pCR4-TOPO) and NotI (in ATXN2) and ligated into pGL2-5A3 prepared with HpaI and NotI. The resulting plasmid included ATXN2 upstream sequence starting at position −5502 from the TSS and was named pGL2-L5A3.
Mutating ATXN2 start codons in pGL2-5A3
Mutation of the second ATXN2 start codon was accomplished by preparing annealed linkers that were then ligated into their corresponding locations in the ATXN2 exon 1 portion of pGL2-5A3, replacing the wild-type start codon. To change the first ATG to CTG, we prepared a 137 bp double-stranded linker by annealing the two oligonucleotides ATG2_sense (5′-CCTCACCcTGTCGCTGAAGCCCCAGC-3′) and ATG1_antisense (5′-TCGAGCTGGGGCTTCAGCGACAgGGTGAGGGGCC-3′). Annealing these oligonucleotides created ApaI and XhoI sticky ends ready to ligate into pGL2.5A3 between the same two restriction sites. To mutate the first ATG to CTG in pGL2-5A3, we utilized a custom mutagenesis service (GenScript). Plasmids were verified by sequencing and were named pGL2.5A3(mATG1) and pGL2.5A3 (mATG2).
Cloning upstream deletions of pGL2-5A3
Deletions of the upstream end of the ATXN2 sequence were accomplished by timed exonuclease III digestion. We first modified pGL2-5A3 by the insertion of an oligonucleotide linker upstream of the sequence to be deleted introducing two new restriction sites including NsiI on the upstream side, producing an exonuclease III resistant 3′ overhang, and SpeI on the downstream side, producing an exonuclease III susceptible 5′ overhang. The linker was made by annealing the two oligonucleotides EAB-A (5′-GATCATGCATCTCGAGACTAGTAGCT-3′) and EAB-B (5′-AGCTACTAGTCTCGAGATGCATGATC-3′). The linker was then ligated into the HpaI restriction site located just the upstream of the ATXN2 upstream sequence insert in pGL2-5A3. We performed timed exonuclease III digestions of NsiI and SpeI double-digested plasmid using the Erase-A-Base (Promega) kit following the vendor's protocol. Twenty-five different deletions in pGL2-5A3 were verified by sequencing, of which 12 were used in the study, designated PD1–PD12.
Cloning of IPDs in pGL2-5A3
IPDs were made by two methods. IPD1–IPD19 were made by a two-step PCR method where each deletion was ∼100 bp in length. In the first step for any particular deletion, two PCRs were conducted. In one reaction, the reverse primer consisted of a ∼30 bp hybrid primer made of ∼15 bp sequence located on either side of the 100 bp region to be deleted and a forward primer across a unique restriction site for cloning (either HpaI or BamHI). In the other reaction, the forward primer consisted of a ∼30 bp hybrid primer also made of the ∼15 bp sequence located on either side of the 100 bp region to be deleted and a reverse primer across a unique restriction site for cloning (either BamHI or XhoI). These two reactions resulted in two amplicons with 15 bp shared sequence that would prime one another in the PCR performed in the second step, each with unique restriction sites on their extreme ends for cloning. To perform the second step, we first gel purified the amplicons from the first step using the Qiaex II Gel Extraction Kit (Qiagen) and then used 1 µl (2 ng) of each in the second-step PCR including the two primers that prime at the unique restriction sites that were also used in the first-step PCRs. This second step resulted in a single amplicon containing the 100 bp deletion with unique restriction sites for cloning on each end. The amplicon from the second step was double-digested (with either HpaI and BamHI, BamHI and XhoI or HpaI and XhoI), gel purified using Qiaex II and ligated into pGL2-5A3 prepared with the same two enzymes. Deletions were verified by sequencing. The primers used to prepare each IPD clone of pGL2-5A3 are provided in Supplementary Material, Table S1, and the combinations how they were used in step 1 PCRs are in Supplementary Material, Table S2.
The remaining IPD clones (IPD20–IPD31) were made by annealing oligonucleotide pairs designed to create short ATXN2 promoter fragments containing the desired deletions flanked by cohesive ends ready to ligate to pGL2-5A3 prepared by double-restriction digestion (either BamHI and PmlI, PmlI and NotI, NotI and SacI or SacI and ApaI). The oligonucleotides and the corresponding combination of restriction enzymes used to create each of these IPDs are provided in Supplementary Material, Table S3.
Cloning of IPDs in pPrl-luc
We cloned a short fragment of the ATXN2 5′-UTR into pPrl-luc, a plasmid expressing luciferase from a rat minimal prolactin promoter. The ATXN2 fragment was 58 bp long with the ETS1-binding site at its center. The fragment corresponded to ATXN2 +72 to +129bp. Inserts were prepared by annealing sense and antisense oligonucleotides with or without deletions (Supplementary Material, Table S4). Annealing created a SacI site upstream and a BglII site downstream. Inserts were ligated into pPrl-luc between the SacI and BglII sites located between the prolactin minimal promoter and the luciferase.
