Abstract
Strigolactones (SLs) control lateral branching in diverse species by regulating transcription factors orthologous to Teosinte branched1 (Tb1). In maize (Zea mays), however, selection for a strong central stalk during domestication is attributed primarily to the Tb1 locus, leaving the architectural roles of SLs unclear. To determine how this signaling network is altered in maize, we first examined effects of a knockout mutation in an essential SL biosynthetic gene that encodes CAROTENOID CLEAVAGE DIOXYGENASE8 (CCD8), then tested interactions between SL signaling and Tb1. Comparative genome analysis revealed that maize depends on a single CCD8 gene (ZmCCD8), unlike other panicoid grasses that have multiple CCD8 paralogs. Function of ZmCCD8 was confirmed by transgenic complementation of Arabidopsis (Arabidopsis thaliana) max4 (ccd8) and by phenotypic rescue of the maize mutant (zmccd8::Ds) using a synthetic SL (GR24). Analysis of the zmccd8 mutant revealed a modest increase in branching that contrasted with prominent pleiotropic changes that include (1) marked reduction in stem diameter, (2) reduced elongation of internodes (independent of carbon supply), and (3) a pronounced delay in development of the centrally important, nodal system of adventitious roots. Analysis of the tb1 zmccd8 double mutant revealed that Tb1 functions in an SL-independent subnetwork that is not required for the other diverse roles of SL in development. Our findings indicate that in maize, uncoupling of the Tb1 subnetwork from SL signaling has profoundly altered the balance between conserved roles of SLs in branching and diverse aspects of plant architecture.
Strigolactones (SLs) are a recently recognized class of plant hormones that inhibit bud outgrowth (Gomez-Roldan et al., 2008; Umehara et al., 2008). Together with auxin and cytokinin, the SLs and their derivatives mediate the remarkable plasticity of plant architecture that results from outgrowth of axillary branches (Beveridge et al., 2009; Dun et al., 2012). In addition to having a hormonal role in plants, SLs stimulate development of arbuscular mycorrhizae and are essential for establishment of these symbioses in maize (Zea mays; Gomez-Roldan et al., 2007). SLs also induce seed germination in a group of destructive, parasitic, flowering plants, including Striga species that target maize and other crops (Xie et al., 2010).
Key constituents of the SL biosynthetic pathway and signaling network have emerged through analysis of branching mutants in multiple plant species (Goulet and Klee, 2010). These include the more axillary growth (max) mutants of Arabidopsis (Arabidopsis thaliana; Sorefan et al., 2003; Booker et al., 2004), ramosus (rms) mutants of pea (Pisum sativum; Foo et al., 2005), decreased apical dominance (dad) mutants of petunia (Petunia hybrida; Snowden et al., 2005; Drummond et al., 2009), and the dwarf (d) and high tillering dwarf (htd) mutants of rice (Oryza sativa; Zou et al., 2006; Arite et al., 2007). That SLs are derived from carotenoids was first established by analyses of maize carotenoid-deficient mutants (Matusova et al., 2005). Essential steps in the SL biosynthetic pathway have been identified by MAX1, MAX3, and MAX4 mutants of Arabidopsis (Sorefan et al., 2003; Booker et al., 2004) and rice D27 (Lin et al., 2009). MAX1 encodes a cytochrome P450 monooxygenase assigned to a late step of the SL biosynthesis pathway (Booker et al., 2005). Orthologs of MAX3 encode CAROTENOID CLEAVAGE DIOXYGENASE7 (CCD7; Booker et al., 2004), whereas MAX4 orthologs encode a second carotenoid cleavage dioxygenase (CCD8; Sorefan et al., 2003). Rice D27 encodes a carotenoid isomerase (Lin et al., 2009; Alder et al., 2012; Waters et al., 2012a). In addition, an ATP-binding cassette transporter (Pleiotropic Drug Resistance1 [PDR1]) is implicated in SL transport (Kretzschmar et al., 2012). Recent biochemical analyses have resolved the early steps in the biosynthetic pathway in plastids. D27 catalyzes isomerization of all trans β-carotene to 9-cis β-carotene (Alder et al., 2012). The 9-cis carotenoid precursor is then cleaved by CCD7 to yield a C27-aldehyde intermediate that is the substrate of CCD8. A complex cyclization reaction catalyzed by CCD8 produces carlactone, which contains the D-ring and the enol ether bridge of SLs (Alder et al., 2012). The function of CCD8 thus emerges as a committed step in SL biosynthesis.
The mechanism of SL signal transduction has been illuminated by studies of Arabidopsis MAX2 (Stirnberg et al., 2002) and its orthologs RMS4 and D3 that encode an F-box Leu-rich repeat protein. SLs are proposed to regulate F-box-directed turnover of downstream target proteins. Orthologs of the rice D14 gene encode a conversed hydrolase also required for SL perception, though the precise role of this factor remains unclear (Arite et al., 2009, Gao et al., 2009; Liu et al., 2009; Waters et al., 2012b). Genetic studies in Arabidopsis, pea, and rice implicate orthologs of the maize Teosinte Branched1 (Tb1) transcription factor (Doebley et al., 1997) as a key downstream target of SL signaling. In all three species, mutants of Tb1 orthologs, Atbrc1 (for branched1) and Atbrc2 of Arabidopsis (Aguilar-Martínez et al., 2007; Finlayson, 2007), Psbrc1 of pea (Braun et al., 2012), and fine culm1 (fc1) of rice (Minakuchi et al., 2010), cause increased branching phenotypes. In Arabidopsis, AtBRC1 and AtBRC2 function as integrators for phytochrome regulation of branching (Finlayson et al., 2010). Placement of Tb1 orthologs downstream of SL signaling is based on two observations. First, brc and fc1 mutants do not respond to SL treatment (Brewer et al., 2009; Minakuchi et al., 2010; Braun et al., 2012). Second, double mutant combinations constructed in diverse species, including Atbrc1 max3, Atbrc1 max4, Psbrc1 rms1, and fc1 d17 (Aguilar-Martínez et al., 2007; Minakuchi et al., 2010; Braun et al., 2012), consistently show branching phenotypes that resemble the corresponding SL-deficient single mutant. Additional support for this hypothesis is also available from Arabidopsis and pea, where expression of AtBRC1 and PsBRC1 is reduced in SL mutants (Aguilar-Martínez et al., 2007; Finlayson, 2007; Braun et al., 2012) and PsBRC1 expression is enhanced by SL treatment (Braun et al., 2012). However, in rice, SL regulation of FC1 expression is not observed, and overexpression of FC1 in the d3 mutant only partially rescues the branching phenotype (Minakuchi et al., 2010). Action of SLs in rice may thus involve an alternate pathway that does not require function of the Tb1 ortholog, FC1, to inhibit outgrowth of branches (Minakuchi et al., 2010).
Selection for increased apical dominance that was central to domestication of maize from ancestral teosinte acted mainly on the Tb1 locus (Doebley et al., 1997). Expression of the dominant maize Tb1 allele is about 2-fold greater than the recessive teosinte allele (Doebley et al., 1997; Hubbard et al., 2002, Studer et al., 2011). A critical regulatory region is located between 58.7 and 69.5 kb upstream of the Tb1 coding sequence (Clark et al., 2006). Recent evidence has shown that two transposable element insertions in this essential region of the Tb1 gene are responsible for elevated expression of the maize Tb1 allele (Studer et al., 2011; Zhou et al., 2011). In maize, plant architecture derives from two types of branches that affect its agronomic utility. The first are basal tillers that arise acropetally from nodes near the ground, and like the main stalk, usually bear a terminal male inflorescence, or tassel. The second type of branch is positioned laterally in a basipetal gradient and bears the female inflorescence, i.e. the ear. The Tb1 gene is required for inhibiting outgrowth of both types of branches as well as initiating development of the female inflorescence (Doebley et al., 1997; Clark et al., 2006; Studer et al., 2011). Maize, which is a host of the parasitic plant, Striga spp., as well as an arbuscular mycorrhizae symbiont, produces at least two SLs (5-deoxy-strigol and sorghumol; Awad et al., 2006). Despite the prominent role of SL signaling in control of branching in diverse species, SL pathway genes have thus far not emerged in genetic studies of branching control in maize. Hence, the role of SL signaling in maize architecture remains unclear.
Although excessive branching is the hallmark of SL mutants in multiple species, pleiotropic phenotypes that point to broader roles of SL signaling in plant development are gradually gaining attention. Recent studies in Arabidopsis and rice have combined genetic analysis and digital morphometric measurements to identify subtle growth phenotypes affecting hypocotyls, mesocotyls, and roots (Shen et al., 2007; Tsuchiya et al., 2010; Ruyter-Spira et al., 2011; Koltai, 2011). Other nonbranching phenotypes include delayed seed germination under specific light conditions (Shen et al., 2007), reduced elongation of hypocotyls (Shen et al., 2007; Tsuchiya et al., 2010), and mesocotyls (Hu et al., 2010), as well as altered root-system architecture (Ruyter-Spira et al., 2011; Koltai, 2011). Chemical genetic screens in Arabidopsis also suggest that SLs can participate in light signaling, thereby sensitizing seeds and seedlings to light-adapted development (Tsuchiya et al., 2010). Some pleiotropic effects of SLs are attributed to interaction with auxin signaling and nutrient status. Auxin up-regulates CCD7 and CCD8 genes in Arabidopsis (Hayward et al., 2009), pea (Brewer et al., 2009), and rice (Arite et al., 2007). SL signaling in turn regulates polar auxin transport in stems and thereby acts as a shoot branching inhibitor (Crawford et al., 2010). In addition, SLs can either inhibit or stimulate lateral root development depending on phosphate availability and auxin status (Ruyter-Spira et al., 2011). Moreover, SL signaling is required for auxin-dependent stimulation of secondary growth in plants (Agusti et al., 2011). Ancient roles for SLs in plants predate evolution of vascular species and are evident in algae (Delaux et al., 2012) and moss (Physcomitrella patens), where SLs regulate branching of the protonema and have been implicated as quorum sensing-like signals (Proust et al., 2011).
Here, we show that ZmCCD8, a single-copy maize gene required for biosynthesis of SLs, has broad pleiotropic effects on architecture of the plant but comparatively mild effects on apical dominance and bud outgrowth. The genetic interactions of tb1 and zmccd8 mutants reveal independent functions of SL signaling and Tb1. Specifically, (1) we describe pleiotropic effects of SL signaling on plant architecture that do not require Tb1, and (2) we show that Tb1 has evolved independence from SL signaling.
