Abstract
Understanding seasonality and longevity is a major challenge in tree biology. In woody species, growth phases and dormancy follow one another consecutively. In the oldest living individuals, the annual cycle may run for more than 1,000 years. So far, however, not much is known about the processes triggering reactivation from dormancy. In this study, we focused on wood rays, which are known to play an important role in tree development. The transition phase from dormancy to flowering in early spring was compared with the phase of active growth in summer. Rays from wood samples of poplar (Populus × canescens) were enriched by laser microdissection, and transcripts were monitored by poplar whole-genome microarrays. The resulting seasonally varying complex expression and metabolite patterns were subjected to pathway analyses. In February, the metabolic pathways related to flower induction were high, indicating that reactivation from dormancy was already taking place at this time of the year. In July, the pathways related to active growth, like lignin biosynthesis, nitrogen assimilation, and defense, were enriched. Based on “marker” genes identified in our pathway analyses, we were able to validate periodical changes in wood samples by quantitative polymerase chain reaction. These studies, and the resulting ray database, provide new insights into the steps underlying the seasonality of poplar trees.
Perennial woody plants dominate many natural land ecosystems. Their major difference, compared with annual herbs, is their long life cycle, which, in trees, may span several centuries. During the annual cycle of trees, growth periods are followed by dormancy and vice versa. These seasonal variations are associated with nutrient uptake from the soil together with nutrient remobilization from the bark and wood as well as deposition within these storage compartments. Trees achieve longevity in an ever-changing environment by taking advantage of this intraorganismic nutrient depot in combination with a variety of other concurrent adaptation strategies. This ability to transfer nutrients between alternative deposition sites depends on dynamic metabolite translocation between tissues and on the storage of nutrients in vegetative organs. Furthermore, wood formation is required to ensure durable mechanical stability and as a long-distance pipeline for the transport of water and electrochemical signals (for review, see Ache et al., 2010).
In temperate climates, trees grow synchronously with the seasons and are able to endure periods that are unfavorable for growth by a phase shift from active growth toward dormancy (Okubo, 2000; Rohde and Bhalerao, 2007). It is generally accepted that during growth, cell cycles, cell expansion, and associated biochemical mechanisms are the dominant processes. According to Lang (1987), dormancy is the temporary absence of visible growth of any plant structure containing a meristem. Dormancy can also be regarded as a developmental process rather than a single static state and may be thought of as consisting of many interrelated subprocesses that are active at different points through its various stages. Following entry into dormancy, for example, drought resistance and frost tolerance increase in buds and cambium (Druart et al., 2007; Ruttink et al., 2007). In deciduous trees, growth cessation in response to reduced hours of daylight is characterized by the termination of cambial activity, shedding of leaves, and completion of bud set (Nitsch, 1957; Critchfield, 1960; Rohde et al., 2011a).
In poplar (Populus × canescens), during the period between late February and early March, flowering represents a visual marker for the reactivation of the tree and the initiation of bud break. These processes are dependent upon an increase in hours of daylight and temperature (Corbesier and Coupland, 2006). At this stage, in general, just a few warm days are required for the inception of flowering (Heide, 1993), but it is to be expected that at the onset of reactivation, “silent” biochemical steps predate the visible steps from dormancy into active growth. The active processes in early spring, therefore, seem to await definite signals for remobilizing the nutrient stores (Rohde and Bhalerao, 2007).
Poplar wood consists of various cell types, such as water-transporting vessels and stabilizing fibers, as well as parenchyma cells that function in short-distance transport and the storage of starch, fat, and proteins (Sauter and van Cleve, 1994). Wood rays provide for the solute demands of cambial cell division and proliferation as well as wood differentiation. Ray parenchyma cells function as a radial route for nutrient distribution. In trees, the vertical xylem parenchyma is connected with the horizontal ray parenchyma to form a three-dimensional storage system. With poplar in particular, axial xylem parenchyma cells are rare; therefore, the storage function of the stem is restricted to ray cells (Ache et al., 2010). Rays play entirely different roles during active growth (nutrient supply) compared with dormancy (nutrient storage) and could thus constitute sites from which gross transcriptional restructuring is initiated and regulated (Ache et al., 2010). It is unclear what the nature of these initiatory signals is and what the precise chain of events is that they trigger. In this study, we test the hypothesis that molecular mechanisms controlling dramatic changes in poplar physiology, in particular the wood ray mechanism, underpin seasonal phase shifts in growth-dormancy biochemistry.
Previous studies on seasonality have focused on hormonal and structural aspects of cambium reactivation, cambial activity, and cessation (Lachaud, 1989; Catesson, 1994; Savidge, 1996; Uggla et al., 1996; Baba et al., 2011), bud set (Rohde et al., 2011b), and frost hardening (Ko et al., 2011). However, the molecular basis of such seasonal behavior, the related signaling system, and the repercussions on nutrient cycles in trees remain largely unknown.
Before any cytological or anatomical changes can occur in the interphase between growth and dormancy, metabolism and gene expression profiles must undergo major reconstruction. It thus comes as no surprise that, when comparing the “summer and winter gene expression” of trees, thousands of transcripts appear differentially regulated (Ko et al., 2011).
In this study, therefore, we used a genomic and metabolomic approach to study poplar ray seasonality. In order to handle the expected large set of data generally associated with such approaches, we applied two types of filters. The first filter was set on the tissue level so that only the rays in question were addressed. For this filter, rays were enriched using the laser microdissection and pressure catapulting (LMPC) technique. This is a powerful tool for the isolation of distinct cell types from complex tissues. The second filter used was a bioinformatic one. To pinpoint general trends in, for example, seasonality-related expression profiles, pathway analyses represent the method of choice. This state-of-the-art approach enabled us to study the poplar ray transcript profiles associated with the remodeling phases linked to reactivation from late dormancy in early spring and to active growth in summer. The goal of our study, therefore, was to identify key processes in the transition phase of winter reactivation compared with summer metabolism in wood rays and to confirm the involvement of rays in this seasonal remodeling by studying the annual dynamics of selected marker genes from the identified pathways. Such knowledge is mandatory for further genome-wide analysis and the interpretation of the physiological dynamics within trees.
RESULTS
Poplar Ray Morphology Clearly Differs between Seasons
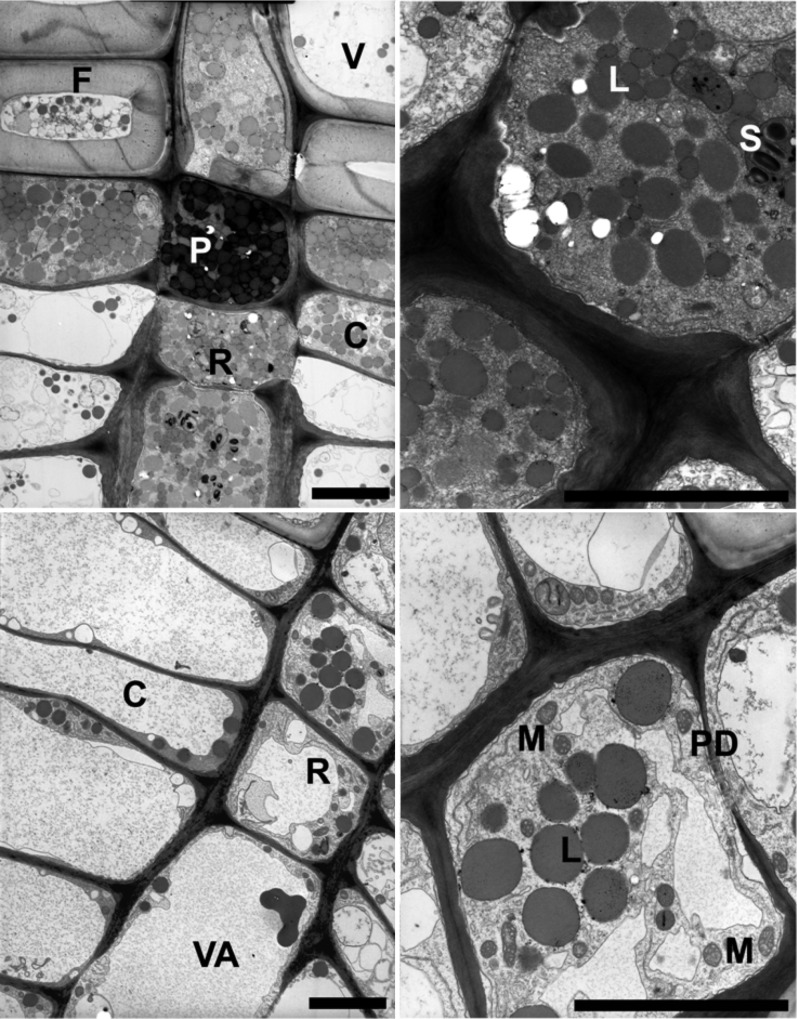
To study the two major physiological states associated with rays, we initially focused on the main growth period at the end of July (summer) and the onset of reactivation in February (early spring). The different stages of poplar wood rays in early spring and summer were documented by electron microscopy of twig cross sections. Poplar ray cells in the cambial and wood formation zones exhibited striking cytological differences between the period of remobilization for flowering in early spring and the period of continuous growth in summer (Fig. 1). In February, ray cells were packed with storage compounds such as lipids, protein, and starch; in July, ray cells were characterized by prominent vacuoles and the strong presence of mitochondria, as typically found during wood production. Based on our microscopic analyses, ray cells appeared to be metabolically highly active in July but still in dormancy in February. To explore whether early metabolic activity was already initiated in February, genome expression profiles of ray samples were analyzed by microarrays.
Figure 1.
Seasonal differences in ray contents as indicated by electron microscopy of the cambial and wood formation zone (cross sections). Top row, early spring: ray cells show a very dense cytoplasm and are packed with storage compounds, such as lipids (L), starch (S), and protein bodies (P). Bottom row, summer: ray cells show a prominent vacuole (VA) and present many mitochondria (M). Intercellular exchange is facilitated by plasmodesmata (PD). C, Typical cambial cell; F, (xylem) fiber; R, typical ray cell; V, vessel. Left panels show overview, and right panels show enlarged different sectional planes of the cells depicted in the left panels. Bars = 20 µm.