Addition of CAG tracts into pGL2-5A3
We modified pGL2-5A3 by addition of different CAG tracts including 108 bp of ATXN2 exon 1 sequence downstream of the CAGs. Because pGL2-5A3 has a CAG1 tract but excludes the 108 bp exon 1 sequence downstream, we first made plasmid pGL2-5Aa3, identical to pGL2-5A3, but including the extra 108 bp exon 1 sequence. pGL2-5Aa3 was created by first preparing an insert made by annealing oligoCAG1sense (5′-CCTCACCATGTCGCTGAAGCCCCAGCCGCCGCCCGCGGCTGCCAATGTCCGCAAGCCCGGCGGCAGCGGCCTTCTAGCGTCGCCCGCCGCCGCGCCTTCGCCGTCCTCGTCCTCGGTCTCCTCGTCCTCGGCCC-3′) with oligoCAG1antisense (5′-TCGAGGGCCGAGGACGAGGAGACCGAGGACGAGGACGGCGAAGGCGCGGCGGCGGGCGACGCTAGAAGGCCGCTGCCGCCGGGCTTGCGGACATTGGCAGCCGCGGGCGGCGGCTGGGGCTTCAGCGACATGGTGAGGGGCC-3′). Annealing these oligonucleotides created ApaI blunt and XhoI sticky ends ready to ligate into pGL2.5A3 between the same two restriction sites. In order to create longer CAG tracts, we used a PCR approach: Using primers SacI-AA (5′-GGCAGAGCTCGCCTCCCTCCGCCTCAGAC-3′) and XhoI-BB (5′-GCATCTCGAGGGCCGAGGACGAGGAGAC-3′), we amplified ATXN2 exon 1 fragments including CAG tracts of lengths 22, 58 and 102 CAGs. Template plasmids are described in Ng et al. (44). These inserts were ligated into pGL2-5A3 prepared by SacI and XhoI digestion. The resultant plasmids were named pGL2-5B3, pGL2-5C3 and pGL2-5D3 (containing 22, 58 and 102 CAGs, respectively) and were sequenced to verify the CAG number. The CAG tract in pGL2-5B3 was the common normal length allelic form (CAG)8CAA(CAG)4CAA(CAG)8, while the CAG tracts in pGL2-5C3 and pGL2-5D3 did not include CAA interruptions.
Addition of TK-Renilla cassettes into pGL2-5Aa3, pGL2-5B3, pGL2-5C3 and pGL2-5D3
We modified the plasmids pGL2-5Aa3, pGL2-5B3, pGL2-5C3 and pGL2-5D3 by the addition of a fragment containing TK-Renilla luciferase. We amplified the TK-Renilla-polyA insert from the plasmid pRL-TK (Promega) with primers TKRenSalA (5′-CTGGCGTCGACGATCTAAATGAGTCTTC-3′) and TKRenSalB (5′-CAAGGTCGACGCCACCTGGATCCTTATC-3′), prepared the insert by SalI digestion and ligated it into the SalI site in each of the target plasmids. Sequencing confirmed the selection of plasmids with TK-Renilla oriented in the opposite direction compared with ATXN2-luc. The resultant plasmids were named pGL2-5Aa3-TK-rLuc, pGL2-5B3-TK-rLuc, pGL2-5C3-TK-rLuc and pGL2-5D3-TK-rLuc. These plasmids were used in the qPCR experiment shown in Fig. 2C.
Full-length ATXN2 expression plasmids
We first acquired a 4 kb ATXN2 upstream sequence (US) by PCR from human BAC clone RP11-798L15 using primers HindIII-Fwd (5′-TATAAAGCTTAAGCCTTCTGAAGTCC-3′) and NotI-Rev (5′-GCTGCGGCCGCTGAGCGCATCGGAGGGCG-3′). The insert was ligated between the HindIII and NotI sites of the plasmid pAdTrack, also containing a CMV-enhanced green florescent protein (EGFP) cassette, creating the plasmid pAdTrack-ATXN2US. The resultant plasmid was then modified by adding ATXN2 cDNAs with varying CAG repeat lengths and bovine growth hormone polyadenylation (BGH PolyA) signal by acquiring inserts from pCMV-Flag-ATXN2(CAG)n plasmids by PCR using NotI primers ATX-Fwd (5′-TCAGCGGCCGCAGCTCCTCGGAGTCCC-3′) and BGH-Rev (5′-TTTGCGGCCGCTTCCCCAGCATGCCTGCTATTG-3′). The inserts were digested with NotI and ligated into NotI digested pAdTrack-ATXN2US to create the pAdTrack-ATXN2(CAG)n plasmids, expressing ataxin-2 C-terminally tagged with the Flag epitope. The pCMV-Flag-ATXN2(CAG)n plasmids were previously cloned by amplifying inserts from corresponding pEGFP-SCA2(CAG)n plasmids (45) using primers HindIII2-Fwd (5′-GGGAAGCTTATGCGCTCAGCGGCCGCAGCTCC-3′) and SalI-Rev (5′-AAAGTCGACCAACTGCTGTTGGTGGTGGGCTTG-3′) and ligating the HindIII/SalI digested inserts into a pCMV-3Tag-8 vector (Stragagene) digested with the same two restriction enzymes. Plasmid sequences were verified by sequencing.