RESULTS
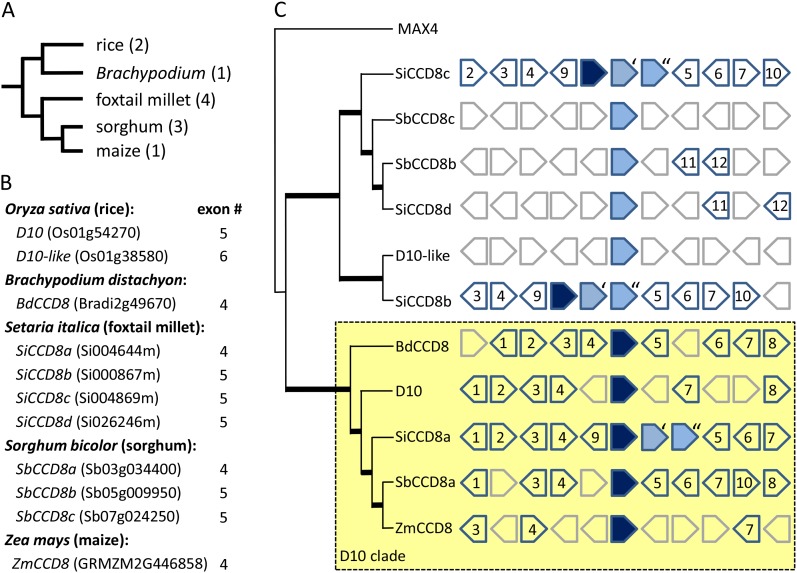
Identification of a Single CCD8 Gene in the Maize Genome
To facilitate genetic analysis of SL signaling in maize, we focused on identifying steps in the maize SL pathway that were encoded by single genes. Although there are two CCD8 genes in the rice genome (D10 and D10-like), mutations in only one gene (d10) are sufficient to increase branching (Arite et al., 2007). To identify D10 and D10-like homologs in maize, we searched the available maize cDNA, EST, GSS, HTGS, and genome sequence databases by TBLASTN (http://www.maizesequence.org/index.html, http://maizegdb.org, www.phytozome.net, and National Center for Biotechnology Information). In addition to the complete B73 genome sequence, the search included the draft genome sequence of the Palomero Toluqueño landrace (Vielle-Calzada et al., 2009). A single D10/MAX4 homolog was found and designated ZmCCD8 (GRMZM2G446858; Fig. 1). The ZmCCD8 sequence was confirmed to be a single copy by Southern-blot analysis (Supplemental Fig. S1), and the genetic map position of ZmCCD8 on the long arm of chromosome 3 (Supplemental Fig. S2) was confirmed using the IBM94 mapping kit (Maize Functional Genomics Project, University of Missouri, Columbia). The four exons of ZmCCD8 comprise a 1719-bp open reading frame that encodes a protein of 572 residues. A 2089-bp cDNA (GenBank accession no. FJ957946) was cloned by reverse transcription (RT)-PCR from roots of B73 seedlings to verify the structure of ZmCCD8. The ZmCCD8 protein sequence shares 84.9%, 59.7%, and 58.9% amino acid identity with rice D10, D10-like, and Arabidopsis MAX4, respectively. The ChloroP 1.1 program predicted a 50-amino acid, N-terminal transit peptide, consistent with plastid localization of CCD8 (Auldridge et al., 2006). Amino acid alignment of ZmCCD8 with CCD8s from different species showed that amino acids implicated in enzyme activity are well conserved in ZmCCD8 (Messing et al., 2010; Supplemental Fig. S3A).
Figure 1.
Structure of the CCD8 gene family in grasses. A, Phylogenetic relationships of grass species with completely sequenced genomes. Number of CCD8 gene copies per genome is in parentheses. B, List of CCD8 genes with number of exons in each homolog. C, Phylogenetic relationships of protein sequences for genes in B (left) and a schematic summary of local-order synteny for genes in the vicinity of CCD8 homologs (right). Gene phylogeny was determined with Clustal and rooted with Arabidopsis MAX4. Nodes depicted with thick lines have bootstrap values >90%. The clade of putative D10 orthologs is highlighted by a shaded rectangle enclosed by a dashed line. Data are adapted from the Phytozome synteny database at www.phytozome.net. The CCD8 homologs are shown as filled boxes positioned at the center of each graphic, along with the nearest five upstream and downstream neighboring genes. Numbers in boxes indicate families of homologous genes. Putative orthologs of rice D10 are shaded dark, CCD8 paralogs shaded light, and genes lacking synteny with other genomes in gray unnumbered boxes. Where sets of tandem CCD8 gene copies occur in the foxtail millet genome (Si000867 and Si004869), these are respectively labeled with single (‘) and double (“) quotation marks. [See online article for color version of this figure.]
Analysis of CCD8 Gene Phylogeny and Synteny in Grass Genomes
To explore variation in CCD8 gene copy number and to determine orthology relationships of CCD8 genes in grasses, we analyzed the phylogeny and synteny of CCD8 genes identified in completely sequenced grass genomes. Figure 1, A and B, shows that, in contrast with other grasses, the maize and Brachypodium spp. genomes contain only a single copy of CCD8. The absence of CCD8 paralogs in Brachypodium spp. was confirmed by TBLASTN searches of the available genome, EST, and cDNA databases. A phylogenetic tree of grass CCD8 homologs was created using ClustalX (Fig. 1C). Both ZmCCD8 and BdCCD8 group in a distinct clade with rice D10. Synteny surrounding CCD8 gene copies in grass genomes was explored using tools at Phytozome (www.phytozome.net) and CoGe (http://genomevolution.org/CoGe/index.pl). All synteny relationships were confirmed by alignments in CoGe, and combined results are summarized in the Phytozome schematic format as shown in Figure 1C. The presence of syntenic blocks surrounding ZmCCD8, BdCCD8, rice D10, and sorghum (Sorghum bicolor) SbCCD8a confirmed that these genes occupy orthologous positions (Fig. 1C). The D10 orthologs of all grasses except rice have four exons (Fig. 1B). Rice D10 has five exons, whereas the eudicot orthologs, including Arabidopsis MAX4, pea RMS1, and petunia DAD1, have six exons. Compared to ZmCCD8, the rice D10 gene contains an additional 106-bp intron that is inserted in the region corresponding to the 2nd exon of ZmCCD8. The additional intron does not affect the structure of the D10 protein. Interestingly, the D10 locus in Setaria spp. includes three tandem CCD8 copies, the first of which, SiCCD8a, is in the same clade as D10 and, therefore, probably its ortholog. The middle copy in the Setaria spp. cluster (SiCCD8c) groups with an unlinked paralog in the same genome (SiCCD8d) and two paralogs in sorghum (SbCCD8b and SbCCD8c). The third gene in the Setaria spp. cluster (SiCCD8b) is most closely related to rice D10-like, whereas sorghum and maize do not have genes in the D10-like clade. That SbCCD8b and SiCCD8d are orthologs is supported by local synteny of two nearby genes. Hence, SbCCD8b was most likely present in the common ancestor of foxtail millet (Setaria italica), sorghum, and maize and subsequently lost in the maize lineage. Using CoGe, we failed to detect synteny between sorghum and maize in the regions surrounding SbCCD8b and SbCCD8c, indicating that genes near these paralogs are also absent from the maize genome (data not shown).
ZmCCD8 Complementation of Arabidopsis max4
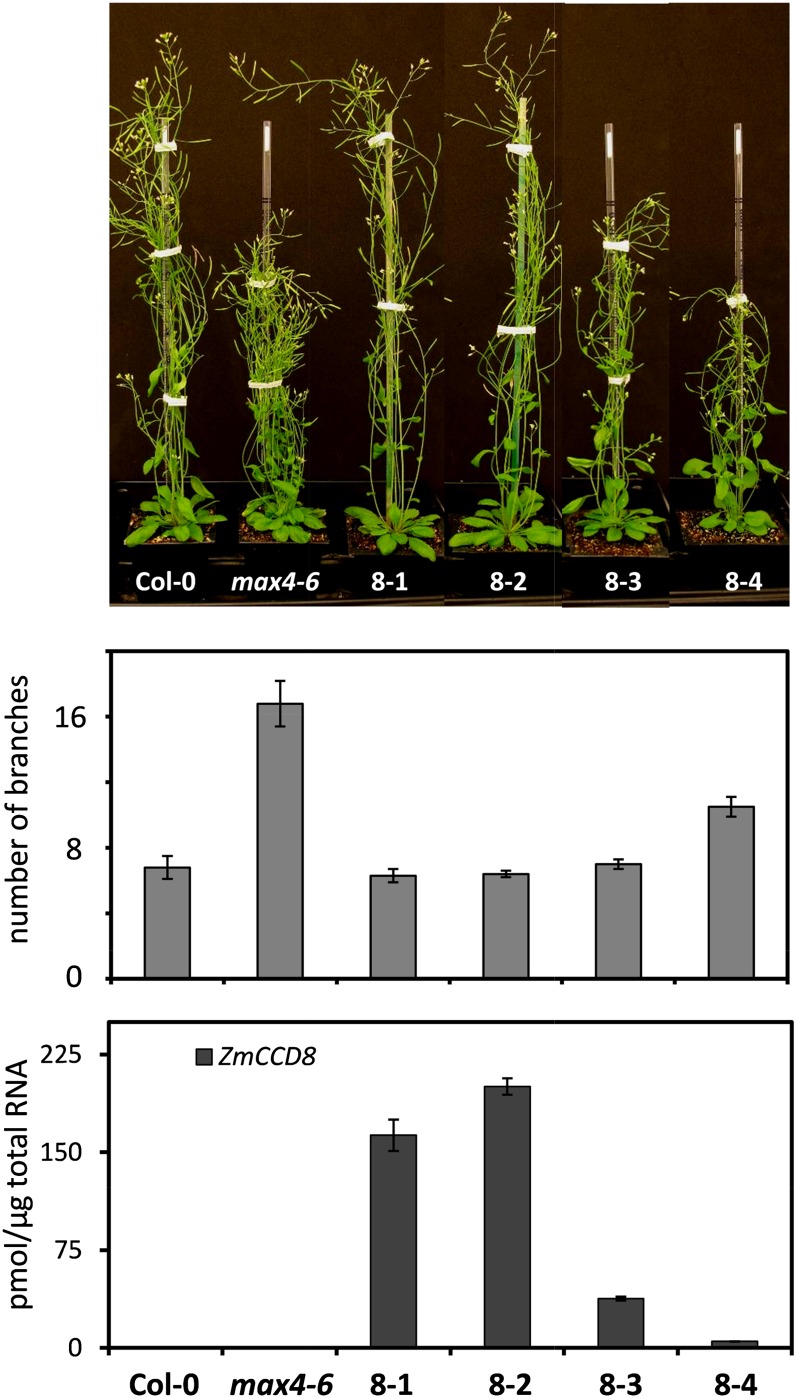
To confirm the biological function of ZmCCD8, a Pro-35S::ZmCCD8 transgene was introduced into the Arabidopsis max4-6 mutant by floral dip (Desfeux et al., 2000). Four independent transgenic lines were selected for phenotypic appraisal and quantification of ZmCCD8 mRNA abundance. As shown in Figure 2, three of four ZmCCD8 transgenic lines had elevated levels of ZmCCD8 mRNA expression, and their branching phenotypes were indistinguishable from those of wild-type plants. These results confirmed ZmCCD8 as the functional ortholog of MAX4. In the remaining line (8-4), lower ZmCCD8 expression correlated with an intermediate number of branches, consistent with a dosage-dependent, quantitative effect of ZmCCD8 on branching.
Figure 2.
Complementation of the Arabidopsis max4-6 mutant by a Pro-35S::ZmCCD8 transgene. The number of rosette branches for each independent transgenic line is shown in the bar graph below their images. Values are means ± se (n = 6). Transcript levels of the overexpressed ZmCCD8 for each line were determined by quantitative RT-PCR. Values are means ± se (n = 3). Col-0, Columbia-0. [See online article for color version of this figure.]