Poplar Ray Cells Were Highly Enriched by Laser Microdissection
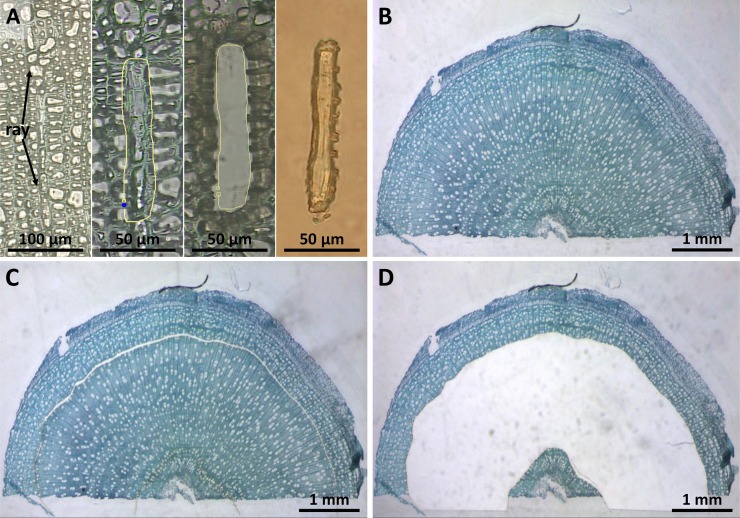
In a first approach (Fig. 2A), ray cell samples from poplar wood were harvested by LMPC and RNA was extracted. When compared with Arabidopsis (Arabidopsis thaliana) phloem samples prepared via LMPC (Deeken et al., 2008), the quality of the RNA gained was poor and not suitable for microarray analysis.
Figure 2.
LMPC of ray cells from poplar wood. A, First LMPC attempt of rays. From left to right: selected ray, signed ray, remains after ray dissection, catapulted ray. RNA yield and quality from these samples were not adequate for microarray hybridization. B to D, Inverse LMPC. B, Cross section of a poplar twig. C, Dissection of the area between the xylem differentiation zone and the central section. D, Remains after removal of ray-enriched wood. [See online article for color version of this figure.]
In considering how to circumvent this problem, we took into account that mature poplar wood contains 10% to 14% ray cells and less than 1% parenchyma cells as living constituents as well as 24% to 44% vessels and 56% to 63% fibers as nonliving cell types (Panshin and De Zeeuw, 1980; Wagenführ, 2007). Supplemental Figure S1 unequivocally shows that the wood samples used in this study contained neither axial parenchyma cells nor living fibers. Therefore, we redefined our LMPC parameters to exclude the xylem differentiation zone (as the outer limit for LMPC) and the central pith section in the sample twig (as the inner limit for LMPC). In this way, we were able to obtain samples that were highly enriched in ray cell material (Fig. 2, B–D). RNA extracted from these samples was of high quality and thus well suited for hybridization studies with microarrays.
Ray Cell Microarrays Revealed about 7,000 Seasonally Regulated Genes
We assumed that microarray analysis of ray cells would provide us with the key elements that regulate the cellular processes essential for the transition from early spring to active growth. The first analysis of the expression profiles for the entire set of 61,251 GeneChip Poplar Genome Array probe sets revealed that 6,879 genes were expressed differentially (Benjamini-Hochberg-adjusted P ≤ 0.05 and fold change ≥ 2). Out of these, 3,459 (5.63% of the total chip genes) appeared up-regulated in summer samples and 3,420 (5.57%) in early-spring samples. Applying a fold-change threshold filter of three still resulted in 2,353 (3.83%) genes up-regulated in summer and 1,842 (3.00%) in early spring, respectively. In total, 8,688 (14.1%) genes appeared significantly regulated when only the adjusted P value cutoff of 5% was considered. This relatively large number of differentially expressed genes may reflect the fact that two different functional stages of the rays were compared.
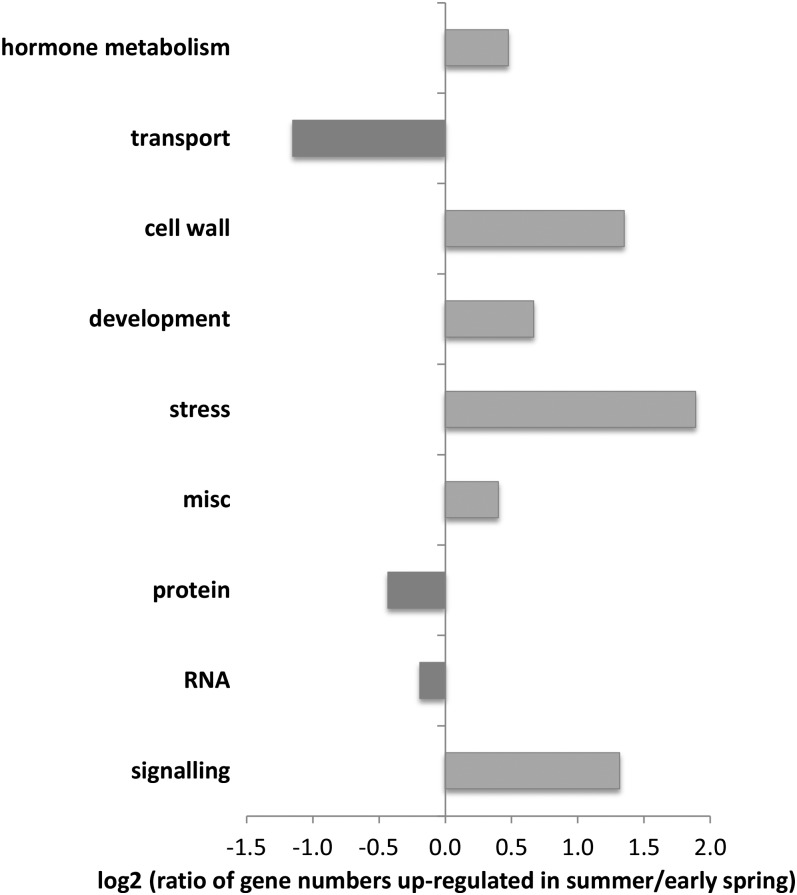
First, in order to obtain a general overview, the 500 most differentially regulated genes (chosen based on the adjusted P values), 294 up-regulated in summer (with base 2 log fold changes [logFC] ≥ 2.0) and 206 up-regulated in early spring (with logFC ≤ −1.7), were subjected to MapMan analysis using the best match with Arabidopsis (Usadel et al., 2009; Fig. 3; Supplemental Fig. S2).
Figure 3.
MapMan analysis of the 500 most significantly regulated mapped genes (chosen based on the adjusted P values) from summer versus early-spring samples. These genes were imported into MapMan 3.5.1 and classified accordingly. Presented clusters were restricted to those pathways containing 10 or more genes. Since the group of unassigned genes was also not included, only 193 (65.7%) of the 294 genes up-regulated in summer and 115 (55.8%) of the 206 genes up-regulated in early spring are shown. Negative values indicate clusters with higher induction in early spring; positive values indicate clusters with higher induction in summer.
Clusters with genes related to stress, signaling, cell wall synthesis, development, and hormone metabolism were more abundant in summer ray samples than in early spring (Fig. 3). This result highlights that wood production is of prime importance in summer and that this production process is supported by an array of genes related to growth hormones, cell differentiation, and cell wall development. It is furthermore notable that genes involved in defense and stress responses were also up-regulated, suggesting that growth processes need to be protected against summer environmental factors such as microbial attacks or drought. In contrast, early-spring samples revealed gene clusters related to RNA metabolism together with protein synthesis and transportation. This profile indicates that, notwithstanding the obvious visible dormancy (Fig. 1), remobilization processes have already been initiated in February.
Pathway Analyses Identified Key Elements of Seasonal Regulation
Having thus validated our sampling at a general level, we sought a more detailed insight into the key elements of seasonal regulation. For gross differential analysis of the gene expression data set obtained in this study, the large number of regulated genes prohibited functional interpretations at the single-gene level. Advanced approaches in microarray analysis, however, enable functional annotation of gene sets to metabolic pathways. Bioinformatic databases, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg), MapMan, and Gene Ontology (GO; Ashburner et al., 2000; http://www.geneontology.org/), provide a broad collection of functional gene sets for many organisms that can be used for gene set enrichment analysis (GSEA). The use of these algorithms requires a functional annotation of the genes present on a chip. Unfortunately, large proportions of the GeneChip Poplar Genome Array are not yet fully annotated. Therefore, we had to apply a homology-based strategy to exploit the wealth of information harbored in the poplar ray transcriptions via the well-annotated Arabidopsis genome database at The Arabidopsis Information Resource (http://www.arabidopsis.org). For this application, we mapped all probe sets with the poplar chip to their corresponding Arabidopsis Genome Initiative (AGI) codes using BLAST mapping from the PLEXdb database (Dash et al., 2012). With this approach (at a BLAST E-value cutoff of 1e-4), we identified Arabidopsis homologs for 69.4% (43,057) of all poplar genes, corresponding to 15,365 different AGI codes. Focusing on this “70%” gene set, we found a total of 4,485 (29.19%) genes differently regulated (BH-adjusted P ≤ 0.05) between the seasons, with 2,189 (14.25%) genes up-regulated in summer and 2,296 (14.94%) up-regulated in the early-spring samples.
Based on the 115 Arabidopsis pathways present in the KEGG database, we retrieved 101 pathways with 2,063 poplar homolog genes out of 2,707 original Arabidopsis genes that are annotated in KEGG pathways in total. Thus, our annotation constitutes 76% of all Arabidopsis genes in KEGG covering 13% of the probe sets present on the poplar arrays (Supplemental Fig. S3).
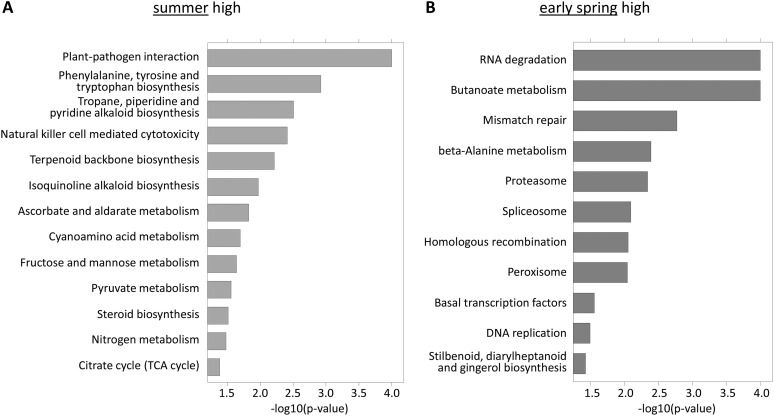
The filtered data set was then analyzed by two state-of-the-art approaches to identify differentially regulated pathways through GSEA: the so-called “self-contained” approach (ROAST; Wu et al., 2010) and the “competitive” approach (ROMER; Majewski et al., 2010). The self-contained analysis compares pathway genes between the defined conditions, whereas the competitive approach compares different gene sets not only between conditions but also against each other. A first approach using the self-contained ROAST analysis identified 32 up-regulated pathways in summer and 20 pathways up-regulated in early spring. The relatively high number of regulated pathways (52 out of 101) reflects the strong differences in cellular physiology between summer and early-spring states. To spot the most strongly regulated pathways, we then performed a competitive enrichment analysis. Using the ROMER approach, we obtained 13 pathways up-regulated in summer and 11 pathways up-regulated in early-spring samples, which were also in the set of pathways identified by the ROAST analysis (Fig. 4).
