Abstract
To further characterize the genetic basis of primary biliary cirrhosis (PBC), we genotyped 2426 PBC patients and 5731 unaffected controls from three independent cohorts using a single nucleotide polymorphism (SNP) array (Immunochip) enriched for autoimmune disease risk loci. Meta-analysis of the genotype data sets identified a novel disease-associated locus near the TNFSF11 gene at 13q14, provided evidence for association at six additional immune-related loci not previously implicated in PBC and confirmed associations at 19 of 22 established risk loci. Results of conditional analyses also provided evidence for multiple independent association signals at four risk loci, with haplotype analyses suggesting independent SNP effects at the 2q32 and 16p13 loci, but complex haplotype driven effects at the 3q25 and 6p21 loci. By imputing classical HLA alleles from this data set, four class II alleles independently contributing to the association signal from this region were identified. Imputation of genotypes at the non-HLA loci also provided additional associations, but none with stronger effects than the genotyped variants. An epistatic interaction between the IL12RB2 risk locus at 1p31and the IRF5 risk locus at 7q32 was also identified and suggests a complementary effect of these loci in predisposing to disease. These data expand the repertoire of genes with potential roles in PBC pathogenesis that need to be explored by follow-up biological studies.
INTRODUCTION
Primary biliary cirrhosis (PBC) is an autoimmune liver disease affecting 1 in 1000 women over the age of 40 and attributed to complex interactions of multiple environmental and genetic risk factors (1). The only available medical therapy, ursodeoxycholic acid, is ineffective in about half of all cases, but development of more widely efficacious therapies has been hindered by limited understanding of disease pathogenesis (2). At least 90% of PBC patients express anti-mitochondrial antibodies (AMA) reactive with the ubiquitously expressed E2 subunit of the pyruvate dehydrogenase complex (PDC-E2) (3). Despite the broad cellular expression of PDC-E2, the pathogenic immune inflammatory response in PBC appears to be targeted to the biliary epithelial cells (BECs) lining the small interlobular bile ducts. Among their many properties, BECs are thought to play important roles in maintaining immune homeostasis in the local cellular environment (4). The mechanisms whereby the biliary epithelia is specifically targeted for immunologic damage in PBC are not well understood, but it has been suggested that an inability of BECs to glutathiolate PDC-E2 during cell apoptosis may result in aberrant persistence of immunogenic PDC-E2 antigens and downstream priming and activation of T cells in the draining lymph nodes (5). Microbial mimicry and xenobiotic modification have also been proposed as mechanisms driving the loss of tolerance underpinning PBC (6).
As for most other autoimmune diseases, underlying genetic variation is considered key to the etiology of PBC (7). This possibility is supported by many lines of evidence, including familial clustering and high sibling risk for PBC, disease concordance among monozygotic twins, concomitant occurrence of PBC with other autoimmune diseases and data from numerous studies of animal models (8). Early efforts to identify PBC genes were hampered by the rarity and late onset of disease, which precluded familial linkage studies. Candidate gene approaches identified a number of associated gene variants, but only a few, most notably variants in the human leukocyte antigen (HLA) and CTLA-4 genes, proved to be replicable across independent studies (9). More recently, through genome-wide association surveys (GWAS) and fine-mapping studies, we identified six non-HLA loci reaching genome-wide significance (P < 5 × 10−8) for association with PBC susceptibility (10–12). A subsequent UK-based GWAS confirmed these associations and identified an additional 15 risk loci (13). Among the PBC loci identified to date, most have also been implicated in risk for other autoimmune diseases and contain genes with key roles in immune cell signaling and/or development. However, functional significance of these associations and the disease-causal alleles within the risk loci remain unknown. We therefore sought to further characterize the known PBC loci and search for additional risk loci, by genotyping three independent PBC patient cohorts with a customized SNP array, the Immunochip, designed to interrogate 186 autoimmune disease loci primarily identified in prior GWAS of PBC and other autoimmune diseases (14).
RESULTS
Study design, patient cohorts and quality control
Our study design involved a meta-analysis of the Immunochip data sets derived from genotyping three independent cohorts of PBC patients and population controls, all of European descent and collected, respectively, from Canada, the USA and Italy. In total, 2426 PBC patients and 5731 unaffected controls were genotyped for 196 524 genetic variants using the Immunochip platform. Quality control measures, including the exclusion of population outliers, were performed separately for each cohort. Prior to meta-analysis, markers not present in all three populations, as well as those demonstrating a significant minor allele frequency (MAF) difference (P < 0.001) between PBC patients from the Canadian and US cohorts were removed. Following these procedures, 109 812 markers were available for study in 2216 PBC patients and 5594 unaffected controls. An overview of the study design, quality control filters and patient cohorts, as well as demographics of retained patients and controls is provided in Supplementary Material, Figure S1 and Supplementary Material, Table S1.
Associations detected at previously identified PBC risk loci
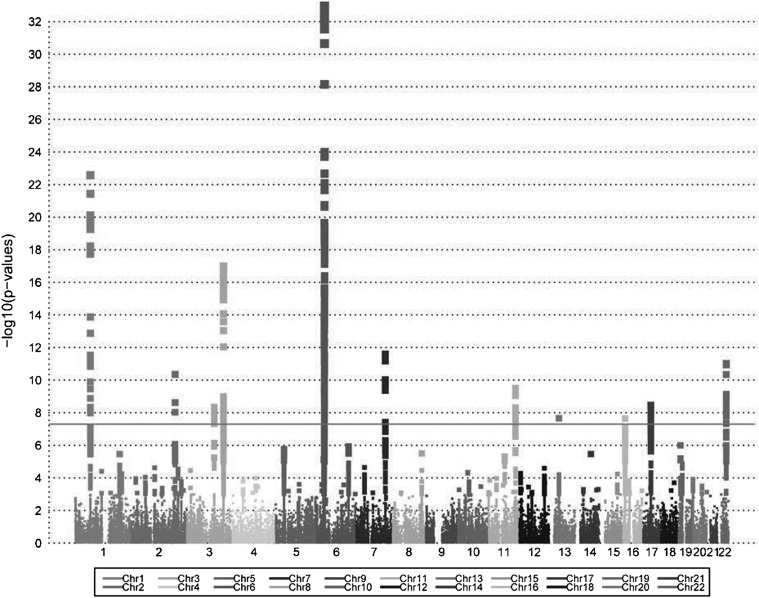
Meta-analysis encompassing all three individually analyzed data sets (Fig. 1) was performed using METAL (15), assuming a fixed effects model. Among the seven risk loci initially identified in studies of Canadian and Italian PBC patients (10–12), five harbored SNPs achieving genome-wide significance in the meta-analysis (1p31, 3q25, 6p21, 7q32 and 17q12), with strong support for association across each individual cohort (Table 1 and Supplementary Material, Table S2). Regarding the other two loci, Immunochip coverage for one, the 19q13 locus, was sparse and provided no good proxy for the SNP previously shown to be associated with risk (rs3745516). This locus was, however, strongly replicated in the UK GWAS (13) and our recent fine-mapping study (16), providing compelling evidence for its involvement in risk for PBC. In contrast, despite dense coverage of the 1p36 region encompassing the MMEL1 gene, evidence for this region's association with disease was weak in the meta-analysis, the individual cohort analysis and also in the prior UK GWAS (13). While the peak SNP identified in our original report of this association (rs3748816) (11) differs from those observed in the UK (rs10752747) and in this study (rs10910108), these three SNPs are in strong LD (pairwise r2 ≥ 0.85), and thus the relevance of this locus to risk for PBC is unclear.
Figure 1.
Manhattan plot illustrating the results of a meta-analysis of Immunochip association studies of three independent cohorts of PBC patients and unaffected controls.
Table 1.
