Abstract
Peroxisome proliferator-activated receptors (PPARs) are ligand-mediated transcription factors, which control both lipid and energy metabolism and inflammation pathways. PPARγ agonists are effective in the treatment of metabolic diseases and, more recently, neurodegenerative diseases, in which they show promising neuroprotective effects. We studied the effects of the pan-PPAR agonist bezafibrate on tau pathology, inflammation, lipid metabolism and behavior in transgenic mice with the P301S human tau mutation, which causes familial frontotemporal lobar degeneration. Bezafibrate treatment significantly decreased tau hyperphosphorylation using AT8 staining and the number of MC1-positive neurons. Bezafibrate treatment also diminished microglial activation and expression of both inducible nitric oxide synthase and cyclooxygenase 2. Additionally, the drug differentially affected the brain and brown fat lipidome of control and P301S mice, preventing lipid vacuoles in brown fat. These effects were associated with behavioral improvement, as evidenced by reduced hyperactivity and disinhibition in the P301S mice. Bezafibrate therefore exerts neuroprotective effects in a mouse model of tauopathy, as shown by decreased tau pathology and behavioral improvement. Since bezafibrate was given to the mice before tau pathology had developed, our data suggest that bezafibrate exerts a preventive effect on both tau pathology and its behavioral consequences. Bezafibrate is therefore a promising agent for the treatment of neurodegenerative diseases associated with tau pathology.
INTRODUCTION
Peroxisome proliferator-activated receptors (PPARs) are a group of nuclear receptor proteins that act as ligand-dependent transcription factors. PPARα, PPARβ and PPARγ are the three known PPAR isotypes. PPARα is predominantly expressed in the liver, kidney, muscle, adipose tissue, heart and, to a lesser extent, brain, whereas PPARβ is found in the brain, adipose tissue and skin and PPARγ is expressed ubiquitously (1). These transcription factors have been linked to lipid transport, metabolism and inflammation pathways (1). Because of this, synthetic PPAR agonists have been generated as therapeutic agents for the treatment of diabetes and metabolic diseases (2,3). PPARs have effects on metabolism and inflammation in both the central nervous system and peripheral tissues, suggesting that they may also play a role in the pathogenesis of neurodegenerative diseases, such as Alzheimer's disease (AD) (4).
Prior reports demonstrated beneficial effects of PPARγ agonists, such as thiazolidinediones (5), in models of AD (6–9), Parkinson's disease (PD) (10), amyotrophic lateral sclerosis (ALS) (11,12) and Huntington's disease (HD) (13,14). Fibrates, such as fenofibrate (15), are another class of PPAR agonists that primarily target the PPARα pathway, with smaller effects on PPARβ and PPARγ (16–18). Fenofibrate has shown promising neuroprotective effects in models of neurodegenerative diseases including PD (19) and brain injury (20). Interestingly, the neuroprotective effects of PPAR agonists occur through mechanisms involving a reduction in oxidative stress and inflammation (6–9,20).
Increased phosphorylation and accumulation of tau within neurons are important pathologic hallmarks of AD and tauopathies. Neurofibrillary tangles are more strongly linked to the cognitive impairment occurring in AD than is the deposition of β-amyloid (Aβ) (21,22). Previous reports showed that in vivo PPARγ agonists reduce Aβ and tau phosphorylation in mouse models of AD (23,24). In CHOtau4R cells, a model of tauopathy, administration of troglitazone, also reduced tau phosphorylation (25). In the present study, we investigated whether the pan-PPAR agonist bezafibrate exerts beneficial effects in the P301S transgenic mouse model of tauopathy. Bezafibrate is similar to other fibrates in that it predominantly activates PPARα, but also acts on PPARβ and PPARγ (26). Although PPAR agonists have been linked to the activation of PGC1α and mitochondrial biogenesis, the activation of 5' adenosine monophosphate-activated protein kinase produces fatty acid (FA) oxidation by activating both PPARα and PGC1α and PGC1β in cardiac muscle, which maintains mitochondrial substrate oxidation and respiration (27,28). We recently showed that bezafibrate had neuroprotective effects in a mouse model of HD (29). The P301S transgenic mice, which express the human tau gene with the P301S mutation, develop progressive tau pathology and accompanied by microglial activation (30,31), synaptic damage (31) and behavioral impairments (32,33). We treated these mice with 0.5% bezafibrate in the diet from 1 to 10 months of age and assessed its effects on tau pathology, markers of inflammation, lipid metabolism and behavior.
RESULTS
Bezafibrate treatment reduced tau pathology and tau hyperphosphorylation in P301S mice
To assess tau pathology in the brains of P301S mice, we used two mouse monoclonal antibodies. MC1 is an anti-human tau antibody and an indicator of early tau pathology related to conformational changes prior to the appearance of paired helical filamentous (PHF) tau (N-terminal conformational change, exon 10, amino acids 5–15 and 312–322). MC1 immunoreactivity appears as non-filamentous cytoplasmic staining in pre-tangle bearing neurons (34,35). AT8 is another anti-human tau antibody that detects PHF-like tau. The epitope of AT8 (around residue 200) is located outside the region of internal repeats and requires the phosphorylation of serines 199 and/or 202 (pSer202/Thr205) (36). An increase in such markers has been previously described in AD and used to evaluate tau pathogenesis (37,38).
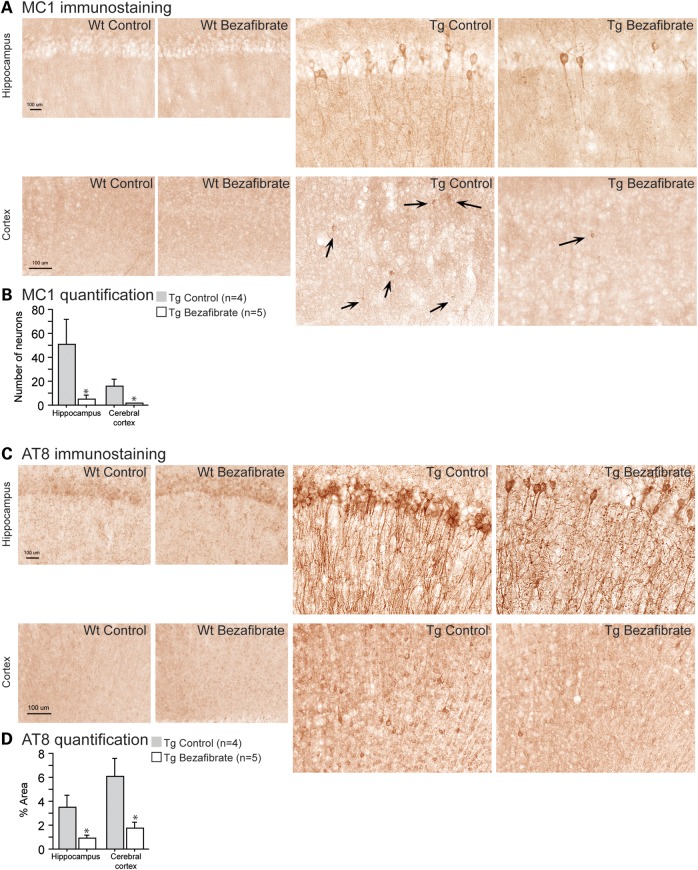
As wild-type mice did not show any tau pathology, we quantified MC1 and AT8 immunostaining only in the P301S mice. In 10-month-old P301S mice, MC1 and AT8 immunoreactivity was markedly increased in the hippocampus and cerebral cortex when compared with their wild-type littermates (Fig. 1). Bezafibrate treatment improved both early tau pathological conformational changes and tau hyperphosphorylation, measured by MC1 (Fig. 1A and B) and AT8 (Fig. 1C and D), respectively.
Figure 1.
Bezafibrate reduced tau pathology and tau hyperphosphorylation in P301S mice. (A) Immunohistochemical staining with the MC1 antibody in the hippocampus and cerebral cortex (scale bar: 100 μm). (B) Number of MC1-positive neurons in the hippocampus and cerebral cortex of P301S mice fed a control diet (Tg Control, n = 4) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 5). (C) Immunohistochemical staining with the AT8 antibody to phosphorylated tau in the hippocampus and cerebral cortex (scale bar: 100 μm). (D) Percent area covered by AT8 immunoreactivity in the hippocampus and cerebral cortex of P301S mice fed a control diet (Tg Control, n = 4) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 5). Administration of bezafibrate in P301S mice significantly reduced tau pathology and phosphorylation (unpaired t-tests, *P < 0.05).
Bezafibrate treatment affected GSK3β phosphorylation in P301S mice
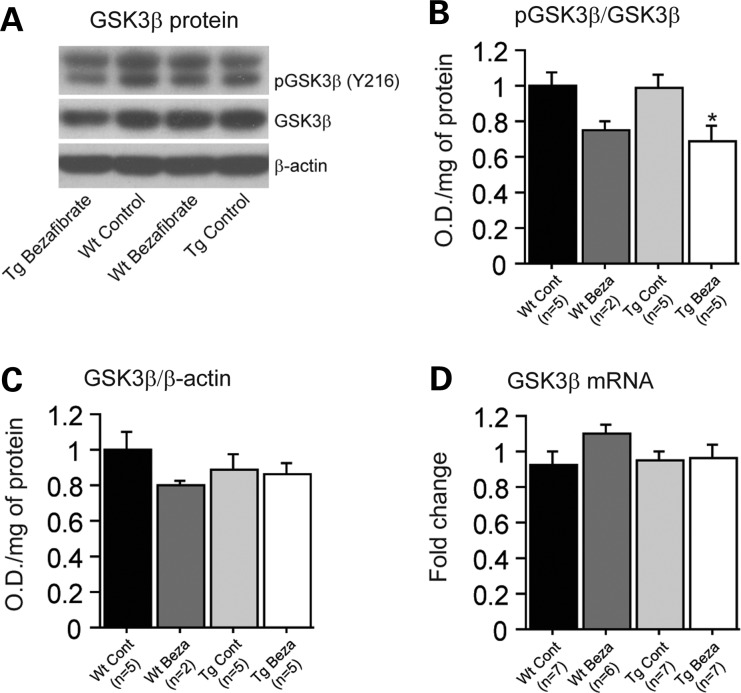
We studied the expression of glycogen synthase kinase-3-beta (GSK3β), a well-characterized serine/threonine protein kinase, which phosphorylates cellular substrates such as the protein tau and is thought to play a major role in tau phosphorylation in vivo (39–41). Administration of bezafibrate significantly decreased protein levels of phospho-GSK3β in P301S mice (Fig. 2A and B). Furthermore, we verified that these effects were not due to an overall inhibition of GSK3β by measuring total GSK3β mRNA and protein levels, which were not altered by bezafibrate treatment in both P301S mice and their wild-type littermates (Fig. 2C and D). It should be noted that no significant changes were seen between P301S mice and their wild-type littermates at baseline regarding levels of GSK3β.
Figure 2.
Bezafibrate treatment affected GSK3β phosphorylation in P301S mice. (A) Western blots of phospho-GSK3β and total GSK3β, and (B and C) quantification by optical densities in wild-type mice fed a control diet (Wt Cont, n = 5), wild-type fed a bezafibrate (Wt Beza, n = 2), P301S mice fed a control diet (Tg Cont, n = 5) and P301S mice fed a bezafibrate diet (Tg Beza, n = 5). (C) Levels of total GSK3β mRNA in wild-type mice fed a control diet (Wt Cont, n = 7), wild-type fed a bezafibrate (Wt Beza, n = 6), P301S mice fed a control diet (Tg Cont, n = 7) and P301S mice fed a bezafibrate diet (Tg Beza, n = 7). Bezafibrate treatment affected the phosphorylation of GSK3β levels in P301S mice, without affecting total GSK3β mRNA and protein levels (Fisher's PLSD, *P < 0.05).