Figure 4.
Pathway enrichment analysis (competitive test). In total, 101 KEGG pathways have been included in the analysis. All significant pathways (P < 0.05) are represented by the negative decadic logarithm of the enrichment P value (i.e. longer bars represent more significant pathways). A, Up-regulated pathways (induced in summer). TCA, Trichloroacetic acid. B, Down-regulated pathways (induced in early spring).
Overall, this KEGG analysis revealed pathways of particular significance in the summer samples, similar to the top 500 analysis (Fig. 3), which could be categorized into three groups. The largest group included pathways involved in biotic and abiotic stress response and defense, namely, pathways linked to plant-pathogen and plant-herbivore interactions: tropane, piperidine, isoquinoline, and pyridine alkaloid biosynthesis (Iriti and Faoro, 2009). In addition, proteins involved in cytotoxicity, steroid biosynthesis (Bishop and Koncz, 2002), terpenoid backbone biosynthesis (Iriti and Faoro, 2009), and antioxidative metabolism (Noctor, 2006) belong to this class. The second group included pathways of sugar, energy, and nitrogen metabolism, all essential for cell proliferation during active growth. Steroid metabolism, a member of the first group, can also be coupled to this group (e.g. as a precursor of membrane components and brassinosteroids that are involved in the modulation of growth, reproduction, and development; Fujioka and Yokota, 2003; Haubrick and Assmann, 2006; Cheon et al., 2010). These groups interact with the third one, which is characterized by pathways involved in phenolic amino acid synthesis that are important not only for protein formation but also for the production of phytohormones, as well as lignin components and numerous other secondary metabolites (Bonawitz and Chapple, 2010).
Early-spring samples also confirmed the top 500 results. Pathways involved in reproduction and protein synthesis were preferentially up-regulated. This population included, for example, pathways for RNA degradation, DNA replication, proteasome and spliceosome synthesis, and mismatch repair. In addition, the induction of β-Ala metabolism implies enhanced spermine and spermidine synthesis, since these are both β-Ala precursors. Particularly in trees, spermine and spermidine are involved in bud break, leaf development, and shoot growth (Fromm, 1997; Santanen and Simola, 2007). Stilbenoids, diarylheptanoids, and gingerols are part of the phytoalexin biosynthesis and, thus, belong to the plant’s antimicrobial defense arsenal (Sobolev et al., 2011).
Subsequent to the KEGG analysis, data were evaluated using GO terms to achieve more complete gene coverage. Whereas KEGG pathways are strongly related to animals and humans, GO terms encompass more complete coverage of plant-specific pathways. Despite restriction to “biological processes,” 13,655 genes were found with AGI matches in 3,216 pathways, amounting to almost 90% coverage of all mapped genes on the chip. Before final analysis data were filtered, GO parent terms including more than 200 genes, which were relatively unspecific (e.g. metabolic process), were removed, as were terms with less than 10 genes, which were classified as too specific. A total of 1,075 terms, comprising 6,196 genes, were thereby obtained for further analysis (Supplemental Fig. S4).
The GO-based analysis indicated that 115 terms in summer and 99 terms in early spring were significantly enriched (at P ≤ 0.05). Among the 75 most enriched terms in the summer samples, 31 were related to defense and immunity and 30 to general metabolism; in early spring, 15 terms were related to RNA degradation, DNA replication, and proteasome synthesis, providing features similar to the KEGG analysis. Since GO analysis considers plant-specific pathways, the most strongly enriched GO terms in the early-spring samples also included those related to photoperiodism/flowering (seven) and seed development and gametogenesis (22). Thus, GO analysis supports the contention suggested by the earlier analyses that dormancy had already been broken by the time of sampling the early-spring probes and that metabolic reactivation and, indeed, flower induction had already started.
To test if the genes that were significantly regulated in our early-spring and summer samples were overlapping with those identified in previous studies on wood formation in active or dormant cambium, eNorthern analyses were conducted employing poplar EST libraries of different tissues and developmental stages (Sjödin et al., 2009). Only a moderate number of our genes clustered in the cambium libraries (Supplemental Fig. S5). The overlap was less than 13% with genes of the cambial zone and even lower in the specific stages relevant for our study, dormant (Supplemental Fig. S5A) and active cambium (Supplemental Fig. S5B). These results indicate that large differences exist between the behaviors of cambial and ray cells in the analyzed seasonal stages. Interestingly, about 7% of the early-spring genes formed a cluster overlapping with the floral bud library (Supplemental Fig. S5A), suggesting a link between gene activation in rays and flower formation.
Metabolite Spectra Mirror Expression Patterns
To study if the changes in transcript profiles were translated into physiological state shifts, wood samples originating from the same twig sections previously used for the collection of ray cells by LMPC were also subjected to metabolite analysis. Prominent changes in metabolism between early-spring and summer samples were detected for sugar compounds. Abundances of Fru, Glc, and maltotriose were higher in the early-spring compared with the summer samples (Fru, 3.1-fold; Glc, 9.1-fold; maltotriose, 2-fold), whereas levels of myoinositol and Suc were lower in the early-spring compared with the summer samples (myoinositol, 8.5-fold; Suc, 2.7-fold; Table I).
Table I. Metabolite abundances in poplar wood samples determined by GC-MS.
Values represent means ± se (n = 3–6) of relative metabolite abundance per g fresh weight. All samples were taken in early spring (es) and summer (su). Statistical significances between early-spring and summer samples were analyzed by a paired Student’s t test. P values were adjusted by the Benjamini-Hochberg method. n.d. = not detected.
| Class | Compound | Early Spring | Summer | Fold Change (es/su) | P Value | Adjusted P Value |
|---|---|---|---|---|---|---|
| Amino acids | 4-Aminobutyric acid | 0.107 ± 0.039 | 0.218 ± 0.029 | −2.04 | 0.046 | 0.094 |
| Ethanolamine | 0.128 ± 0.019 | 0.065 ± 0.021 | 1.96 | 0.037 | 0.094 | |
| l-Glu | 0.098 ± 0.032 | 0.163 ± 0.007 | −1.67 | 0.335 | 0.396 | |
| Orn | 0.362 ± 0.034 | 0.248 ± 0.101 | 1.46 | 0.369 | 0.399 | |
| Carbohydrates | Fru | 2.54 ± 0.53 | 0.809 ± 0.416 | 3.14 | 0.024 | 0.079 |
| Glc | 4.92 ± 0.57 | 0.539 ± 0.287 | 9.14 | 0.004 | 0.019 | |
| Maltotriose | 0.385 ± 0.132 | 0.197 ± 0.025 | 1.95 | 0.056 | 0.094 | |
| Myoinositol | 1.91 ± 0.53 | 16.3 ± 4.9 | −8.50 | 0.001 | 0.008 | |
| Suc | 8.52 ± 0.87 | 23.3 ± 9.6 | −2.74 | 0.125 | 0.18 | |
| Glycerol and derivatives | Galactosylglycerol | 0.308 ± 0.068 | n.d. | n.d. | n.d. | n.d. |
| Glycerol | 11.5 ± 2.8 | 0.305 ± 0.152 | 37.60 | 0.001 | 0.008 | |
| Organic acids | Citric acid | 3.61 ± 0.34 | 3.03 ± 1.18 | 1.19 | 0.156 | 0.203 |
| Malic acid | 2.05 ± 0.56 | 2.32 ± 0.38 | −1.13 | 0.559 | 0.559 | |
| Sugar alcohols | Galactinol | 0.377 ± 0.062 | n.d. | n.d. | n.d. | n.d. |
| Galactitol | 0.461 ± 0.111 | 0.185 ± 0.009 | 2.48 | 0.058 | 0.094 | |
| Threitol | 0.108 ± 0.024 | n.d. | n.d. | n.d. | n.d. |
The glycerol level was 37.6-fold higher in the early-spring relative to summer samples. The sugar alcohols galactinol and threitol were not detectable in summer samples but were found in samples from early spring. Moreover, the galactitol level was 2.5-fold higher in the early-spring compared with summer samples (Table I).
Using the coupled gas chromatography-mass spectrometry (GC-MS) approach, only a few amino compounds were detectable. In order to get a more detailed picture about shifts in nitrogen metabolism, amino acids were quantified by ultra-performance liquid chromatography (UPLC; Table II). In early-spring samples, concentrations of Gln, Glu, Asp, and γ-aminobutyric acid were lower compared with summer samples. Ser and Gly were nondetectable in early spring but accumulated to detectable concentrations in summer. Arg was by far the most abundant amino acid in early-spring samples but was not detectable in summer.
Table II. Amino acids and ammonium determined by UPLC.
Data (µmol g−1 fresh weight) indicate means ± sd (n = 3). Compounds were assigned to biosynthesis pathways according to Ehlting et al. (2007). Statistical significances between early-spring and summer samples were analyzed by a paired Student’s t test. P values were adjusted by the Benjamini-Hochberg method. AABA, α-Aminobutyric acid; GABA, γ-aminobutyric acid. n.d. = not detected.
| Biosynthesis Pathway | Compound | Early Spring | Summer | P Value | Adjusted P Value |
|---|---|---|---|---|---|
| Asp group | l-Asp | 0.16 ± 0.010 | 0.23 ± 0.050 | 0.079 | 0.105 |
| l-Thr | n.d. | 0.03 ± 0.010 | n.d. | n.d. | |
| l-Lys | 0.03 ± 0.002 | n.d. | n.d. | n.d. | |
| Glu group | l-Gln | 0.05 ± 0.010 | 0.41 ± 0.210 | 0.040 | 0.064 |
| l-Arg | 3.9 ± 1.290 | n.d. | n.d. | n.d. | |
| l-Glu | 0.17 ± 0.010 | 0.28 ± 0.050 | 0.021 | 0.042 | |
| GABA | 0.05 ± 0.016 | 0.25 ± 0.010 | 8.78E-05 | 7.02E-04 | |
| AABA | 0.01 ± 0.004 | n.d. | n.d. | n.d. | |
| Pyruvate group | l-Ala | 0.03 ± 0.010 | 0.03 ± 0.010 | 0.803 | 0.803 |
| Ser group | l-Ser | n.d. | 0.11 ± 0.020 | n.d. | n.d. |
| Gly | n.d. | 0.02 ± 0.001 | n.d. | n.d. | |
| Shikimate group | l-Phe | 0.03 ± 0.003 | n.d. | n.d. | n.d. |
| Others | Ammonium | 0.12 ± 0.040 | 0.08 ± 0.040 | 0.344 | 0.393 |
| Ethanolamine | 0.14 ± 0.020 | 0.07 ± 0.003 | 0.001 | 0.004 | |
| Total | 4.83 ± 1.320 | 1.56 ± 0.30 | 0.014 | 0.037 |
Time-Course Analyses of Marker Transcripts Revealed Seasonal Switch Points
In order to obtain a more detailed, quantitative view of the seasonal differences in transcript abundance in the rays, the expression of key transcripts in wood samples was analyzed by quantitative PCR (qPCR). The particular transcripts selected for qPCR analysis were informed by our GO pathway analysis.