Results of meta-analysis of three independent PBC case–control cohorts at previously reported PBC risk loci
| Locus | GWASa | Immunochip |
Peak SNP | Locationc | RA | OR (95% CI) | P-value | Gene(s)d | Location | |
|---|---|---|---|---|---|---|---|---|---|---|
| Region (Mb) | Coverageb | |||||||||
| 1p31 | Canada1 | 67.57–67.96 | Dense | rs72678531 | 67798445 | C | 1.68 (1.51–1.86) | 2.66 × 10−23 | IL12RB2 | Intron |
| 1p36 | Canada2 | 2.40–2.79 | Dense | rs10910108 | 2539532 | G | 1.15 (1.05–1.25) | 1.81 × 10−3 | MMEL1 | Intron |
| 3q25 | Canada1 | 159.55–159.82 | Dense | rs9877910 | 159730819 | T | 1.47 (1.34–1.60) | 1.02 × 10−17 | IL12A/IFT80 | Intergenic |
| 6p21 | Canada1 | 27.96–34.07 | Dense | rs7775055 | 32657916 | C | 3.71 (3.00–4.59) | 1.11 × 10−33 | HLA-DQB1/HLA-DQA2 | Intergenic |
| 7q32 | Canada2 | 128.55–128.78 | Dense | rs10488631 | 128594183 | C | 1.56 (1.38–1.77) | 2.52 × 10−12 | IRF5/TNPO3 | Intergenic |
| 17q12 | Canada2 | 37.39–38.25 | Dense | rs907091 | 37921742 | C | 1.29 (1.19–1.41) | 3.43 × 10−9 | IKZF3 | UTR |
| 19q13 | Italy3 | 50.03–51.80 | Sparse | rs2546551 | 50896360 | T | 1.13 (1.04–1.24) | 3.80 × 10−3 | POLD | Intron |
| 1q31 | UK4 | 197.31–197.94 | Dense | rs1539414 | 197743506 | A | 1.26 (1.14–1.39) | 3.46 × 10−6 | DENND1B | Intron |
| 2q32 | UK4 | 191.85–192.01 | Dense | rs3024921 | 191943272 | A | 1.75 (1.48–2.07) | 4.45 × 10−11 | STAT4 | Intron |
| 3p24 | UK4 | 16.74–17.09 | Sparse | rs1025818 | 16965021 | G | 1.20 (1.10–1.31) | 3.53 × 10−5 | PLCL2 | Intron |
| 3q13 | UK4 | 119.11–119.33 | Dense | rs1131265 | 119222456 | G | 1.42 (1.26–1.60) | 4.49 × 10−9 | TIMMDC1 | Exon (syn) |
| 4q24 | UK4 | 103.42–103.65 | Sparse | rs7665090 | 103551603 | G | 1.18 (1.09–1.28) | 1.00 × 10−4 | NFKB1/MANBA | Intergenic |
| 5p13 | UK4 | 35.77–36.03 | Dense | rs700172 | 35934157 | A | 1.27 (1.15–1.40) | 1.63 × 10−6 | CAPSL | Intron |
| 7p14 | UK4 | 37.36–37.47 | Dense | rs17259795 | 37398273 | T | 1.24 (1.13–1.37) | 1.19 × 10−5 | ELMO1 | Intron |
| 11q13 | UK4 | 64.09–64.21 | Sparse | rs694739 | 64097233 | A | 1.18 (1.08–1.29) | 2.85 × 10−4 | PRDX5/CCDC88B | Intergenic |
| 11q23 | UK4 | 118.34–118.77 | Dense | rs7117261 | 118741157 | C | 1.46 (1.30–1.64) | 3.18 × 10−10 | DDX6/CXCR5 | Intergenic |
| 12p13 | UK4 | 6.41–6.53 | Dense | rs1860545 | 6446777 | T | 1.20 (1.10–1.30) | 5.27 × 10−5 | TNFRSF1A | Intron |
| 14q24 | UK4 | 68.75–69.12 | Sparse | rs911263 | 68753593 | A | 1.25 (1.14–1.37) | 3.43 × 10−6 | RAD51L1 | Intron |
| 14q32 | UK4 | 102.98–103.86 | Sparse | rs12148050 | 103263788 | G | 1.09 (1.00–1.19) | 0.0491 | TRAF3 | Intron |
| 16p13 | UK4 | 10.90–11.53 | Dense | rs413024 | 11354091 | T | 1.31 (1.19–1.44) | 2.29 × 10−8 | SOCS1/TNP2 | Intergenic |
| 16q24 | UK4 | 85.92–86.03 | Dense | rs35703946 | 86021505 | G | 1.27 (1.11–1.45) | 6.10 × 10−4 | IRF8/FOXF1 | Intergenic |
| 22q13 | UK4 | 39.66–39.78 | Dense | rs715505 | 39751251 | C | 1.41 (1.28–1.60) | 9.58 × 10−12 | SYNGR1 | Intron |
RA, risk allele; OR, odds ratio; 95%CI, 95% confidence interval.
aGWAS reference: (1) Hirschfield (10); (2) Hirschfield (11); (3) Liu (12); 4. Mells (13); italics denotes unconfirmed finding from meta-analysis. bApproximate coverage density of markers on the Immunochip with Dense denoting >100 markers in region and Sparse denoting <20 markers in region. cGRCh37 assembly. dGene in which the peak signal is located in, or genes flanking peak signal if intergenic.
The 15 PBC risk loci newly reported in the recent UK GWAS (13) have not yet been validated in independent studies. Of these, five achieved genome-wide significance in our meta-analysis (2q32, 3q13, 11q23, 16p13 and 22q13) and another nine (1q31, 3p24, 4q24, 5p13, 7p14, 11q13, 12p13, 14q24 and 16q24) demonstrated at least suggestive evidence for association (i.e. P < 0.001), despite sparse coverage at some of the loci (Table 1). Although the peak SNPs identified by meta-analysis at most of these loci did not correspond to those previously reported, these associations were generally evident in all three cohorts studied and did show moderate to strong LD with the peak SNP identified previously at each locus (data not shown). The one locus reported in the UK GWAS that was not replicated here, 14q32, was too poorly covered by the Immunochip to be assessed. However, together our observations do confirm 19 of the previously reported associations with the risk of PBC.
Identification of a novel PBC risk locus at 13q14, and suggestive evidence of PBC association at six additional loci
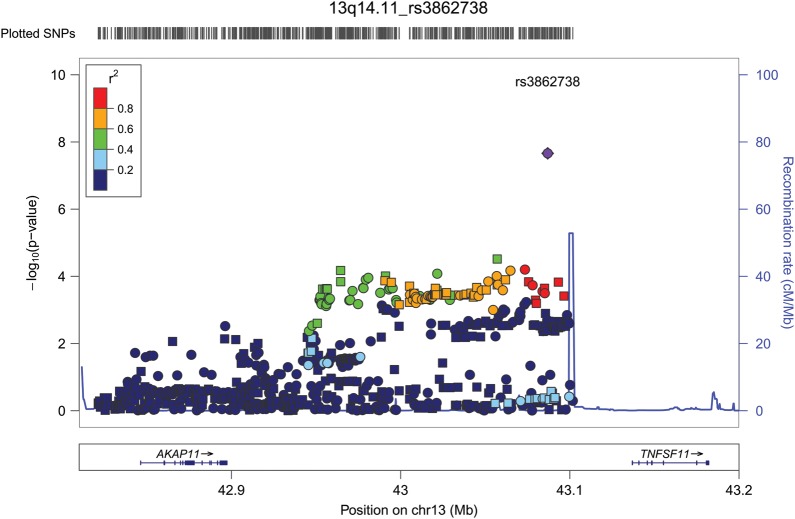
Although many of the associations detected in our screen derive from SNPs in previously reported PBC risk loci, the quantile–quantile (Q-Q) plot of the meta-analysis results also indicate association signals that are not accounted for by the previously identified loci (Fig. 2). We took a conservative approach to the identification of novel PBC risk loci from our data, considering only those regions with dense coverage on the Immunochip and requiring a suggestive level of significance (P < 5 × 10−5) by meta-analysis, the same direction of effect across all three cohorts and supportive (P < 0.05) evidence for association in at least two of the groups. Using this approach, one novel association achieving genome-wide significance was identified at 13q14 and six additional loci (2q12, 6q23, 8q24, 11q13, 12q24 and 19p13) were found to have suggestive evidence for association (Table 2 and Supplementary Material, Table S3). Within the 13q14 locus, the most strongly associated SNP, rs3862738 (Pmeta = 2.18 × 10−8), lies 50 kb upstream of the TNFSF11 gene (Fig. 3) in a region also associated with risk for Crohn's disease (CD) (17) and regulation of bone mineral density (BMD) (18,19). Evidence for this association was strong in the meta-analysis, with the same direction of effect detected in each cohort with strong effect in the Canadian and US cohorts [Canada odds ratio (OR) = 1.41, P = 1.55 × 10−6; US OR = 1.40, P = 4.08 × 10−4], but a weaker effect seen in the Italian cohort (Italy OR = 1.33, P = 0.087). Among the six loci showing suggestive association, five have previously been associated with one or more autoimmune diseases and the sixth, 11q13, has been associated with population differences in vitamin D levels (20,21). Considering that PBC affects primarily women and that AMA are found to be present in the majority of patients, we also performed subgroup analyses, limiting the patient population to either those who were confirmed AMA positive or who were of female gender. These subgroup analyses revealed ORs and P-values at previously known and novel reported risk loci to be essentially unchanged compared with those derived in the full cohort analyses (Supplementary Material, Table S4), suggesting little effect of AMA status and gender on these associations.