Bezafibrate treatment reduced inflammation in P301S mice
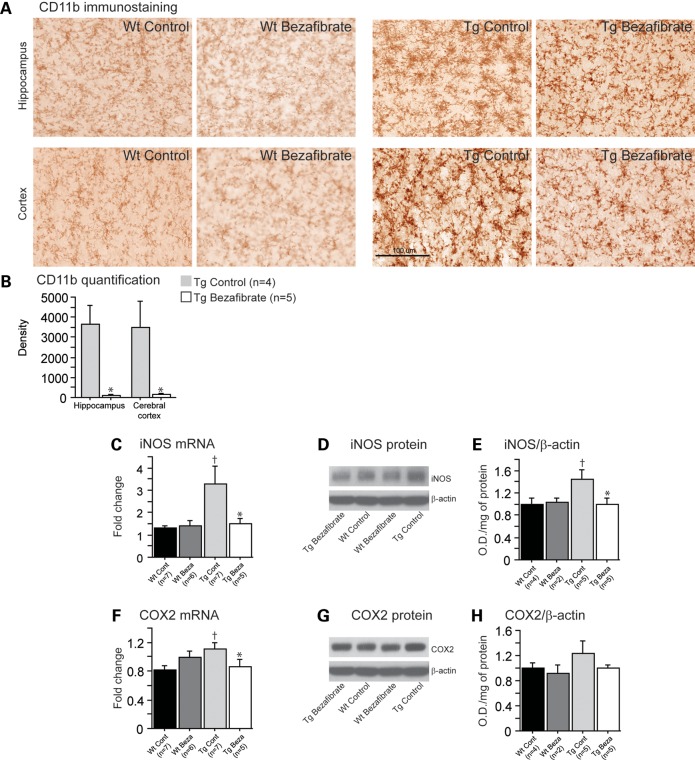
Previous reports showed that P301S mice develop early microglial activation when compared with their wild-type littermates (30,31). In this study, we observed increased microglial activation in 10-month-old P301S mice relative to non-transgenic littermates, as evidenced by elevated cluster of differentiation 11b (CD11b) immunoreactivity in the hippocampus and cerebral cortex (Fig. 3A). Bezafibrate treatment significantly decreased CD11b staining intensity associated with activated microglia in P301S mice (Fig. 3B).
Figure 3.
Bezafibrate treatment reduced inflammation in P301S mice. (A) Immunohistochemical staining with the CD11b antibody in the hippocampus and cerebral cortex. (B) Intensity (density) of CD11b in P301S mice fed a control diet (Tg Control, n = 4) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 5; scale bar: 100 μm). Administration of bezafibrate significantly reduced microglial activation in the brains of P301S mice (unpaired t-tests, *P < 0.05). Levels of iNOS (C) and COX2 (F) mRNA in wild-type mice fed a control diet (Wt Control, n = 7), wild-type mice fed a bezafibrate diet (Wt Bezafibrate, n = 6), P301S mice fed a control diet (Tg Control, n = 7) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 5). Western blots of iNOS (D) and COX2 (G) protein and their quantifications by optical densities normalized to β-actin (E and H) in wild-type mice fed a control diet (Wt Control, n = 4), wild-type mice fed a bezafibrate diet (Wt Bezafibrate, n = 2), P301S mice fed a control diet (Tg Control, n = 5) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 5). P301S mice had significantly elevated inflammation relative to their wild-type littermates (Fisher's PLSD, †P < 0.05). After bezafibrate treatment, both iNOS and COX2 mRNA and protein expression were significantly decreased in P301S mice (Fisher's PLSD, *P < 0.05).
PPAR activation is known to inhibit the transcriptional regulation of several key inflammatory genes (4). Also, considering the important role of inducible nitric oxide synthase (iNOS) and cyclooxygenase 2 (COX2) in the pro-inflammatory responses of microglia, we measured their levels after bezafibrate treatment. Both iNOS mRNA and protein levels were significantly reduced in P301S mice fed a bezafibrate diet relative to P301S mice fed a control diet (Fig. 3C–E). Similarly, COX2 mRNA and protein levels were decreased after bezafibrate treatment in the P301S mice (Fig. 3F–H).
Bezafibrate treatment improved behavioral deficits in P301S mice
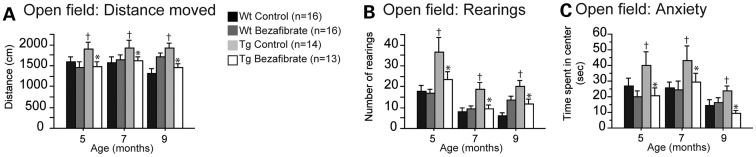
The open-field test was used to assess locomotion and exploration. In this test, P301S mice were hyperactive relative to their wild-type littermates at 5, 7 and 9 months of age (Fig. 4A and B). P301S mice were also disinhibited as evidenced by the increased time spent in the central area of the apparatus (Fig. 4C). Both locomotor and anxiety-related abnormalities were improved in P301S mice fed a bezafibrate diet when compared with P301S mice fed a control diet (Fig. 4).
Figure 4.
Bezafibrate treatment improved behavioral deficits in P301S mice. Distance moved (A), rearings (B) and anxiety (time spent in the center) (C) of the open field in wild-type mice fed a control diet (Wt Control, n = 16), wild-type mice fed a bezafibrate diet (Wt Bezafibrate, n = 16), P301S mice fed a control diet (Tg Control, n = 14) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 13). P301S mice were significantly hyperactive and disinhibited in the open-field test when compared with wild-type littermates (Fisher's PLSD, †P < 0.05). Bezafibrate treatment significantly reduced behavioral abnormalities in P301S mice (Fisher's PLSD, *P < 0.05).
Bezafibrate treatment affected oxidative stress markers and energy metabolism in P301S mice
We have previously shown that P301S mice have increased protein carbonyl levels starting at 7 months of age (32). Here, we assessed oxidative stress by measuring levels of malondialdehyde, a marker of lipid peroxidation (Supplementary Material, Fig. S1A), ratios of oxidized glutathione disulfide (GSSG) to reduced glutathione (GSH; Supplementary Material, Fig. S1C) and protein carbonyls in the brains of P301S mice (Supplementary Material, Fig. S1B). Levels of both malondialdehyde and protein carbonyls were increased in 10-month-old P301S mice relative to their wild-type littermates (Supplementary Material, Fig. S1A and B). However, bezafibrate treatment did not affect lipid peroxidation (Supplementary Material, Fig. S1A). Both protein carbonyls and glutathione levels in P301S mice fed a bezafibrate diet were significantly reduced when compared with P301S mice fed the control diet (Supplementary Material, Fig. S1B and C), although levels of glutathione were not significantly increased between P301S and wild-type littermates at baseline.
In order to understand the mechanisms in which bezafibrate affected oxidative stress, we examined energy metabolism pathways. First, we analyzed mitochondrial DNA (mtDNA) copy number. There was a trend toward a decrease in mtDNA copy number in P301S mice when compared with their wild-type littermates. We found that this level was elevated after bezafibrate treatment, suggesting that bezafibrate may have induced mitochondrial biogenesis and energy metabolism (Supplementary Material, Fig. S1D). To confirm this hypothesis, we assessed the gene expression of PPAR downstream targets involved in energy metabolism, such as sirtuin 1 (Sirt1), PPARγ coactivator 1-α (PGC1α), nuclear respiratory factor 1 (NRF1) and mitochondrial transcription factor A (Tfam). No differences were found in these genes at baseline between P301S mice and their wild-type littermates (Supplementary Material, Fig. S1E). However, after bezafibrate treatment, mRNA levels of Sirt1 and Tfam were significantly increased in P301S mice (Supplementary Material, Fig. S1E).
We also evaluated whether this could be attributed to a mitochondrial antioxidant-mediated response. Bezafibrate treatment did not affect the enzymatic activities of either glutathione reductase or superoxide dismutase (Supplementary Material, Fig. S2A). We also checked enzymatic activities of other mitochondrial enzymes, such as aconitase, citrate synthase, complex I, succinate dehydrogenase, isocitrate dehydrogenase and malic enzyme (Supplementary Material, Fig. S2B), and protein levels of ATPase (Supplementary Material, Fig. S2C). We did not observe any significant differences in these mitochondrial enzymes with or without bezafibrate treatment in either P301S mice or their wild-type littermates (Supplementary Material, Fig. S2).
Bezafibrate treatment increased FA β-oxidation in the P301S brains
In the brains of P301S mice, there was a trend for an increase in both PPARα and PPARβ gene expression (Fig. 5A). In addition, we examined their downstream targets which are involved in FA β-oxidation pathways, such as peroxisomal acyl-coenzyme A oxidase 1 (ACOX1), carnitine palmitoyltransferase 1A (CPT1A) and 3-hydroxy-3-methylglutaryl-coenzyme A synthase 2 (HMGCS2). After bezafibrate treatment, mRNA levels of HMGCS2 were significantly increased in P301S mice (Fig. 5B).
Figure 5.
Bezafibrate treatment increased FA metabolism in P301S brains. Expression of PPAR (A) and fatty β-oxidation genes (B) in the brains of wild-type mice fed a control diet (Wt Control, n = 6), wild-type mice fed a bezafibrate diet (Wt Bezafibrate, n = 6), P301S mice fed a control diet (Tg Control, n = 6) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 6). P301S mice had reduced mRNA levels of HMGCS2 (Fisher's PLSD, †P < 0.05). After bezafibrate treatment, there was a trend toward an increase in PPARα and PPARβ, and there was a significant increase in HMGCS2 mRNA levels in P301S mouse brains (Fisher's PLSD, *P < 0.05).
To determine whether these effects had functional consequences on FA content, we studied types and levels of free FAs in the brains of P301S mice (Supplementary Material, Table S1). At baseline, we did not detect any significant differences in saturated, mono-unsaturated or total FA levels in P301S mice when compared with their wild-type littermates. However, P301S mice had elevated levels of poly-unsaturated FAs when compared with wild-type mice, as evidenced by a significant increase in linoleic acid (C18:2; Supplementary Material, Table S1). After bezafibrate treatment, the level of linoleic acid was significantly reduced in both wild-type and P301S mice (Supplementary Material, Table S1). There was also a trend toward a decrease in other poly-unsaturated FAs in P301S mice fed the bezafibrate diet relative to P301S mice fed a control diet, such as linolenic acid (C18:3) and eicosapentaenoic acid (C20:5).