Site-directed mutagenesis of pGL2-5A3
We identified a 26 bp region 75 bp upstream of the start codon important for supporting ATXN2 expression and possessing predictions for binding by multiple TFs. Mutations in pGL2-5A3 in this region were accomplished by using the QuickChange Lightning Site-Directed Mutagenesis Kit (Stratagene). The mCGA mutation, a TCC→CGA mutation in the ATXN2 promoter, was created using the mutagenesis oligonucleotides MutR1A (5′-CCGTCTGACCCCTCCGACTCGAGGTAAAGAGTCCC-3′) and MutR1B (5′-GGGACTCTTTACCTCGAGTCGGAGGGGTCAGACGG-3′). The D14 mutation, a 14 bp deletion in this region of the ATXN2 promoter, was created using the mutagenesis oligonucleotides MutR2A (5′-CCGTCTGACCCCTCGAGTCCCTATCCGC-3′) and MutR2B (5′-GCGGATAGGGACTCGAGGGGTCAGACGG-3′). Mutagenesis was accomplished following the vendor's protocol.
TF expression plasmids
The expression plasmid for ETS1 was provided by Barbara Graves (University of Utah). To create an expression plasmid for ELF2, we ligated the ELF2 cDNA between the BamHI and XbaI sites of pcDNA3.1(+). The ELF2 cDNA was obtained by PCR using primers ELF2Bam-A (5′-CAGCGGATCCATAAACATGGCGACGTCTC-3′) and ELF2Xba-B (5′-GGAGCTTCTAGATTATTTCTCACATGTCACTAGT-3′) using template pBluescript-ELF2. An insert for the dn-ETS1 fragment including ETS1 amino acids 307–441 (46) was obtained by PCR with primers ETS1DN-A (5′-CCAGGGATCCATGTATGTGCGGGACCGTGCTGAC-3′) and ETS1DN-B (5′-AGTGCTCGAGTCACTCGTCGGCATCTGGCTTGA-3′) and ligated between the BamHI and XhoI sites of pcDNA3.1/Hygro(+) (Invitrogen).
Luciferase assays
HEK293 or SH-SY5Y neuroblastoma cells were grown in Dulbecco's Modified Eagle Medium with 10% fetal bovine serum and 1× penicillin/streptomycin. Cells were transfected using Metafectene (Biontex Laboratories) or Xfect (Clontech). Transfections were conducted in triplicate wells of a 24-well plate and typically consisted of 125 ng of pGL2-5A3 plasmid (mutant or wild-type) and 40 ng of pRL-SV40 or pRL-TK (Promega). Dose-wise transfections including ETS1 plasmids were balanced with an empty vector to ensure consistent total DNA quantities. Assays were performed in triplicate per transfection after 48 h transfection using the Dual-Glo Luciferase Assay System (Promega), recording relative light units (RLUs) from firefly luciferase and Renilla luciferase on a multimode plate reader (Beckman DT880). Values were reported as the mean ± SD of the ratios of RLUfirefly/RLURLuc (referred to in the text as ‘relative luciferase activity’), with n = 3 transfections for the calculation of SD. Each experiment was repeated at least three times.
Immunoblotting
Proteins were separated on precast polyacrylamide gels (Bio-Rad), transferred to Hybond (Amersham) and detected by ECL (Amersham). Antibodies included goat anti-luciferase (Rockland Immunochemicals), β-actin mAb (AC-40, Sigma), anti-ETS-1(C-4, Santa Cruz Biotechnology), anti-FLAG antibody (Sigma) and anti-GFP antibody (Invitrogen). Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories.
TF-binding site prediction
Predictions for TF-binding sites were made using MatInspector (Genomatix) (47) and TESS (48).