Figure 2.
Quantile–quantile plot of observed versus expected P-values for the meta-analysis. Black circles specify the results of the complete Immunochip, gray circles specify the results after all known loci (as identified by previous GWAS) are removed. The genomic control (λGC = 1.29) was estimated after removal of the known loci. The gray shaded region of the expectation line represents the 95% confidence interval for the expected range of P-values.
Table 2.
Results of meta-analysis of three independent PBC case–control cohorts at novel and suggestive PBC risk loci
| Risk locus | Immunochip content |
Peak SNP | Locationb | RA | OR (95% CI) | P-value | Gene(s)c | Type | |
|---|---|---|---|---|---|---|---|---|---|
| Region (Mb) | Coveragea | ||||||||
| 13q14 | 42.83–43.20 | Dense | rs3862738 | 43086907 | G | 1.33 (1.20–1.47) | 2.18 × 10−8 | AKAP11/TNFSF11 | Intergenic |
| 2q12 | 102.30–103.21 | Dense | rs10186746 | 102866377 | G | 1.21 (1.11–1.33) | 2.40 × 10−5 | IL1RL2/IL1RL1 | Intergenic |
| 6q23 | 137.88–138.29 | Dense | rs6920220 | 138006504 | A | 1.29 (1.16–1.43) | 1.17 × 10−6 | OLIG3/TNFAIP3 | Intergenic |
| 8q24 | 129.13–129.30 | Dense | rs2608029 | 129170126 | C | 1.23 (1.13–1.34) | 3.14 × 10−6 | PVT1/GSDMC | Intergenic |
| 11q13 | 70.95–71.36 | Dense | rs10898201 | 71201527 | A | 1.31 (1.17–1.48) | 4.91 × 10−6 | NADSYN1 | Intron |
| 12q24 | 111.70–113.04 | Dense | rs7309325 | 111849515 | G | 1.26 (1.13–1.41) | 2.54 × 10−5 | SH2B3 | Intron |
| 19p13 | 18.17–18.44 | Dense | rs73003205 | 18370790 | A | 1.35 (1.18–1.54) | 1.43 × 10−5 | KIAA1683 | Intron |
RA, risk allele; OR, odds ratio; 95% CI, 95% confidence interval.
aApproximate coverage density of markers on the Immunochip with Dense denoting > 100 markers in region and Sparse denoting <20 markers in region. bGRCh37/hg19 assembly. cGene in which the peak signal is located in, or genes flanking peak signal if intergenic.
Figure 3.
Association results from meta-analysis of three independent cohorts for the AKAP11/TNFSF11 region on chromosome 13q14.11. Genotyped SNPs are illustrated as circles and imputed SNPs as squares. The peak SNP, rs3862738, is shown in purple and pairwise correlation (r2) with this SNP is indicated by a color gradient ranging from red (high LD) to dark blue (low LD).
Identification of multiple independent association signals at four PBC risk loci
To further refine the association signals, the contributions of individual SNPs to the signal at each of the 11 loci achieving genome-wide significance in the meta-analysis were evaluated by logistic regression analyses conditioning on the most significant SNP(s) in the region. Four loci harboring multiple association signals that independently achieved genome-wide significance were identified, including 2q32 (independent signals tagged by rs3024921 and rs7568275), 16p13 (rs413024 and rs12928537), 3q25 (rs9877910, rs34913294 and rs582537) and 6p21 (rs7775055, rs9271588, rs1042544 and rs707929) (Supplementary Material, Table S5). Haplotype analyses were then carried out to evaluate whether the regional effects are primarily attributable to individual variants or are better represented by haplotypic effects. These analyses revealed little or no differences in haplotype and individual SNP effects at 2q32 and 16p13, suggesting separate allelic effects at these loci (Supplementary Material, Table S6). In contrast, the haplotype analyses suggested the presence of cis-acting elements or a single effect tagged specifically by multiple variants at 3q25 and 6p21. At 3q25, for example, two haplotypes were more strongly associated with disease (rs34913294, rs582537, rs9877910; CAT: OR = 1.61, P = 4.10 × 10−23 and TCT: OR = 2.10, P = 7.06 × 10−21) than any of the individual SNPs at this locus (top SNP rs9877910: OR = 1.47, P = 1.02 × 10−17).
As shown in Figure 4, the two most significant and independent association signals at the 2q32 locus lie in close proximity within the same intron of STAT4, but the LD of the peak SNP extends into the regulatory region of the neighboring STAT1 gene (illustrated in Supplementary Material, Fig. S2), suggesting possible effects of either or both genes. The two independent signals at 16p13 appear quite separated with one localizing in the CLEC16A coding region and the other mapping upstream of a gene cluster including SOCS1, raising the possibility of effects from multiple genes. The three independent association signals at 3q25 flank the IL12A gene, and based on the haplotype analysis, appear to have a complex effect on risk that may also reflect added effects of the nearby SCHIP1 gene.
Figure 4.
Association results from meta-analysis of three independent cohorts illustrating independent association signals identified by conditional analyses at 2q32, 16p13 and 3q25. The peak SNPs are indicated by large dark blue circles and correlated SNPs (r2 ≥ 0.5) as medium-sized light blue circles. The peak SNP remaining after the first round of conditioning (SNP2) is indicated by large red circle and correlated SNPs as medium-sized pink circles. For 3q25, the peak and correlated SNPs representing the third signal remaining after the second round of conditioning are indicated by dark and light gold circles.
As expected, the strongest associations in this study were with 6p21 HLA region polymorphisms. Due to the complexity of this region and likelihood of strong haplotype effects, we tried to further resolve this locus by imputing classical HLA alleles from the SNP genotypes (22,23). Meta-analysis of the results obtained from the three independent cohorts identified a total of eight HLA class II alleles HLA DRB1 (*0801, *1101 and *1501), HLA DQA1 (*0501) and HLA DQB1 (*0301, *0302 and *0402) achieving genome-wide significance levels (Table 3). Independence of these HLA allele associations was then evaluated by logistic regression analyses conditioning on the most significant allele(s). This analysis provided evidence for four largely independent association signals among the eight alleles, specifically (i) DQB1*0402:DRB1*0801 (risk), (ii) DQB1*0301:DRB1*1101:DQA1*0501 (protective), (iii) DRB1*1501 (risk) and (iv) DQB1*0302 (risk) (Table 3), a result that correlates with the SNP association data. HLA allele frequencies and scale of the observed associations were quite similar between the Canadian and US cohorts, with direction and magnitude of the effects generally consistent in the Italian cohort, despite large allele frequency differences in the Italian relative to the other two populations (Supplementary Material, Table S7).
