Abstract
The crucial role of carbohydrate in plant growth and morphogenesis is widely recognized. In this study, we describe the characterization of nana, a dwarf Arabidopsis (Arabidopsis thaliana) mutant impaired in carbohydrate metabolism. We show that the nana dwarf phenotype was accompanied by altered leaf morphology and a delayed flowering time. Our genetic and molecular data indicate that the mutation in nana is due to a transfer DNA insertion in the promoter region of a gene encoding a chloroplast-located aspartyl protease that alters its pattern of expression. Overexpression of the gene (oxNANA) phenocopies the mutation. Both nana and oxNANA display alterations in carbohydrate content, and the extent of these changes varies depending on growth light intensity. In particular, in low light, soluble sugar levels are lower and do not show the daily fluctuations observed in wild-type plants. Moreover, nana and oxNANA are defective in the expression of some genes implicated in sugar metabolism and photosynthetic light harvesting. Interestingly, some chloroplast-encoded genes as well as genes whose products seem to be involved in retrograde signaling appear to be down-regulated. These findings suggest that the NANA aspartic protease has an important regulatory function in chloroplasts that not only influences photosynthetic carbon metabolism but also plastid and nuclear gene expression.
Plants synthesize carbohydrates through photosynthesis. The subsequent partitioning of carbohydrates between the site of their production, the chloroplast, and other cellular components is regulated by complex mechanisms to meet the diverse needs of the plant in terms of carbon availability (Rolland et al., 2006; Stitt et al., 2010). Sugars formed during the day are a prerequisite for plant growth, as they represent both cellular building blocks and substrates for mitochondrial respiration. During the night, when sugars are no longer produced by photosynthesis, metabolism and growth are largely dependent on transitory starch reserves that are accumulated during the day and broken down at night (Smith and Stitt, 2007; Graf et al., 2010). This complex time-tuned mechanism of carbon partitioning among organelles, tissues, and organs can be easily disrupted by the mutation of genes encoding components of sugar metabolism or regulatory networks. Unless plants are grown under very-long-photoperiod conditions so that photoassimilates are almost always available, mutations affecting enzymes involved in starch synthesis or degradation result in a phenotype of reduced growth. This demonstrates the importance of leaf carbohydrate reserves for the whole plant’s growth and development (Gibon et al., 2004; Smith and Stitt, 2007; Usadel et al., 2008). Defects in starch metabolism clearly limit the supply of energy and building blocks for growth at night (Yazdanbakhsh and Fisahn, 2011). However, it remains unclear whether the reduced growth of starch mutants is entirely attributable to such limitations or if other factors also play a role. Sugars can indeed also act as osmoregulatory or signaling molecules, with effects on plant growth and development throughout the plant’s life cycle, from germination right through the floral transition and senescence (Gibson, 2000, 2004, 2005; Koch, 2004).
The ability of plants to sense sugars might play an important role in carbon partitioning and allocation in source and sink tissues (Smith and Stitt, 2007). In particular, the expression of genes involved in photosynthate accumulation, mobilization, and storage is regulated by Glc and Suc (Koch, 1996) and by light- and circadian clock-mediated signaling mechanisms (Harmer et al., 2000; Neff et al., 2000). This regulatory network maintains an optimal dynamic carbohydrate status, integrating the synthesis and the consumption of carbohydrates in response to environmental changes and in response to the availability of other nutrients, such as nitrogen (Coruzzi and Bush, 2001).
Sugar homeostasis is also tightly linked to the regulation of photosynthesis and chloroplast physiology, with the expression of photosynthesis-related genes being modulated by light intensity, temperature, and CO2 availability (Jang and Sheen, 1997; Rolland et al., 2002, 2006; Stitt et al., 2010). For example, in conditions of high carbohydrate demand and nonlimiting light availability, plants increase the production and export of photosynthate by increasing the expression of genes involved in photosynthesis (Koch, 1996). Conversely, when photosynthates are not immediately required, genes involved in starch synthesis are activated to maintain a balance between photosynthate supply, demand, and storage (Rook et al., 2001).
Therefore, chloroplasts play a central role in primary metabolism, supporting the growth and differentiation of plant cells. Chloroplasts are thought to have originated from a cyanobacterium-like endosymbiont, but during evolution most genes of cyanobacterial origin were transferred from the chloroplast genome to the nucleus (Mayfield et al., 1995; Martin et al., 2002; Dyall et al., 2004; Ajjawi et al., 2010; Myouga et al., 2010). Relatively few photosynthesis-related genes remain in the chloroplast genome, and chloroplast development and functionality are dependent on the import of the nucleus-encoded plastid proteins (Woodson and Chory, 2008; Inaba, 2010).