ZmCCD8 Expression in Reproductive Organs and Vegetative tissues
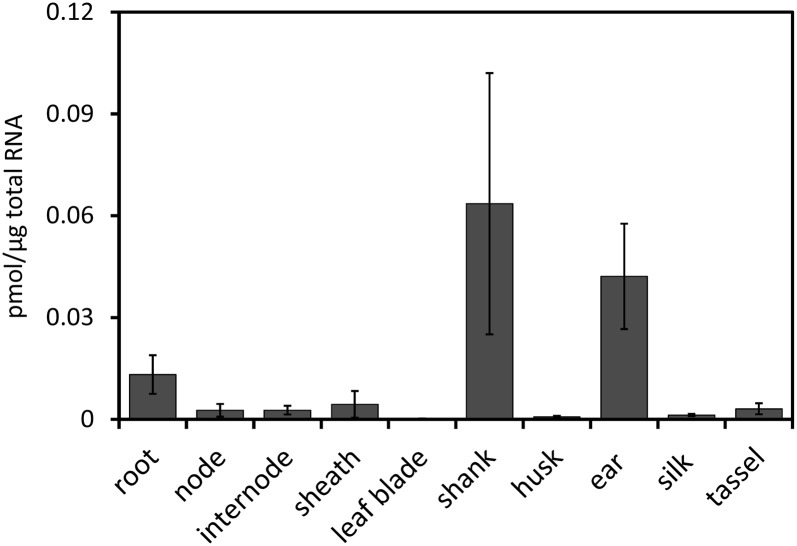
At the time of flowering (tassel emergence), ZmCCD8 mRNA was present in all vegetative tissues (Fig. 3) except the leaf blade. Expression in roots was about 3-fold higher than in sheaths, which showed highest expression among aerial vegetative tissues, including nodes, internodes, sheathes, and leaf blades. ZmCCD8 mRNA levels were elevated in shanks and ear shoots of the female inflorescence, whereas the expression in tassels was only about 7% of ear levels.
Figure 3.
Expression of ZmCCD8 in organs of mature maize plants (B73 inbred). Samples were harvested at tassel emergence. Bars show pmol ZmCCD8 mRNA per microgram total RNA. Values are means ± se (n = 3).
Characterization of a ZmCCD8 Mutant
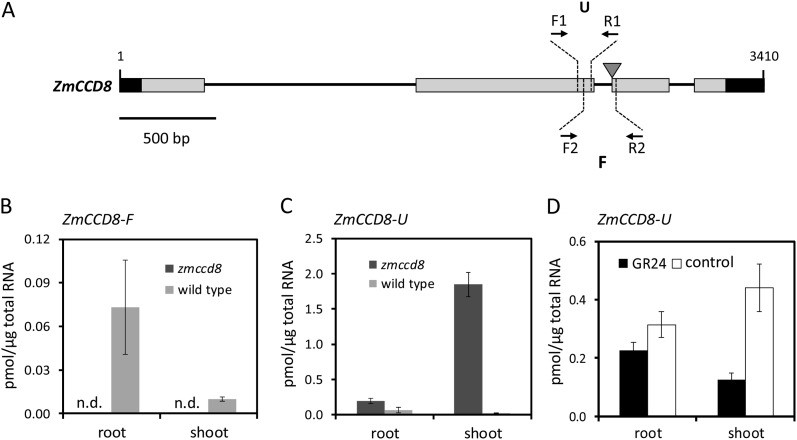
To test the hypothesis that SL regulation of branching is conserved in maize, we analyzed a ZmCCD8 knockout mutant. A Ds transposon insertion (zmccd8::Ds), initially reported in this line by Vollbrecht et al. (2010) (Ds accession B.W06.1407), was reconfirmed here by PCR and sequencing. A Ds6-like element was located exactly at the junction of the 2nd intron and 3rd exon of ZmCCD8 (Fig. 4A). In quantitative RT-PCR experiments, primers (Fig. 4A, F2/R2) flanking the insertion site efficiently amplified ZmCCD8 transcripts from tissues of wild-type plants but not from the mutant (Fig. 4B), indicating that the zmccd8 mutant did not produce an intact transcript that would include coding sequence for the highly conserved, C-terminal region of the CCD8 enzyme (Messing et al., 2010). Using primers located upstream of the insertion site (Fig. 4A, primers F1 and R1), transcripts (ZmCCD8-U) were detected in both mutant and wild-type RNAs, consistent with production of a truncated mRNA in the mutant (Fig. 4C).
Figure 4.
Structure and expression of the zmccd8::Ds insertion mutant. A, Exon-intron structure of ZmCCD8 with its four exons (solid rectangles) and three introns (horizontal lines). The 5′ and 3′ untranslated regions are shown in black. The inverted triangle indicates the site at which the Ds6-like transposable element is inserted in the zmccd8::Ds mutant allele. Primer locations for quantitative RT-PCR are indicated by arrowheads and dashed lines. The F fragment of ZmCCD8 spans the insertion site amplified by the F2/R2 pair. The U fragment of ZmCCD8 is an exon sequence located upstream of the Ds insertion site and is amplified by the F1/R1 primer pair. B, Quantification of ZmCCD8-F-containing transcripts in roots and shoots of 14-d-old zmccd8 and wild-type seedlings. C, Up-regulation of a truncated nonfunctional transcript that includes ZmCCD8-U in roots and shoots of the zmccd8 mutant. D, Suppression of ZmCCD8-U transcripts by GR24 (SL analog) treatment. Seedlings in D were grown hydroponically with 1 μm GR24 or DMSO (control). Bars show pmol mRNA per microgram total RNA. Values are means ± se (n = 3), and n.d. indicates not detected.
Feedback Regulation of ZmCCD8 by SL Signaling
As shown in Figure 4C, levels of a truncated, nonfunctional ZmCCD8 transcript were strongly elevated in shoot tissues of zmccd8 seedlings, consistent with the loss of feedback repression of CCD8 seen in SL-deficient mutants of other species (Foo et al., 2005; Snowden et al., 2005; Arite et al., 2007; Minakuchi et al., 2010). Interestingly, up-regulation of the truncated mRNA was less prominent in roots, indicating a degree of tissue specificity to feedback control in maize. To test whether the up-regulation observed here was due to SL deficiency, zmccd8 seedlings were grown in hydroponic solution and treated with the synthetic SL, GR24. As shown in Figure 4D, GR24 treatment decreased levels of ZmCCD8-U mRNA in shoots by 3.5-fold, confirming the hypothesis that ZmCCD8 is negatively regulated by SL signaling (Fig. 4D). The response in roots was less marked, consistent with a modest effect of the mutation on ZmCCD8-U mRNA abundance in that tissue. The influence of GR24 treatments on wild-type seedlings was also tested. As expected, ZmCCD8 expression in shoots was slightly down-regulated by GR24 treatments (Supplemental Fig. S4).
The Mild Branching Phenotype of zmccd8
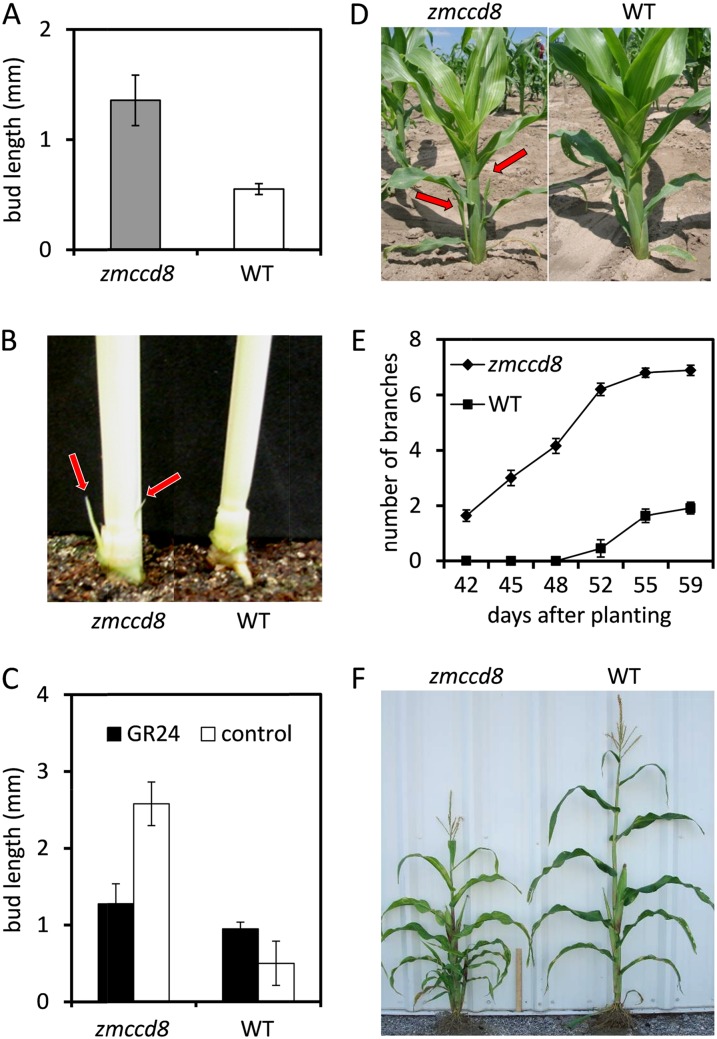
To evaluate the role of SLs in repression of lateral bud outgrowth in maize, the branching phenotype of zmccd8 was analyzed at different growth stages. In 14-d-old wild-type seedlings, buds were typically not visible in the axil of the first leaf, whereas buds were clearly evident at that position on a majority (about 70%) of zmccd8 seedlings (data not shown). In instances where visible buds were present, the average length was 2.5-fold greater on zmccd8 seedlings (Fig. 5A). Figure 5B shows that elongation of lateral buds on zmccd8 seedlings was clearly evident at 21 d after germination but not visible in wild-type plants. These observations indicate that SLs inhibit branch outgrowth from maize plants. In zmccd8 mutant seedlings grown hydroponically, the outgrowth of buds in the axil of the second leaf was suppressed by treatment with the synthetic SL, GR24 (Fig. 5C), establishing that the branching phenotype of the zmccd8 mutant is caused by SL deficiency. In Arabidopsis and rice, ccd8 mutants produce 3- and 7-fold more branches (Auldridge et al., 2006; Arite et al., 2007), respectively, with extensive elongation of primary branches and production of secondary or tertiary branches (branches that form on branches). By contrast, the overall effect of zmccd8 on plant architecture was comparatively modest. Analysis of branching in field-grown maize plants revealed that although axillary branch outgrowth had initiated by 6 weeks after planting in zmccd8 mutant plants (Fig. 5D; about 2 weeks earlier than in wild-type plants [Fig. 5E]), elongation of lateral branches on mutant plants was limited in comparison to the height of the main stem (compare also the relative branch elongation of Arabidopsis max4 in Fig. 2). The total number of lateral branches in mature zmccd8 plants (3 weeks after anthesis) was about 2-fold greater than on wild-type plants (Fig. 5F; Table I) due primarily to the initial outgrowth of lateral branches located below the ear nodes. Secondary or tertiary branches were not observed.