To have a better understanding of the impact of bezafibrate diet on brain lipid metabolism, we also conducted a comprehensive lipidomic analysis of the brain tissue derived from wild-type and P301S mice. We found that neither the P301S transgene expression nor the bezafibrate diet caused any major changes in the levels of various lipid classes (Supplementary Material, Fig. S3A). However, two types of alterations were observed in the metabolism of sphingolipids: first, a significant increase in the levels of monohexyl ceramide (MHCer) was found in the P301S brains from mice treated with a control diet, but not with the bezafibrate diet; second, the wild-type mice on the bezafibrate diet showed an increase in the levels of sulfatide (Supplementary Material, Fig. S3A). Interestingly, while no major differences were found in the average carbon lengths and levels of unsaturated fatty acyl groups associated with glycerolipids and phospholipids (Supplementary Material, Fig. S3B and C), several alterations were noted in specific molecular species of various lipid families (Supplementary Material, Fig. S4). Of particular interest were brain lipid changes that correlate with the phenotypic rescue of P301S mice treated with the bezafibrate diet (see the fourth lane in the heat map of Supplementary Material, Fig. S4). These include decreases in various ether phosphatidylcholine (ePC) species (ePCs 34:0, 36:0, 36:1 and 38:4), diacylglycerol (DGs 34:2, 36:2 and 36:3) and triglycerides (TGs 52:2. 54:3, 58:9, 60:7 and 60:9). The case of DG is particularly interesting as these very same species of DG (i.e. 34:2, 36:2 and 36:3) are increased by the expression of the P301S transgene (see the second lane in the heat map of Supplementary Material, Fig. S4), indicating that the bezafibrate diet reversed lipid alterations that occur in the brains of mice with the P301S tauopathy. Overall, these data indicate that the brain lipidome is differentially affected by the expression of the P301S transgene, the bezafibrate diet and the two together.
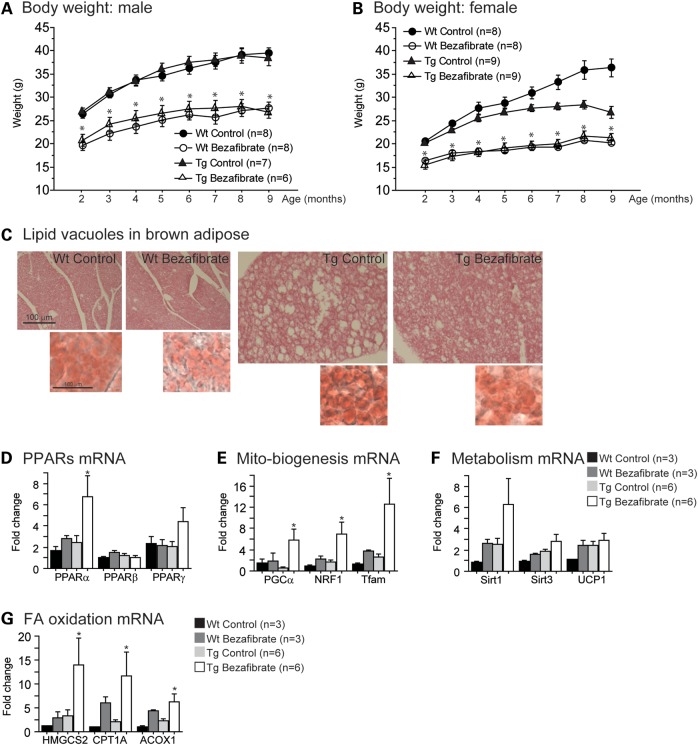
Bezafibrate treatment had beneficial effects in the brown adipose tissue of P301S mice
Body weight and lipid metabolism in the brown adipose tissue were also studied (Fig. 6). Previous reports showed that bezafibrate reduces body weight (42). In our study, both wild-type and P301S mice fed the bezafibrate diet had lower body weights than those fed the control diet (approximately a 30% reduction; Fig. 6A and B). Interestingly, in the brown adipose tissue, P301S mice had an increase in the size and the number of lipid vacuoles when compared with their wild-type littermates, using both hematoxylin and oil red O staining (Fig. 6C). This phenomenon was not due to the presence of human tau protein, since the human tau protein was not detected using either immunohistochemistry or western blotting with the HT7 antibody (an anti-human tau antibody; Supplementary Material, Fig. S6). However, this pathology was improved after bezafibrate treatment, as evidenced by a reduction in size and number of lipid vacuoles in the brown adipose tissue of P301S mice, similar to our observations in the R6/2 transgenic mouse model of HD (29).
Figure 6.
Bezafibrate treatment prevented lipid vacuoles and activated mitochondrial biogenesis and β-oxidation genes in brown adipose tissue of P301S mice. Body weight of male (A) and female (B) wild-type mice fed a control diet (Wt Control, total n = 16), wild-type mice fed a bezafibrate diet (Wt Bezafibrate, total n = 16), P301S mice fed a control diet (Tg Control, total n = 16) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, total n = 15). Bezafibrate-treated mice had significantly lower body weight than control-treated mice (Fisher's PLSD, *P < 0.05). (C) Hematoxylin–eosin staining of brown adipose tissue in wild-type mice fed a bezafibrate diet (Wt Bezafibrate, n = 5), P301S mice fed a control diet (Tg Control, n = 5) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 5). (C) Oil red O staining in bottom right panels was used to verify the presence of lipids. P301S mice had increased size and number of lipid vacuoles, pathology which was improved after bezafibrate treatment. Expression of PPARs (D) and energy metabolism-related genes (E and F) in wild-type mice fed a control diet (Wt Control, n = 3), wild-type mice fed a bezafibrate diet (Wt Bezafibrate, n = 3), P301S mice fed a control diet (Tg Control, n = 6) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 6). There were significant increases in PPARα, PGC1α, NRF1 and Tfam in the brown adipose of P301S mice after bezafibrate treatment (Fisher's PLSD, *P < 0.05). Also, there was a trend toward an increase in PPARγ and Sirt1. (G) Expression of FA β-oxidation genes in wild-type mice fed a control diet (Wt Control, n = 3), wild-type mice fed a bezafibrate diet (Wt Bezafibrate, n = 3), P301S mice fed a control diet (Tg Control, n = 6) and P301S mice fed a bezafibrate diet (Tg Bezafibrate, n = 6). There was a significant increase in HMGCS2, CPT1A and ACOX1 in the brown adipose of P301S mice after bezafibrate treatment (Fisher's PLSD, *P < 0.05).
To determine how bezafibrate exerts beneficial effects in the brown adipose tissue of P301S mice, we measured the gene expression of PPARs and their downstream targets involved in energy metabolism as previously done in the brain, including Sirt1, Sirt3, uncoupling protein 1 (UCP1), PGC1α, NRF1 and Tfam (Fig. 6D–F). It should be noted that there was a trend toward a decrease in PGC1α mRNA in the brown adipose tissue of P301S mice relative to their wild-type littermates, which may play a role in the lipid pathology. Bezafibrate administration increased both PPARα and PPARγ in the brown adipose tissue of P301S mice (Fig. 6D). Levels of Sirt1, PGC1α, NRF1 and Tfam were significantly increased after bezafibrate treatment, suggesting an increase in energy metabolism.
Bezafibrate treatment increased FA β-oxidation in the brown adipose tissue of P301S mice
We also measured the expression of PPAR downstream target genes involved in FA β-oxidation, including ACOX1, CPT1A and HMGCS2 (Fig. 6G). After bezafibrate treatment, the levels of ACOX1, CPT1A and HMGCS2 were significantly increased (Fig. 6G).
To investigate the effects of bezafibrate on free FAs in the brown adipose tissue of P301S mice, we generated another cohort of mice and administered 0.5% bezafibrate by intraperitoneal injection in a single dose (Supplementary Material, Table S2). Mice were sacrificed 4 h after the injection. Levels of three saturated FAs were decreased in P301S mice when compared with their wild-type littermates: docosanoic acid (C22:0), tetracosanoic acid (C24:0) and hexacosanoic acid (26:0). After bezafibrate treatment, levels of saturated FAs were increased. In particular, levels of hexacosanoic acid were significantly increased in the P301S mice treated with bezafibrate (Supplementary Material, Table S2). Total levels of free FAs did not change after bezafibrate treatment. As in the brains, there was a decrease in all poly-unsaturated FAs in the brown adipose tissues of P301S mice injected with bezafibrate relative to P301S mice injected with a vehicle control (Supplementary Material, Table S2). We conducted a comprehensive lipidomics analysis for the brown adipose tissue and found that two classes of phospholipids, phosphatidylinositols (PIs) and phosphatidylserines (PSs), were significantly down-regulated in the P301S mice treated with bezafibrate, while there is a trend for an elevation of DG in both wild-type and P301S animals treated with bezafibrate (Supplementary Material, Fig. S3D). A further examination of the levels of individual lipid species showed significant changes in the P301S animals treated with bezafibrate including decreases in multiple species of PI and PS containing polyunsaturated FAs and increases in multiple species of DG and TG (the third lane of the heat map of Supplementary Material, Fig. S5). A FA analysis of the phospholipids showed that the acute bezafibrate treatment produced a significant increase in short chain phospholipids relative to longer chain phospholipids (Supplementary Material, Fig. S3E), as well as an increase in saturated and monosaturated phospholipids relative to polyunsaturated phospholipids.
DISCUSSION
There is a large body of evidence demonstrating the importance of PPARs in lipid metabolism, energy metabolism and inflammation. Several groups have investigated the role of PPARs in the central nervous system and effects of PPARγ agonists, such as pioglitazone and rosiglitazone in models of neurodegenerative diseases (43,44). We previously showed that administration of pioglitazone extended survival and attenuated neuronal loss, gliosis and oxidative damage in a transgenic mouse model of ALS (11), as did another group of investigators (12). Interestingly, neuroprotective effects were also found in transgenic mouse models of AD (45). PPARγ agonists reduced Aβ levels and inflammation (6), as well as cerebrovascular dysfunction (8), and behavioral deficits in transgenic mouse models of AD (46). PPARγ agonists also enhance mitochondrial biogenesis (47).
Several clinical trials in AD patients have been initiated using PPARγ agonists. In one study, there was an increase in cerebral glucose metabolism after rosiglitazone treatment in the early stages of the disease (48). In both a pilot trial and a phase II clinical trial, the use of pioglitazone improved memory and cognition in AD patients who did not have apolipoprotein E4 alleles (49). However, in large phase III trials, cognition was not significantly improved in AD patients (48,50–52), suggesting that the mechanism of the action of PPARγ agonists in animal models of amyloid deposition may differ from those in humans or that therapeutic intervention once the AD pathology is fully developed may not be efficacious (53). Also, neurofibrillary tangles are not present in most of the animal models of amyloid deposition.
In fact, in AD and tauopathies, very little is known about whether PPARs play any role in the pathologic abnormalities which occur in the tau protein. Tauopathies are a group of diseases in which the predominant pathology is neurofibrillary tangles and hyperphosphorylated tau, including fronto-temporal dementia, cortico-basal degeneration and progressive supranuclear palsy, as well as AD. Tau pathology has been closely linked to the dementia which occurs in AD (21,22). This pathology is also reflective of tau oligomer species which have been shown to be released in the interstitial space as well as cerebrospinal fluid (54). Tau oligomers appear to propagate and produce neurotoxic effects (55–57).
In addition to PPARγ, other PPAR agonists have also been tested as potential therapeutic agents for the treatment of neurodegenerative diseases. PPARα agonists, such as fenofibrate, showed promising effects in both 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine and 6-hydroxydopamine mouse models of PD (19). Here, we investigated the role of a pan-PPAR agonist on tau-related pathology. We chose bezafibrate, a well-known PPAR agonist that has been used for more than 25 years in the treatment of elevated TGs and cholesterol. Bezafibrate primarily activates PPARα, but also has some activity on both PPARβ and PPARγ (26,58). Recently, Wenz et al. (59) showed that all three isotypes of PPARs were elevated in the muscle of mice treated with bezafibrate, while another study showed an increase in PPARα and PPARβ (60).