Most of our knowledge of chloroplastic proteins is focused on the enzymes and transporters involved in photosynthesis and starch metabolism, but other biochemical processes in the chloroplast require diverse enzymatic activities, such as proteases. Proteolysis has several important functions in chloroplasts (Adam, 1996; Andersson and Aro, 1997; Adam et al., 2001), such as processing of precursor proteins, degradation of incomplete proteins, and removal of damaged proteins (Nair and Ramaswamy, 2004). Aspartic proteinases (APs; Enzyme Commission 3.4.23) are one of the major catalytic classes of proteases in plants as well as in vertebrates, nematode parasites, bacteria, fungi, and viruses (Timotijević et al., 2010) and are involved in many aspects of plant physiology and development. APs contain two Asp residues that are crucial for catalytic activity. They are most active under acidic conditions (pH 2–6) and are specifically inhibited by pepstatin A (Mutlu and Gal, 1999; Milisavljevic et al., 2008). Several functions have been proposed for APs, including processing and degradation of storage proteins, protein degradation during organ senescence and cell death, autolysis during the formation of tracheary elements, prey digestion by carnivorous plants, and adhesion-mediated proteolytic mechanisms in pollen recognition and growth (Simões and Faro, 2004). In tobacco (Nicotiana tabacum) chloroplasts, an AP named CND41 (for 41 kD Chloroplast Nucleoid DNA-binding protein) is involved in degradation of the Rubisco holoprotein during leaf senescence (Murakami et al., 2000; Kato et al., 2004). CND41 homologs have been identified in Arabidopsis (Arabidopsis thaliana), and their role in the regulation of Rubisco turnover and senescence has also been confirmed in this species (Kato et al., 2005a, 2005b; Diaz et al., 2008). Moreover, CND41 accumulation is negatively correlated with chloroplast transcript levels in tobacco cells (Nakano et al., 1997), indicating that CND41 may function as a negative regulator of chloroplast gene expression.
In this study, we identified and characterized a dwarf Arabidopsis mutant bearing a T-DNA insertion in a gene encoding a chloroplast-located AP. We named the mutant nana, which means “dwarf” in Italian. NANA plays an important role in carbohydrate homeostasis, as demonstrated by the severe alteration in carbohydrate metabolism resulting from misexpression or overexpression of this AP.
RESULTS AND DISCUSSION
Isolation and Morphological Characterization of the nana Mutant
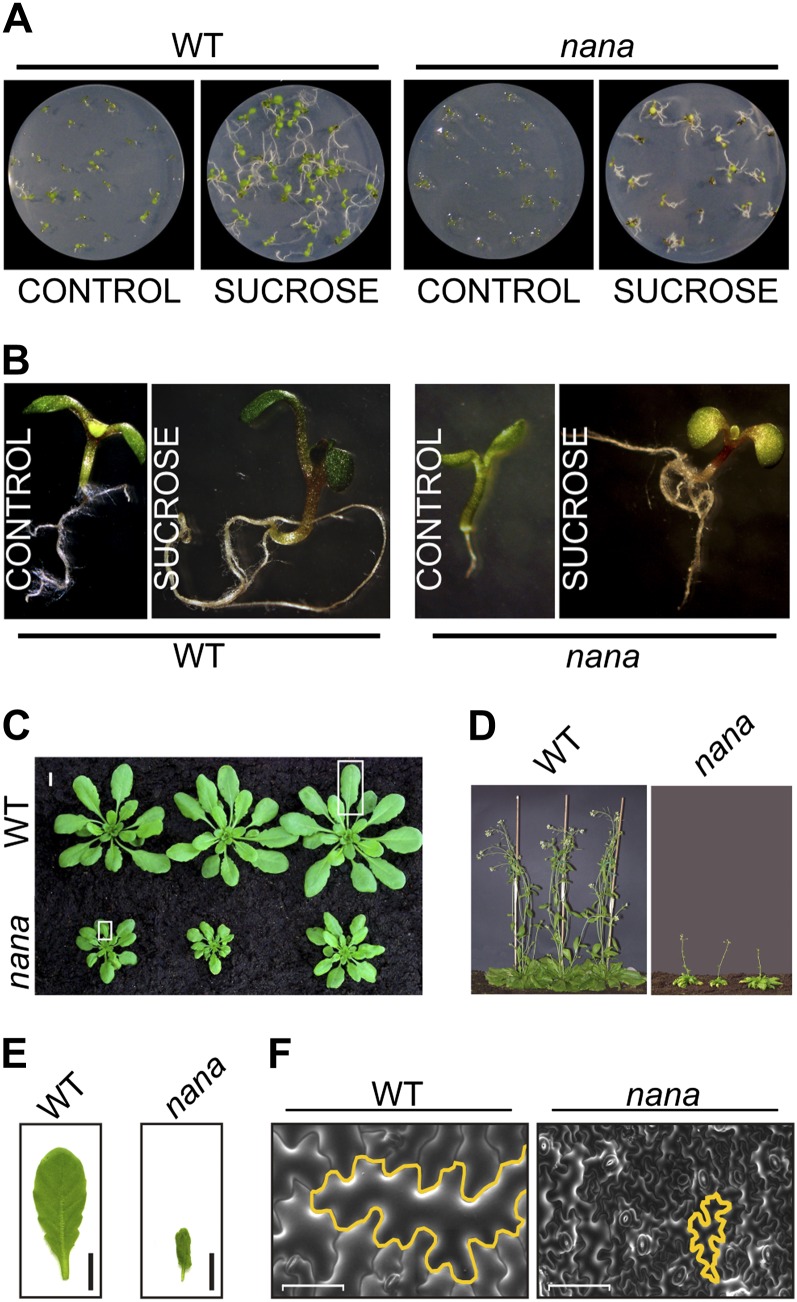