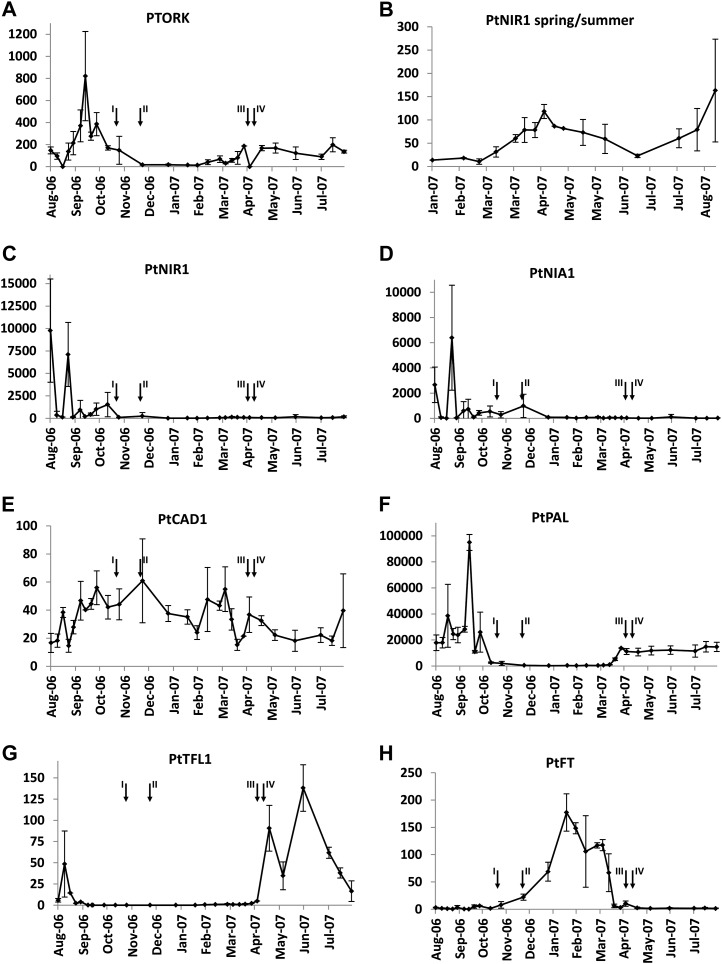
The potassium channel, PTORK, is known to be engaged in potassium transfer from the leaves to the wood, as well as in tracheid development, and was used as a control (Langer et al., 2002; Arend et al., 2005). This potassium channel is preferentially expressed in late autumn and thus serves as a marker for the finalization of tracheid development during late wood production and the transition to leaf abscission. In fact, qPCR data document that PTORK was barely expressed in winter, increased before the onset of the growth phase (March/April), and reached a maximum before leaf abscission in autumn (September/October).
As potential summer markers, we analyzed transcript levels of nitrate and nitrite reductase (NIA1 and NIR1), which are key enzymes in nitrogen assimilation. Their transcripts were highly enriched in the summer rays (logFC = 5.9 for NIA1 and NIR1). In qPCR analyses, the seasonal expression of both NIA1 and NIR1 showed an almost identical pattern to the expected increase at the beginning of the growth period, as shown in detail for NIR1 (Fig. 5B), as well as a strong increase during late wood development between August and November (Fig. 5, C and D). This general increase in NIA1 and NIR1 transcript levels in summer compared with early spring validated the microarray data in relation to nitrogen assimilation. Maximum NIA1 and NIR1 transcript abundances were observed in late August.
Figure 5.
Seasonal time course of key transcript abundances from qPCR analyses. All values represent numbers of molecules per 10,000 molecules of actin. Black arrows are as follows: I, first visible signs of leaf senescence; II, completion of leaf abscission in 2006; III, start of (leaf) bud break; IV, end of bud break in 2007 (according to Wildhagen et al., 2010). n = 3 ± sd.
As additional summer markers, we analyzed transcript abundances of phenylalanine ammonia-lyase (PAL) and cinnamyl alcohol dehydrogenase (CAD), which are key enzymes for defense metabolism (Bhuiyan et al., 2009) and lignin biosynthesis (Boerjan et al., 2003). PAL has already been shown to be involved in poplar lignin biosynthesis (Emiliani et al., 2011). In our analysis, transcript abundances of both PAL and CAD1 in rays were found to be enhanced in the summer arrays (logFC = 4.0 and 1.9). CAD1 mRNAs, however, fluctuated throughout the year without any obvious seasonal specificity. The observed difference of CAD1 expression in early-spring and summer ray arrays thus very likely originated from such a fluctuation. In contrast, PAL showed low expression in winter and high expression during the growth phase when lignification took place.
The most striking up-regulation of pathways observed in the arrays in early spring was related to reproductive processes. Therefore, we analyzed the seasonal transcript abundance of genes related to flower induction and development. In poplar, this includes Flowering Locus T (FT), as a phloem-mobile signal, and its antagonist Terminal Flower1 (TFL1; Böhlenius et al., 2006; Mohamed et al., 2010; Hsu et al., 2011). While FT was highly induced in the microarrays in early spring (logFC = −6.7), TFL1 was absent in rays. Seasonal qPCR analyses of debarked wood samples showed that TFL1 expression appeared directly after flowering at the beginning of April. Transcript abundance increased to maximum levels in June and then decreased until the end of August, before disappearing completely during FT expression (Fig. 5G). FT transcript abundance, in contrast, increased continuously from the onset of dormancy and, after reaching a maximum just before leaf-bud break, completely disappeared immediately before flowering (Fig. 5H).
DISCUSSION
Throughout the seasons, trees restructure their wood physiology in general and that of rays in particular. This study has sought to investigate season-specific physiology and biochemistry in poplar at the seasonal interphases in autumn and spring. As might be expected, ray transcription profiles from samples collected during the dormancy/spring interphase showed a large number of differentially expressed genes when compared with summer samples. Since monitoring major changes in whole-genome gene expression results in data sets of high complexity, in order to distinguish season-related pathways associated with wood formation from background noise, we focused on gene expression and metabolism of rays. In contrast to classical reductionist approaches, which concentrate on the differential expression of single genes only, we used a systematic state-of-the-art pathway analysis method to provide a global overview of gene regulation on the level of higher order cellular processes. Using this approach, we were able to identify key genes/pathways in rays that then allowed us to relate the transcriptome and the metabolite profiles of wood cells to their physiological activities through the course of the seasons. Based on these data sets, we were able to highlight certain modules within biochemical remodeling processes during the step from dormancy to reactivation. Previous transcriptomic analyses focused on cambial function and wood formation (Hertzberg et al., 2001; Andersson et al., 2004; Schrader et al., 2004; Ruttink et al., 2007; Hsu et al., 2011; Ko et al., 2011). Our analysis, in which we mined the available EST libraries, uncovered significant differences between gene profiles in cambial and ray cells. This might have been expected because these tissues have different functions, but this highlights the need for sophisticated cell type-specific analyses.
Comparing the gene expression profiles in summer and early-spring rays showed that almost all major pathways were subject to changes, thus indicating that rays play a fundamental role in the initiation of seasonal adaptation in tree physiology. In order to pinpoint the switchboard for steps from dormancy to growth, we had to identify the pathways that become active in the interphase between winter and early spring, when processes such as flowering and bud break are primed. The ROMER analysis thus served to directly extend the classical gene-wise Student’s t test to the analysis of entire pathways, and this greatly facilitated the biological interpretation of the obtained results. Furthermore, using a homology-based mapping of all poplar array genes to the well-annotated Arabidopsis genome, we were able to obtain a high coverage of pathway genes (Supplemental Fig. S3). The results suggest that these strategies, when successfully combined, could be generally applicable to complex expression profiles of plants lacking a thorough functional genome annotation.
Our analysis of summer samples identified transcript profiles associated with a lifestyle struggling with episodes of biotic and abiotic stress (Lawrence et al., 2006; Ralph et al., 2006; Rennenberg and Schmidt, 2010). Previous comparisons between summer and winter wood have identified stress-regulated genes mainly in deep winter (Ko et al., 2011). The composition of the stress-related clusters in winter, however, differed from those found here in summer. In winter, the genes required for dehydration protection and freezing tolerance (e.g. raffinose synthase, DREBA transcription factor, etc.) were increased (Ko et al., 2011), whereas in summer, pathways linked to plant-pathogen and plant-herbivore interactions were up-regulated. Our study also revealed no enrichment of defense and stress in early spring, underlining that, at this time, ray physiology is in transition in order to trigger reproductive structures.
The pathways dominating summer physiology identify photosynthesis-dependent sugar metabolism and the production of lignin precursors as major biochemical processes. The early-spring data confirmed the prediction that poplar is reactivating stored nutrients and global gene expression well before flowering. The dramatic differences in pathways active in the two interseason snapshots were found to be mirrored in cytological changes in the rays. In line with these interphase pathways, tree-specific metabolite sets confirmed predictions from the annual variations in gene activity.
In the remaining sections of this discussion, we will integrate the potassium transport, nitrogen assimilation, carbon metabolism, and lignin biosynthesis processes into our understanding of the seasonality of poplar physiology.
K+ Transport
At the transition from active growth to dormancy, a massive reallocation of nutrients from the leaves to the wood takes place via the rays, and vice versa, in spring during growth reactivation (Langer et al., 2002; Arend et al., 2005). In those prior studies, seasonally regulated K+ transporters were found in the phloem, differentiating fibers, and rays. In particular, the K+ release channel, PTORK, was highly expressed in contact cells (i.e. where a ray cell neighbors a vessel). PTORK appeared to be polarly expressed only in the membrane between both cells, where they seemed to be engaged in ion allocation into the wood vessels observed in the autumn. Based on our analysis, however, the expression of PTORK at the beginning of the growth phase suggests that PTORK is also associated with wood reactivation, channeling potassium ions from rays to the cambial region. There, potassium is required for turgor formation upon the transition from dormancy (cell division and differentiation) to expansion and growth (cell proliferation).