In the present study, we used transgenic mice with the P301S mutation, which causes fronto-temporal dementia in human patients (61–63). These mice develop progressive tau pathology and neurodegeneration with early synaptic deficits, as well as inflammation (31). The P301S mice and their wild-type littermates were administered 0.5% bezafibrate (∼800 mg/kg/day) in the diet from 1 to 10 months of age. This is a relatively high dose, but we wished to achieve the best possible brain levels to fully assess its effects. Pharmacodynamic analyses in rodents have been conducted with lower doses of bezafibrate, such as 10–100 mg/kg/day, the most clinically relevant dose being 10 mg/kg/day for patients (64–66). However, many groups studying the in vivo effects of bezafibrate in mice used higher doses, such as 0.2% (320 mg/kg) or 0.5% (800 mg/kg) (59,60,67–69). Since fibrates cross the blood brain barrier slowly (70) and we wished to achieve therapeutic effects in the brain, we utilized 0.5% bezafibrate, which is the highest dose that had previously been shown to be tolerable and efficacious in mice (29,59).
After bezafibrate administration, immunoreactivity of both MC1, which immunostains non-filamentous cytoplasmic tau with a conformational change, and AT8, which immunostains phosphorylated tau, were decreased by 77 and 65%, respectively, in the brains of P301S mice. This demonstrates that bezafibrate reduces filamentous and non-filamentous tau pathology. To understand the mechanism by which bezafibrate improves tau pathology, we studied its effects on GSK3β. Phosphorylation levels of tau are regulated by a number of kinases and phosphatases (71,72). However, several reports have shown that GSK3β is a specific therapeutic target for PPARs. Activation of PPARα by overexpression or treatment with fenofibrate inhibited the phosphorylation of GSK3β (73). Sharma et al. (74) similarly demonstrated that the activation of PPARγ in stable AML-12 hepatocyte cell lines down-regulated GSK3β kinase activity. Even though no major changes were seen between P301S mice and their wild-type littermates at baseline, we found that bezafibrate significantly reduced phospho-GSK3β levels.
In addition to tau pathology, P301S mice show early activation of microglia (31). PPAR agonists are well known to reduce inflammation by inhibiting the transcriptional regulation of pro-inflammatory genes such as the nuclear factor kappa-light-chain-enhancer of activated B cells pathway (75), and we therefore studied microglia activation after bezafibrate treatment. CD11b immunoreactivity, which is a marker for activated microglia, was significantly reduced by bezafibrate treatment in the brains of P301S mice. We assessed levels of iNOS and COX2, which are two critical mediators of inflammation and which are repressed by PPAR activation. Bezafibrate treatment inhibited both iNOS and COX2 expression in the brains of P301S mice. In a model of neuro-inflammation produced by intracerebral injection of lipopolysaccharide in mice, the pharmacological activation of PPARα decreased expression of tumor necrosis factor α (TNFα), COX2 and iNOS as well as other markers of inflammation (76). A PPARβ/PPARδ agonist also decreased iNOS after septic shock (77). In transgenic AD mice, pioglitazone reduced glial inflammation by decreasing levels of iNOS and COX2 (6). Recently, Escribano et al. found that rosiglitazone inhibited microglial activation by decreasing the expression of TNFα and COX2 in transgenic AD mice. Consistent with these data, rosiglitazone also elevated the expression of the gene CD36 (24). The authors concluded that the PPARγ agonist rosiglitazone induced a switch of microglia activation from the classic (M1) to the alternative phenotype (M2), which could contribute to a reduction in tau accumulation in transgenic AD mice (24).
The beneficial effects of bezafibrate in P301S mice were associated with an improvement in behavioral deficits at various ages. P301S mice are known to be hyperactive and disinhibited when compared with wild-type mice (32,33). In this study, bezafibrate treatment improved both hyperactivity and disinhibition at 5, 7 and 9 months of age. We however did not determine the effects of bezafibrate on cognition. Chronic administration of rosiglitazone prevents early cognitive impairment in young transgenic AD mice and reversed memory decline in aged transgenic AD mice, as shown using the object recognition test (24,46). Further investigation to determine whether bezafibrate has effects on cognition would therefore be worthwhile in future studies.
To confirm that PPAR activation occurred in the brains of our mouse model, we assessed gene expression levels of the three PPAR isotypes (PPARα, PPARβ and PPARγ) and their downstream targets. In bezafibrate-treated P301S mice, there was an increase in both PPARα and PPARβ in the brain. Consistent with PPARα activation being involved in FA β-oxidation (78,79), we found that bezafibrate treatment increased gene expression of HMGCS2. This effect was associated with a reduction in polyunsaturated free FA levels in the brains of P301S mice. Our data are consistent with a previous report showing that in rats, levels of mitochondrial HMG-coenzyme A (CoA) synthase, ACOX and medium chain acyl-CoA dehydrogenase were increased after ciprofibrate administration (80).
To further examine the effects of bezafibrate on the brain lipid metabolism, we conducted a comprehensive lipidomic analysis. Unlike the dramatic changes in subclass lipid composition present in familial AD mouse models (81), we only observed two alterations of sphingolipids with increased levels of MHCer in the untreated P301S mice and sulfatide in wild-type mice in the brain, following treatment with bezafibrate. However, numerous other lipid species were differentially regulated, the most interesting being an elevation of various DG species in the P301S mice followed by its reduction with bezafibrate treatment. This may have important implications since accumulation of DG, a major bioactive lipid, may lead to overactivation of protein kinase C, a biochemical signaling pathway that impairs working memory in rats (82).
We also examined the effects of bezafibrate on energy metabolism and oxidative stress. Indeed, PPARs are known to play a role in the induction of mitochondrial biogenesis via the PGC1α pathway (83,84). Bezafibrate treatment decreased oxidative stress as shown by decreased levels of both protein carbonyls and GSSG/GSH in P301S mice. This effect of bezafibrate was not due to an increase in the activity of mitochondrial electron transport enzymes as they remained unchanged after bezafibrate treatment. Interestingly, bezafibrate augmented gene expression of the Tfam in the brains of P301S mice, which was accompanied by an increase in mtDNA copy number. Although no major changes were observed between P301S mice and their wild-type littermates regarding energy metabolism, it is possible that some of the beneficial effects of bezafibrate may be secondarily mediated by increased mitochondrial biogenesis, which leads to a reduction in oxidative stress (85). This finding may seem paradoxical since bezafibrate improved pathways that did not appear damaged in untreated P301S mice.
We found that body weights of mice treated with the bezafibrate diet were markedly reduced. Even though tau pathology is found mostly in the brain in the P301S mice, we studied the effect of bezafibrate in brown adipose tissue, since it is a tissue which develops marked increases in lipid vacuoles in response to a reduction in PGC1α. Brown adipose tissue, which is used to generate body heat in mammals, contains small lipid droplets also called lipid vacuoles and large numbers of mitochondria (86).
In our study, long-term bezafibrate treatment improved brown adipose tissue pathology in P301S mice, by reducing the size and the number of lipid droplets. To better understand these beneficial effects, we examined the expression of PPARs and their downstream targets in the brown adipose tissue of P301S mice. Both PPARα and PPARγ mRNA levels were increased in the brown adipose tissue. In isolated adipocytes, the activation of PPARα with bezafibrate reduces adipocyte hypertrophy and elevates both adipogenic and FA oxidation-related genes, such as peroxisomal ACOX and CPT1 (87), similar to our observations in the P301S mice. Using a chromatin immunoprecipitation assay, bezafibrate treatment resulted in the recruitment of PPARα to the promoter regions of both adipogenic and FA oxidation-related genes (87). In a mouse model of mitochondrial myopathy, bezafibrate treatment induced FA oxidation in the liver, as evidenced by increases in both the FA translocase CD36 and ACOX1 (69). Similar effects occurred in the muscle of mice treated with bezafibrate (60).
Since we observed a reduction in lipid droplets, we hypothesized that bezafibrate treatment may induce β-oxidation of FAs and lipid oxidation of TG, the major lipid component of brown adipose tissue and/or other lipid classes. To investigate this possibility, we examined the lipidome of brown adipose tissue after acute bezafibrate treatment by intraperitoneal injections. Similar to the brains of P301S mice undergoing long-term treatment of bezafibrate, levels of poly-unsaturated free FAs were reduced in acutely treated mice. These data are consistent with previous data published by Tremblay-Mercier et al. (88) showing that bezafibrate lowered plasma levels of free FAs in patients with hypertriglyceridemia as a result of increased FA β-oxidation in linoleic acid. In our study, further examination of the lipidome revealed that only PS and PI were significantly decreased in P301S mice treated with bezafibrate. However, there was a clear shift toward phospholipids with shorter FA chain length and a lower degree of saturation, thereby suggesting that significant lipid remodeling is taking place due to bezafibrate treatment. Interestingly, there was also a trend for an increase in DG in bezafibrate-treated mice, suggesting the potential activation of lipolysis of TG, even within this small time window.
In the brown adipose tissue, beneficial effects of bezafibrate in P301S mice may also be attributed to its ability to increase the expression of energy metabolism genes and mitochondrial biogenesis, such as PGC1α, NRF1, Tfam and Sirt1. These effects of PPAR activation on transcription factors which regulate mitochondrial biogenesis were also observed in other models, such as in the muscle of bezafibrate-treated mice with a mitochondrial myopathy (89) and in the brains of mice treated with rosiglitazone (47). We also found that bezafibrate increased the brain and muscle mitochondrial biogenesis in the R6/2 transgenic mouse model of HD (29). However, in studies of other mice with different myopathies, bezafibrate treatment had no effect on mitochondrial biogenesis (60,69). The effects of bezafibrate on mitochondrial biogenesis therefore appear to vary with the background strains of the mice being studied or with the origin of the mitochondrial dysfunction or its metabolic context.
In conclusion, we showed that bezafibrate exerts beneficial effects in both the central nervous system as well as brown adipose tissue in the P301S transgenic mice. To our knowledge, this is the first in vivo evidence showing that bezafibrate ameliorates tau pathology and behavioral deficits in diseases that manifest tauopathy. The beneficial effects of bezafibrate are associated with its ability to reduce inflammation and stimulate lipid metabolism. Importantly, bezafibrate was administered prior to the appearance of any tau pathology, which supports a possible preventive role of bezafibrate. The present results therefore show that drugs such as bezafibrate may be useful for the treatment of patients with tauopathies or other diseases such as AD, in which tau pathology plays a major role, both administered during symptoms but especially when given during pre-symptomatic stages of the disease.
MATERIALS AND METHODS
Animals and treatment
Animals were generated by breeding P301S transgenic male mice with wild-type female mice, under the original C57BL/6 × C3H background obtained from Jackson Laboratory (Bar Harbor, ME, USA). Offspring were genotyped by polymerase chain reaction (PCR) of tail DNA. P301S transgenic mice and their wild-type littermates were randomly assigned to receive either control diet (LabDiet 5002) or 0.5% bezafibrate diet (Sigma, St Louis, MO, USA) from 1 to 10 months of age ad libitum. The chow was pelleted by Purina-Mills (Richmond, IN, USA).
Behavioral analyses were performed at 5, 7 and 9 months of age. Histopathological and biochemical analyses were conducted at 10 months of age on the same animals. All experiments were approved by the Weill Cornell Medical College Institutional Animal Care and Use Committee.
Behavior
Body weight was recorded once a month from 2 to 9 months of age. Locomotor activity and exploration were assessed in the open-field test as described previously (90). Briefly, mice were placed in the apparatus for a 5 min trial. Distance traveled and rearing frequency were recorded using a video tracking system (Ethovision 3.1, Noldus Technology, Attleborough, MA, USA). As an indicator of exploration and anxiety, the time spent in the periphery and the center of the apparatus was recorded.