Figure 5.
Branching phenotypes and GR24 (SL analog) rescue of the zmccd8::Ds mutant. A, Axial bud length of zmccd8 seedlings at 14 d after germination. Mutant buds are significantly longer (P < 0.05, ANOVA). Values are means ± se (n = 7 for zmccd8; n = 4 for the wild type). B, Outgrowth of the two, lowermost axial buds (arrows) in zmccd8 seedlings at 21 d after germination. C, Suppression of bud outgrowth in zmccd8 seedlings by GR24. Seedlings were grown hydroponically and supplied with 1 μm GR24 or DMSO (control). Bud length was measured at 21 d after planting (corresponding to emergence of the 5th leaf). Mutant buds treated by GR24 are significantly shorter (P < 0.05, ANOVA). Values are means ± se (n = 4). D, Outgrowth of the axial buds in third and fourth position from the base (red arrows) of 6-week-old zmccd8 plants grown in the field. E, Time course of visible outgrowth by axial branches appearing above subtending leaf sheaths of zmccd8 mutant (diamonds) or wild-type plants (squares). The zmccd8 mutants had statistically more axillary branches (P < 0.001, ANOVA) at 42 d after planting. Values are means ± se (n = 25 for zmccd8; n = 14 for the wild type). F, Morphology of a mature zmccd8 mutant (left) compared to a wild-type plant (right) at 3 weeks after pollination. Ruler = 30 cm. WT, wild type.
Table I. Axillary branch numbers in zmccd8 mutant and wild-type plants.
Data were collected at 3 weeks after pollination. Values are means ± se.
| Genotype | Tillers | Lateral Branches | Total Branches |
|---|---|---|---|
| zmccd8 | 3.3 ± 0.2 | 8.3 ± 0.3a | 11.6 ± 0.5a (n = 53) |
| Wild type | 2.5 ± 0.1 | 3.5 ± 0.1 | 6.0 ± 0.2 (n = 35) |
P < 0.01 (ANOVA).
Interpretation of these results depends on whether the Ds allele is a complete knockout mutation. Although no intact transcripts could be detected by RT-PCR, we also considered the possibility that rare functional transcripts might still be created due to splicing of the Ds sequence from the mRNA. We tested this hypothesis by evaluating the phenotype of a stable, derivative allele (zmccd8::trDs) created by imperfect excision of Ds from zmccd8::Ds. The zmccd8::trDs derivative was identified by PCR in a screen of Ac-active progeny for germinal excision derivatives of zmccd8::Ds (data not shown). Sequences of RT-PCR products amplified by primers flanking the Ds insertion site revealed a 7-bp insertion in the transcript (Supplemental Fig. S5A), which was attributed to incomplete removal of the 8-base, host site duplication flanking the Ds insertion. The resulting frame shift creates a protein of 568 residues in which the wild-type sequence located downstream of the insertion site is replaced with a 142-amino acid sequence unrelated to ZmCCD8 (Supplemental Fig. S5B). Importantly, the phenotype of the zmccd8::trDs plants was indistinguishable from the original Ds allele (Supplemental Fig. S6). These results rule out the possibility that Ds-mediated splicing events could allow functionally significant but undetectable levels of a ZmCCD8 transcript. Thus, zmccd8::Ds is a full, loss-of-function mutation.
Short Stature, Inflorescence Size, and Narrow-Stalk Phenotypes of the zmccd8 Mutant
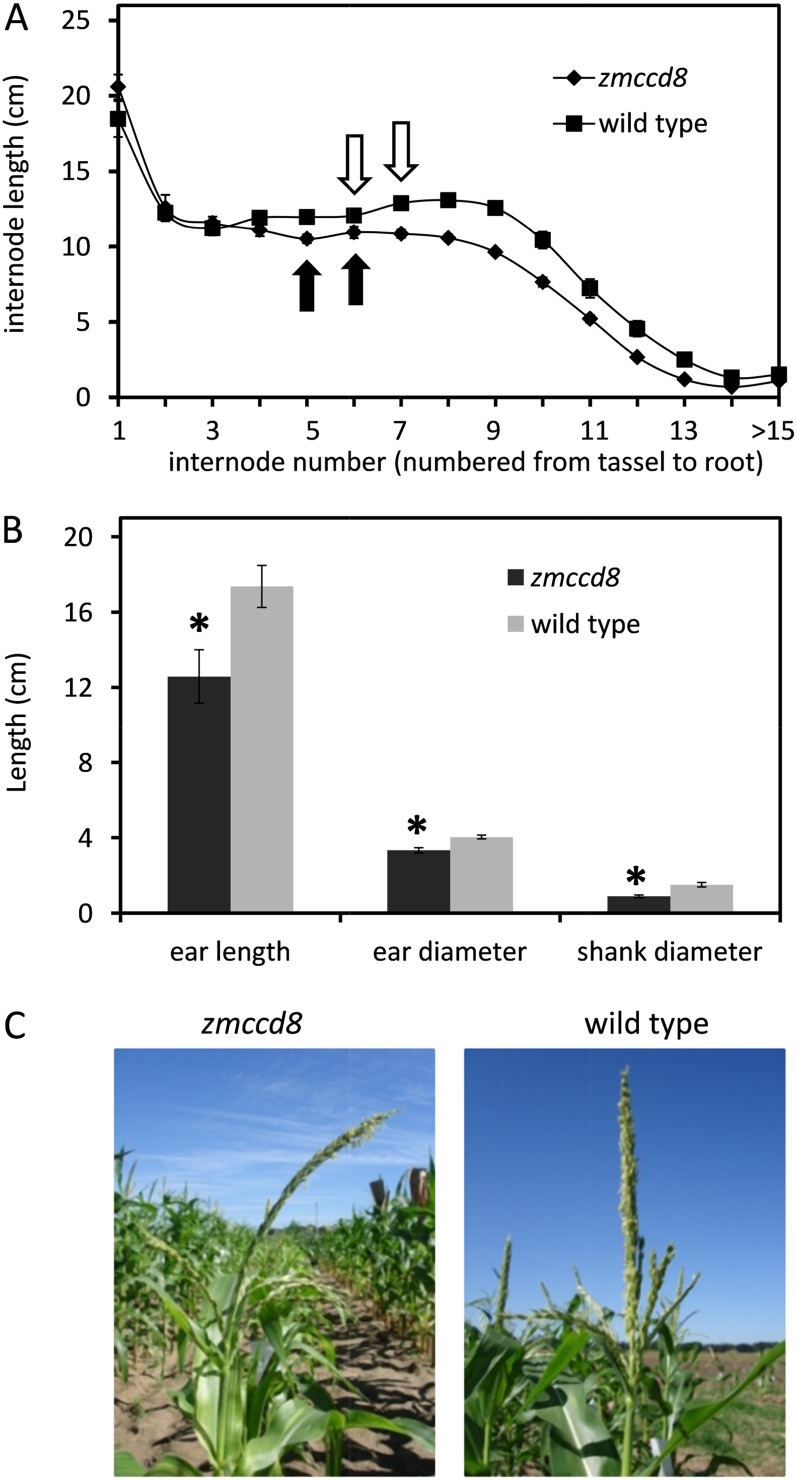
In addition to the mild branching phenotype, zmccd8 plants had shorter stature, small ears, and narrow stalks. As shown in Figure 5F, the zmccd8 plants were about 10% (P < 0.001) shorter than the wild type (151.13 ± 3.47 cm versus 167.05 ± 3.02 cm, n = 8). The difference in plant stature could be detected in seedlings as early as 10 d after planting (Supplemental Fig. S7). All internodes of zmccd8 stems below the ear node were shorter than corresponding wild-type internodes (Fig. 6A). We tested the hypothesis that the effect of SL deficiency on plant stature was due to an altered growth rate. This was indeed the case and was confirmed by data showing that the growth rate of zmccd8 was 45% less throughout the elongation stage (from 42 to 59 d after planting; Supplemental Fig. S8). The role of SL signaling during reproductive development is unclear. In the petunia dad1 mutant, flower dry weight is reduced by 40% (Snowden et al., 2005). As shown in Figure 6B, ear length and ear diameter of the zmccd8 mutant were 28% and 18% less, respectively, and both differences were significant. In addition, the shank diameter of zmccd8 ears was reduced by 41%. The structural effects were consistent with the relatively strong expression of ZmCCD8 in female inflorescences (refer back to Fig. 3). In contrast with shorter internodes of the main stalk, tassels of zmccd8 were slightly but significantly longer than those of wild-type plants (approximately 7%; P < 0.01, ANOVA; 31.3 ± 0.46 cm versus 29.3 ± 0.45 cm, n = 22). Tassel length thus comprised about 16% of total plant height (from ground to tassel tip) in zmccd8 plants compared to about 13% in wild-type plants. The zmccd8 tassels also tended to droop (Fig. 6C), probably because tassel internodes of mutant plants were longer and narrower than those of wild-type plants (Supplemental Fig. S9). At the ear node, the internode diameter was 23% less in the zmccd8 mutant (P < 0.001, ANOVA; 1.37 ± 0.02 cm, n = 53, versus 1.79 ± 0.03 cm, n = 34; Supplemental Fig. S9).
Figure 6.
Salk, ear, and tassel phenotypes of zmccd8 mutants. A, Internode length in zmccd8 mutant and wild-type plants. Internodes were numbered from tassel to base. In zmccd8, internodes between positions 5 and 14 were significantly shorter (P < 0.01, ANOVA). The internodes immediately above ear nodes are marked with arrows. Values are means ± se (n = 10). B, Statistical analysis of ear phenotypes of zmccd8 mutant and the wild type. Data were collected from dry mature ear. Asterisks indicate statistically significant differences between ears of mutant and the wild type (P < 0.0001, ANOVA). Values are means ± se (n = 20 to approximately 50). C, The drooping-tassel phenotype of the zmccd8 mutant is shown at anthesis. [See online article for color version of this figure.]