Carbon Metabolites
Metabolite profiling revealed increased abundances of Fru, Glc, and maltotriose, but decreased abundances of myoinositol and Suc, in early-spring compared with summer samples (Table I). Maltotriose represents a substrate for Glc biosynthesis and is produced, for example, during starch degradation. An elevated concentration of this trisaccharide suggests enhanced starch catabolism leading to subsequent degradation of maltotriose. The latter may have resulted in the release of Glc, which is required to meet the plant’s increased energy demand in spring. This view is supported by the expression in early spring of genes encoding enzymes engaged with starch and Suc metabolism, such as Suc synthases, Suc phosphate synthases, invertases, and amylases (Supplemental Table S1). Similarly, Druart et al. (2007) observed an induction of Suc catabolism at the transcriptional level in the cambial meristem cells of aspen (Populus spp.) in spring. In summer, the Suc content of the rays increased because this disaccharide is produced in the leaves and transported via the phloem and the rays to the sink tissues (Konishi et al., 2004, Ache et al., 2010).
The strongly increased glycerol levels in early-spring compared with summer samples (Table I) suggests that in addition to the catabolism of stored carbohydrates, the degradation of triacylglycerols fuel the energy demand in early spring. An accelerated catabolism of triacylglycerols results in an increased production of glycerol and fatty acids. Fatty acid catabolism is linked to the glyoxylate cycle, which itself has a gluconeogenic function and is important for anaplerotic reactions. Our microarray analyses revealed 97 genes involved in fatty acid metabolism. Of these, 59 remained unregulated, 18 were up-regulated in summer (up to 11-fold), and 20 were up-regulated in early spring. In the latter group, two isocitrate lyases appeared 40-fold up-regulated, which points to high glyoxylate cycle activity during this season (Supplemental Table S1). Along with other genes encoding enzymes of β-oxidation and the glyoxylate cycle, isocitrate lyase was highly expressed in cambial cells of aspen in spring (Druart et al., 2007). Metabolite profiles also support the notion of increased metabolism of storage fats during reactivation in spring (Druart et al., 2007). Additionally, the expression of genes encoding enzymes of the β-oxidation pathway was up-regulated in poplar bark in late winter (Park et al., 2008).
The increased concentrations of polyalcohols in early-spring samples are most likely linked to cryoprotection, since below-freezing-point temperatures are still occurring frequently during this time of the year. In line with these findings, expression of GolS2, a galactinol synthase that catalyzes the production of UDP-Gal from myoinositol, was highly induced in early spring (Supplemental Table S1). Previous studies reported an increase of galactinol in cambial meristem cells upon the establishment of dormancy (Druart et al., 2007) and an increased expression of galactinol synthase genes in the bark of poplar in early winter (Park et al., 2008).
Nitrogen Assimilation
In poplar, similar to annual plants but different from most forest trees, nitrogen assimilation takes place not only in roots but also in leaves. The partitioning of nitrogen assimilation between roots and the shoot is controlled by environmental factors (Black et al., 2002; Gessler et al., 2004). Nitrogen fertilization, for example, increases the allocation of photosynthates in the shoot (Ingestad and Agren, 1988; Pregitzer et al., 1990). As a matter of fact, in early summer, as crown growth and leaf numbers are increasing (Ibrahim et al., 1997), most of the nitrate required for growth originates from the soil. In contrast, as our study has shown, nitrate allocation for high protein demand during the time of late wood formation in August is reflected in the extremely high expression of NIA as well as NIR (Fig. 5). This suggests that, even in wood, the enzymes required for nitrogen assimilation are present. Since rays of twigs also contain chloroplasts (Goué et al., 2008), reduction of nitrate to ammonium may also take place.
In late summer, amino acids derived from nitrogen assimilation might already be directed into the synthesis of storage proteins. However, in the bark of field-grown poplar, total protein concentrations and transcript abundances of BSP1 and -2, essential for the formation of the bark storage proteins, do not increase before the onset of visible leaf senescence (Wildhagen et al., 2010). Thus, the expression peaks of NIA1 and NIR1 observed in late August do not correlate with those of BSPs. This indicates that increased NIA and NIR transcript abundances are not directly related to BSP synthesis.
In early spring, reduced concentrations of major amino acids (e.g. Gln, Glu, and Asp) indicated that the nitrogen assimilation pathway has not yet been released from its dormancy-related down-regulation. This notion is supported by the strong decline of transcript abundance for Gln metabolism enzymes, including the Gln synthetase GS2 (Supplemental Table S1). Moreover, a large number of transcripts for nitrate and nitrite reductase, and nitrate transporters, are down-regulated in early spring (Supplemental Table S1). Arg was by far the most abundant amino acid in early-spring samples (Table II), which is consistent with earlier studies showing that this nitrogen-rich amino acid is dominant in the xylem sap (Schneider et al., 1994), phloem exudates, and bark (Sagisaka 1974; Wildhagen et al., 2010) of poplar during winter until bud break. Since Arg is considered an important nitrogen storage compound (Rennenberg et al., 2010), we conclude that nitrogen metabolism remains adjusted to nitrogen storage in early spring, approximately 2 months before bud break occurs. The fact that total protein concentrations in the bark of field-grown poplars are not yet decreased in early spring tends to support this conclusion (Wildhagen et al., 2010).
Lignin Biosynthesis
Lignin biosynthesis in angiosperms is based on the formation of coniferyl and sinapyl alcohol from Phe and on the polymerization of the monolignol building blocks. PAL is the entry point for the production of secondary metabolites, whereas the final dedicated step is catalyzed by CAD1. The increase in lignin biosynthesis in summer is likely to be related to an induction of PAL expression (Fig. 5). The drastic increase of PAL in autumn before dormancy may reflect an enhanced demand for lignification during this time of the year. Since early wood has large vessels and thin cell walls, whereas late wood contains small vessels and thick cell walls (Arend and Fromm, 2007), the amount of lignin required for incorporation increases toward autumn. Further increases in lignin occur in poplar wood after growth cessation (Luo et al., 2008) but not throughout winter (Bugos et al., 1991). The observed seasonal changes in PAL expression, therefore, trace these patterns in lignification.
Flowering
Flower induction in Arabidopsis is based on a regulatory network containing light-, temperature-, and hormone-dependent elements (for review, see Corbesier and Coupland, 2006). In Arabidopsis, FT is a phloem-mobile signal, which together with its antagonist TFL1 determines the flowering time dependence on the daylength (Kotake et al., 2003; Turck et al., 2008). FT and TFL1 have also been shown to represent key elements for flowering in poplar (Böhlenius et al., 2006; Mohamed et al., 2010; Hsu et al., 2011). Therefore, it is to be expected that an early-flowering species like poplar would exhibit seasonality in certain network components different from those of an annual herbaceous plant, which flowers at the end of its life cycle. Indeed, the expression of FT, in contrast to TFL1, was induced in the early spring compared with summer. In the qPCR of wood samples, only FT expression continuously increased from the onset of dormancy to the induction of flowering. TFL1 transcript levels, in contrast, occurred strictly inversely to FT throughout the year. On the one hand, as an antagonist to FT, this behavior of TFL1 was expected. On the other hand, TFL1 was absent in rays and seems, therefore, to be expressed in other tissues during the growth period. Moreover, FT has been discussed as a mobile flowering initiation component (Turck et al., 2008). In early spring, when leaves are absent, rays represent the ideal origin for such a mobile signal. In fact, FT1 and FT2 have been shown to switch poplars between vegetative and sexual growth (Hsu et al., 2011). We found similar expression patterns as described for FT1 from Populus deltoides (Hsu et al., 2011). Our FT expression profile, together with the antagonistic TFL1 expression levels, suggest that the FT found in poplar rays might represent FT1. Here, we show that the antagonistic regulation of FT1 and TFL1 is not confined to vegetative and reproductive buds but appears also in ray cells.
Timing of Ray Elements Integrated in Poplar Seasonality
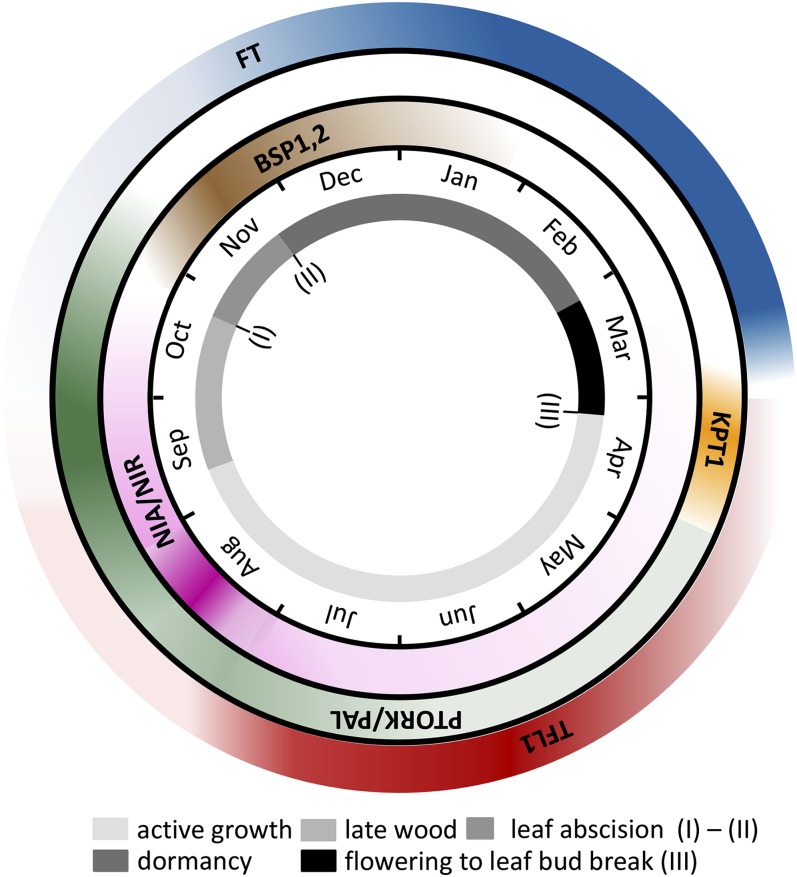
In this study, we show that annual cycles are not only characterized by pronounced variations of most of the transcripts but appear also to be synchronized in chronological order. For example, mRNAs of NIA and NIR, required for nitrogen assimilation, peaked in the second half of August. Expression of the potassium channel PTORK, which is associated with K+ retrieval and tracheid development (Arend et al., 2005), was most pronounced around the middle of September, when late wood production is characterized by increased PAL mRNAs and lignin formation. Taking these findings (Fig. 5) together, and combining them with previously published data, it is possible to provide an account of the seasonal cycle of processes in poplar (Fig. 6).