Table 3.
Results of association study and conditional analyses of imputed HLA alleles
| Independent signal | HLA allele | OR (95% CI) | P-value observeda | P-value cond. DQB1*0402b | P-value cond. + DQB1*0301c | P-value cond. + DRB1*1501d |
|---|---|---|---|---|---|---|
| 1 | DQB1*0402 | 3.93 (3.17–4.87) | 7.76 × 10−36 | 1.00 | – | – |
| 1 | DRB1*0801 | 3.99 (3.19–5.00) | 2.78 × 10−33 | 0.4278 | – | – |
| 2 | DQB1*0301 | 0.54 (0.47–0.61) | 1.81 × 10−20 | 3.39 × 10−16 | 1.00 | – |
| 2 | DRB1*1101 | 0.49 (0.41–0.59) | 4.11 × 10−14 | 2.00 × 10−12 | 0.0023 | – |
| 2 | DQA1*0501 | 0.66 (0.59–0.75) | 6.59 × 10−11 | 3.54 × 10−7 | 0.1979 | – |
| 3 | DRB1*1501 | 0.62 (0.53–0.73) | 7.66 × 10−9 | 2.99 × 10−6 | 1.04 × 10−8 | 1.00 |
| 4 | DQB1*0302 | 1.51 (1.31–1.75) | 2.73 × 10−8 | 1.23 × 10−10 | 1.71 × 10−7 | 1.22 × 10−5 |
OR, odds ratio; 95%CI, 95% confidence interval.
aFrom meta analysis of individual cohort association results, bafter conditioning on DQB1*0402, cafter adding DQB1*0301 to the conditioning model, dafter additionally adding DRB1*1501 to the conditioning model.
Fine mapping and functional annotation of PBC risk loci
To facilitate fine mapping and functional annotation efforts, we next imputed additional genotypes across the non-HLA PBC risk loci validated or identified in this study, using 1000 Genomes. We found little difference in P-values of the peak genotyped versus peak imputed SNPs (Supplementary Material, Table S8), consistent with the comprehensive coverage provided by the Immunochip (Supplementary Material, Fig. S2). Association signals with strong LD (r2 > 0.8) extending across multiple genes were observed at 9 of the 10 non-HLA loci reaching genome-wide significance in this study, the exception being for 1p31 where the signal localized to a single gene (IL12RB2).
Functional annotation was then carried out for all observed and imputed SNPs with meta-analysis P ≤ 10−4 and/or in LD (r2 ≥ 0.5) with a peak SNP, using SNPnexus and NCBI's GTEx expression quantitative trait loci (eQTL) browser (Table 4). This analysis identified five non-synonymous SNPs in a total of four genes (TMEM39A, IL7R, ZPBP2 and GSDMB), four SNPs in conserved promoter regions of three genes (STAT1, TMEM39A and IKZF3) and 27 SNPs representing eQTL for four genes (IRF5, C16orf75, ORMDL3 and SYNGR1). While signal localization and functional annotation data do not conclusively identify disease-relevant variants, these findings highlight a number of compelling gene candidates for follow-up biological studies.
Table 4.
Results of functional annotation of PBC risk loci
| Locus | Implicated gene | SNPa | r2 with peak SNP | SNP type | P-value | Note |
|---|---|---|---|---|---|---|
| 2q32 | STAT1 | rs4138244* | – | Promoter | 5.01 × 10−10 | DNAse I hypersensitivity |
| 3q13 | TMEM39A | rs2282171* | – | Promoter | 3.90 × 10−5 | DNAse I hypersensitivity |
| rs1132200 | 0.776 | Coding, A487T | 1.20 × 10−6 | SIFT tolerated | ||
| 5p13 | IL7R | rs6897932 | 0.728 | Coding, T244I | 5.08 × 10−6 | SIFT tolerated |
| 7q32 | IRF5 | rs3807306* | – | eQTL | 4.93 × 10−6 | eQTL P = 7.46 × 10−22 |
| rs4728142 | 0.119 | eQTL | 1.13 × 10−5 | eQTL P = 7.58 × 10−19 | ||
| 16p13 | C16orf75 | rs243325 | 0.940 | eQTL | 1.56 × 10−7 | eQTL P = 7.28 × 10−27 |
| rs243323 | 0.880 | eQTL | 9.07 × 10−8 | eQTL P = 8.70 × 10−26 | ||
| Multiple | Varies | eQTL | Varies | 5 additional SNPs eQTL P < 1 × 10−10 | ||
| 17q12 | IKZF3 | rs1453559* | – | Promoter | 2.71 × 10−6 | DNAse I hypersensitivity, CpG island |
| rs2941522 | – | Promoter | 2.44 × 10−6 | DNAse I hypersensitivity, CpG island | ||
| ZPBP2 | rs11557467 | 0.931 | Coding, S151I | 1.14 × 10−7 | SIFT tolerated | |
| GSDMB | rs2305479 | 0.902 | Coding, G282R | 3.82 × 10−8 | SIFT tolerated | |
| rs2305480 | 0.729 | Coding, P289S | 3.82 × 10−7 | SIFT tolerated | ||
| rs11078929 | 0.792 | Splice site | 4.30 × 10−7 | Intron retention? | ||
| ORMDL3 | rs8067378 | 0.946 | eQTL | 5.09 × 10−9 | eQTL P = 5.90 × 10−17 | |
| rs12936231 | 0.946 | eQTL | 6.73 × 10−9 | eQTL P = 8.69 × 10−17 | ||
| Multiple | Varies | eQTL | Varies | 7 additional SNPs eQTL P < 1 × 10−10 | ||
| 22q13 | SYNGR1 | rs909685* | – | eQTL | 7.90 × 10−8 | eQTL P = 1.72 × 10−28 |
| rs11704319 | 0.363 | eQTL | 1.50 × 10−6 | eQTL P = 9.52 × 10−18 | ||
| Multiple | Varies | eQTL | Varies | 7 additional SNPs eQTL P < 1 × 10−10 |
All observed or imputed SNPs at the 22 known and 1 novel PBC risk loci with meta-analysis P < 1.0 × 10−4 and/or in LD with the peak SNP at one of the loci (r2 > 0.5) were subjected to functional annotation. Non-synonymous coding SNPs, promoter region SNPs and SNPs located in consensus splice acceptor or donor sequences were identified using SNPnexus (www.snp-nexus.org). SNPs associated with eQTL in lymphoblastoid cell lines were identified using NCBI's GTEx eQTL browser (www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi).
aimputed SNP denoted by *; eQTL, expression quantitative trait loci.