Real-time PCR
Real-time PCR (qPCR) was conducted by the SYBR Green method with standard curves on the iCycler (Bio-Rad) in 96-well plates in quadruplicate. RNAs were isolated from 30 mg tissue using the RNeasy Kit (Qiagen). cDNAs were generated using the QuantiTect Reverse Transcription kit (Qiagen). Primers for luc were GL2luc-2F (5′-ATCCGGAAGCGACCAACGCC-3′) and GL2luc-2R (5′-GTCGGGAAGACCTGCCACGC-3′) (212 bp product, 60°C annealing temperature). Primers for mouse ATXN2 were ATXN-m5a (5′-AAGATACAGACTCCAGTTATGCACGG-3′) and ATXN-m6b (5′-GCTCCAGGTCCTTCTCCTTGTGC-3′) (96 bp product, 60°C annealing temperature). Primers for human ATXN2 were HAtxn2-fwd (5′-AAGATATGGACTCCAGTTATGCAAA-3′) and HAtxn2-rev (5′-CAAAGCCTCAAGTTCCTCAT-3′) (143 bp product, 60°C annealing temperature). Primers for Renilla luciferase were RenA (5′-GGCCATGATTGGGGTGCTTGTT-3′) and RenB (5′-CGGGATTTCACGAGGCCATGA-3′) (321 bp product, 55°C annealing temperature). Reactions were 20 μl consisting of 15 ng of cDNA, 2 μl of each primer (0.3 μm final) and 10 μl of SYBR Green Master Mix (Bio-Rad). Cycling parameters included a 95°C denaturation for 10 s, an incubation at the annealing temperature for 20 s and a 40 s incubation at 72°C. Each plate included a standard curve using cerebellar RNA prepared from multiple pGL2-5A3 transgenic mice. Single amplicons were verified by denaturation analysis and gel electrophoresis.
Electromobility shift assays
Nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce) from HEK293 or SH-SY5Y cells, following the vender's protocol. Protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad). EMSA was performed using the non-radioactive LightShift Chemiluminescent EMSA kit (Pierce) following the vendor's protocol starting with 3 μg nuclear protein per assay. For ETS and STAT EMSA supershift experiments, the oligonucleotide probe sets included WT-sense (5′-TCCGACTTCCGGTAAAGAGTC-3′) and WT-antisense (5′-GACTCTTTACCGGAAGTCGGA-3′), mCGA-sense (5′-TCCGACTCGAGGTAAAGAGTC-3′) and mCGA-antisense (5′-GACTCTTTACCTCGAGTCGGA-3′) and RANDOM-sense (5′-CTAGCAGCTGATCTCAGCTGA-3′) and RANDOM-antisense (5′-TCAGCTGAGATCAGCTGCTAG-3′). For forkhead EMSA supershift experiments, the oligonucleotide probe sets included SO7-A (5′-ACCCCTCCGACTTCCGGTAAAGAGTCCCTA-3′) and SO7-B (5′-TAGGGACTCTTTACCGGAAGTCGGAGGGGT-3′) and SO7m-A (5′-ACCCCTCCGACTTCCGGTATTCACTCCCTA-3′) and SO7m-B (5′-TAGGGAGTGAATACCGGAAGTCGGAGGGGT-3′). Oligonucleotides were synthesized by the University of Utah Sequencing Core either unlabeled or biotinylated on the 3′ end. Annealing was accomplished by mixing sense and antisense oligonucleotides of each set at equimolar concentrations in annealing buffer (0.1 mm Tris–HCl, pH 8), heating to 95°C in a heat block, then allowing the block to slowly cool to room temperature over 60 min. EMSAs were 20 μl consisting of 1 × reaction buffer, 5 μg/μl poly(dI.dC), 10% glycerol, 2.5 mm MgCl2, 25 mm KCl, 1 mm ethylenediaminetetraacetic acid and 20 fmol biotinylated probe. Supershift assays were accomplished by including 4 μg antibody. Antibodies use for supershift assays included ETS-1(C-4)X, NERF(C-20)X, GABPα(C-20)X, ELF1(C-20)X, FKHRL1(N-16)X, FKHR(H-128)X, STAT1 p84/p91(E-23)X, STAT2(C-20)X, STAT3(C-20)X and STAT5(C-17)X (Santa Cruz Biotechnology).