Figure 6.
Timetable of seasonal transcript abundance. The inner ring shows seasons throughout the year with unique events as indicated: I, onset of leaf senescence; II, end of leaf abscission; III, start of (leaf) bud break. The outer rings show activity windows of seasonal expression patterns. Color strengths correspond to expression levels, where the darkest colors indicate peak times of marker transcripts.
We will enter the cycle during late wood formation, which is characterized by increased nitrogen uptake and gross metabolic activity. As this phase progresses, wood differentiation is completed, followed by lignification and a nitrogen storage phase. Concomitantly, leaf senescence processes come to an end when, before abscission, potassium and nutrients are recycled from the primary photosynthetic organ and are redirected via the rays toward the wood. In this last phase of the annual cycle, the expression levels of BSP genes in the bark reflect enhanced synthesis of storage proteins (Wildhagen et al., 2010). TFL1 expression in wood samples diminishes until the end of August. In line with its antagonistic function, at the same time, low levels of FT appear. From the end of November, during dormancy, FT expression starts to increase. Peak FT transcript abundances are recorded in early February and remain high until the onset of flowering, at the beginning of March (Fig. 5). Apparently synchronized with FT rundown, TFL1 transcripts increase again. Before bud swelling, the expression of the potassium uptake channel, KPT1, rises. Potassium is taken up from the stores in the rays and the wood. As a consequence, the turgor of leaf cells increases and drives elongation growth and finally bud opening (Langer et al., 2004). Now the annual cycle enters the active growth phase, which ends with the production of late wood.
CONCLUSION
The roadmap of the annual cycle of poplar offered here shows that, by using pathway analyses together with metabolite and periodic transcript profiling, one can pinpoint processes that are ongoing during interphases, especially those between dormancy and the major growth phase. Instead of analyzing gene by gene, we have introduced and exemplified a strategy that will help to associate entire and complex metabolic pathways to distinct phases of remodeling. To enable a closer look into seasonal remodeling, we monitored the expression of marker genes within the pathways, recognized in the arrays, throughout the entire year. This set of single-gene analyses showed that most marker transcripts undergo pronounced annual fluctuations accompanying the seasonal changes in wood biology.
Rays represent platforms for the distribution of nutrients in summer, while they operate as a store for nutrients and reserve proteins in winter. It is self-evident that from array experiments with ray samples from two critical windows of the annual cycle, one cannot get a complete picture of the seasonal restructuring of poplar trees. The tools developed here, however, enabled us to identify pathways, as well as single genes, that are significant for the reactivation of poplar growth in spring. Using the marker genes to analyze rays along the annual cycle, we can also support the notion that this cell layer plays a central role in the growing season in summer as well as the interphase between dormancy and reactivation in early spring.
MATERIALS AND METHODS
Plant Material and Sample Preparation for LMPC
Twigs from the six field-grown, approximately 10-year-old poplar (Populus × canescens trees; Institut National de la Recherche Agronomique clone 717 1B4) located in a Forest Botanical Garden in Goettingen, Germany (51°33′ N, 9°57′ E, 293 m above sea level) were taken on February 12, 2007 (early-spring samples: mean air temperature in the week before harvest, −1.1°C to +2.1°C; long-term mean in February, 1.0°C) and on July 29, 2008 (summer samples: mean air temperature in the week before harvest, +11.5°C to +24.6°C; long-term mean in July, 17.1°C). The early-spring samples showed completely closed buds and appeared to be in dormancy, while the summer samples were fully leaved, healthy, and obviously in the active growth phase. From the harvested twigs, the part between the last two nodes was used as a sample and cut into approximately 5-mm pieces. These samples were decorticated with a razor blade. For fixation, the wood pieces were transferred in at least 10 volumes of ethanol:acetic acid (5:1, v/v). Following vacuum infiltration, the fixation solution was exchanged, and the material was stored at 4°C overnight. Samples were then dehydrated with 70% ethanol (20 min, room temperature) and 96% ethanol (two times, 20 min, room temperature), followed by an ethanol-Roticlear series (Carl Roth), 3:1, 1:1, 3:1 (30 min each, room temperature), and finally pure Roticlear (30 min, 30°C). The material was then transferred to Paraplast regular (Sigma-Aldrich), melted in 42°C Roticlear, and stored at 42°C overnight. The next day, the Paraplast/Roticlear was changed to pure melted Paraplast (56°C), which was replaced several times until the Roticlear was completely removed. With this procedure, the Paraplast was vacuum infiltrated using a preheated metal block as described previously (Deeken et al., 2008). Samples were placed in embedding forms, slowly cooled down to room temperature, then to 4°C, and finally stored at −80°C.
Cross sections of 10 µm of the embedded material were cut on a rotary microtome (RM2165; Leica) and placed on membrane glass slides (MembraneSlide NF 1.0 PEN; Carl Zeiss MicroImaging) treated with SigmaCote (Sigma-Aldrich). The sections were allowed to melt on the slides for at least 10 min on a 56°C heating plate and were then stored at −20°C under dehydrating conditions using plastic boxes half-filled with silica gel, with the membrane slides placed on a perforated plastic plate on top.
For time-course analyses, samples were taken from poplar trees (Institut National de la Recherche Agronomique clone 717 1B4) growing in the tree nursery of the Institute of Forest Botany and Tree Physiology in Freiburg, Germany (48°01′ N, 7°50′ E, 236 m above sea level). Details of the cultivation of these trees are described by Wildhagen et al. (2010). Wood samples were collected from August 2006 to September 2007. Weekly harvests were carried out in the transition phases in spring and autumn, whereas larger sampling intervals were chosen in summer and winter. To maximize intervals between two consecutive harvests of each tree, three to four trees were rotationally chosen out of a total of 16 trees. All harvests were carried out at 10 am. Wood from the twig section between leaves 5 and 15 was collected by separating it from the bark of one twig of three trees on each sampling date. Samples were immediately frozen in liquid nitrogen and stored at −80°C until further analysis.
LMPC
For LMPC, the cross sections on the membrane slides were deparaffinized several times with pure Histoclear (National Diagnostics) followed by 3-fold washing with 96% ethanol. The cross sections were stained using a saturated toluidine blue solution in ethanol. The slides were dried at 37°C on a heating plate. Slides were covered with PALM Liquid CoverGlass N (Carl Zeiss MicroImaging) to keep the cross sections flat and in place.
LMPC was carried out using the PALM MicroBeam System (Carl Zeiss MicroImaging). The cross-section outer differentiating zone and inner pith region were separated as described in Figure 2. The woody part was removed from the membrane slide using RNase-free forceps. The pieces were collected in a 1.5-mL low-binding tube (Eppendorf) on dry ice and stored at −80°C. On average, 80 pieces were used for one RNA extraction.
Total RNA Extraction
LMPC-Derived Samples
Total RNA was extracted using the RNeasy Micro Kit (Qiagen). Material collected via LMPC was topped with a 350-µL RLT buffer containing 1% β-mercaptoethanol and placed in a rotary mixer (Hartenstein) for 20 to 30 min and RNA was extracted according to the manufacturer’s advice. RNA quality was tested using the Experion Automated Electrophoresis System (Bio-Rad Laboratories). The purified mRNA was stored at −80°C. All extraction steps were carried out using low-binding tubes and tips (Eppendorf and Biozym, Hess) to avoid the loss of low-amount RNAs.
Samples for Time-Course Analyses
Wood prepared from the same samples previously described for bark preparation (Wildhagen et al., 2010) was ground in liquid nitrogen. Approximately 100 mg was transferred in 1.5-mL reaction tubes containing two RNase-free ceramic balls (Precellys Keramik-Kit, 1.4 mm; Peqlab). Under cooling with liquid nitrogen, the plant material was further homogenized in a mixer mill (MM300; Retsch). Total RNA was extracted using the E.Z.N.A. Plant RNA Kit (Ω Bio-tek) according to the manufacturer’s advice. A total of 2.5 µg of RNA was DNase digested using RNase-free DNase I (Fermentas). Following precipitation, RNA was reverse transcribed using Moloney murine leukemia virus RNase H− Reverse Transcriptase (Promega).
Electron Microscopy
Small wood sections of poplar twigs were cut with a razor blade and immediately immersed for 4 h in a fixation medium containing 1% (w/v) formaldehyde, 1 mm EGTA, 50 mm cacodylate buffer, and 5% glutaraldehyde. Subsequently, the tissue was postfixed with 2% (w/v) osmium tetroxide overnight at room temperature, stained with 3% (w/v) uranyl acetate in 20% ethanol for 1 h, dehydrated using a graded series of ethanol, and embedded in Spurr’s epoxy resin (Spurr, 1969). For detailed methodological information, see Arend and Fromm (2003). Ultrathin sections were cut to a thickness of 70 to 80 nm using a diamond knife on an ultramicrotome (Reichert-Jung; Ultracut E), transferred onto copper grids coated with Formvar (polyvinyl formal; Plano), and stained with lead citrate. Sections were examined using a Philips CM 12 transmission electron microscope at 80 kV.
Metabolite Profiling by GC-MS and UPLC
Wood samples representing the samples used for LMPC (GC-MS data) or a subset of the samples used for time-course analysis of marker transcripts (UPLC data) were homogenized in liquid nitrogen with mortar and pestle. For extraction (GC-MS analysis), 1.5 mL of 87% (v/v) methanol containing ribitol (0.2 mg mL−1) was added to approximately 50 mg of frozen homogenate. ribitol served as an internal standard. After extraction for 15 min at 70°C under agitation, the samples were centrifuged (5 min, 12,000g, 4°C). Aliquots (500 µL) of the supernatant were vacuum dried and thereafter dissolved in 25 μL of methoxamine hydrochloride (20 mg mL−1 pyridine) and incubated at 30°C for 90 min under continuous agitation. Then, 40 μL of N-methyl-N-trimethylsilyl trifluoroacetamide was added at 37°C for 30 min to derivatize polar functional groups. After the addition of 25 µL of alkane standard (Sigma-Aldrich), 1-µL aliquots were analyzed on a GC-quadrupole MS system (gas chromatograph, 7890A; mass spectrometer, 5975C; Agilent Technologies) as described by Fiehn (2006). For data deconvolution, peak identification and peak area determination the AMDIS software (http://chemdata.nist.gov/mass-spc/amdis/) and the Golm Metabolome Database (Kopka et al., 2005) were used. To account for differences in derivatization efficiency, peak areas of identified compounds were related to the internal standard peak area. Finally, the peak area ratio was related to the amount of plant material used for extraction. Detailed quantification of amino acids was performed by UPLC as described by Luo et al. (2009).