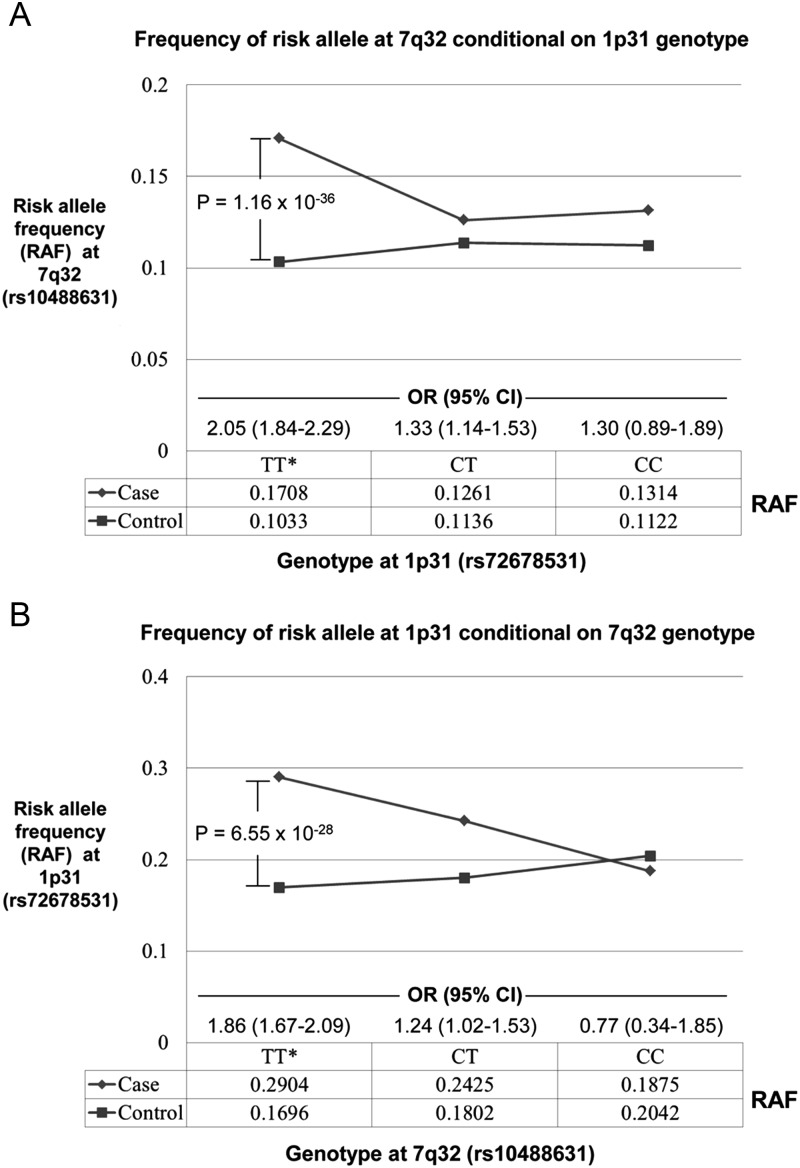
Evidence of an epistatic interaction between the 1p31 (IL12RB2) and 7q32 (IRF5) loci
Potential interactions between associated loci were also explored using the Epistasis module in PLINK v1.07 (24) to test for epistasis between all pairs of the most strongly associated SNPs at each locus. Meta-analysis of the individual cohort results revealed one significant interaction involving rs72678531 (1p31, IL12RB2) and rs10588631 (7q32, IRF5) (Pmeta = 2.06 × 10−5, P = 0.0048 after Bonferroni correction). Risk for PBC was increased for all joint genotypes incorporating risk alleles (Table 5), but most notably by the combination of rs72678531 homozygous risk genotype ‘CC’ and rs10488631 heterozygous risk genotype ‘CT’ (OR = 6.31, P = 2.20 × 10−12). To further characterize this interaction, risk allele frequency for each SNP was plotted conditional on the alternate genotype, and logistic regression analysis was used to estimate the genotype-specific effects of the risk alleles under a log-additive model (Fig. 5). This analysis revealed the epistasis to largely reflect excess risk alleles in patients having non-risk genotypes (i.e. ‘TT’) at the interacting locus. Specifically, the allelic ORs are elevated and P-values highly significant only in the context of the ‘TT’ genotype at the alternate locus. This observation suggests complementary effects of the risk alleles at 1p31 and 7q32 on PBC susceptibility.
Table 5.
Results of the Cochran–Mantel–Haenszel test of genotypes at interacting loci 1p31 and 7q32
| 1p31 | 7q32 | Meta Analysis |
Canada | USA | Italy | |||
|---|---|---|---|---|---|---|---|---|
| Frequency |
OR (95%CI) | P-value | ||||||
| rs72678531a | rs10488631a | Cases | Controls | P-value | P-value | P-value | ||
| TT | TT | 0.3612 | 0.5507 | 1 | – | – | – | – |
| CT | TT | 0.3016 | 0.2265 | 1.94 (1.71–2.21) | 6.07 × 10−24 | 1.17 × 10−15 | 2.06 × 10−7 | 3.06 × 10−4 |
| TT | CT | 0.1521 | 0.1249 | 2.00 (1.70–2.35) | 2.12 × 10−17 | 4.79 × 10−13 | 3.40 × 10−4 | 5.48 × 10−3 |
| CT | CT | 0.0849 | 0.0580 | 2.13 (1.74–2.61) | 1.84 × 10−13 | 2.10 × 10−14 | 5.50 × 10−3 | 0.5165 |
| CC | TT | 0.0587 | 0.0224 | 3.67 (2.77–4.87) | 1.47 × 10−19 | 1.13 × 10−11 | 3.30 × 10−6 | 3.66 × 10−6 |
| TT | CC | 0.0140 | 0.0082 | 3.66 (2.35–5.69) | 8.13 × 10−9 | 7.79 × 10−6 | 0.1400 | 0.0909 |
| CC | CT | 0.0199 | 0.0048 | 6.31 (3.77–10.5) | 2.20 × 10−12 | 4.34 × 10−11 | 0.0254 | 9.89 × 10−4 |
| CT | CC | 0.0072 | 0.0038 | 3.30 (1.60–6.82) | 1.23 × 10−3 | 5.02 × 10−3 | 0.2061 | NA |
| CC | CC | 0.0005 | 0.0007 | NA | NA | NA | NA | NA |
| Not TT | Not TT | 0.6388 | 0.4493 | 2.15 (1.92–2.38) | 3.6 × 10−44 | 2.20 × 10−16 | 3.98 × 10−11 | 4.45 × 10−7 |
OR, odds ratio; 95% CI, 95% confidence interval.
aThese SNPs represent the peak signal at the respective loci in the meta-analysis; C is the risk allele for each.
Figure 5.
Risk allele frequencies of interacting SNPs at 1p31 (IL12RB2) and 7q32 (IRF5/TNPO3) stratified by the genotype at the alternate locus. (A) The rs10488631 risk allele ‘C’ at 7q32 is highly associated with PBC only in patients with the non-risk rs72678531 genotype ‘TT’ at 1p31. Conversely (B), the rs72678531 risk allele ‘C’ at 1p31 is highly associated with PBC only in patients with rs10488631 ‘TT’ genotypes. These findings suggest the possibility of a complementary effect between the risk alleles at these two loci. Results from logistic regression analyses in three independent cohorts under a log-additive model; combined by meta-analysis.
DISCUSSION
The results of this genetic association study identify a novel risk locus for PBC near the TNFSF11 gene at 13q14 and provide suggestive evidence for PBC association with six other loci not previously implicated in PBC. The data also replicate 14 of 15 non-MHC risk loci identified in a recent UK-based PBC GWAS, but do not provide robust support for an association previously identified at 1p36 (MMEL1). Our conditional analyses also reveal the presence of multiple independent association signals at four of the validated risk loci, and the functional annotation of associated SNPs identifies a number of potentially interesting candidate genes for future study. While the available eQTL data were limited by cell type and activation status, and may not have captured all relevant candidates, these findings identify a number of new candidate genes for PBC that require further molecular studies. Our data also provide new evidence for epistatic effects of SNPs within the 1p31 (IL12RB2) and 7q32 (IRF5) loci, an interaction characterized primarily by a reciprocal overrepresentation of risk alleles in the context of the non-risk genotype at the alternate locus and thus likely reflecting complementary effects of these loci on the risk for PBC.
The novel PBC risk locus identified at 13q14 is positioned upstream of the RANKL-encoding TNFSF11 gene, a region also associated with CD (17) and BMD (18,19). Critical roles for RANKL in immunoregulation have been revealed by the absence of lymph nodes in RANKL-deficient mice (25) and by data showing RANKL involvement in the CD4+CD25+ regulatory T (Treg) cell development relevant to colitis and dermal immunity (26,27). Circulating RANKL levels have also been shown to be decreased in PBC patients relative to controls (28), with numbers of circulating CD4+CD25+ Tregs reduced in both PBC patients and their daughters and sisters (29); and the ratio of CD8+T cells to FoxP3+Tregs increased in PBC livers, relative to other chronic liver conditions. RANKL also regulates osteoclast development and function (30) and may be relevant to the decreased BMD and osteoporosis seen frequently in PBC patients (31), and previously ascribed to impaired vitamin D metabolism (32). Further replication and functional studies are required, however, to interpret the potential relevance of genetic variation in TNFSF11 to PBC and its complications.