The Small Root Architecture Phenotype of the zmccd8 Mutant
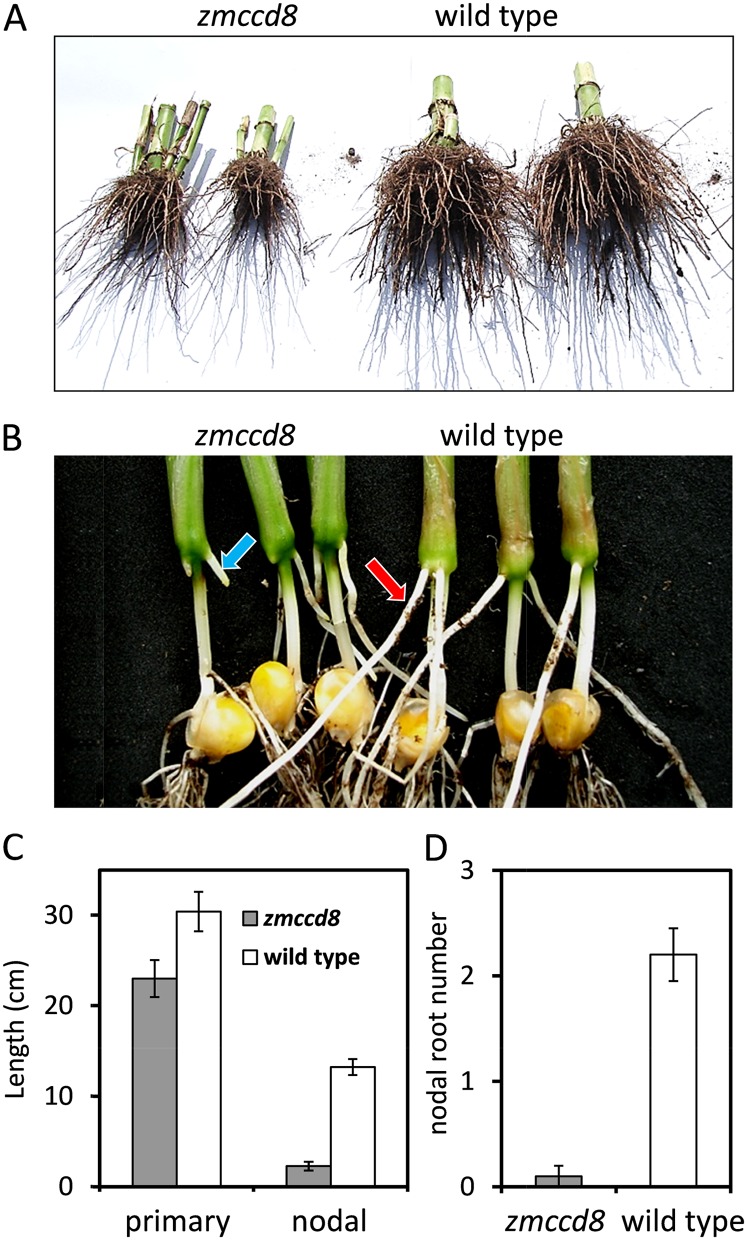
In addition to the aboveground phenotypes, zmccd8 plants had a smaller root system. As shown in Figure 7A, the root system of zmccd8 mutants was small compared to that of wild-type plants, indicative of defects in development. To determine how early the root architecture phenotype was evident, primary root length and nodal (crown) root number and length were measured in seedlings. At 14 d, the length of nodal roots on wild-type seedlings averaged 13.25 cm compared with 2.3 cm for the mutant (Fig. 7, B and C). In addition, the primary root of zmccd8 mutants was 24% shorter than that of wild-type seedlings (Fig. 7C). By 10 d after germination, all wild-type seedlings had at least two nodal roots emerging from the junctions between coleoptiles and mesocotyls, whereas most mutant seedlings had no detectable roots at these nodes (Fig. 7D). Results indicate that development of the nodal root system is delayed in the zmccd8 mutant.
Figure 7.
Root phenotype of the zmccd8 mutant. A, The central root system of field-grown zmccd8 and wild-type plants. Central root systems were excavated by severing them about 30 cm below and away from the base of the plant and gently shaking the root mass to remove the soil. B, Delayed outgrowth of nodal roots from 14-d-old zmccd8 seedlings. Nodal roots are indicated by arrows for zmccd8 and the wild type. C, Quantitative analysis of primary root lengths and nodal root length at 14 d after planting. Plants were grown in a container (0.5 L) with soil media, and root systems were exposed by submerging the entirely loosening soil in water. D, Numbers of nodal roots >5 mm were determined for zmccd8 and wild-type seedlings 10 d after planting. Values are means ± se (n = 10). [See online article for color version of this figure.]
Tb1 Expression in the zmccd8 Mutant
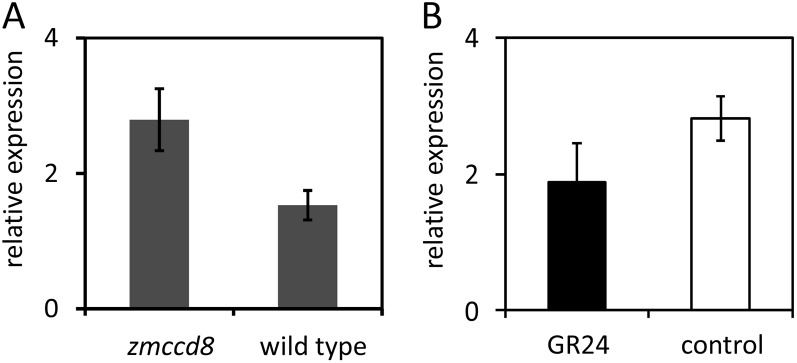
Studies in other species indicate that orthologs of the Tb1 transcription factor (AtBRC1, AtBRC2, and PsBRC1) act downstream of SL signaling and are regulated by SLs (Aguilar-Martínez et al., 2007; Braun et al., 2012). We hypothesized that one reason why branching in maize may be less dependent on ZmCCD8 than has been observed elsewhere is that maize domestication is associated with an unusual, dominant, gain-of-function mutation in the Tb1 gene (Doebley et al., 1997). To determine whether Tb1 is regulated by the SL pathway, as reported in Arabidopsis and pea (Aguilar-Martínez et al., 2007; Braun et al., 2012), we analyzed Tb1 expression in the zmccd8 mutant. Contrary to the SL-dependent regulation of Tb1 orthologs observed in eudicots, level of Tb1 expression was elevated about 80% (P < 0.05, ANOVA) in zmccd8 mutant shoots compared to those of wild-type plants (Fig. 8A). To further test if Tb1 transcription was regulated by SLs, Tb1 mRNAs were quantified in seedlings grown hydroponically either with or without GR24. No significant effect on Tb1 expression was observed (Fig. 8B; P = 0.23, ANOVA). These results indicate that the dominant Tb1 allele associated with maize domestication is not regulated by SL signaling. Recent genetic studies indicate that Grassy tiller1 (Gt1) operates downstream of Tb1 (Whipple et al., 2011). Consistent with those results, we found that Gt1 expression is also not affected in the zmccd8 mutant (data not shown). To test whether Tb1 is required for the SL feedback regulation of ZmCCD8 observed in Figure 4C, we constructed a tb1 zmccd8 double mutant and compared ZmCCD8 mRNAs in shoots of tb1 single mutant and tb1 zmccd8 double mutant seedlings. Expression of the wild-type ZmCCD8 allele was unaltered in the tb1 single mutant (Supplemental Fig. S10A). Furthermore, levels of the truncated ZmCCD8-U mRNAs in the tb1 zmccd8 double mutant and Tb1 zmccd8 single mutant were similar (Supplemental Fig. S10B; P = 0.39, ANOVA), indicating that activity of Tb1 is not required for feedback regulation of ZmCCD8.
Figure 8.
Expression of Tb1 in shoots of zmccd8 and wild type. A, Levels of Tb1 mRNA in shoots of zmccd8 mutants and wild-type seedlings. Shoot tissues (2.5 cm from the base of the coleoptile) were harvested from 14-d-old seedlings. Values are means ± se (n = 6). B, Levels of Tb1 mRNA in GR24-treated seedlings. Wild-type seedlings were grown hydroponically and treated with 1 μm GR24 (SL analog) or DMSO (control). Values are means ± se (n = 3). The expression of 18S rRNA was used as a reference.
The Genetic Interaction of tb1 and zmccd8
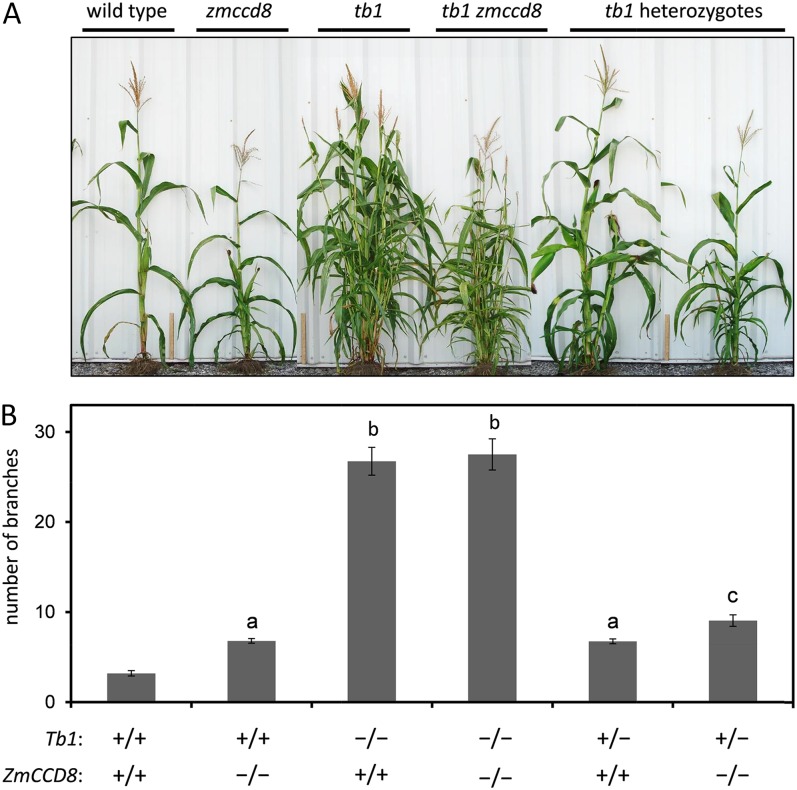
To further test the hypothesis that Tb1 is uncoupled from the regulation of SL signaling in maize, the genetic interaction between tb1 and zmccd8 was analyzed. As shown in Figure 9A, the overall effect of zmccd8 on plant architecture was markedly less severe compared to the dramatic bushy phenotype of the tb1 mutant. In sharp contrast with the zmccd8 mutant, branches of the tb1 mutant elongated extensively, formed secondary and tertiary branches, and invariably terminated with male inflorescences rather than forming ears. In addition, tb1 did not reduce plant height. The contrast between the zmccd8 and tb1 single mutants indicated that branching is still substantially inhibited in SL-deficient, zmccd8 plants. If this SL-independent component of branching control is due to activity of Tb1, then we would expect branching to be fully activated in the tb1 zmccd8 double mutant. To test this hypothesis, we compared tb1 zmccd8 double mutants to their single mutant siblings in an F2 family (Fig. 9). Although the double mutant (tb1 zmccd8; Fig. 9A, middle right) had a short stature similar to that of the zmccd8 single mutant (zmccd8; Fig. 9A, left second), the number of branches was indistinguishable between the double mutant and its tb1 single mutant sibling (Fig. 9A, middle left; 26.8 ± 1.5 versus 27.5 ± 1.7; Fig. 9B). Moreover, like the tb1 single mutant, double mutant plants exhibited extensive branch elongation, outgrowth of secondary and tertiary branches, and presence of terminal male inflorescences. Analysis of heterozygous siblings (Fig. 9A, right) indicated that a single dose of Tb1 conferred the expected, intermediate branching phenotype. In such plants, loss of ZmCCD8, and hence SL signaling, significantly increased branch numbers (6.8 ± 0.3 in ZmCCD8 +/+ Tb1 +/− compared to 9.1 ± 0.6 in zmccd8 −/− Tb1 +/− plants; Fig. 9B). These results are consistent with an additive effect of SL signaling under a limiting dosage of Tb1. In summary, the double mutant analysis, together with the expression analyses (Fig. 8), indicates that the mild branching phenotype of the zmccd8 mutant is due to activity of Tb1. Conversely, the pleiotropic short stature and narrow stalk phenotypes of the zmccd8 mutant are independent of Tb1.