Chromatin immunoprecipitation PCR
Chromatin was prepared using the Pierce Chromatin Prep Module (Pierce). Formaldehyde was added to the cells (2 × 106cells/ChIP reaction) at a final concentration of 1%. Cells were incubated at room temperature for 10 min. Then excess formaldehyde was neutralized by the addition of glycine (1×) and incubating at room temperature for 5 min. After washing twice with cold phosphate buffered saline (PBS), the cells were scraped into 1 ml of cold PBS plus 1 × Halt Cocktail. The cells were pelleted and then suspended in 100 µl of lysis buffer 1 plus 1 × Halt Cocktail. After incubating 10 min on ice, the sample was centrifuged at 9000g and the supernatant was discarded. Isolated nuclei were suspended in MNase Digestion Buffer containing 1 mm dithiothreitol. Cross-linked DNA was fragmented by Micrococcal Nuclease (10 U/µl) at 37°C for 15 min, and the reaction was stopped by the addition of MNase Stop Solution. The solution was then centrifuged at 9000g to recover the nuclei. Digested chromatin was resuspended in 50 µl of lysis buffer 2 plus 1 × Halt Cocktail and incubated on ice for 15 min. The sample was centrifuged at 900g and chromatin was recovered in supernatant. ChIP assay was carried out using Pierce Agarose ChIP Kit (Pierce). Diluted chromatin was incubated with 10 μl anti-ETS1 antibody (diluted to 1 mg/ml, Santa Cruz ETS-1(C-4)X), 10 μl normal rabbit IgG (1 mg/ml stock) or 10 μl antibody diluent, at 4°C overnight. Complexes were column purified using protein A/G-agarose (incubated at 4°C for 1 h) with multiple washes using IP wash buffer 2 and IP wash buffer 3. Samples were eluted in 150 µl of IP elution buffer for 40 min at 65°C with intermittent vortexing. Reverse cross-linked DNAs were purified by using a kit provided DNA Clean-up Columns. Two microliters of each of the purified DNA was used as a template for 30 cycles of PCR amplification using primers C-ATXN2A3 (5′-AGGAAGGCGGATCCGGGTAG-3′) and C-ATXN2B2 (5′-GGGAGCGGAGGTGCGGATAG-3′). ChIP reactions were also evaluated by qPCR and binding ratios were calculated as (starting quantity for anti-ETS1)/(starting quantity for rabbit IgG).
ETS1 knockdown
ETS1 knockdown was accomplished using an ETS1 shRNA-8236 expression plasmid provided by Barbara Graves known to the knockdown expression of both the 42 and 51 kDa ETS1 splicing isoforms (referred to as ‘A’ in 49). The sequence targeted in ETS1 is 5′-AGGTGTAGACTTCCAGAAG-3′. A control shRNA targeting EGFP was also used (#RHS4459, Open Biosystems).
Transgenic mice
We made transgenic mice using an ATXN2-luc fragment excised from pGL2-5A3 with HpaI and AfeI. The mice were created by pronuclear injection at the University of Utah Transgenic Mouse Core using C57BL/6J mice. Founder screening was accomplished by tail-tip PCR identifying eight founders. Each founder was back-crossed to wild-type mice and one F1 littermate from each line was evaluated by assaying for luciferase. To do so, we homogenized ∼100 mg cerebellum in PBS, added an equal volume of Bright-Glo Luciferase Assay Reagent (Promega) and determined RLUs on a multimode plate reader (Beckman DT880). We saved two transgenic lines with the highest ATXN2-luc expression determined as RLU/mg cerebellum tissue. Lines were designated L74 and L75. Only data on the highest expressing line L75 are presented.
Optical imaging
Optical imaging was performed using a Xenogen IVIS 100 imaging system. Mice were IP injected with 50 mg/kg of d-luciferin diluted in PBS (10 μl/g body weight of 5 mg/ml d-luciferin). After 5 min, mice were anesthetized with isoflurane and were either directly imaged or tissues were excised for imaging. Imaging was conducted for a period of 1 min at 10 min after injection of substrate. Imaging of whole mice was preceded by hair removal using Nair.
IF labeling
The OB from C57Bl/6 mice was frozen mounted in OCT and sectioned in 18 μm sections on a cryostat. Sections were fixed in 4% paraformaldehyde for 1 h then were permeabilized and blocked in blocking buffer (PBS, 0.3% Triton X-100, 5% milk) for 5 h. For labeling of HEK293 cells, cells transfected with ETS1 were grown on coverslips 48 h then fixed in 4% paraformaldehyde for 20 min. The primary antibody in antibody diluent (PBS, 0.05% Triton X-100, 5% milk) was incubated on tissues overnight at 4°C. Primary antibodies were polyclonal rabbit anti-ataxin-2-1080 (5 μg/ml), Atxn-2 mAb (BD Transduction Laboratories 611378, 2.5 μg/ml), interneuron marker antibody GAD65/67 (SC-75-13, Santa Cruz Biotechnology, 4 μg/ml) and polyclonal rabbit anti-ETS-1(C-4, Santa Cruz Biotechnology, 1:10:000). Cells were washed in wash buffer (PBS, 0.01% Triton X-100, 1% milk) then incubated with secondary antibodies (1 μg/ml) diluted in secondary antibody diluent (PBS, 3% normal goat serum) for 2 h at room temperature. Secondary antibodies were anti-rabbit-Dylight 488 (611-741-127), anti-mouse-Dylight 549 (610-742-124) and anti-goat-549 Dylight (605-742-125) (Rockland Inc., Gilbertsville, PA, USA). Cells were washed in wash buffer, incubated in 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI)/PBS for 5 min and mounted using Prolong Gold (Invitrogen). Standard fluorescent images were collected using a Nikon C1 fluorescent microscope (NIS Elements software) with a 488/561 nm filter set (for DyLight 488 and 549) or 408 nm filter (for DAPI) and a 408/488/561dichroic mirror.