Western blotting
After behavioral testing, half of the mice in each group were sacrificed by decapitation. The brains were collected, dissected, snap frozen in liquid nitrogen and stored at −80°C for biochemical studies.
Tissues were homogenized in radioimmunoprecipitation assay buffer with protease and phosphatase inhibitors (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Equal amounts of protein were electrophoresed through 4–12% Tri–Bis NuPage gels (Invitrogen, Carlsbad, CA, USA). After transfer to polyvinylidene fluoride, membranes were blocked in 5% non-fat dry milk in phosphate buffer saline with 0.05% Tween 20 and exposed overnight to the primary antibody at 4°C. Horseradish peroxidase (HRP)-conjugated secondary antibody binding was visualized with enhanced chemiluminescence (Pierce, Rockford, IL, USA).
Primary antibodies and concentrations used for western blotting were: mouse monoclonal anti-GSK3β (1:500, Abcam, Cambridge, MA, USA), rabbit monoclonal anti-phospho-GSK3β (Y216; 1:1000, Cell Signaling, Danvers, MA, USA), rabbit polyclonal anti-COX2 (1:200, Abcam), rabbit polyclonal anti-iNOS (1:500, Santa Cruz Biotechnology) and mouse monoclonal anti-β-actin (1:10 000, Sigma). Quantitative analysis was performed using NIH-based Scion Image software (Scion Corp., Frederick, MD, USA). Statistical analysis was performed using ratios of the densitometric value of each band normalized to β-actin as loading control.
Immunohistochemistry and histology
The remaining mice from each group were deeply anesthetized using sodium pentobarbital and transcardially perfused with ice-cold 0.9% sodium chloride and 4% paraformaldehyde. The brains were collected, dissected, post-fixed in 4% paraformaldehyde followed by gradient sucrose (15 and 30%) and stored in cryoprotectant for immunohistochemical studies.
Sections were cut at 50 μm thickness and stained with the following antibodies: MC1 mouse monoclonal anti-human tau (N-terminal conformational change, Exon 10) (1:500, gift from Dr Peter Davies), AT8 mouse monoclonal anti-human tau pSer202/Thr205 (1:500, Thermo Fisher Scientific, Rockford, IL, USA) and rat monoclonal anti-CD11b (1:1000, AbD Serotec, Raleigh, NC, USA). Immunolabeling was detected by the streptavidin-HRP method and visualized after diaminobenzidine incubation (Vector, Burlingame, CA, USA). Quantification was done using five 50 μm serial non-adjacent sections per animal (300 μm apart, from bregma −1.34 through bregma −2.84 through the cerebral cortex and hippocampus). The percentage area occupied by AT8 and the intensity (optical density) of CD11b were measured using Scion Image (Scion Corp.). For the cerebral cortex, measures were determined within a 0.9 mm2 area encompassing the primary (M1) and secondary (M2) motor cortex. The threshold was set at 140. For the hippocampus, the percent area occupied by AT8-labeled structures in the CA1 was determined. The intensity of CD11b-stained microglia in the hippocampus was measured within a 0.9 mm2 area using the dentate gyrus as an anatomical landmark. The threshold was set at 140. The number of neurons intensely stained with MC1 were counted using Scion Image (Scion Corp.). There were few MC1 immunoreactive neurons in both the cerebral cortex and the hippocampus. For this reason, stereological analysis was not used because this approach requires a larger number of countable cells by applying the random paradigm. For the cerebral cortex, the numbers of MC1-positive neurons were counted within a 0.9 mm2 area encompassing the primary (M1) and secondary (M2) motor cortex. For consistency, the cingulum was used as a landmark. For the hippocampus, the numbers of MC1-positive neurons were counted in the CA1 region of the hippocampus. Results are expressed as the mean of the numbers of MC1-positive neurons/section.
For the histology, perfused and fixed brown adipose tissues were cut at 16 μm thickness and mounted on slides. Sections were processed for hematoxylin–eosin and oil red O staining to visualize lipid vacuoles in the brown adipose.
Gene expression by quantitative real-time PCR
Fresh frozen tissues stored at −80°C were processed for RNA extraction (Qiagen kit, Valencia, CA, USA). Quantitative real-time PCR was performed at the Weill Cornell Medical College Microarray Core Facility using SyberGreen assays with the ABI Prism 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA). The following genes were analyzed: PPARα, PPARα, PPARα, GSK3α, ACOX1, CPT1A, HMGCS2, PGC1α, NRF1, Tfam, UCP1, Sirt1, Sirt3, COX2, iNOS and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as control. The primer sequences are listed in the Supplementary Material, Method.
Mitochondria characterization
Sample preparation
Dissected, non-perfused frontal lobe samples (∼30–55 mg) were stored frozen at −80°C until assaying. Before assays, tissue samples were thawed on ice and homogenized with Dounce-type 2 ml homogenizer (glass vessel/glass pestle). The homogenate was centrifuged at 1000g × 5 min to get rid of nuclear fraction and cell debris; the resulting supernatant was centrifuged at 14 000g × 5 min. The pellet was collected and centrifuged again at 14 000g × 5 min; the final pellet obtained in this step was resuspended in 20 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.8) and used for all assays.
Immunoblot analysis: mitochondria-enriched fraction
The protein lysates containing equal amounts of protein were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis, electroblotted onto a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA, USA) and immunoreacted with an appropriate primary antibody (see below), followed by HRP-conjugated secondary antibodies (Kierkegaard Perry Labs Inc., Gaithersburg, MD, USA). Immunoreactive proteins were visualized by incubating blots in chemiluminescence substrate (Pierce). Quantitative analysis was performed using NIH ‘Image J’ software. Statistical analysis was performed using ratios of the densitometric value of each band normalized to β-actin as a loading control.
Assays
All samples were assayed for the following: complex I activity (Nicotinamide adenine dinucleotide:CoQ reductase, rotenone-sensitive) (91), malic enzyme activity [malate:nicotinamide adenine dinucleotide phosphate (NADP) reductase], isocitric dehydrogenase activity (threo-Ds-isocitrate:NADP+ reductase), succinate dehydrogenase activity (succinate:CoQ:DCIP reductase, TTFA-sensitive) (92), citrate synthase activity (93), aconitase activity (‘Aconitase Assay Kit’, Cayman Chemical, Ann Arbor, MI, USA), glutathione reductase activity (‘Glutathione Reductase Assay Kit’, Cayman Chemical), superoxide dismutase activity (‘Superoxide Dismutase Assay Kit’, Cayman Chemical), ATPase subunit expression (immunoblotting with anti-ATPase ATP5A1 subunit 1:1000, Invitrogen) and protein carbonyls by 2,4-Dinitrophenol derivatization followed by immunoblotting (‘Oxyblot’ kit, Cayman Chemical). All activities and content values were normalized by protein content (Bicinchoninic acid protein assay, Thermo Scientific, FL, USA).
mtDNA copy number
Frozen tissues stored at −80°C were processed for DNA extraction according to the manufacturer's protocol (Qiagen kit). The relative mtDNA copy number was determined by quantitative real-time PCR on an ABI PRISM 7900H Sequence Detection System (Applied Biosystems) using the TaqMan® Universal PCR Master mix and predeveloped TaqMan® Gene Expression Assay primers/probes (Applied Biosystems) for mitochondrial cytochrome oxidase 2 and β-actin (nuclear DNA control). Results were calculated from the threshold cycle values and expressed as the 2−ΔCT of cytochrome oxidase 2 to β-actin.
Lipidomics-lipid extraction
Lipid extraction was performed using a modified Bligh and Dyer protocol and spiked with an internal standard mixture as described previously (81). Mass spectrometry analyses were done with an Agilent Technologies 6490 Ion Funnel liquid chromatography/mass spectrometry (LC/MS) Triple Quadrupole system with front end 1260 Infinity high-performance liquid chromatography (HPLC). Phospholipids and sphingolipids were analyzed by a normal-phase HPLC, while neutral lipids were analyzed using a reverse-phase HPLC. For normal-phase analysis, lipids were separated on a Phenomenex Luna silica column (i.d. 2.0 × 150 mm) using a gradient consisting of A: chloroform/methanol/ammonium hydroxide (90:9.5:0.5) and B: chloroform/methanol/water/ammonium hydroxide (55:39:5.5:0.5), starting at 5% and changing to 70% over a 40 min period as described previously (94). Neutral lipids were separated on an Agilent Zorbax XDB-C18 column (i.d. 4.6 × 100 mm) using an isocratic mobile-phase chloroform:methanol:0.1 m ammonium acetate (100:100:4) at a flow rate of 300 µl/min (94,95). The instrument capillary voltage, sheath gas flow rate, temperature and nebulizer pressure were set to 3000 V, 12 l/min, 300°C and 35 psi, respectively. Multiple reaction monitoring transitions were set-up for quantitative analysis of different lipid subclasses as described previously (81). The solvents employed for sample extractions and liquid chromatography were LC/MS or LC grade when the LC/MS grade was not available and were purchased from Sigma.
Free FA levels
Unesterified FA concentrations were determined for the brain and brown adipose samples using negative electrospray ionization-MS and the selected ion recording mode exactly as described by Clugston et al. (96). All measurements were carried out on a Waters Xevo TQ MS ACQUITY UPLC system (Waters, Milford, MA, USA). The system was controlled by Mass Lynx Software version 4.1. (Waters). The solvents employed for sample extractions and liquid chromatography were the LC/MS or LC grade when the LC/MS grade was not available and were purchased from Thermo Fisher (Pittsburgh, PA, USA).
Oxidative stress markers (HPLC)
Malondialdehyde levels
The HPLC determination of malondialdehyde (MDA) was carried out by a method modified from a previous report by Agarwal and Chase (97). Fresh tissues were homogenized in 40% ethanol solution; 50 μl of sample homogenate or MDA standard was prepared in 40% ethanol; 50 μl of 0.05% butylated hydroxytoluene, 400 μl of 0.44 m H3PO4 and 100 μl of 0.42 mm 2-thiobarbituric acid (TBA) were added to each. Samples were then vortexed, heated for 1 h at 100°C and immediately cooled with ice water to stop the derivative reaction. The MDA-TBA derivative was extracted by adding 250 μl n-butanol, followed by vortexing and centrifugation. About 50 μl of n-butanol extract was used for the HPLC assay. The HPLC mobile phase used acetonitrile buffer (20:80, v/v, buffer 50 mm KH2PO4, pH 6.8). The column was an ESA of 150 × 3 mm C18 column with particle size of 3 μm (ESA, Inc., Bedford, MA, USA). Fluorescence detectors were set at an excitation wavelength of 515 nm and emission wavelength of 553 nm. MDA was eluted from the column in 2 min. Data were normalized by protein content (Bio-Rad Protein Assay Kit).
GSH and GSSG levels
Brain tissues were homogenized and centrifuged in chilled 0.1 m perchloric acid. Supernatants were used for reduced (GSH) and oxidized (GSSG) glutathione measurements by HPLC, as described previously (98). Briefly, 15 μl of supernatant was isocratically eluated through a 4.6 × 150 mm C18 column (ESA, Inc., Chelmsford, MA, USA) with a mobile phase containing 50 mm LiH2PO4, 1.0 mm 1-octanesulfonic acid and 1.5% (v/v) methanol. The two-channel Coulochem III electrochemical detector (ESA, Inc., Chelmsford) was set with guard cell potential 950 mV, channel 1 potential 500 mm for GSH detection and channel 2 potential 880 mV for GSSG detection. Concentrations of both GSH and GSSG are expressed as nmol per mg of protein. Protein concentrations of tissue homogenates were measured using the Bio-Rad protein protocol (Bio-Rad Laboratories) and Perkin Elmer Bio Assay Reader (Norwalk, CT, USA).