Figure 9.
Phenotypes of zmccd8 and tb1 single and double mutants. A, Morphology of F2 progeny segregating for zmccd8, tb1, and their double mutant genotypes. Plants were imaged approximately 3 weeks after anthesis. A 30-cm ruler is included between plants. B, Axillary branch production in tb1 and zmccd8 F2 progeny. Branch emergence was determined by visibility of the bud tip above the subtending sheath. Genotypes of F2 progeny are indicated at the bottom of the graph. Branches were counted at 3 weeks after anthesis. Values are means ± se (n = 15 to approximately 23). Values with the same lowercase letter are not significantly different from one another (P < 0.05, ANOVA).
DISCUSSION
Mutants in SL biosynthesis and signaling increase branching in diverse plant species, including Arabidopsis, pea, and petunia (for review, see Goulet and Klee, 2010), as well as tomato (Solanum lycopersicum; Vogel et al., 2010), kiwifruit (Actinidia deliciosa; Ledger et al., 2010), and rice (Wang and Li, 2011). In contrast with the prevailing model, which places SLs upstream of Tb1 orthologs in a linear pathway of branching control, our results indicate that Tb1 mediates a branching-specific subnetwork that has become independent of SL signaling in maize. The relative importance of SL signaling to suppression of branching in maize is thus reduced relative to other species, including rice. This fundamental alteration in the relationship between SL signals and Tb1 function accentuates other nonbranching functions of SL signaling in maize architecture that are independent of Tb1. Our results suggest that relative to other grasses, selection for reduced branching during maize domestication may have been constrained to the Tb1 locus by pleiotropic effects of SL genes, combined with loss of CCD8 paralogs during recent evolutionary history of the maize genome.
Loss of CCD8 Paralog Diversity in Maize Evolution
The gene phylogeny analysis reveals dynamic gain and loss of CCD8 paralogs in the course of grass evolution (Barker et al., 2001). In contrast with its panicoid cousins, Setaria spp. and sorghum, the maize genome contains a single CCD8 homolog (ZmCCD8; Fig. 1). Sorghum has two CCD8 paralogs that are not present in maize, including at least one gene that is common to Setaria spp. and thus probably present in the most recent common ancestor of maize and sorghum (approximately 12 million years ago). It is unknown whether the loss of CCD8 paralogs and nearby genes predated the whole-genome duplication that occurred after divergence of maize from sorghum (Blanc and Wolfe, 2004). Orthology between the single CCD8 in maize (ZmCCD8) and the rice D10 gene is supported by phylogenetic relationships and synteny. In Setaria spp., three of four CCD8 copies, including the apparent D10 ortholog, occur in a tandem gene cluster. The other two genes in the Setaria spp. cluster are closely related to CCD8 paralogs that occupy unlinked locations in the sorghum and rice genomes. A parsimonious interpretation is that the Setaria spp. gene cluster arose early in grass evolution, prior to the divergence of rice (pooids) and panicoids. Subsequent duplication and/or transposition of genes in the cluster could have given rise to unlinked paralogs in Setaria spp., rice, and sorghum, whereas those paralogs were lost independently in the Brachypodium spp. and maize lineages. The remaining centrally positioned gene in the Setaria spp. cluster, SiCCD8c, would then have been duplicated in the lineage that led to sorghum, while the D10-like paralog shared by Setaria spp. and rice was apparently lost in the sorghum/maize lineage. Tandem CCD8 duplications similar to the Setaria spp. cluster have evidently arisen independently in eudicot lineages leading to Eucalyptus spp. and Citrus spp. (www.phytozome.net).
Conserved Function and Regulation of ZmCCD8
The function of ZmCCD8 in SL biosynthesis was confirmed by transgenic complementation of the max4 mutant of Arabidopsis (Fig. 2). In this analysis, the degree of phenotypic rescue was correlated with the level of ZmCCD8 mRNA, which is intriguing given the generally low expression of MAX4 in Arabidopsis (Auldridge et al., 2006). This dosage dependence indicates that CCD8 is potentially a rate-limiting step in biosynthesis of SL, a suggestion consistent with the specialized function of CCD8 in SL biosynthesis (Alder et al., 2012).
Although studies in eudicot species have emphasized SL biosynthesis in roots (Foo et al., 2005; Snowden et al., 2005; Auldridge et al., 2006); our analysis of ZmCCD8 expression (Figs. 3 and 4B) together with the expression profile of D10 in rice (Arite et al., 2007) suggest that shoot tissues of grasses have significant capacity for SL biosynthesis. In maize, CCD8 expression is especially high in shanks and developing ears of female inflorescences. Interestingly, in maize, negative feedback regulation of ZmCCD8 by SLs is much stronger in shoots than in roots (Fig. 4C), implying that SL synthesis in aboveground organs is highly regulated by SL supply. This was confirmed by showing that treatment with GR24 partially suppressed accumulation of the ZmCCD8-U transcript in shoots (Fig. 4D). The overall mechanism for feedback regulation of SL biosynthesis is evidently conserved in diverse species (Foo et al., 2005; Snowden et al., 2005; Arite et al., 2007; Mashiguchi et al., 2009; Minakuchi et al., 2010).
Establishing that zmccd8::Ds is a full, loss-of-function mutation is critical to interpretation of its complex phenotype. Analysis of ZmCCD8 mRNA transcripts produced by the mutant allele (zmccd8::Ds; Fig. 4B) indicates that zmccd8 is null for expression of transcripts that have potential to encode a functional protein. No transcript is detected using primers flanking the insertion site (Fig. 4B, ZmCCD8-F) or in exon sequences downstream of the Ds insertion (data not shown). This conclusion is further supported by our finding that a derivative, frame-shift allele (zmcccd8::trDs) has a phenotype indistinguishable from the original mutant (zmccd8::Ds; Supplemental Fig. S6). Due to the frame shift, the predicted mutant protein lacks highly conserved residues that are essential for the structure of a functional enzyme (Messing et al., 2010). Consistent with this conclusion, we note that the d10-2 mutation, a strong allele of rice D10, is due to a termination codon in the fifth exon that results in a 47-amino acid truncation from the C terminus (Arite et al., 2007). Several other ccd8 mutant alleles are due to point mutations, including pea rms1-7 (Foo et al., 2005), petunia dad1-2 (Snowden et al., 2005), and rice d10-1 (Arite et al., 2007). Based on these observations, we exclude the possibility that the mild branching phenotype of zmccd8 can be attributed to leaky expression of ZmCCD8 or residual activity of ZmCCD8 protein. However, we cannot formally exclude the possibility of an alternative pathway for SL biosynthesis in plants (Delaux et al., 2012).
Pleiotropic zmccd8 Phenotypes Reveal Diverse Roles of SL Signaling in Maize
The comparatively modest increase in branching caused by the zmccd8::Ds mutation contrasts with the suite of other pronounced, nonbranching aspects of the phenotype, including short stature, narrow stem diameter, and delayed development of the adventitious nodal root system. These diverse effects highlight the pleiotropic nature of phenotypes that have been attributed to SL signaling mutants in other systems. Although thus far all known SL-deficient and signaling mutants are associated with dwarf phenotypes (Foo, et al., 2005; Snowden et al., 2005; Zou et al., 2006; Arite et al., 2007; Finlayson et al., 2010), the role of SL signaling in determining plant height is unclear. In the rice ccd7 mutant (htd1/d17), for example, the dwarf phenotype is reversed by removal of axillary buds, indicating that the dwarfism of the htd1 mutant could be a consequence of resources allocated to the excessive tiller production rather than a direct effect of SL signaling (Zou et al., 2006). However, due to the SL-independent action of Tb1 in maize (discussed below), zmccd8 plants do not have the excessive level of branching comparable to that of the rice d10 or htd1 mutants. Competition between tillers and the main stem for resources is thus unlikely to be the basis for reduced stature. An indirect growth effect due to a smaller root system remains a possibility (Fig. 7). Recent studies of rice mesocotyl growth in darkness suggest that SLs negatively regulates cell division but not cell elongation (Hu et al., 2010). While this is consistent with the reduced growth rate of zmccd8 plants observed in this study (Supplemental Fig. S8), our data do not resolve effects due to cell number and cell size.
Together, the plant height and narrow-stem phenotypes of SL mutants in grasses (maize and rice) highlight an interesting parallel in roles of SL signaling in monocot and eudicot development. In eudicots, the thickening of stems and roots results from secondary growth, which is due to cell proliferation activity in the vascular cambium. Agusti et al. (2011) showed that SL signaling interacts with auxin to stimulate the activity of vascular cambium in Arabidopsis and pea. In grasses, elongation and thickening of the stem is primarily due to activity of intercalary meristems located in nodes of the culm. In maize, reduced stature of the zmccd8 mutant is mainly attributable to reduced elongation of internodes located below the ear node (Fig. 6A). A similar semidwarf phenotype due to shortened lower internodes (Knöller et al., 2010) results from dysfunction of the maize Brachytic2 gene, which encodes an ATP-binding cassette type B auxin transporter. It is tempting to speculate that SL signaling interacts with auxin signaling to control elongation and thickening of the stem in maize. The comparatively strong expression of ZmCCD8 in ear tissues is consistent with the reduced size of the ear (Fig. 6B), whereas the tassel is slightly enlarged (Fig. 6C), highlighting the possibility of differential roles for SL in development of female and male inflorescences in monoecious maize.
The root architecture of maize has two components: the seminal (seed) root system and the nodal (crown) root system. Robust development of the nodal root system after germination and seedling emergence is essential for plant survival and reproduction under field conditions. The slow development of nodal roots on zmccd8 mutant plants (Fig. 7) indicates that the transition from dependence on the seminal system to the nodal root system is delayed. The nodal roots of maize are adventitious roots, and recent work in Arabidopsis and pea has implicated SLs in suppressing this class of roots (Rasmussen et al., 2012). However, in maize, SL signaling evidently has an opposite effect; SL-deficient zmccd8 seedlings have fewer and shorter nodal roots (Fig. 7). Because the formation of adventitious roots involves interactions with other plant hormones (Rasmussen et al., 2012), the contrasts between species may point to fundamental differences in interactions among signaling pathways of auxin, cytokinin, and SLs. The small root system phenotype of zmccd8 is consistent with the differences of root morphology between Striga spp.-resistant and -susceptible maize lines (Amusan et al., 2008). The resistant inbred line has fewer nodal roots.