Statistical analysis
Unpaired two-tailed t-tests were conducted for selected pairwise comparisons. When multiple tests were performed, it was accomplished by one-way ANOVA and we reported probabilities from Bonferroni post-tests.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health (R01NS033123 to S.M.P. and RC4NS073009 to S.M.P. and D.R.S.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Barbara Graves for contributing to the interpretation of data and for providing ETS1 expression plasmid and rat prolactin minimal promoter reporter plasmid rPrl-luc, and Mary Lucero for interpretations on OB IHC. We also thank Stephen Lessnick for providing access to the Xenogen IVIS100 instrument for in vivo imaging of ATXN2-luc in mice. We thank Karla Figueroa, Seth Christensen, Patrick Gordon and Sharan Paul for their contributions.
Conflict of Interest statement. None declared.
References
- 1.Pulst S.M. Inherited ataxias: an introduction. In: Pulst S.M., editor. Genetics of Movement Disorders. Amsterdam: Elsevier, Inc.; 2003. pp. 19–34. [Google Scholar]
- 2.Kim J.M., Hong S., Kim G.P., Choi Y.J., Kim Y.K., Park S.S., Kim S.E., Jeon B.S. Importance of low-range CAG expansion and CAA interruption in SCA2 Parkinsonism. Arch. Neurol. 2007;64:1510–1518. doi: 10.1001/archneur.64.10.1510. [DOI] [PubMed] [Google Scholar]
- 3.Ross O.A., Rutherford N.J., Baker M., Soto-Ortolaza A.I., Carrasquillo M.M., Dejesus-Hernandez M., Adamson J., Li M., Volkening K., Finger E., et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum. Mol. Genet. 2011;20:3207–3212. doi: 10.1093/hmg/ddr227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corrado L., Mazzini L., Oggioni G.D., Luciano B., Godi M., Brusco A., D'Alfonso S. ATXN-2 CAG repeat expansions are interrupted in ALS patients. Hum. Genet. 2011;130:575–580. doi: 10.1007/s00439-011-1000-2. [DOI] [PubMed] [Google Scholar]
- 5.Elden A.C., Kim H.J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Damme P., Veldink J.H., van Blitterswijk M., Corveleyn A., van Vught P.W., Thijs V., Dubois B., Matthijs G., van den Berg L.H., Robberecht W. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76:2066–2072. doi: 10.1212/WNL.0b013e31821f445b. [DOI] [PubMed] [Google Scholar]
- 7.Lajoie P., Snapp E.L. Formation and toxicity of soluble polyglutamine oligomers in living cells. PLoS One. 2011;5:e15245. doi: 10.1371/journal.pone.0015245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynh D.P., Del Bigio M.R., Ho D.H., Pulst S.M. Expression of ataxin-2 in brains from normal individuals and patients with Alzheimer's disease and spinocerebellar ataxia 2. Ann. Neurol. 1999;45:232–241. doi: 10.1002/1531-8249(199902)45:2<232::aid-ana14>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Huynh D.P., Yang H.T., Vakharia H., Nguyen D., Pulst S.M. Expansion of the polyQ repeat in ataxin-2 alters its Golgi localization, disrupts the Golgi complex and causes cell death. Hum. Mol. Genet. 2003;12:1485–1496. doi: 10.1093/hmg/ddg175. [DOI] [PubMed] [Google Scholar]
- 10.Duvick L., Barnes J., Ebner B., Agrawal S., Andresen M., Lim J., Giesler G.J., Zoghbi H.Y., Orr H.T. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamanaka T., Nukina N. Transcription factor sequestration by polyglutamine proteins. Methods Mol. Biol. 2010;648:215–229. doi: 10.1007/978-1-60761-756-3_14. [DOI] [PubMed] [Google Scholar]
- 12.Koshy B., Matilla T., Burright E.N., Merry D.E., Fischbeck K.H., Orr H.T., Zoghbi H.Y. Spinocerebellar ataxia type-1 and spinobulbar muscular atrophy gene products interact with glyceraldehyde-3-phosphate dehydrogenase. Hum. Mol. Genet. 1996;5: 1311–1318. doi: 10.1093/hmg/5.9.1311. [DOI] [PubMed] [Google Scholar]