The nana mutant was isolated while searching for mutants altered in their sugar-sensing abilities. The growth of the hypocotyl and/or root was indeed reduced in nana seedlings compared with the wild type in various growth conditions, either in the presence or absence of Suc (Fig. 1, A and B; Supplemental Table S1). When grown on medium without Suc, the length of the root in nana seedlings was shorter than in the wild type, when kept in the presence of light, while in darkness, hypocotyl elongation was severely inhibited in nana (Supplemental Table S1). When grown in soil, adult nana plants maintained a dwarf phenotype, with a reduced diameter of the rosette and a diminished height of the inflorescence axis (Fig. 1, C and D; Table I). The nana mutant also exhibited small leaves, which were more lobed than wild-type leaves and slightly curled, particularly during the early vegetative stages of development (Fig. 1E). We did not observe differences in leaf number between nana and wild-type plants (Table I). At the generative stage, bolting was significantly delayed in nana compared with wild-type plants (Fig. 1D; Table I). Epidermal cells were substantially smaller in the mutant compared with wild-type plants (Fig. 1F). However, in contrast to other mutants with altered epidermal cell size, the nana mutant did not show any differences in the degree of endoreduplication (Supplemental Fig. S1).
Figure 1.
Phenotypes of nana. A, Effects of treatments with Suc on seedling growth. Seedlings of nana and the wild type (WT) grown in vitro for 10 d on agar plates containing Murashige and Skoog medium in the absence (control) or presence of Suc (90 mm) are shown. B, Effects of treatments with Suc on hypocotyl and root growth. Details of one seedling from the plates in A are shown. C, Representative growth phenotypes of nana and wild-type plants grown on soil for 30 d. D, Phenotypes of 6-week-old mutant and wild-type plants. E, Photographs of 4-week-old leaves from wild-type and nana plants. Bars = 1 cm. F, Cryo-scanning electron microscopy images of frozen-hydrated leaf freeze fractures in wild-type and nana plants. Representative leaf cells are highlighted in yellow. Bars = 50 μm.
Table I. Comparison of wild-type and nana mutant phenotypes.
Morphological parameters were measured in 4-week-old adult plants.
| Parameter | Wild Type | nana |
|---|---|---|
| Leaf number | 20.1 ± 1.65 | 19.1 ± 1.80 |
| Leaf length (mm) | 18.6 ± 1.85 | 7.4 ± 0.55a |
| Leaf width (mm) | 12.5 ± 1.52 | 6.7 ± 1.06a |
| Petiole length (mm) | 13.8 ± 1.70 | 6.2 ± 1.15a |
| Rosette size (cm) | 8.74 ± 0.28 | 4.02 ± 0.37a |
| Bolting time (d after germination) | 29.6 ± 1.70 | 36.7 ± 1.55a |
| Inflorescence height (cm) | 34.1 ± 4.51 | 8.3 ± 2.31a |
| Inflorescence number | 10.2 ± 1.58 | 5.1 ± 1.25a |
| Silique number | 50.3 ± 9.24 | 16.2 ± 1.52a |
Significantly different (P < 0.001) as calculated using a Student’s t test.