Regulation of Tb1 Is Uncoupled from SL Signaling in Maize
An emerging consensus of studies in Arabidopsis, rice, and pea is that Tb1 orthologs (i.e. BRC1 and FC1) act downstream of SL signaling to control branching (Aguilar-Martínez et al., 2007; Minakuchi et al., 2010; Braun et al., 2012). The BRC1 (ortholog of Tb1) is proposed to act as an integrator of branching signals that responds to different growth conditions in Arabidopsis (Aguilar-Martínez et al., 2007; Finlayson et al., 2010). In Arabidopsis and pea, the BRCs are transcriptionally regulated by SLs, as evidenced by reduced expression of AtBRC1 and PsBRC1 in SL mutants (Aguilar-Martínez et al., 2007; Finlayson, 2007; Braun et al., 2012) and by induction of PsBRC1 expression by SL treatments (Braun et al., 2012). However, in contrast with Arabidopsis and pea, FC1 is not induced in rice seedlings by SL treatments nor is its expression reduced in SL-deficient mutants of rice (Minakuchi et al., 2010). Thus, these results indicate that FC1 expression may be coupled less directly to SL signaling in rice. Our analyses of gene expression confirm that regulation of Tb1 transcript level is uncoupled from SL signaling in maize, where we find that expression of Tb1 is not reduced in zmccd8 mutants or up-regulated by GR24 treatment (Fig. 8). Moreover, expression of ZmCCD8 is not affected by Tb1 (Supplemental Fig. S10).
SL-Independent Activity of Tb1 Suppresses Branching in zmccd8 Mutant Maize
Our finding that regulation of Tb1 is uncoupled from SL signaling suggested that SL-independent activity of Tb1 might account for the muted branching phenotype of the zmccd8 mutant. This hypothesis is confirmed by analysis of the tb1 zmccd8 double mutant (Fig. 9). Notably, the significant inhibition of branching evident in the zmccd8 single mutant is abolished by the introduction of tb1 into the SL-deficient zmccd8 background. Double mutant plants closely resemble the tb1 single mutant in key branching characteristics that distinguish the zmccd8 and tb1 single mutants, including branch number, extensive branch elongation, formation of secondary and tertiary branches, and exclusive formation of branches that have terminal male inflorescences. Although the observation that tb1 is epistatic to zmccd8 with respect to branching implies that the effect of SL signaling on branching requires activity of Tb1, an additive effect of SL deficiency and Tb1 dosage is observed in comparison of zmccd8 impact on branch number in Tb1 Tb1 homozygous and Tb1 tb1 heterozygous backgrounds. One possibility is that branching is already maximized in homozygous tb1 plants, leaving little or no capacity for an additive zmccd8 effect in the double mutant. Alternatively, an additive effect could result if SL signaling and Tb1 interact with a common downstream target. Conversely, analysis of the double mutant has shown that in contrast to the branching phenotype, the effects of zmccd8 on plant height, stem diameter, and nodal root development, as well as the SL-mediated negative feedback regulation of ZmCCD8, are independent of Tb1 function. Hence, our results delineate a discrete subnetwork of Tb1-dependent functions in maize.
Evolution of an Altered SL/Tb1 Network in Grasses and Implications for Domestication
Based on the above observations, we propose a model for the interaction between Tb1 and SL signaling in maize (Fig. 10). Whether the uncoupling of Tb1 from SL signaling is specific to maize or extends more broadly in related grasses is an open question. While, FC1 and SL signaling are also at least partially independent, SLs are clearly major regulators of branching in rice (Minakuchi et al., 2010). In maize, the SL/Tb1 system has evolved still further toward an SL independence of the subnetwork that controls branching. This contrast may have implications for the genetics of domestication in grasses. Selection for a strong central stem through increased apical dominance has occurred multiple times during the domestications of foxtail millet, sorghum, and maize from highly branched, wild ancestors (Doust et al., 2004; Doust, 2007; de Alencar Figueiredo et al., 2008, Remigereau et al., 2011). In maize, increased apical dominance is attributed principally to selection at the Tb1 locus. However, multiple quantitative trait loci are implicated in reduced branching associated with domestication of foxtail millet (Doust et al., 2004), indicating that the genetic potential for altered branching in related grasses is not limited to Tb1. We note that the CCD8 gene cluster in foxtail millet is linked to a branching quantitative trait locus on chromosome 5 (Doust et al., 2004). Hence, the loss of CCD8 paralogs in maize may have narrowed the potential for genetic variation in the SL biosynthetic pathway in a way that constrained the path of domestication relative to other grasses.
Figure 10.
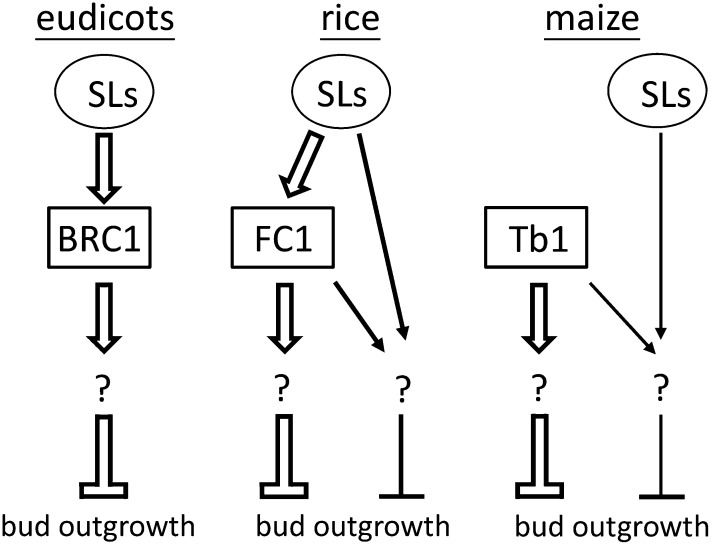
Models for SL regulation of axillary bud outgrowth in eudicots, rice, and maize. In eudicots, SL promotes expression of BRC1 and acts strictly through BRC1 to suppress axillary bud outgrowth. In rice, SL signaling acts partially through FC1 to inhibit outgrowth of axial buds. In maize, SL acts primarily on a downstream component of the Tb1 pathway to regulate bud outgrowth. The open arrows indicate a major effect on the suppression of bud outgrowth.
In conclusion, our results suggest that a Tb1 subnetwork that principally controls branching has evolved toward independence from SL signaling in maize. The uncoupling of Tb1 from the SL network has enabled a clearer delineation of diverse roles of SL signaling in maize architecture. Key nonbranching phenotypes that are genetically independent of Tb1 include narrow stem diameter, short stature, and delayed development of the adventitious nodal root system. The evolution of SL-independent regulation of branching in maize and variation in CCD8 paralog diversity in grass genomes suggest that innovation and functional diversification of the SL signaling network have had significant roles in the evolution of grasses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Maize (Zea mays) inbreds B73 and W22 were used in this study. The zmccd8::Ds insertion line was developed by Vollbrecht et al. (2010), and seeds were obtained from the Boyce Thompson Institute (Ithaca, NY). The lines were maintained in a W22 genetic background. The wild type used in this study was derived from a nonmutant sibling of the homozygous zmccd8::Ds insertion isolate. The tb1 reference mutant (117D) was obtained from the Maize Genetics Cooperation Stock Center. For work with seedlings, maize seeds were sown in plastic pots with unfertilized soil and grown in a walk-in growth chamber at 24°C with a 16/8-h light/dark cycle. For developmental analyses of larger plants, greenhouse-grown materials were used during winters of 2009 and 2010 and compared to field-grown individuals from the University of Florida research farm in Citra, Florida during the spring and fall of 2010. Arabidopsis (Arabidopsis thaliana) Columbia-0 wild-type and max4-6 mutants were grown under the same growth chamber conditions described above.
Cloning of ZmCCD8
Total RNA was extracted from roots of 14-d-old B73 seedlings using Trizol reagent (Invitrogen). First-strand cDNA was synthesized from purified poly(A)+ RNA primed with oligo(dT) by using SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). The poly(A)+ RNA was isolated by using Oligotex Direct mRNA Mini kit (Qiagen) according to the manufacturer’s instructions. A full-length fragment was amplified using high-fidelity Pfx50 DNA polymerase (Invitrogen). The primers used for PCR are listed as follows: 5′-GTACACACCACACTCCCCCGGCTAGCACC-3′ and 5′-CTATGGGCTCGCTCA-CATGAGCTAGCTCTG-3′ for ZmCCD8. The PCR fragments were gel purified by GFX PCR DNA and Gel Band Purification kits (GE Healthcare) and then cloned into the pCR4-TOPO vector using the TOPO TA cloning kit for sequencing analysis (Invitrogen).
Complementation Analyses of ZmCCD8 in Arabidopsis max4-6 Mutant
To generate constructs for complementation analysis in Arabidopsis, the ZmCCD8 coding region was amplified by PCR and then cloned into SpeI and BstEII sites of the pCAMBIA1305.1 vector, placing it downstream of the Cauliflower mosaic virus 35S promoter. The primers are as listed (restriction enzyme recognition sites are underlined): 5′-ACTAGTATGTCTCCCACTATGGCTTCG-3′ and 5′-AGGTAACCCTACGCGTCTGAGGTACTCCTAGG-3′ for ZmCCD8. The constructs were transformed into Agrobacterium tumefaciens strain GV3101 and used to transform the Arabidopsis max4-6 using the floral dip method (Desfeux et al., 2000). The T2 seeds were selected using hygromycin (30 μg/mL) in one-half-strength Murashige and Skoog plates and then transferred to soil and grown in a growth chamber as described above. Wild-type (Columbia-0) control plants and max4-6 seeds were germinated without antibiotics under the same culture conditions. The number of rosette branches was recorded at 3 weeks after flowering. For expression analyses, leaves from each independent line were harvested at 3 weeks after planting. Total RNA was extracted from leaves and purified using the plant RNeasy kit (Qiagen). The RQ1 RNase-free DNase (Promega) was used for removal of genomic DNA.
Hydroponic Culture and GR24 Treatment
The hydroponic solution was adapted according to Motta et al. (2001). Seeds were sterilized for 10 min in 3% (v/v) sodium hypochlorite/deionized water, rinsed thoroughly under running water, and germinated for 2 d in the dark inside wet filter paper rolls at 25°C. Germinated seedlings were then incubated in a growth chamber for 1 d. Seedling were grown in aerated hydroponic solution [8 mm KNO3, 2 mm Ca(NO3)2, 2 mm KH2PO4, 2 mm MgSO4, 46 μm H3BO3, 10 μm MnSO4, 0.32 μm CuSO4, 0.77 μm ZnSO4, 0.58 μm MoO3, and 0.25 μm NH4VO3] with a photoperiod of 16 h light (150 μmol of photons m−2 s−1) and 8 h dark at 22°C. The GR24 was synthesized and kindly provided by Kinga Chojnacka at the University of Florida (Chojnacka et al., 2011). A 100-mm GR24 stock solution was prepared in dimethyl sulfoxide (DMSO). Stock solution was added to the hydroponic media until a concentration of 1 μm (±) GR24 was reached. An equal volume of DMSO was added to controls. For gene expression analysis, seedlings were grown for 11 d and then harvested for total RNA preparation. For rescue experiments, seedlings were grown continually in hydroponic solution for 3 weeks and then harvested for bud length measurement. The hydroponic solution was refreshed each week.
Southern-Blot Analysis
Ten micrograms of genomic DNA (B73 inbred) was digested by restriction enzymes as indicated, resolved in a 1.0% agarose gel, and then transferred onto nylon membrane. A genomic fragment (approximately 3,900 bp) containing the ZmCCD8 gene was used as a probe. Hybridization was performed as described previously (Settles et al., 2004).