- 13.Burke J.R., Enghild J.J., Martin M.E., Jou Y.S., Myers R.M., Roses A.D., Vance J.M., Strittmatter W.J. Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nat. Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 14.Nonhoff U., Ralser M., Welzel F., Piccini I., Balzereit D., Yaspo M.L., Lehrach H., Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata H., Huynh D.P., Pulst S.M. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 16.Ciosk R., DePalma M., Priess J.R. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- 17.Satterfield T.F., Pallanck L.J. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 18.Nonis D., Schmidt M.H., van de Loo S., Eich F., Dikic I., Nowock J., Auburger G. Ataxin-2 associates with the endocytosis complex and affects EGF receptor trafficking. Cell Signal. 2008;20:1725–1739. doi: 10.1016/j.cellsig.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Tang T.S., Tu H., Nelson O., Herndon E., Huynh D.P., Pulst S.M., Bezprozvanny I. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. J. Neurosci. 2009;29:9148–9162. doi: 10.1523/JNEUROSCI.0660-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pulst S.M., Santos N., Wang D., Yang H., Huynh D., Velazquez L., Figueroa K.P. Spinocerebellar ataxia type 2: polyQ repeat variation in the CACNA1A calcium channel modifies age of onset. Brain. 2005;128:2297–2303. doi: 10.1093/brain/awh586. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto A., Lucas J.J., Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 22.Zu T., Duvick L.A., Kaytor M.D., Berlinger M.S., Zoghbi H.Y., Clark H.B., Orr H.T. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J. Neurosci. 2004;24:8853–8861. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boy J., Schmidt T., Wolburg H., Mack A., Nuber S., Bottcher M., Schmitt I., Holzmann C., Zimmermann F., Servadio A., et al. Reversibility of symptoms in a conditional mouse model of spinocerebellar ataxia type 3. Hum. Mol. Genet. 2009;18:4282–4295. doi: 10.1093/hmg/ddp381. [DOI] [PubMed] [Google Scholar]
- 24.Alves S., Nascimento-Ferreira I., Dufour N., Hassig R., Auregan G., Nobrega C., Brouillet E., Hantraye P., Pedroso de Lima M.C., Deglon N., et al. Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3? Hum. Mol. Genet. 2010;19:2380–2394. doi: 10.1093/hmg/ddq111. [DOI] [PubMed] [Google Scholar]
- 25.Kiehl T.R., Nechiporuk A., Figueroa K.P., Keating M.T., Huynh D.P., Pulst S.M. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem. Biophys. Res. Commun. 2006;339:17–24. doi: 10.1016/j.bbrc.2005.10.186. [DOI] [PubMed] [Google Scholar]
- 26.Aguiar J., Santurlidis S., Nowok J., Alexander C., Rudnicki D., Gispert S., Schulz W., Auburger G. Identification of the physiological promoter for spinocerebellar ataxia 2 gene reveals a CpG island for promoter activity situated into the exon 1 of this gene and provides data about the origin of the nonmethylated state of these types of islands. Biochem. Biophys. Res. Commun. 1999;254:315–318. doi: 10.1006/bbrc.1998.9929. [DOI] [PubMed] [Google Scholar]
- 27.Hallen L., Klein H., Stoschek C., Wehrmeyer S., Nonhoff U., Ralser M., Wilde J., Rohr C., Schweiger M.R., Zatloukal K., et al. The KRAB-containing zinc-finger transcriptional regulator ZBRK1 activates SCA2 gene transcription through direct interaction with its gene product, ataxin-2. Hum. Mol. Genet. 2011;20:104–114. doi: 10.1093/hmg/ddq436. [DOI] [PubMed] [Google Scholar]
- 28.Hollenhorst P.C., Shah A.A., Hopkins C., Graves B.J. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes. Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crespo-Barreto J., Fryer J.D., Shaw C.A., Orr H.T., Zoghbi H.Y. Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet. 2010;6:e1001021. doi: 10.1371/journal.pgen.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seattle, WA: Allen Institute for Brain Science; 2009. Allen Mouse Brain Atlas [Internet] http://mouse.brain-map.org . [Google Scholar]
- 31.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 32.Lastres-Becker I., Brodesser S., Lutjohann D., Azizov M., Buchmann J., Hintermann E., Sandhoff K., Schurmann A., Nowock J., Auburger G. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum. Mol. Genet. 2008;17:1465–1481. doi: 10.1093/hmg/ddn035. [DOI] [PubMed] [Google Scholar]