The six suggestive loci identified in our study also harbor genes involved in signaling pathways implicated in pathogenesis of PBC. The 2q12 locus is associated with ulcerative colitis (UC), CD and celiac disease (17,33,34), and harbors a cluster of genes encoding key effectors of toll-like receptor (TLR) signaling. The 6q23 locus associated with celiac disease, rheumatoid arthritis (RA), systemic lupus erythematosus, psoriasis and UC (34–38) contains TNFAIP3, an inhibitor of NF-κB. The 8q24 locus maps upstream of PVT1, a region associated with multiple sclerosis (MS), celiac disease and IgA deficiency (38–40) which is enriched in putative immuno-active micro RNAs (41). The 12q24 locus includes the SH2B3 gene associated with type 1 diabetes, celiac disease and RA (38,42,43) and coding for LNK, a regulator of cytokine signaling and the 19p13 locus harbors IL12RB1, a component in the IL12 signaling pathway already implicated in PBC. Finally, the 11q13 locus is associated with population differences in levels of vitamin D (20,21), a molecule with integral roles in immunity (44).
Our study corroborates the majority of previously reported PBC risk loci and although the peak SNPs detected at many of these loci in this study differ from those reported in prior GWAS analyses, the ORs for peak associations detected here were similar to those previously reported. While these peak signals were often in strong LD with other SNPs extending across multiple genes or across the whole of the genotyped region, our data did illuminate multiple independent association signals within a number of loci. Thus, for example, the detection of independent signals emanating from the STAT4 gene at 2q32 and a second SNP in the region that exists within a haplotype extending into the STAT1 gene promoter identifies STAT1 as well as STAT4 as a possible player in PBC pathogenesis. The detection of two strong independent signals at the CLEC16A and SOCS1 genes, respectively, in the 16p13 locus is also consistent with our previous data implicating both of these genes in PBC (16) and with other data implicating the two genes in MS (45). Our functional annotation results identify c16orf75 (Table 4) as another possible contributor to risk from this region, suggesting the possible complexity of this association and reinforcing the potential for multiple sources of signals from single loci. Similarly, the detection and positions of three independent association signals flanking the IL12A gene at 3q25 suggest that at least one other gene in the regions SCHIP1 may contribute to risk for PBC.
Our data also identify a significant interaction involving the 1p31 (IL12RB2) and 7q32 (IRF5) PBC risk loci, suggesting a complementary or balancing effect of these alleles on PBC risk. Further studies are required to validate this finding and test its relevance to disease, but these data are consistent with the contribution of both the candidate genes at these loci, IL12RB2 and IRF5, to the immune response downstream of TLR activation (46).
In summary, the data presented here identify TNFSF11 as a new risk locus for PBC, provide suggestive evidence for six additional immune-related gene loci and corroborate almost all associations previously identified from PBC GWAS data sets. The dense coverage of targeted loci afforded by the Immunochip array also enabled definition of multiple independent associations at a number of loci, the validation by imputation of classical HLA alleles of a number of HLA associations variably detected in other studies and the demonstration that risk alleles at the IL12RB2 and IRF5 susceptibility loci have complementary effects on risk. Together, these findings contribute new insights into the genetic etiology of PBC that will inform future genetic and functional studies of PBC pathogenesis.
MATERIALS AND METHODS
Ethics
All aspects of this research project were approved by the local institutional review boards or ethics committees at each contributing institution or were obtained from outside genotype databases with the appropriate approval processes.
Subjects
The Toronto-based ‘Canadian’ collection consisted of PBC patients of European descent collected from centers in Canada, USA and Europe. Controls for the Canadian cohort were drawn from the WTCCC shared control initiative. The PBC patients and controls for the US Cohort were all participants in the Mayo Clinic PBC Genetic Epidemiology Registry and Biospecimen repository (47). This resource consists primarily of current or former Mayo Clinic PBC patients (plus a small number of self-referrals) and recruits controls during or shortly after annual preventative maintenance visits at the Mayo Clinic Division of General Internal Medicine. The Italian PBC cases were collected from multiple centers in Italy including Milano, Bologna, Padova, Torino, Napoli, Pisa, Udine, Cuneo, Roma, Firenze, Genova, Bari and Palermo. Italian population controls with no history of autoimmune diseases were derived from three sources: (i) 117 control blood samples were collected at IRCCS Istituto Clinico Humanitas (Rozzano, Italy), (ii) 195 control DNAs were provided from the HYPERGENES project, and (iii) 315 Immunochip genotypes (performed at Kiel, Germany) of confirmed Italian controls were obtained from Drs Fabrizio Bossa (Division of Gastroenterology, IRCCS-CCS Hospital, San Giovanni Rotondo, Italy) and Andre Franke (University of Kiel, Kiel, Germany). All PBC patients were diagnosed according to the guidelines set forth by the American Association for the Study of Liver Disease (1).
Genotyping
Immunochip genotyping was performed at the Mount Sinai Hospital/University Health Network Gene Profiling Facility for the Canadian PBC patients at Christian Albrechts University, Kiel, Germany for the US PBC patients and controls and at three centers, Feinstein Institute for Medical Research, North Shore LIJ Health System (440 cases and 205 controls), Manhasset, New York; the University of Toronto (240 cases and 127 controls), and Christian Albrechts University, Kiel, Germany (315 controls), for the Italian subjects. Bead intensity data were processed, normalized and genotypes were called within the individual populations using GenomeStudio.
Quality control
Initial quality control efforts were performed separately for the three independent cohorts used in this study. Samples were removed from the analysis if (i) call rate was <95%, (ii) gender was discrepant to expectation, (iii) unexpected relatedness was detected (estimated identity by descent > 20%), or (iv) individuals with greater than 15% non-European ancestry were identified using STRUCTURE v2.3 (48,49), a Bayesian clustering algorithm. For the STRUCTURE analyses, we used a set of 883 independent SNPs (r2 < 0.1) that were common to each data set and available in the reference populations. The reference populations included at least 75 individuals for each of the following groups: European, East Asian, sub-Saharan Africa, South Asian, and Amerindian. The STRUCTURE analyses were performed under K (number of clusters) =4, using the prior population information. The runs were performed using >100 000 resamplings and >50 000 burn-in cycles under the admixture model with the λ = 1 option, where λ estimates the prior probability of the allele frequency and is based on the Dirichlet distribution of allele frequencies. In addition, we excluded samples that were outliers (>4 standard deviations) in principal component analyses (PCA) (see below). Following quality control 2216 PBC patients and 5594 controls were available for study. Of these, 399 PBC patients and 199 controls from the Italian cohort and 414 PBC patients from the Canadian cohort had been included in the discovery panels of two earlier PBC GWAS publications (10,12). SNPs were removed from the analysis if (i) the call rate was <95%, (ii) the test of the Hardy–Weinberg equilibrium was P < 1 × 10−5, (iii) MAF was <1%, or (iv) any discrepant alleles were detected among replicated samples (21 replicates in US data, 20 replicates in Italian data). For the seven novel loci reported, the minimum HWE in a single control group was found to be 0.1279, so these novel loci would not have disappeared with the use of a more stringent cut-off. For the remainder of the specifically reported SNPs at known loci, only two had HWE P-values <0.01, both in a single population (rs7665090, HWE P = 0.007 in Canada controls and rs9271588, HWE P = 0.002 in US controls). Prior to meta-analysis, markers not available in all three populations or where MAF difference between US and Canada cohort controls were significantly different (P < 0.001 by Chi-square test) were removed. Following quality control 109 812 SNPs were available. Cluster plots for all SNPs presented in Tables 1 and 2 were visually inspected, and confirmed to be of high quality.