Gene Expression Analysis by Quantitative Real-Time RT-PCR
For analysis of expression profiles, tissues from field-grown plants (B73 inbred) at the stage of flowering (tassel emergence) were harvested separately. For expression analyses performed at seedling stage, tissues were taken from seedlings at 14 d after germination or 21 d after germination as indicated. All samples were frozen in liquid nitrogen immediately and then kept at a −80°C freezer until used. Tissues were ground in liquid nitrogen. Total RNA was extracted as indicated and purified using the plant RNeasy kit (Qiagen). RNAs were quantified using a NanoDrop 1000 (Thermo Fisher Scientific), and 5.5 μg of RNA from each sample was treated with the RQ1 RNase-free DNase (Promega). Negative RT controls were used to confirm that there was no carryover of genomic DNA. For quantitative PCR, a Power SYBR green RNA-to-CT 1-step kit (Applied Biosystems) was used with an iCycler iQ real-time PCR detection system (Bio-Rad). For analysis of ZmCCD8 expression, absolute quantitative methods were used. 3H-labeled RNA transcripts for standard curves (synthesized as described in Vogel et al., 2010). The forward and reverse primer used for ZmCCD8-U was 5′-CGATCATCGCCGACTGCTGCGAGC/TGGAAGAGCGCAGGTTCTG-3′ and for ZmCCD8-F was 5′-CTCCAGAACCTGCGCTCTTCCA/CGTCCAGCGGGATCCTGAAG-3′. For analysis of Tb1 expression, relative quantitative methods were used following the mathematical model of Pfaffl (2001). The 18S rRNA was used an internal reference for determining expression of Tb1. The primer sequences used were adapted from Whipple et al. (2011). The forward and reverse primer sequence for Tb1 was 5′-TTCCCTCAACGTGAGCTTCT/TTCATCGTCACACAGCCAAT-3′ and for 18S rRNA was 5′-ATTCTATGGGTGGTGGTGCAT/TCAAACTTCGCGGCCTAAA-3′. ANOVA was used to determine statistical significance of expression level differences.
Morphological Analyses of tb1 and zmccd8 Double Mutants
A heterozygous zmccd8 mutant was crossed to the tb1mutant, followed by genotyping of the F1 progeny and growth of selected individuals in the winter green house. The F2 and F3 progeny were grown in spring and fall field seasons at the University of Florida research farm in Citra, Florida. The genotype of zmccd8 alleles was identified by PCR and confirmed by genetic analyses. In F2 progeny, plants homozygous and heterozygous at the tb1 locus were identified by phenotype, and their genotype was confirmed by progeny tests. The branches produced from nodes at or below ground level were considered tillers, whereas those originating from nodes further up the stalk were classified as lateral branches or ear branches. Nodes having a leaf tip visible at the junction between leaf sheath and stalk were counted as having a branch.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number FJ957946.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Southern-blot analysis of ZmCCD8 (B73 inbred).
Supplemental Figure S2. Linkage map of ZmCCD8.
Supplemental Figure S3. Amino acid alignment of ZmCCD8 with rice D10, petunia DAD1, pea RMS1 and Arabidopsis MAX4.
Supplemental Figure S4. Suppression of ZmCCD8-F transcripts by GR24 treatment in shoots of wild-type seedlings (W22).
Supplemental Figure S5. Identification of the zmccd8::trDs frameshift allele.
Supplemental Figure S6. The branching phenotype of zmccd8::Ds and zmccd8::trDs.
Supplemental Figure S7. Quantitative analysis of plant stature differences in mutant and wild-type seedlings.
Supplemental Figure S8. Growth rate of zmccd8 and wild-type stalks.
Supplemental Figure S9. The narrow stalk phenotype of the zmccd8 mutants.
Supplemental Figure S10. Expression analyses of ZmCCD8 in shoots of zmccd8 and tb1 single mutants or tb1 zmccd8 double mutants.
Supplementary Material
Acknowledgments
We thank the Maize Genetics Cooperative Stock Center for providing the tb1 seed stock. GR24 was kindly provided by Kinga Chojnacka in the laboratory of Nigel G.J. Richards (Department of Chemistry, University of Florida).
Glossary
- SL
strigolactone
- RT
reverse transcription
- DMSO
dimethyl sulfoxide
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, Greb T. (2011) Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci USA 108: 20242–20247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amusan IO, Rich PJ, Menkir A, Housley T, Ejeta G. (2008) Resistance to Striga hermonthica in a maize inbred line derived from Zea diploperennis. New Phytol 178: 157–166 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ. (2006) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45: 982–993 [DOI] [PubMed] [Google Scholar]
- Awad A, Sato D, Kusumoto D, Kamioka H, Takeuchi Y, Yoneyama K. (2006) Characterization of strigolactones, germination stimulants for the root parasitic plants Striga and Orobanche, produced by maize, millet and sorghum. Plant Growth Regul 48: 221–227 [Google Scholar]
- Barker NP, Clark LG, Davis JI, Duvall MR, Guala GF, Hsiao C, Kellogg EA, Linder HP, Mason-Gamer RJ, Mathews SY, et al. (2001) Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Mo Bot Gard 88: 373–457 [Google Scholar]
- Beveridge CA, Dun EA, Rameau C. (2009) Pea has its tendrils in branching discoveries spanning a century from auxin to strigolactones. Plant Physiol 151: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot J-P, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacka K, Santoro S, Awartani R, Richards NGJ, Himo F, Aponick A. (2011) Synthetic studies on the solanacol ABC ring system by cation-initiated cascade cyclization: implications for strigolactone biosynthesis. Org Biomol Chem 9: 5350–5353 [DOI] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J. (2006) A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet 38: 594–597 [DOI] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O. (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913 [DOI] [PubMed] [Google Scholar]
- de Alencar Figueiredo LF, Calatayud C, Dupuits C, Billot C, Rami JF, Brunel D, Perrier X, Courtois B, Deu M, Glaszmann JC. (2008) Phylogeographic evidence of crop neodiversity in sorghum. Genetics 179: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux PM, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N. (2012) Origin of strigolactones in the green lineage. New Phytol 195: 857–871 [DOI] [PubMed] [Google Scholar]
- Desfeux C, Clough SJ, Bent AF. (2000) Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral-dip method. Plant Physiol 123: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Doust AN. (2007) Architectural evolution and its implications for domestication in grasses. Ann Bot (Lond) 100: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN, Devos KM, Gadberry MD, Gale MD, Kellogg EA. (2004) Genetic control of branching in foxtail millet. Proc Natl Acad Sci USA 101: 9045–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RSM, Martínez-Sánchez NM, Janssen BJ, Templeton KR, Simons JL, Quinn BD, Karunairetnam S, Snowden KC. (2009) Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol 151: 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson SA. (2007) Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol 48: 667–677 [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. (2010) Phytochrome regulation of branching in Arabidopsis. Plant Physiol 152: 1914–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA. (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Qian Q, Liu X, Yan M, Feng Q, Dong G, Liu J, Han B. (2009) Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol Biol 71: 265–276 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 180–194 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Roux C, Girard D, Bécard G, Puech-Pagés V. (2007) Strigolactones: promising plant signals. Plant Signal Behav 2: 163–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet C, Klee HJ. (2010) Climbing the branches of the strigolactones pathway one discovery at a time. Plant Physiol 154: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yan H, Yang J, Yamaguchi S, Maekawa M, Takamure I, Tsutsumi N, Kyozuka J, Nakazono M. (2010) Strigolactones negatively regulate mesocotyl elongation in rice during germination and growth in darkness. Plant Cell Physiol 51: 1136–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S. (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162: 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöller AS, Blakeslee JJ, Richards EL, Peer WA, Murphy AS. (2010) Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. J Exp Bot 61: 3689–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H. (2011) Strigolactones are regulators of root development. New Phytol 190: 545–549 [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E. (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344 [DOI] [PubMed] [Google Scholar]
- Ledger SE, Janssen BJ, Karunairetnam S, Wang T, Snowden KC. (2010) Modified CAROTENOID CLEAVAGE DIOXYGENASE8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytol 188: 803–813 [DOI] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, Wang Y. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wu C, Fu Y, Hu G, Si H, Zhu L, Luan W, He Z, Sun Z. (2009) Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230: 649–658 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano T, Yoneyama K, Suzuki Y, Asami T. (2009) Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci Biotechnol Biochem 73: 2460–2465 [DOI] [PubMed] [Google Scholar]
- Matusova R, Rani K, Verstappen FW, Franssen MCR, Beale MH, Bouwmeester HJ. (2005) The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol 139: 920–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing SAJ, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM. (2010) Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S, Kyozuka J. (2010) FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 51: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta A, Basso B, Dell’Orto M, Briat J-F, Soave C. (2001) Ferritin synthesis in response to iron in the Fe-inefficient maize mutant ys3. Plant Physiol Biochem 39: 461–465 [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust H, Hoffmann B, Xie X, Yoneyama K, Schaefer DG, Yoneyama K, Nogué F, Rameau C. (2011) Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138: 1531–1539 [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, Brewer PB, Herold S, Agusti J, Geelen D, Greb T, Goormachtig S, Beeckman T, Beveridge CA. (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol 158: 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigereau MS, Lakis G, Rekima S, Leveugle M, Fontaine MC, Langin T, Sarr A, Robert T. (2011) Cereal domestication and evolution of branching: evidence for soft selection in the Tb1 orthologue of pearl millet (Pennisetum glaucum [L.] R. Br.). PLoS ONE 6: e22404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, Verstappen F, Bouwmeester H. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles AM, Latshaw S, McCarty DR. (2004) Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res 32: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E. (2007) The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol 145: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. (2005) The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, Leyser O. (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43: 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P. (2010) A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat Chem Biol 6: 741–749 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Martínez de la Vega O, Hernández-Guzmán G, Ibarra-Laclette E, Alvarez-Mejía C, Vega-Arreguín JC, Jiménez-Moraila B, Fernández-Cortés A, Corona-Armenta G, Herrera-Estrella L, et al. (2009) The Palomero genome suggests metal effects on domestication. Science 326: 1078. [DOI] [PubMed] [Google Scholar]
- Vogel JT, Walter MH, Giavalisco P, Lytovchenko A, Kohlen W, Charnikhova T, Simkin AJ, Goulet C, Strack D, Bouwmeester HJ, et al. (2010) SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J 61: 300–311 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Duvick J, Schares JP, Ahern KR, Deewatthanawong P, Xu L, Conrad LJ, Kikuchi K, Kubinec TA, Hall BD, et al. (2010) Genome-wide distribution of transposed Dissociation elements in maize. Plant Cell 22: 1667–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J. (2011) Branching in rice. Curr Opin Plant Biol 14: 94–99 [DOI] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. (2012a) The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol 159: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. (2012b) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP. (2011) grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA 108: E506–E512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K. (2010) The strigolactone story. Annu Rev Phytopathol 48: 93–117 [DOI] [PubMed] [Google Scholar]
- Zhou L, Zhang J, Yan J, Song R. (2011) Two transposable element insertions are causative mutations for the major domestication gene teosinte branched 1 in modern maize. Cell Res 21: 1267–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. (2006) The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48: 687–698 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.