Statistical analysis
For behavioral data, we used analysis of variance with repeated measurements followed by post hoc Fisher's PLSD tests for multiple comparisons (comparing four groups: wild-type mice fed a control diet, wild-type mice fed a bezafibrate diet, P301S mice fed a control diet and P301S mice fed a bezafibrate diet). To analyze any other data, we used two-tailed unpaired t-tests (comparing two groups: P301S mice fed a control diet and P301S mice fed a bezafibrate diet) and post hoc Fisher's PLSD tests for multiple comparisons (comparing four groups: wild-type mice fed a control diet, wild-type mice fed a bezafibrate diet, P301S mice fed a control diet and P301S mice fed a bezafibrate diet; Statview 5.0.1, SAS Institute Inc., Cary, NC, USA). All presented data were expressed as means ± standard errors of the means.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Tau Consortium and the National Institute of Aging (AG014930) to M.F.B. and by National Institute of Health (NIH) (P50 AG008702) to G.P. (PI: Michael Shelanski).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Peter Davies for providing the tau MC1 and DA9 antibodies. We also thank Dr William Blaner (Columbia University Medical Center) for his analysis of free fatty acids using LC/MS. We are also grateful to Serge Cremers and Tiffany Thomas (Columbia University Medical Center) as well as Bryan Lavery and Douglas Postl (Agilent Technologies) for help setting up the lipidomics core at Columbia University.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bensinger S.J., Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454:470–477. doi: 10.1038/nature07202. doi:10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- 2.Schulman I.G. Nuclear receptors as drug targets for metabolic disease. Adv. Drug Deliv. Rev. 2010;62:1307–1315. doi: 10.1016/j.addr.2010.07.002. doi:10.1016/j.addr.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y.X. PPARs: diverse regulators in energy metabolism and metabolic diseases. Cell Res. 2010;20:124–137. doi: 10.1038/cr.2010.13. doi:10.1038/cr.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandrekar-Colucci S., Landreth G.E. Nuclear receptors as therapeutic targets for Alzheimer's disease. Expert Opin. Ther. Targets. 2011;15:1085–1097. doi: 10.1517/14728222.2011.594043. doi:10.1517/14728222.2011.594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaundal R.K., Sharma S.S. Peroxisome proliferator-activated receptor gamma agonists as neuroprotective agents. Drug News Perspect. 2010;23:241–256. doi: 10.1358/dnp.2010.23.4.1437710. doi:10.1358/dnp.2010.23.4.1437710. [DOI] [PubMed] [Google Scholar]
- 6.Heneka M.T., Sastre M., Dumitrescu-Ozimek L., Hanke A., Dewachter I., Kuiperi C., O'Banion K., Klockgether T., Van Leuven F., Landreth G.E. Acute treatment with the PPARgamma agonist pioglitazone and ibuprofen reduces glial inflammation and Abeta1-42 levels in APPV717I transgenic mice. Brain. 2005;128:1442–1453. doi: 10.1093/brain/awh452. doi:10.1093/brain/awh452. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Q., Heneka M., Landreth G.E. The role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in Alzheimer's disease: therapeutic implications. CNS Drugs. 2008;22:1–14. doi: 10.2165/00023210-200822010-00001. doi:10.2165/00023210-200822010-00001. [DOI] [PubMed] [Google Scholar]
- 8.Nicolakakis N., Aboulkassim T., Ongali B., Lecrux C., Fernandes P., Rosa-Neto P., Tong X.K., Hamel E. Complete rescue of cerebrovascular function in aged Alzheimer's disease transgenic mice by antioxidants and pioglitazone, a peroxisome proliferator-activated receptor gamma agonist. J. Neurosci. 2008;28:9287–9296. doi: 10.1523/JNEUROSCI.3348-08.2008. doi:10.1523/JNEUROSCI.3348-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolakakis N., Hamel E. The nuclear receptor PPARgamma as a therapeutic target for cerebrovascular and brain dysfunction in Alzheimer's disease. Front. Aging Neurosci. 2010;2:1–10. doi: 10.3389/fnagi.2010.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulusoy G.K., Celik T., Kayir H., Gursoy M., Isik A.T., Uzbay T.I. Effects of pioglitazone and retinoic acid in a rotenone model of Parkinson's disease. Brain Res. Bull. 2011;85:380–384. doi: 10.1016/j.brainresbull.2011.05.001. doi:10.1016/j.brainresbull.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Kiaei M., Kipiani K., Chen J., Calingasan N.Y., Beal M.F. Peroxisome proliferator-activated receptor-gamma agonist extends survival in transgenic mouse model of amyotrophic lateral sclerosis. Exp. Neurol. 2005;191:331–336. doi: 10.1016/j.expneurol.2004.10.007. doi:10.1016/j.expneurol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Schutz B., Reimann J., Dumitrescu-Ozimek L., Kappes-Horn K., Landreth G.E., Schurmann B., Zimmer A., Heneka M.T. The oral antidiabetic pioglitazone protects from neurodegeneration and amyotrophic lateral sclerosis-like symptoms in superoxide dismutase-G93A transgenic mice. J. Neurosci. 2005;25:7805–7812. doi: 10.1523/JNEUROSCI.2038-05.2005. doi:10.1523/JNEUROSCI.2038-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalonia H., Kumar P., Kumar A. Pioglitazone ameliorates behavioral, biochemical and cellular alterations in quinolinic acid induced neurotoxicity: possible role of peroxisome proliferator activated receptor-Upsilon (PPARUpsilon) in Huntington's disease. Pharmacol. Biochem. Behav. 2010;96:115–124. doi: 10.1016/j.pbb.2010.04.018. doi:10.1016/j.pbb.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 14.Quintanilla R.A., Jin Y.N., Fuenzalida K., Bronfman M., Johnson G.V. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in the pathogenesis of Huntington disease. J. Biol. Chem. 2008;283:25628–25637. doi: 10.1074/jbc.M804291200. doi:10.1074/jbc.M804291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakhshandehroo M., Knoch B., Muller M., Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res., 2007. 2010:1–13. doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abourbih S., Filion K.B., Joseph L., Schiffrin E.L., Rinfret S., Poirier P., Pilote L., Genest J., Eisenberg M.J. Effect of fibrates on lipid profiles and cardiovascular outcomes: a systematic review. Am. J. Med. 2009;122:962.e1–968.e1. doi: 10.1016/j.amjmed.2009.03.030. doi:10.1016/j.amjmed.2009.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Munigoti S.P., Rees A. Evidence for use of fibrates in diabetic dyslipidemia: are we looking hard enough? Curr. Opin. Lipidol. 2011;22:76–77. doi: 10.1097/MOL.0b013e328342b0e9. doi:10.1097/MOL.0b013e328342b0e9. [DOI] [PubMed] [Google Scholar]
- 18.Staels B., Maes M., Zambon A. Fibrates and future PPARalpha agonists in the treatment of cardiovascular disease. Nat. Clin. Pract. Cardiovasc. Med. 2008;5:542–553. doi: 10.1038/ncpcardio1278. doi:10.1038/ncpcardio1278. [DOI] [PubMed] [Google Scholar]
- 19.Kreisler A., Duhamel A., Vanbesien-Mailliot C., Destee A., Bordet R. Differing short-term neuroprotective effects of the fibrates fenofibrate and bezafibrate in MPTP and 6-OHDA experimental models of Parkinson's disease. Behav. Pharmacol. 2010;21:194–205. doi: 10.1097/FBP.0b013e32833a5c81. doi:10.1097/FBP.0b013e32833a5c81. [DOI] [PubMed] [Google Scholar]
- 20.Chen X.R., Besson V.C., Palmier B., Garcia Y., Plotkine M., Marchand-Leroux C. Neurological recovery-promoting, anti-inflammatory, and anti-oxidative effects afforded by fenofibrate, a PPAR alpha agonist, in traumatic brain injury. J. Neurotrauma. 2007;24:1119–1131. doi: 10.1089/neu.2006.0216. doi:10.1089/neu.2006.0216. [DOI] [PubMed] [Google Scholar]
- 21.Arriagada P.V., Growdon J.H., Hedley-Whyte E.T., Hyman B.T. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer's disease. Neurology. 1992;42:631–639. doi: 10.1212/wnl.42.3.631. doi:10.1212/WNL.42.3.631. [DOI] [PubMed] [Google Scholar]
- 22.Medeiros R., Baglietto-Vargas D., LaFerla F.M. The role of tau in Alzheimer's disease and related disorders. CNS Neurosci. Ther. 2011;17:514–524. doi: 10.1111/j.1755-5949.2010.00177.x. doi:10.1111/j.1755-5949.2010.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.d'Abramo C., Massone S., Zingg J.M., Pizzuti A., Marambaud P., Dalla Piccola B., Azzi A., Marinari U.M., Pronzato M.A., Ricciarelli R. Role of peroxisome proliferator-activated receptor gamma in amyloid precursor protein processing and amyloid beta-mediated cell death. Biochem. J. 2005;391:693–698. doi: 10.1042/BJ20050560. doi:10.1042/BJ20050560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escribano L., Simon A.M., Gimeno E., Cuadrado-Tejedor M., Lopez de Maturana R., Garcia-Osta A., Ricobaraza A., Perez-Mediavilla A., Del Rio J., Frechilla D. Rosiglitazone rescues memory impairment in Alzheimer's transgenic mice: mechanisms involving a reduced amyloid and tau pathology. Neuropsychopharmacology. 2010;35:1593–1604. doi: 10.1038/npp.2010.32. doi:10.1038/npp.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.d'Abramo C., Ricciarelli R., Pronzato M.A., Davies P. Troglitazone, a peroxisome proliferator-activated receptor-gamma agonist, decreases tau phosphorylation in CHOtau4R cells. J. Neurochem. 2006;98:1068–1077. doi: 10.1111/j.1471-4159.2006.03931.x. doi:10.1111/j.1471-4159.2006.03931.x. [DOI] [PubMed] [Google Scholar]