- 33.McCann C., Holohan E.E., Das S., Dervan A., Larkin A., Lee J.A., Rodrigues V., Parker R., Ramaswami M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc. Natl Acad. Sci. USA. 2011;108:E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velazquez-Perez L., Fernandez-Ruiz J., Diaz R., Gonzalez R.P., Ochoa N.C., Cruz G.S., Mederos L.E., Gongora E.M., Hudson R., Drucker-Colin R. Spinocerebellar ataxia type 2 olfactory impairment shows a pattern similar to other major neurodegenerative diseases. J. Neurol. 2006;253:1165–1169. doi: 10.1007/s00415-006-0183-2. [DOI] [PubMed] [Google Scholar]
- 35.Laffita-Mesa J.M., Bauer P.O., Kouri V., Pena Serrano L., Roskams J., Almaguer Gotay D., Montes Brown J.C., Martinez Rodriguez P.A., Gonzalez-Zaldivar Y., Almaguer Mederos L., et al. Epigenetics DNA methylation in the core ataxin-2 gene promoter: novel physiological and pathological implications. Hum. Genet. 2011;131:625–638. doi: 10.1007/s00439-011-1101-y. [DOI] [PubMed] [Google Scholar]
- 36.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A., et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broda M., Kierzek E., Gdaniec Z., Kulinski T., Kierzek R. Thermodynamic stability of RNA structures formed by CNG trinucleotide repeats. Implication for prediction of RNA structure. Biochemistry. 2005;44:10873–10882. doi: 10.1021/bi0502339. [DOI] [PubMed] [Google Scholar]
- 38.Dansithong W., Wolf C.M., Sarkar P., Paul S., Chiang A., Holt I., Morris G.E., Branco D., Sherwood M.C., Comai L., et al. Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features. PLoS One. 2008;3:e3968. doi: 10.1371/journal.pone.0003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobczak K., Krzyzosiak W.J. CAG repeats containing CAA interruptions form branched hairpin structures in spinocerebellar ataxia type 2 transcripts. J. Biol. Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
- 40.Gerhauser I., Alldinger S., Ulrich R., Baumgartner W. Spatio-temporal expression of immediate early genes in the central nervous system of SJL/J mice. Int. J. Dev. Neurosci. 2005;23:637–649. doi: 10.1016/j.ijdevneu.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Miyake T., Aoki M., Osako M.K., Shimamura M., Nakagami H., Morishita R. Systemic administration of ribbon-type decoy oligodeoxynucleotide against nuclear factor kappaB and ets prevents abdominal aortic aneurysm in rat model. Mol. Ther. 2011;19:181–187. doi: 10.1038/mt.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Souissi I., Najjar I., Ah-Koon L., Schischmanoff P.O., Lesage D., Le Coquil S., Roger C., Dusanter-Fourt I., Varin-Blank N., Cao A., et al. A STAT3-decoy oligonucleotide induces cell death in a human colorectal carcinoma cell line by blocking nuclear transfer of STAT3 and STAT3-bound NF-kappaB. BMC Cell Biol. 2011;12:14. doi: 10.1186/1471-2121-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander J.H., Hafley G., Harrington R.A., Peterson E.D., Ferguson T.B., Jr, Lorenz T.J., Goyal A., Gibson M., Mack M.J., Gennevois D., et al. Efficacy and safety of edifoligide, an E2F transcription factor decoy, for prevention of vein graft failure following coronary artery bypass graft surgery: PREVENT IV: a randomized controlled trial. JAMA. 2005;294:2446–2454. doi: 10.1001/jama.294.19.2446. [DOI] [PubMed] [Google Scholar]
- 44.Ng H., Pulst S.M., Huynh D.P. Ataxin-2 mediated cell death is dependent on domains downstream of the polyQ repeat. Exp. Neurol. 2007;208:207–215. doi: 10.1016/j.expneurol.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huynh D.P., Figueroa K., Hoang N., Pulst S.M. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 2000;26:44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- 46.Holterman C.E., Franovic A., Payette J., Lee S. ETS-1 oncogenic activity mediated by transforming growth factor alpha. Cancer Res. 2010;70:730–740. doi: 10.1158/0008-5472.CAN-09-2090. [DOI] [PubMed] [Google Scholar]
- 47.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr. Protoc. Bioinformatics. 2008;Chapter 2:Unit 2.6. doi: 10.1002/0471250953.bi0206s21. [DOI] [PubMed] [Google Scholar]
- 49.Hollenhorst P.C., Chandler K.J., Poulsen R.L., Johnson W.E., Speck N.A., Graves B.J. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.