Statistical methodology
Association analysis
For each of the data sets, PCA was used to control for population stratification; performed using EIGENSTRAT software (50) and the genotypes from a set of 12 579 SNPs (with low linkage disequilibrium (r2 < 0.5) distributed over all autosomal chromosomes (see Supplementary Material is available at HMG Online). The population substructure was defined by PCs 1, 2, and 4 in the Canadian data set, the first 2 PCs in the US data set and 0 PCs in the Italian data set (due to strong homogeneity of the Italian subjects selected for Immunochip genotyping). The PCA results were used as covariates in the association analyses, which were performed using logistic regression in PLINK v1.07 (log-additive model) (24). Cochran's test for heterogeneity between studies was performed and was not significant (P > 0.05). Thus, subsequent meta-analysis was carried out using METAL software (15) assuming a fixed effects model. For all associated SNPs reported (P < 0.05), the direction of the risk allele was the same in the three cohorts. In addition to the primary meta-analysis, subset meta-analyses restricting the patient population to either AMA positive patients (n = 1994) or female patients (n = 2083) and controls (n = 2974) were also performed.
Conditional analysis
To search for independent association signals at PBC risk loci, we performed logistic regression, conditioning on the most significant SNP in each region using PLINK. For loci with SNPs demonstrating genome-wide significance (P < 5.0 × 10−8) after conditioning, the most significant SNP was added to the model and additional rounds of analysis were performed. Residual SNPs achieving P < 5 × 10−7 were considered to represent an independent signal but were not taken forward for further analysis.
Haplotype analysis
To evaluate whether or not association could be explained by effects from individual SNPs or the associations were better explained by haplotypic effects, we first used the EM algorithm implemented in PLINK to estimate haplotypic frequencies in cases and controls for each population (Mayo, Italy and Canada) separately. Then, we used R to perform logistic regression adjusting for effects from the populations. We first performed an association analysis jointly testing all haplotypes, versus a model testing effects from each SNP separately. We used a likelihood ratio test to obtain P-values for the omnibus haplotype effect (in which an indicator variable was used to model variant haplotypes against the most common haplotype, which formed the referent) versus the joint effects of SNPs (in which indicator variables were used contrasting the variant SNP alleles jointly at the loci against the more common alleles). Because the tests are not nested, we used Akaike's information criterion to identify the best fitting model.
Imputation
MaCH version 1.0.16 was used to impute SNPs within defined regions using 1000 Genomes release 20101123, build 37, European population. SNPs with MAF < 1%, insertion–deletions or unknown positions in the 1000 Genomes data were excluded for imputation. Only imputation results with R2 values >0.8 or MAF > 1% were retained for the analysis. Imputation of classical HLA alleles from the genotype information was performed using the program HLA*IMP (https://oxfordhla.well.ox.ac.uk/hla/) as described in references (22,23), which includes the 1958 Birth Cohort, HapMap CEU and CEPH CEU samples as the reference set.
Functional annotation
All observed or imputed SNPs at the 22 known and 1 novel PBC risk loci with meta-analysis P < 1.0 × 10−4 and/or in LD with the peak SNP at one of the loci (r2 > 0.5) were taken forward for functional annotation. SNPnexus (51) (www.snp-nexus.org) was used to identify non-synonymous coding SNPs, promoter region SNPs and SNPs located in consensus splice acceptor or donor sequences. SNPs associated with eQTL in lymphoblastoid cell lines were identified using NCBI's GTEx (genotype tissue expression) eQTL browser (www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi).
Epistasis
We tested for evidence of gene–gene interaction by interrogating all possible pairs of the peak SNPs detected by meta-analysis at 21 known (14q32 excluded) and 1 novel PBC risk loci identified in the study. This was accomplished using the Epistasis module in PLINK v1.07. We used the Bonferroni method to correct for the 231 independent tests, establishing the cut-off for significance at P = 2.16 × 10−4. To follow-up a significant interaction finding between the peak SNPs at 1p31 and 7q32, we performed a Cochran–Mantel–Haenszel test procedure, using the most prevalent joint genotype as reference, to evaluate the effect of the joint genotypes on PBC risk while allowing for variability in genotype frequencies among centers. Based on these findings, we constructed plots of the risk allele frequency of each marker in relation to the genotype of the other marker, and compared the frequencies relative to genotype in patients and controls as well as between cases and controls using logistic regression in the individual groups followed by meta-analysis.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Canadian Institutes for Health Research (MOP74621 to G.M.H. and K.A.S.), the Ontario Research Fund (RE01-061 to G.M.H. and K.A.S.), the Canadian Primary Biliary Cirrhosis Society (to G.M.H. and K.A.S.), the US National Institutes of Health (RO1 DK056839 to M.E.G, M.F.S. and C.I.A.), (RO1 DK091823 to M.F.S., M.E.G. and C.I.A.), (RO1 DK80670 to K.N.L), (AR44422 to C.I.A., Y.L. and P.K.G.), (8UL1TR000058 to Virginia Commonwealth University Center for Clinical and Translational Research), the American Gastroenterological Association (to K.N.L.), the A.J. and Sigismunda Palumbo Charitable Trust (to K.N.L.), HYPERGENES (FP7 - HEALTH-F4-2007-201550) and INTEROMICS (MIUR - CNR Italian Flagship Project). K.A.S. holds the Sherman Family Chair in Genomic Medicine and a Tier 1 Canada Research Chair.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the subjects and physicians who provided DNA specimens and clinical data for this study.
Conflict of Interest statement. The authors report no conflicts of interest.
REFERENCES
- 1.Lindor K.D., Gershwin M.E., Poupon R., Kaplan M., Bergasa N.V., Heathcote E.J. Primary biliary cirrhosis. Hepatology. 2009;50:291–308. doi: 10.1002/hep.22906. [DOI] [PubMed] [Google Scholar]
- 2.Corpechot C., Abenavoli L., Rabahi N., Chretien Y., Andreani T., Johanet C., Chazouilleres O., Poupon R. Biochemical response to ursodeoxycholic acid and long-term prognosis in primary biliary cirrhosis. Hepatology. 2008;48:871–877. doi: 10.1002/hep.22428. [DOI] [PubMed] [Google Scholar]
- 3.Invernizzi P., Lleo A., Podda M. Interpreting serological tests in diagnosing autoimmune liver diseases. Semin. Liver Dis. 2007;27:161–172. doi: 10.1055/s-2007-979469. [DOI] [PubMed] [Google Scholar]
- 4.Invernizzi P., Selmi C., Gershwin M.E. Update on primary biliary cirrhosis. Dig. Liver Dis. 2010;42:401–408. doi: 10.1016/j.dld.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odin J.A., Huebert R.C., Casciola-Rosen L., LaRusso N.F., Rosen A. Bcl-2-dependent oxidation of pyruvate dehydrogenase-E2, a primary biliary cirrhosis autoantigen, during apoptosis. J. Clin. Invest. 2001;108:223–232. doi: 10.1172/JCI10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selmi C., De Santis M., Cavaciocchi F., Gershwin M.E. Infectious agents and xenobiotics in the etiology of primary biliary cirrhosis. Dis. Markers. 2010;29:287–299. doi: 10.3233/DMA-2010-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juran B.D., Lazaridis K.N. Update on the genetics and genomics of PBC. J. Autoimmun. 2010;35:181–187. doi: 10.1016/j.jaut.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirschfield G.M., Invernizzi P. Progress in the genetics of primary biliary cirrhosis. Semin. Liver Dis. 2011;31:147–156. doi: 10.1055/s-0031-1276644. [DOI] [PubMed] [Google Scholar]