- 26.Tenenbaum A., Motro M., Fisman E.Z. Dual and pan-peroxisome proliferator-activated receptors (PPAR) co-agonism: the bezafibrate lessons. Cardiovasc. Diabetol. 2005;4:14. doi: 10.1186/1475-2840-4-14. doi:10.1186/1475-2840-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haemmerle G., Moustafa T., Woelkart G., Buttner S., Schmidt A., van de Weijer T., Hesselink M., Jaeger D., Kienesberger P.C., Zierler K., et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat. Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. doi:10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee W.J., Kim M., Park H.S., Kim H.S., Jeon M.J., Oh K.S., Koh E.H., Won J.C., Kim M.S., Oh G.T., et al. AMPK activation increases fatty acid oxidation in skeletal muscle by activating PPARalpha and PGC-1. Biochem. Biophys. Res. Commun. 2006;340:291–295. doi: 10.1016/j.bbrc.2005.12.011. doi:10.1016/j.bbrc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Johri A., Calingasan N.Y., Hennessey T.M., Sharma A., Yang L., Wille E., Chandra A., Beal M.F. Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington's disease. Hum. Mol. Genet. 2011;21:1124–1137. doi: 10.1093/hmg/ddr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellucci A., Westwood A.J., Ingram E., Casamenti F., Goedert M., Spillantini M.G. Induction of inflammatory mediators and microglial activation in mice transgenic for mutant human P301S tau protein. Am. J. Pathol. 2004;165:1643–1652. doi: 10.1016/S0002-9440(10)63421-9. doi:10.1016/S0002-9440(10)63421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshiyama Y., Higuchi M., Zhang B., Huang S.M., Iwata N., Saido T.C., Maeda J., Suhara T., Trojanowski J.Q., Lee V.M. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. doi:10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Dumont M., Stack C., Elipenahli C., Jainuddin S., Gerges M., Starkova N.N., Yang L., Starkov A.A., Beal F. Behavioral deficit, oxidative stress, and mitochondrial dysfunction precede tau pathology in P301S transgenic mice. FASEB J. 2011;25:4063–4072. doi: 10.1096/fj.11-186650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scattoni M.L., Gasparini L., Alleva E., Goedert M., Calamandrei G., Spillantini M.G. Early behavioural markers of disease in P301S tau transgenic mice. Behav. Brain Res. 2010;208:250–257. doi: 10.1016/j.bbr.2009.12.002. doi:10.1016/j.bbr.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Jicha G.A., Bowser R., Kazam I.G., Davies P. Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J. Neurosci. Res. 1997;48:128–132. doi: 10.1002/(sici)1097-4547(19970415)48:2<128::aid-jnr5>3.0.co;2-e. doi:10.1002/(SICI)1097-4547(19970415)48:2<128::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 35.Weaver C.L., Espinoza M., Kress Y., Davies P. Conformational change as one of the earliest alterations of tau in Alzheimer's disease. Neurobiol. Aging. 2000;21:719–727. doi: 10.1016/s0197-4580(00)00157-3. doi:10.1016/S0197-4580(00)00157-3. [DOI] [PubMed] [Google Scholar]
- 36.Biernat J., Mandelkow E.M., Schroter C., Lichtenberg-Kraag B., Steiner B., Berling B., Meyer H., Mercken M., Vandermeeren A., Goedert M., et al. The switch of tau protein to an Alzheimer-like state includes the phosphorylation of two serine-proline motifs upstream of the microtubule binding region. EMBO J. 1992;11:1593–1597. doi: 10.1002/j.1460-2075.1992.tb05204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braak H., Braak E. Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol. Aging. 1995;16:271–278. doi: 10.1016/0197-4580(95)00021-6. discussion 278–284 doi:10.1016/0197-4580(95)00021-6. [DOI] [PubMed] [Google Scholar]
- 38.Mi K., Johnson G.V. The role of tau phosphorylation in the pathogenesis of Alzheimer's disease. Curr. Alzheimer Res. 2006;3:449–463. doi: 10.2174/156720506779025279. doi:10.2174/156720506779025279. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez F., de Barreda E.G., Fuster-Matanzo A., Goni-Oliver P., Lucas J.J., Avila J. The role of GSK3 in Alzheimer disease. Brain Res. Bull. 2009;80:248–250. doi: 10.1016/j.brainresbull.2009.05.017. doi:10.1016/j.brainresbull.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Plattner F., Angelo M., Giese K.P. The roles of cyclin-dependent kinase 5 and glycogen synthase kinase 3 in tau hyperphosphorylation. J. Biol. Chem. 2006;281:25457–25465. doi: 10.1074/jbc.M603469200. doi:10.1074/jbc.M603469200. [DOI] [PubMed] [Google Scholar]
- 41.Soutar M.P., Kim W.Y., Williamson R., Peggie M., Hastie C.J., McLauchlan H., Snider W.D., Gordon-Weeks P.R., Sutherland C. Evidence that glycogen synthase kinase-3 isoforms have distinct substrate preference in the brain. J. Neurochem. 2010;115:974–983. doi: 10.1111/j.1471-4159.2010.06988.x. doi:10.1111/j.1471-4159.2010.06988.x. [DOI] [PubMed] [Google Scholar]
- 42.Fernandes-Santos C., Carneiro R.E., de Souza Mendonca L., Aguila M.B., Mandarim-de-Lacerda C.A. Pan-PPAR agonist beneficial effects in overweight mice fed a high-fat high-sucrose diet. Nutrition. 2009;25:818–827. doi: 10.1016/j.nut.2008.12.010. doi:10.1016/j.nut.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Chaturvedi R.K., Beal M.F. PPAR: a therapeutic target in Parkinson's disease. J. Neurochem. 2008;106:506–518. doi: 10.1111/j.1471-4159.2008.05388.x. doi:10.1111/j.1471-4159.2008.05388.x. [DOI] [PubMed] [Google Scholar]
- 44.Heneka M.T., Landreth G.E. PPARs in the brain. Biochim. Biophys. Acta. 2007;1771:1031–1045. doi: 10.1016/j.bbalip.2007.04.016. doi:10.1016/j.bbalip.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Landreth G. Therapeutic use of agonists of the nuclear receptor PPARgamma in Alzheimer's disease. Curr. Alzheimer Res. 2007;4:159–164. doi: 10.2174/156720507780362092. doi:10.2174/156720507780362092. [DOI] [PubMed] [Google Scholar]
- 46.Escribano L., Simon A.M., Perez-Mediavilla A., Salazar-Colocho P., Del Rio J., Frechilla D. Rosiglitazone reverses memory decline and hippocampal glucocorticoid receptor down-regulation in an Alzheimer's disease mouse model. Biochem. Biophys. Res. Commun. 2009;379:406–410. doi: 10.1016/j.bbrc.2008.12.071. doi:10.1016/j.bbrc.2008.12.071. [DOI] [PubMed] [Google Scholar]
- 47.Strum J.C., Shehee R., Virley D., Richardson J., Mattie M., Selley P., Ghosh S., Nock C., Saunders A., Roses A. Rosiglitazone induces mitochondrial biogenesis in mouse brain. J. Alzheimers Dis. 2007;11:45–51. doi: 10.3233/jad-2007-11108. [DOI] [PubMed] [Google Scholar]
- 48.Tzimopoulou S., Cunningham V.J., Nichols T.E., Searle G., Bird N.P., Mistry P., Dixon I.J., Hallett W.A., Whitcher B., Brown A.P., et al. A multi-center randomized proof-of-concept clinical trial applying [(1)F]FDG-PET for evaluation of metabolic therapy with rosiglitazone XR in mild to moderate Alzheimer's disease. J. Alzheimers Dis. 2010;22:1241–1256. doi: 10.3233/JAD-2010-100939. [DOI] [PubMed] [Google Scholar]
- 49.Hanyu H., Sato T., Sakurai H., Iwamoto T. The role of tumor necrosis factor-alpha in cognitive improvement after peroxisome proliferator-activator receptor gamma agonist pioglitazone treatment in Alzheimer's disease. J. Am. Geriatr. Soc. 2010;58:1000–1001. doi: 10.1111/j.1532-5415.2010.02841.x. doi:10.1111/j.1532-5415.2010.02841.x. [DOI] [PubMed] [Google Scholar]
- 50.Geldmacher D.S., Fritsch T., McClendon M.J., Landreth G. A randomized pilot clinical trial of the safety of pioglitazone in treatment of patients with Alzheimer disease. Arch. Neurol. 2011;68:45–50. doi: 10.1001/archneurol.2010.229. doi:10.1001/archneurol.2010.229. [DOI] [PubMed] [Google Scholar]
- 51.Gold M., Alderton C., Zvartau-Hind M., Egginton S., Saunders A.M., Irizarry M., Craft S., Landreth G., Linnamagi U., Sawchak S. Rosiglitazone monotherapy in mild-to-moderate Alzheimer's disease: results from a randomized, double-blind, placebo-controlled phase III study. Dement. Geriatr. Cogn. Disord. 2010;30:131–146. doi: 10.1159/000318845. doi:10.1159/000318845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrington C., Sawchak S., Chiang C., Davies J., Donovan C., Saunders A.M., Irizarry M., Jeter B., Zvartau-Hind M., van Dyck C.H., et al. Rosiglitazone does not improve cognition or global function when used as adjunctive therapy to AChE inhibitors in mild-to-moderate Alzheimer's disease: two phase 3 studies. Curr. Alzheimer Res. 2011;8:592–606. doi: 10.2174/156720511796391935. [DOI] [PubMed] [Google Scholar]
- 53.Selkoe D.J. Resolving controversies on the path to Alzheimer's therapeutics. Nat. Med. 2011;17:1060–1065. doi: 10.1038/nm.2460. doi:10.1038/nm.2460. [DOI] [PubMed] [Google Scholar]
- 54.Spires-Jones T.L., Kopeikina K.J., Koffie R.M., de Calignon A., Hyman B.T. Are tangles as toxic as they look? J. Mol. Neurosci. 2011;45:438–444. doi: 10.1007/s12031-011-9566-7. doi:10.1007/s12031-011-9566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frost B., Jacks R.L., Diamond M.I. Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. doi:10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meraz-Rios M.A., Lira-De Leon K.I., Campos-Pena V., De Anda-Hernandez M.A., Mena-Lopez R. Tau oligomers and aggregation in Alzheimer's disease. J. Neurochem. 2010;112:1353–1367. doi: 10.1111/j.1471-4159.2009.06511.x. doi:10.1111/j.1471-4159.2009.06511.x. [DOI] [PubMed] [Google Scholar]
- 57.Patterson K.R., Remmers C., Fu Y., Brooker S., Kanaan N.M., Vana L., Ward S., Reyes J.F., Philibert K., Glucksman M.J., et al. Characterization of prefibrillar Tau oligomers in vitro and in Alzheimer disease. J. Biol. Chem. 2011;286:23063–23076. doi: 10.1074/jbc.M111.237974. doi:10.1074/jbc.M111.237974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldenberg I., Benderly M., Goldbourt U. Update on the use of fibrates: focus on bezafibrate. Vasc. Health Risk Manag. 2008;4:131–141. doi: 10.2147/vhrm.2008.04.01.131. doi:10.2147/vhrm.2008.04.01.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wenz T., Diaz F., Spiegelman B.M., Moraes C.T. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. doi:10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Viscomi C., Bottani E., Civiletto G., Cerutti R., Moggio M., Fagiolari G., Schon E.A., Lamperti C., Zeviani M. In vivo correction of COX deficiency by activation of the AMPK/PGC-1alpha axis. Cell Metab. 2011;14:80–90. doi: 10.1016/j.cmet.2011.04.011. doi:10.1016/j.cmet.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baba Y., Baker M.C., Le Ber I., Brice A., Maeck L., Kohlhase J., Yasuda M., Stoppe G., Bugiani O., Sperfeld A.D., et al. Clinical and genetic features of families with frontotemporal dementia and parkinsonism linked to chromosome 17 with a P301S tau mutation. J. Neural. Transm. 2007;114:947–950. doi: 10.1007/s00702-007-0632-9. doi:10.1007/s00702-007-0632-9. [DOI] [PubMed] [Google Scholar]
- 62.Lossos A., Reches A., Gal A., Newman J.P., Soffer D., Gomori J.M., Boher M., Ekstein D., Biran I., Meiner Z., et al. Frontotemporal dementia and parkinsonism with the P301S tau gene mutation in a Jewish family. J. Neurol. 2003;250:733–740. doi: 10.1007/s00415-003-1074-4. doi:10.1007/s00415-003-1074-4. [DOI] [PubMed] [Google Scholar]
- 63.Morris H.R., Khan M.N., Janssen J.C., Brown J.M., Perez-Tur J., Baker M., Ozansoy M., Hardy J., Hutton M., Wood N.W., et al. The genetic and pathological classification of familial frontotemporal dementia. Arch. Neurol. 2001;58:1813–1816. doi: 10.1001/archneur.58.11.1813. doi:10.1001/archneur.58.11.1813. [DOI] [PubMed] [Google Scholar]