The statistical significance of differences between early-spring and summer samples was tested by paired Student’s t tests. P values were adjusted for multiple testing by applying the method of Benjamini and Hochberg (1995).
Microarrays
Microarray analyses were conducted at the Microarray Facility, University of Tübingen. A total of four arrays were hybridized for each summer and early-spring season. All samples were amplified using the One-Cycle Target Labeling Assay (Affymetrix) according to the manufacturer’s protocol and hybridized to the Gene Chip Poplar Genome Array (Affymetrix). Microarrays were scanned using the GCS3000 GeneChip scanner (Affymetrix) and GCOS software, version 1.4. Scanned images were subjected to visual inspection to control for hybridization artifacts and proper grid alignment. Quality control files were generated using the program Expression Console (Affymetrix).
Preprocessing
Data preprocessing was performed using the Bioconductor software (Gentleman et al., 2004) with the statistical programming environment R (R Development Core Team, 2011). Background correction, normalization, and probe set summary were performed using the procedures of robust microarray analysis (Irizarry et al., 2003). Exploratory analysis by hierarchical clustering of the arrays and correspondence analysis (Fellenberg et al., 2001) revealed a strong signal between the summer and early-spring samples (Supplemental Fig. S6), with a high inner group correlation between the biological replicates.
Mapping Poplar to Arabidopsis
Mapping of poplar probe sets was performed based on the BLAST mappings available in PLEXdb (Dash et al., 2012) using the following settings: maximum hits = 1, E value = 1e-4. Out of the 61,251 poplar probe sets on the Affymetrix Poplar Genome Array, 43,057 could be mapped to the corresponding Arabidopsis (Arabidopsis thaliana) genome loci, yielding a total of 15,365 different AGI gene codes (The Arabidopsis Information Resource; www.arabidopsis.org).
Functional Analysis
Differential gene expression between the summer and the early-spring seasons was calculated using the moderated t-statistic approach as implemented in the R package limma (Smyth, 2004), which has been specifically developed for the analysis of small sample size experiments. By exploiting information across genes, this delivers more stable results than a conventional Student’s t test. The P values of all results were corrected for multiple testing by applying the false discovery rate (Benjamini and Hochberg, 1995). Differential expression is reported in logFC throughout the entire paper.
GSEA was carried out using a model-based approach that can be directly integrated into the limma analysis of differential expression. Competitive and self-contained null hypotheses were analyzed using the ROMER (Majewski et al., 2010) and ROAST (Wu et al., 2010) methods as implemented in the limma package. Both hypothesis tests are similar to that of the classical GSEA (Subramanian et al., 2005) approach but have been designed for use with linear models. Instead of permutations, they use a background distribution generated by random rotation of the regression coefficients.
Significant up- or down-regulated genes were mapped to poplar tissue and developmental libraries (Sterky et al., 2004). For this purpose, the Affymetrix gene identifiers were converted to poplar gene identifiers (version 2.0) using the annotation list provided by the Tsai laboratory (http://aspendb.uga.edu/downloads). The lists of up- and down-regulated ray genes were analyzed in PopGenIE V2.0 (Sjödin et al., 2009; http://popgenie.org/) using the tool eNorthern. The following parameters were chosen for clustering: wards and Euclidian distance. The default settings were used for minimum gene frequency and number of genes per intensity unit.
qPCR
For real-time qPCR, cDNA was diluted 1:20 with 10 ng/μL tRNA-water. The qPCR was carried out using the ABsolute QPCR SYBR Green Capillary Mix (ABgene) in a Realplex Mastercycler (Eppendorf). All primers were chosen to amplify fragments not exceeding 500 bp. The following primers were used: PtAct2 (forward: 5′-CCCAGAAGTCCTCTT-3′), PtAct2 (reverse: 5′-ACTGAGCACAATGTTAC-3′), PtCAD1 (forward: 5′-TACAAAGGGTGGATATTC-3′), PtCAD1 (reverse: 5′-GCTAGATCGGATGTATCA-3′), PtFT (forward: 5′-AGAGAATATTTGCACTGG-3′), PtFT (reverse: 5′-CTCTGGCAGTTGAAATAA-3′), PtLFY (forward: 5′-TTATATGAACAGTGTCGT-3′), PtLFY (reverse: 5′-AGAAGTGGCACTATTG-3′), PtMYB58 (forward: 5′-AAAGATTGCCTTGTAGAA-3′), PtMYB58 (reverse: 5′-TTCCTCGTTTAATGTCAG-3′), PtNIA1 (forward: 5′-ATCAGACGATACTCTGG-3′), PtNIA1 (reverse: 5′-GCCTCTCTTTTACCTGTTC-3′), PtNIR1 (forward: 5′-AAAGGAGGGACTATTTCG-3′), PtNIR1 (reverse: 5′-CAATATCGGCAACTTGTA-3′), PtPAL (forward: 5′-AGAGGGTTTTAACTACAG-3′), PtPAL (reverse: 5′-CCAGTGAGCAATCCAGTTC-3′), PtTFL1 (forward: 5′-TAAAGTTGAGGTTCATGG-3′), and PtTFL1 (reverse: 5′-TGGTGTTAAATTTGTCCC-3′). Pt primers were designed using sequences from the Populus trichocarpa genome. Each primer pair was tested for compatibility with poplar prior to qPCR. Each transcript was quantified using an individual standard. Expression levels were normalized to 10,000 molecules of actin cDNA fragments. Due to the currently ambiguous poplar genome annotation, PAL primers might match both PAL1 and PAL3, at least, and therefore are designated here just as PAL.
GenBank accession numbers for transcripts measured by qPCR shown in Figure 5 are as follows: PtPAL (CV271667), PtCAD1 (AI161886), PtFT (AB110612.1), PtTFL1 (AB181183), PtNIA1 (BU816698), PtNIR1 (CV237671), and PTORK (CV277426). The data discussed in this paper have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession no. GSE33977 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33977).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Anatomy of poplar wood.
Supplemental Figure S2. Clustering of poplar differentially expressed genes in rays (summer versus early spring).
Supplemental Figure S3. KEGG pathway coverage.
Supplemental Figure S4. GO enrichment analysis of biological processes (competitive test).
Supplemental Figure S5. Comparison of rays with other seasonal array analyses.
Supplemental Figure S6. Exploratory array analysis by hierarchical clustering and correspondence analysis.
Supplemental Table S1. Differentially regulated transcripts according to seasonal metabolite profiles.
Supplementary Material
Acknowledgments
We thank S. Wolfart (born Elend) for help with sampling.
Glossary
- LMPC
laser microdissection and pressure catapulting
- logFC
base 2 log fold changes
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSEA
gene set enrichment analysis
- GO
Gene Ontology
- AGI
Arabidopsis Genome Initiative
- GC
gas chromatography
- MS
mass spectrometry
- UPLC
ultra-performance liquid chromatography
- qPCR
quantitative PCR
- PAL
phenylalanine ammonia-lyase
References
- Ache P, Fromm J, Hedrich R. (2010) Potassium-dependent wood formation in poplar: seasonal aspects and environmental limitations. Plant Biol (Stuttg) 12: 259–267 [DOI] [PubMed] [Google Scholar]
- Andersson A, Keskitalo J, Sjödin A, Bhalerao R, Sterky F, Wissel K, Tandre K, Aspeborg H, Moyle R, Ohmiya Y, et al. (2004) A transcriptional timetable of autumn senescence. Genome Biol 5: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arend M, Fromm J. (2003) Ultrastructural changes in cambial cell derivatives during xylem differentiation in poplar. Plant Biol 5: 255–264 [Google Scholar]
- Arend M, Fromm J. (2007) Seasonal change in the drought response of wood cell development in poplar. Tree Physiol 27: 985–992 [DOI] [PubMed] [Google Scholar]
- Arend M, Stinzing A, Wind C, Langer K, Latz A, Ache P, Fromm J, Hedrich R. (2005) Polar-localised poplar K+ channel capable of controlling electrical properties of wood-forming cells. Planta 223: 140–148 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Karlberg A, Schmidt J, Schrader J, Hvidsten TR, Bako L, Bhalerao RP. (2011) Activity-dormancy transition in the cambial meristem involves stage-specific modulation of auxin response in hybrid aspen. Proc Natl Acad Sci USA 108: 3418–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bhuiyan NH, Selvaraj G, Wei Y, King J. (2009) Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J Exp Bot 60: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Koncz C. (2002) Brassinosteroids and plant steroid hormone signaling. Plant Cell (Suppl) 14: S97–S110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BL, Fuchigami LH, Coleman GD. (2002) Partitioning of nitrate assimilation among leaves, stems and roots of poplar. Tree Physiol 22: 717–724 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C. (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bugos RC, Chiang VLC, Campbell WH. (1991) cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol Biol 17: 1203–1215 [DOI] [PubMed] [Google Scholar]
- Catesson AM. (1994) Cambial ultrastructure and biochemistry: changes in relation to vascular tissue differentiation and the seasonal cycle. Int J Plant Sci 155: 251–261 [Google Scholar]
- Cheon J, Park S-Y, Schulz B, Choe S. (2010) Arabidopsis brassinosteroid biosynthetic mutant dwarf7-1 exhibits slower rates of cell division and shoot induction. BMC Plant Biol 10: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Coupland G. (2006) The quest for florigen: a review of recent progress. J Exp Bot 57: 3395–3403 [DOI] [PubMed] [Google Scholar]
- Critchfield WB. (1960) Leaf dimorphism in Populus trichocarpa. Am J Bot 47: 699–711 [Google Scholar]
- Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA. (2012) PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res 40: D1194–D1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R. (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Druart N, Johansson A, Baba K, Schrader J, Sjödin A, Bhalerao RR, Resman L, Trygg J, Moritz T, Bhalerao RP. (2007) Environmental and hormonal regulation of the activity-dormancy cycle in the cambial meristem involves stage-specific modulation of transcriptional and metabolic networks. Plant J 50: 557–573 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting B, Dluzniewska P, Dietrich H, Selle A, Teuber M, Hänsch R, Nehls U, Polle A, Schnitzler J-P, Rennenberg H, et al. (2007) Interaction of nitrogen nutrition and salinity in Grey poplar (Populus tremula × alba). Plant Cell Environ 30: 796–811 [DOI] [PubMed] [Google Scholar]