- 9.Invernizzi P. Human leukocyte antigen in primary biliary cirrhosis: an old story now reviving. Hepatology. 2011;54:714–723. doi: 10.1002/hep.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirschfield G.M., Liu X., Xu C., Lu Y., Xie G., Gu X., Walker E.J., Jing K., Juran B.D., Mason A.L., et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N. Engl. J. Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirschfield G.M., Liu X., Han Y., Gorlov I.P., Lu Y., Xu C., Chen W., Juran B.D., Coltescu C., Mason A.L., et al. Variants at IRF5-TNPO3, 17q12–21 and MMEL1 are associated with primary biliary cirrhosis. Nat. Genet. 2010;42:655–657. doi: 10.1038/ng.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Invernizzi P., Lu Y., Kosoy R., Bianchi I., Podda M., Xu C., Xie G., Macciardi F., Selmi C., et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat. Genet. 2010;42:658–660. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mells G.F., Floyd J.A., Morley K.I., Cordell H.J., Franklin C.S., Shin S.Y., Heneghan M.A., Neuberger J.M., Donaldson P.T., Day D.B., et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trynka G., Hunt K.A., Bockett N.A., Romanos J., Mistry V., Szperl A., Bakker S.F., Bardella M.T., Bhaw-Rosun L., Castillejo G., et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirschfield G.M., Xie G., Lu E., Sun Y., Juran B.D., Chellappa V., Coltescu C., Mason A.L., Milkiewicz P., Myers R.P., et al. Association of primary biliary cirrhosis with variants in the CLEC16A, SOCS1, SPIB and SIAE immunomodulatory genes. Genes Immun. 2012;13:328–335. doi: 10.1038/gene.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Styrkarsdottir U., Halldorsson B.V., Gretarsdottir S., Gudbjartsson D.F., Walters G.B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Center J.R., Nguyen T.V., et al. Multiple genetic loci for bone mineral density and fractures. N. Engl. J. Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 19.Rivadeneira F., Styrkarsdottir U., Estrada K., Halldorsson B.V., Hsu Y.H., Richards J.B., Zillikens M.C., Kavvoura F.K., Amin N., Aulchenko Y.S., et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn J., Yu K., Stolzenberg-Solomon R., Simon K.C., McCullough M.L., Gallicchio L., Jacobs E.J., Ascherio A., Helzlsouer K., Jacobs K.B., et al. Genome-wide association study of circulating vitamin D levels. Hum. Mol. Genet. 2010;19:2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T.J., Zhang F., Richards J.B., Kestenbaum B., van Meurs J.B., Berry D., Kiel D.P., Streeten E.A., Ohlsson C., Koller D.L., et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leslie S., Donnelly P., McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am. J. Hum. Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dilthey A.T., Moutsianas L., Leslie S., McVean G. HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–972. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong Y.Y., Yoshida H., Sarosi I., Tan H.L., Timms E., Capparelli C., Morony S., Oliveira-dos-Santos A.J., Van G., Itie A., et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 26.Totsuka T., Kanai T., Nemoto Y., Tomita T., Okamoto R., Tsuchiya K., Nakamura T., Sakamoto N., Akiba H., Okumura K., et al. RANK-RANKL signaling pathway is critically involved in the function of CD4+CD25+ regulatory T cells in chronic colitis. J. Immunol. 2009;182:6079–6087. doi: 10.4049/jimmunol.0711823. [DOI] [PubMed] [Google Scholar]
- 27.Loser K., Mehling A., Loeser S., Apelt J., Kuhn A., Grabbe S., Schwarz T., Penninger J.M., Beissert S. Epidermal RANKL controls regulatory T-cell numbers via activation of dendritic cells. Nat. Med. 2006;12:1372–1379. doi: 10.1038/nm1518. [DOI] [PubMed] [Google Scholar]
- 28.Guanabens N., Enjuanes A., Alvarez L., Peris P., Caballeria L., Jesus Martinez de Osaba M., Cerda D., Monegal A., Pons F., Pares A. High osteoprotegerin serum levels in primary biliary cirrhosis are associated with disease severity but not with the mRNA gene expression in liver tissue. J. Bone Miner. Metab. 2009;27:347–354. doi: 10.1007/s00774-009-0042-1. [DOI] [PubMed] [Google Scholar]
- 29.Lan R.Y., Cheng C., Lian Z.X., Tsuneyama K., Yang G.X., Moritoki Y., Chuang Y.H., Nakamura T., Saito S., Shimoda S., et al. Liver-targeted and peripheral blood alterations of regulatory T cells in primary biliary cirrhosis. Hepatology. 2006;43:729–737. doi: 10.1002/hep.21123. [DOI] [PubMed] [Google Scholar]
- 30.Leibbrandt A., Penninger J.M. RANK/RANKL: regulators of immune responses and bone physiology. Ann. N.Y. Acad. Sci. 2008;1143:123–150. doi: 10.1196/annals.1443.016. [DOI] [PubMed] [Google Scholar]
- 31.Mounach A., Ouzzif Z., Wariaghli G., Achemlal L., Benbaghdadi I., Aouragh A., Bezza A., El Maghraoui A. Primary biliary cirrhosis and osteoporosis: a case-control study. J. Bone Miner. Metab. 2008;26:379–384. doi: 10.1007/s00774-007-0833-1. [DOI] [PubMed] [Google Scholar]
- 32.Nakchbandi I.A., van der Merwe S.W. Current understanding of osteoporosis associated with liver disease. Nat. Rev. Gastroenterol. Hepatol. 2009;6:660–670. doi: 10.1038/nrgastro.2009.166. [DOI] [PubMed] [Google Scholar]
- 33.Hunt K.A., Zhernakova A., Turner G., Heap G.A., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson C.A., Boucher G., Lees C.W., Franke A., D'Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A., et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plenge R.M., Cotsapas C., Davies L., Price A.L., de Bakker P.I., Maller J., Pe'er I., Burtt N.P., Blumenstiel B., DeFelice M., et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham R.R., Cotsapas C., Davies L., Hackett R., Lessard C.J., Leon J.M., Burtt N.P., Guiducci C., Parkin M., Gates C., et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair R.P., Duffin K.C., Helms C., Ding J., Stuart P.E., Goldgar D., Gudjonsson J.E., Li Y., Tejasvi T., Feng B.J., et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A., Adany R., Aromaa A., et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreira R.C., Pan-Hammarstrom Q., Graham R.R., Gateva V., Fontan G., Lee A.T., Ortmann W., Urcelay E., Fernandez-Arquero M., Nunez C., et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat. Genet. 2010;42:777–780. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- 40.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huppi K., Volfovsky N., Runfola T., Jones T.L., Mackiewicz M., Martin S.E., Mushinski J.F., Stephens R., Caplen N.J. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol. Cancer Res. 2008;6:212–221. doi: 10.1158/1541-7786.MCR-07-0105. [DOI] [PubMed] [Google Scholar]
- 42.Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl E.A., Raychaudhuri S., Remmers E.F., Xie G., Eyre S., Thomson B.P., Li Y., Kurreeman F.A., Zhernakova A., Hinks A., et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantorna M.T., Mahon B.D. Mounting evidence for vitamin D as an environmental factor affecting autoimmune disease prevalence. Exp. Biol. Med. (Maywood) 2004;229:1136–1142. doi: 10.1177/153537020422901108. [DOI] [PubMed] [Google Scholar]
- 45.Zuvich R.L., Bush W.S., McCauley J.L., Beecham A.H., De Jager P.L., Ivinson A.J., Compston A., Hafler D.A., Hauser S.L., Sawcer S.J., et al. Interrogating the complex role of chromosome 16p13.13 in multiple sclerosis susceptibility: independent genetic signals in the CIITA-CLEC16A-SOCS1 gene complex. Hum. Mol. Genet. 2011;20:3517–3524. doi: 10.1093/hmg/ddr250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T.W., et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 47.Lazaridis K.N., Juran B.D., Boe G.M., Slusser J.P., de Andrade M., Homburger H.A., Ghosh K., Dickson E.R., Lindor K.D., Petersen G.M. Increased prevalence of antimitochondrial antibodies in first-degree relatives of patients with primary biliary cirrhosis. Hepatology. 2007;46:785–792. doi: 10.1002/hep.21749. [DOI] [PubMed] [Google Scholar]
- 48.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 51.Chelala C., Khan A., Lemoine N.R. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.