- 64.Hoffman A., Lomnicky Y., Luria M.H., Gilhar D., Friedman M. Improved lipid lowering activity of bezafibrate following continuous gastrointestinal administration: pharmacodynamic rationale for sustained release preparation of the drug. Pharm. Res. 1999;16:1093–1097. doi: 10.1023/a:1018900219616. doi:10.1023/A:1018900219616. [DOI] [PubMed] [Google Scholar]
- 65.Nakajima T., Tanaka N., Kanbe H., Hara A., Kamijo Y., Zhang X., Gonzalez F.J., Aoyama T. Bezafibrate at clinically relevant doses decreases serum/liver triglycerides via down-regulation of sterol regulatory element-binding protein-1c in mice: a novel peroxisome proliferator-activated receptor alpha-independent mechanism. Mol. Pharmacol. 2009;75:782–792. doi: 10.1124/mol.108.052928. doi:10.1124/mol.108.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakajima T., Tanaka N., Li G., Hu R., Kamijo Y., Hara A., Aoyama T. Effect of bezafibrate on hepatic oxidative stress: comparison between conventional experimental doses and clinically-relevant doses in mice. Redox Rep. 2010;15:123–130. doi: 10.1179/174329210X12650506623807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peters J.M., Aoyama T., Burns A.M., Gonzalez F.J. Bezafibrate is a dual ligand for PPARalpha and PPARbeta: studies using null mice. Biochim. Biophys. Acta. 2003;1632:80–89. doi: 10.1016/s1388-1981(03)00065-9. doi:10.1016/S1388-1981(03)00065-9. [DOI] [PubMed] [Google Scholar]
- 68.Poirier H., Niot I., Monnot M.C., Braissant O., Meunier-Durmort C., Costet P., Pineau T., Wahli W., Willson T.M., Besnard P. Differential involvement of peroxisome-proliferator-activated receptors alpha and delta in fibrate and fatty-acid-mediated inductions of the gene encoding liver fatty-acid-binding protein in the liver and the small intestine. Biochem. J. 2001;355:481–488. doi: 10.1042/0264-6021:3550481. doi:10.1042/0264-6021:3550481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yatsuga S., Suomalainen A. Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum. Mol. Genet. 2011;21:526–535. doi: 10.1093/hmg/ddr482. [DOI] [PubMed] [Google Scholar]
- 70.Deplanque D., Gele P., Petrault O., Six I., Furman C., Bouly M., Nion S., Dupuis B., Leys D., Fruchart J.C., et al. Peroxisome proliferator-activated receptor-alpha activation as a mechanism of preventive neuroprotection induced by chronic fenofibrate treatment. J. Neurosci. 2003;23:6264–6271. doi: 10.1523/JNEUROSCI.23-15-06264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dolan P.J., Johnson G.V. The role of tau kinases in Alzheimer's disease. Curr. Opin. Drug Discov. Devel. 2010;13:595–603. [PMC free article] [PubMed] [Google Scholar]
- 72.Virshup D.M., Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. doi:10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 73.Li R., Zheng W., Pi R., Gao J., Zhang H., Wang P., Le K., Liu P. Activation of peroxisome proliferator-activated receptor-alpha prevents glycogen synthase 3beta phosphorylation and inhibits cardiac hypertrophy. FEBS Lett. 2007;581:3311–3316. doi: 10.1016/j.febslet.2007.06.017. doi:10.1016/j.febslet.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 74.Sharma C., Pradeep A., Wong L., Rana A., Rana B. Peroxisome proliferator-activated receptor gamma activation can regulate beta-catenin levels via a proteasome-mediated and adenomatous polyposis coli-independent pathway. J. Biol. Chem. 2004;279:35583–35594. doi: 10.1074/jbc.M403143200. doi:10.1074/jbc.M403143200. [DOI] [PubMed] [Google Scholar]
- 75.Li M.D., Yang X. A retrospective on nuclear receptor regulation of inflammation: lessons from GR and PPARs. PPAR Res. 2011;2011:742–785. doi: 10.1155/2011/742785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang G., Namura S. Effects of chronic systemic treatment with peroxisome proliferator-activated receptor alpha activators on neuroinflammation induced by intracerebral injection of lipopolysaccharide in adult mice. Neurosci. Res. 2011;70:230–237. doi: 10.1016/j.neures.2011.02.001. doi:10.1016/j.neures.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kapoor A., Shintani Y., Collino M., Osuchowski M.F., Busch D., Patel N.S., Sepodes B., Castiglia S., Fantozzi R., Bishop-Bailey D., et al. Protective role of peroxisome proliferator-activated receptor-beta/delta in septic shock. Am. J. Respir. Crit. Care Med. 2010;182:1506–1515. doi: 10.1164/rccm.201002-0240OC. doi:10.1164/rccm.201002-0240OC. [DOI] [PubMed] [Google Scholar]
- 78.Nakamura M.T., Cheon Y., Li Y., Nara T.Y. Mechanisms of regulation of gene expression by fatty acids. Lipids. 2004;39:1077–1083. doi: 10.1007/s11745-004-1333-0. doi:10.1007/s11745-004-1333-0. [DOI] [PubMed] [Google Scholar]
- 79.Pyper S.R., Viswakarma N., Yu S., Reddy J.K. PPARalpha: energy combustion, hypolipidemia, inflammation and cancer. Nucl. Recept. Signal. 2010;8:e002. doi: 10.1621/nrs.08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cullingford T.E., Dolphin C.T., Sato H. The peroxisome proliferator-activated receptor alpha-selective activator ciprofibrate upregulates expression of genes encoding fatty acid oxidation and ketogenesis enzymes in rat brain. Neuropharmacology. 2002;42:724–730. doi: 10.1016/s0028-3908(02)00014-x. doi:10.1016/S0028-3908(02)00014-X. [DOI] [PubMed] [Google Scholar]
- 81.Chan R.B., Oliveira T.G., Cortes E.P., Honig L.S., Duff K.E., Small S.A., Wenk M.R., Shui G., Di Paolo G. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J. Biol. Chem. 2012;287:2678–2688. doi: 10.1074/jbc.M111.274142. doi:10.1074/jbc.M111.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Birnbaum S.G., Yuan P.X., Wang M., Vijayraghavan S., Bloom A.K., Davis D.J., Gobeske K.T., Sweatt J.D., Manji H.K., Arnsten A.F. Protein kinase C overactivity impairs prefrontal cortical regulation of working memory. Science. 2004;306:882–884. doi: 10.1126/science.1100021. doi:10.1126/science.1100021. [DOI] [PubMed] [Google Scholar]
- 83.Feige J.N., Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. doi:10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 84.Fernandez-Marcos P.J., Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011;93:884S–8890. doi: 10.3945/ajcn.110.001917. doi:10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmitt K., Grimm A., Kazmierczak A., Strosznajder J.B., Gotz J., Eckert A. Insights into mitochondrial dysfunction: aging, amyloid-beta and tau - a deleterious trio. Antioxid. Redox Signal. 2011;16:1456–1466. doi: 10.1089/ars.2011.4400. [DOI] [PubMed] [Google Scholar]
- 86.Richard D., Picard F. Brown fat biology and thermogenesis. Front. Biosci. 2011;16:1233–1260. doi: 10.2741/3786. doi:10.2741/3786. [DOI] [PubMed] [Google Scholar]
- 87.Goto T., Lee J.Y., Teraminami A., Kim Y.I., Hirai S., Uemura T., Inoue H., Takahashi N., Kawada T. Activation of peroxisome proliferator-activated receptor-alpha stimulates both differentiation and fatty acid oxidation in adipocytes. J. Lipid Res. 2011;52:873–884. doi: 10.1194/jlr.M011320. doi:10.1194/jlr.M011320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tremblay-Mercier J., Tessier D., Plourde M., Fortier M., Lorrain D., Cunnane S.C. Bezafibrate mildly stimulates ketogenesis and fatty acid metabolism in hypertriglyceridemic subjects. J. Pharmacol. Exp. Ther. 2010;334:341–346. doi: 10.1124/jpet.110.166504. doi:10.1124/jpet.110.166504. [DOI] [PubMed] [Google Scholar]
- 89.Wenz T., Rossi S.G., Rotundo R.L., Spiegelman B.M., Moraes C.T. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc. Natl Acad. Sci. USA. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. doi:10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Dumont M., Wille E., Calingasan N.Y., Nathan C., Flint Beal M., Lin M.T. N-iminoethyl-L-lysine improves memory and reduces amyloid pathology in a transgenic mouse model of amyloid deposition. Neurochem. Int. 2010;56:345–351. doi: 10.1016/j.neuint.2009.11.006. doi:10.1016/j.neuint.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 91.Degli Esposti M., Ghelli A., Crimi M., Estornell E., Fato R., Lenaz G. Complex I and complex III of mitochondria have common inhibitors acting as ubiquinone antagonists. Biochem. Biophys. Res. Commun. 1993;190:1090–1096. doi: 10.1006/bbrc.1993.1161. doi:10.1006/bbrc.1993.1161. [DOI] [PubMed] [Google Scholar]
- 92.Arrigoni O., Singer T.P. Limitations of the phenazine methosulphate assay for succinic and related dehydrogenases. Nature. 1962;193:1256–1258. doi: 10.1038/1931256a0. doi:10.1038/1931256a0. [DOI] [PubMed] [Google Scholar]
- 93.Srere P.A. Citrate synthase. Methods Enzymol. 1969;13:3–11. [Google Scholar]
- 94.Shui G., Guan X.L., Gopalakrishnan P., Xue Y., Goh J.S., Yang H., Wenk M.R. Characterization of substrate preference for Slc1p and Cst26p in Saccharomyces cerevisiae using lipidomic approaches and an LPAAT activity assay. PLoS One. 2010;5:e11956. doi: 10.1371/journal.pone.0011956. doi:10.1371/journal.pone.0011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shui G., Guan X.L., Low C.P., Chua G.H., Goh J.S., Yang H., Wenk M.R. Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol. Biosyst. 2010;6:1008–1017. doi: 10.1039/b913353d. doi:10.1039/b913353d. [DOI] [PubMed] [Google Scholar]
- 96.Clugston R.D., Jiang H., Lee M.X., Piantedosi R., Yuen J.J., Ramakrishnan R., Lewis M.J., Gottesman M.E., Huang L.S., Goldberg I.J., et al. Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study. J. Lipid Res. 2011;52:2021–2031. doi: 10.1194/jlr.M017368. doi:10.1194/jlr.M017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Agarwal R., Chase S.D. Rapid, fluorimetric-liquid chromatographic determination of malondialdehyde in biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2002;775:121–126. doi: 10.1016/s1570-0232(02)00273-8. doi:10.1016/S1570-0232(02)00273-8. [DOI] [PubMed] [Google Scholar]
- 98.Melnyk S., Pogribna M., Pogribny I., Hine R.J., James S.J. A new HPLC method for the simultaneous determination of oxidized and reduced plasma aminothiols using coulometric electrochemical detection. J. Nutr. Biochem. 1999;10:490–497. doi: 10.1016/s0955-2863(99)00033-9. doi:10.1016/S0955-2863(99)00033-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.