- Emiliani G, Traversi ML, Anichini M, Giachi G, Giovannelli A. (2011) Transcript accumulation dynamics of phenylpropanoid pathway genes in the maturing xylem and phloem of Picea abies during latewood formation. J Integr Plant Biol 53: 783–799 [DOI] [PubMed] [Google Scholar]
- Fellenberg K, Hauser NC, Brors B, Neutzner A, Hoheisel JD, Vingron M. (2001) Correspondence analysis applied to microarray data. Proc Natl Acad Sci USA 98: 10781–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. (2006) Metabolite profiling in Arabidopsis. Methods Mol Biol 323: 439–447 [DOI] [PubMed] [Google Scholar]
- Fromm J. (1997) Hormonal physiology of wood growth in willow (Salix viminalis L): effects of spermine and abscisic acid. Wood Sci Technol 31: 119–130 [Google Scholar]
- Fujioka S, Yokota T. (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler A, Kopriva S, Rennenberg H. (2004) Regulation of nitrate uptake at the whole-tree level: interaction between nitrogen compounds, cytokinins and carbon metabolism. Tree Physiol 24: 1313–1321 [DOI] [PubMed] [Google Scholar]
- Goué N, Lesage-Descauses MC, Mellerowicz EJ, Magel E, Label P, Sundberg B. (2008) Microgenomic analysis reveals cell type-specific gene expression patterns between ray and fusiform initials within the cambial meristem of Populus. New Phytol 180: 45–56 [DOI] [PubMed] [Google Scholar]
- Haubrick LL, Assmann SM. (2006) Brassinosteroids and plant function: some clues, more puzzles. Plant Cell Environ 29: 446–457 [DOI] [PubMed] [Google Scholar]
- Heide OM. (1993) Daylength and thermal time responses of budburst during dormancy release in some northern deciduous trees. Physiol Plant 88: 531–540 [DOI] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlén M, Teeri TT, Lundeberg J, et al. (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA 98: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CY, Adams JP, Kim HJ, No K, Ma CP, Strauss SH, Drnevich J, Vandervelde L, Ellis JD, Rice BM, et al. (2011) FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc Natl Acad Sci USA 108: 10756–10761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim L, Proe MF, Cameron AD. (1997) Main effects of nitrogen supply and drought stress upon whole-plant carbon allocation in poplar. Can J Res 27: 1413–1419 [Google Scholar]
- Ingestad T, Agren GI. (1988) Nutrient-uptake and allocation at steady-state nutrition. Physiol Plant 72: 450–459 [Google Scholar]
- Iriti M, Faoro F. (2009) Chemical diversity and defence metabolism: how plants cope with pathogens and ozone pollution. Int J Mol Sci 10: 3371–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ko JH, Prassinos C, Keathley D, Han KH. (2011) Novel aspects of transcriptional regulation in the winter survival and maintenance mechanism of poplar. Tree Physiol 31: 208–225 [DOI] [PubMed] [Google Scholar]
- Konishi T, Ohmiya Y, Hayashi T. (2004) Evidence that sucrose loaded into the phloem of a poplar leaf is used directly by sucrose synthase associated with various β-glucan synthases in the stem. Plant Physiol 134: 1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K. (2003) Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol 44: 555–564 [DOI] [PubMed] [Google Scholar]
- Lachaud S. (1989) Participation of auxin and abscisic acid in the regulation of seasonal variations in cambial activity and xylogenesis. Trees Struct Funct 3: 125–137 [Google Scholar]
- Lang GA. (1987) Dormancy: a new universal terminology. HortScience 22: 817–820 [Google Scholar]
- Langer K, Ache P, Geiger D, Stinzing A, Arend M, Wind C, Regan S, Fromm J, Hedrich R. (2002) Poplar potassium transporters capable of controlling K+ homeostasis and K+-dependent xylogenesis. Plant J 32: 997–1009 [DOI] [PubMed] [Google Scholar]
- Langer K, Levchenko V, Fromm J, Geiger D, Steinmeyer R, Lautner S, Ache P, Hedrich R. (2004) The poplar K+ channel KPT1 is associated with K+ uptake during stomatal opening and bud development. Plant J 37: 828–838 [DOI] [PubMed] [Google Scholar]
- Lawrence SD, Dervinis C, Novak N, Davis JM. (2006) Wound and insect herbivory responsive genes in poplar. Biotechnol Lett 28: 1493–1501 [DOI] [PubMed] [Google Scholar]
- Luo Z-B, Calfapietra C, Scarascia-Mugnozza G, Liberloo M, Polle A. (2008) Carbon-based secondary metabolites and internal nitrogen pools in Populus nigra under free air CO2 enrichment (FACE) and nitrogen fertilisation. Plant Soil 304: 45–57 [Google Scholar]
- Luo Z-B, Janz D, Jiang X, Göbel C, Wildhagen H, Tan Y, Rennenberg H, Feussner I, Polle A. (2009) Upgrading root physiology for stress tolerance by ectomycorrhizas: insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiol 151: 1902–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski IJ, Ritchie ME, Phipson B, Corbin J, Pakusch M, Ebert A, Busslinger M, Koseki H, Hu Y, Smyth GK, et al. (2010) Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood 116: 731–739 [DOI] [PubMed] [Google Scholar]
- Mohamed R, Wang CT, Ma C, Shevchenko O, Dye SJ, Puzey JR, Etherington E, Sheng X, Meilan R, Strauss SH, et al. (2010) Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J 62: 674–688 [DOI] [PubMed] [Google Scholar]
- Nitsch JP. (1957) Photoperiodism in woody plants. Proc Am Soc Hortic Sci 70: 526–544 [Google Scholar]
- Noctor G. (2006) Metabolic signalling in defence and stress: the central roles of soluble redox couples. Plant Cell Environ 29: 409–425 [DOI] [PubMed] [Google Scholar]
- Okubo H. (2000) Growth cycle and dormancy in plants. In J Viémont, J Crabbé, eds, Dormancy in Plants. CABI Publishing, Oxon, UK, pp 1–22 [Google Scholar]
- Panshin AJ, De Zeeuw C. (1980) Textbook of Wood Technology, Ed 4. McGraw-Hill, New York [Google Scholar]
- Park S, Keathley DE, Han K-H. (2008) Transcriptional profiles of the annual growth cycle in Populus deltoides. Tree Physiol 28: 321–329 [DOI] [PubMed] [Google Scholar]
- Pregitzer KS, Dickmann DI, Hendrick R, Nguyen PV. (1990) Whole-tree carbon and nitrogen partitioning in young hybrid poplars. Tree Physiol 7: 79–93 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Ralph S, Oddy C, Cooper D, Yueh H, Jancsik S, Kolosova N, Philippe RN, Aeschliman D, White R, Huber D, et al. (2006) Genomics of hybrid poplar (Populus trichocarpa × deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol Ecol 15: 1275–1297 [DOI] [PubMed] [Google Scholar]
- Rennenberg H, Schmidt S. (2010) Perennial lifestyle: an adaptation to nutrient limitation? Tree Physiol 30: 1047–1049 [DOI] [PubMed] [Google Scholar]
- Rennenberg H, Wildhagen H, Ehlting B. (2010) Nitrogen nutrition of poplar trees. Plant Biol (Stuttg) 12: 275–291 [DOI] [PubMed] [Google Scholar]
- Rohde A, Bastien C, Boerjan W. (2011a) Temperature signals contribute to the timing of photoperiodic growth cessation and bud set in poplar. Tree Physiol 31: 472–482 [DOI] [PubMed] [Google Scholar]
- Rohde A, Bhalerao RP. (2007) Plant dormancy in the perennial context. Trends Plant Sci 12: 217–223 [DOI] [PubMed] [Google Scholar]
- Rohde A, Storme V, Jorge V, Gaudet M, Vitacolonna N, Fabbrini F, Ruttink T, Zaina G, Marron N, Dillen S, et al. (2011b) Bud set in poplar: genetic dissection of a complex trait in natural and hybrid populations. New Phytol 189: 106–121 [DOI] [PubMed] [Google Scholar]
- Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A. (2007) A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19: 2370–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagisaka S. (1974) Effect of low temperature on amino acid metabolism in wintering poplar: arginine-glutamine relationships. Plant Physiol 53: 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santanen A, Simola LK. (2007) Polyamine levels in buds and twigs of Tilia cordata from dormancy onset to bud break. Trees Struct Funct 21: 337–344 [Google Scholar]
- Sauter JJ, van Cleve B. (1994) Storage, mobilization, and interrelations of starch, sugars, protein, and fat in the ray storage tissue of poplar trees. Trees (Berl) 8: 297–304 [Google Scholar]
- Savidge RA. (1996) Xylogenesis, genetic and environmental regulation: a review. IAWA J 17: 269–310 [Google Scholar]
- Schneider A, Kreuzwieser J, Schupp R, Sauter JJ, Rennenberg H. (1994) Thiol and amino acid composition of the xylem sap of poplar trees (Populus × canadensis ‘robusta’). Can J Bot 72: 347–351 [Google Scholar]
- Schrader J, Moyle R, Bhalerao R, Hertzberg M, Lundeberg J, Nilsson P, Bhalerao RP. (2004) Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J 40: 173–187 [DOI] [PubMed] [Google Scholar]
- Sjödin A, Street NR, Sandberg G, Gustafsson P, Jansson S. (2009) The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol 182: 1013–1025 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 [DOI] [PubMed] [Google Scholar]
- Sobolev VS, Khan SI, Tabanca N, Wedge DE, Manly SP, Cutler SJ, Coy MR, Becnel JJ, Neff SA, Gloer JB. (2011) Biological activity of peanut (Arachis hypogaea) phytoalexins and selected natural and synthetic stilbenoids. J Agric Food Chem 59: 1673–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH, et al. (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101: 13951–13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Uggla C, Moritz T, Sandberg G, Sundberg B. (1996) Auxin as a positional signal in pattern formation in plants. Proc Natl Acad Sci USA 93: 9282–9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. (2009) A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant Cell Environ 32: 1211–1229 [DOI] [PubMed] [Google Scholar]
- Wagenführ R. (2007) Holzatlas, Ed 6. Fachbuchverlag Leipzig, Leipzig, Germany [Google Scholar]
- Wildhagen H, Dürr J, Ehlting B, Rennenberg H. (2010) Seasonal nitrogen cycling in the bark of field-grown Grey poplar is correlated with meteorological factors and gene expression of bark storage proteins. Tree Physiol 30: 1096–1110 [DOI] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. (2010) ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26: 2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.