Molecular Characterization of the nana Mutation
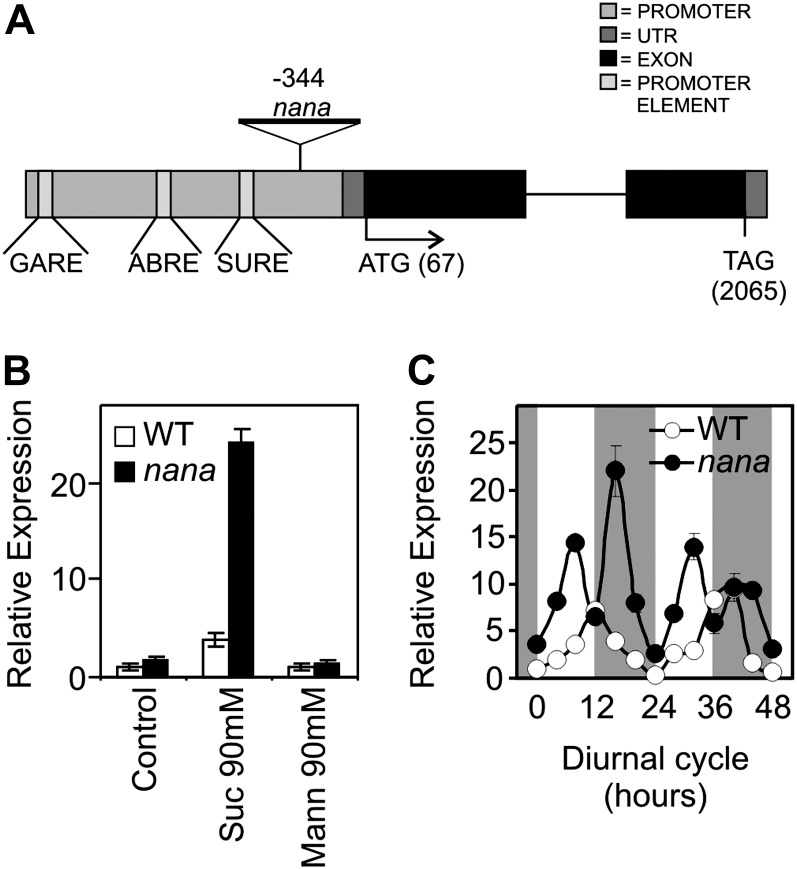
A T-DNA insertion was identified in the nana mutant, 344 nucleotides upstream of the start codon of the coding sequence of the At3g12700 (NANA) gene, which is annotated to encode a member of the aspartyl protease family. This gene is composed of two exons separated by one intron (Fig. 2A). As a result of the T-DNA insertion, the At3g12700 coding sequence is unchanged, but a series of putative regulatory elements (e.g. GARE, SURE, and ABRE elements) in the promoter are displaced away from the 5′ untranslated region and coding sequence (Fig. 2A).
Figure 2.
Structure, Suc inducibility, and expression pattern of At3g12700 and its mutated form. A, Position of the T-DNA insertion identified in chromosome 3 within the promoter region (344 nucleotides upstream of the ATG) of At3g12700 (NANA). GARE, Gibberellin-responsive element; ABRE, abscisic acid-responsive element; SURE, Suc-responsive element, identified by the PLACE database (www.dna.affrc.go.jp/PLACE/); UTR, untranslated region. B, Real-time quantitative PCR analysis of NANA expression in 4-d-old dark-grown wild-type and nana seedlings incubated for an additional 24 h on 0.5× Murashige and Skoog medium (control), 90 mm Suc (Suc 90mM), or 90 mm mannitol (Mann 90mM). Expression levels are indicated in relative units, assuming as unitary the value of the wild-type control. Each value is the mean ± sd of three independent measurements. White bars show the wild type (WT), and black bars show nana. C, Relative expression pattern of the NANA gene in the wild type (white circles) and nana mutant (black circles) during a time course of 48 h (12-h-light/12-h-dark photoperiod, light irradiance of 80 µmol photons m−2 s−1). At3g12700 expression levels in wild type and nana mutant rosettes, grown in a hydroponic system, are expressed as relative units, assuming as unitary the value of the wild type at the beginning of the day (light input). Each value is the mean ± sd of three independent measurements. White and dark gray background shading show the light period and the night, respectively.
Suc slightly induced the expression of At3g12700 in wild-type plants, and the effect was enhanced in the mutant (Fig. 2B). In wild-type adult rosette leaves, At3g12700 was found to be expressed with a pronounced diurnal rhythm characterized by a peak at the end of the day (Fig. 2C). In nana plants, the level of expression of the gene was not only higher than in wild-type plants at almost all times of day but also peaked earlier in the light period (8 h instead of 12 h) and showed a second peak in the middle of the night (Fig. 2C). This indicates that the T-DNA insertion in the promoter region leads to misexpression of At3g12700 in nana plants.
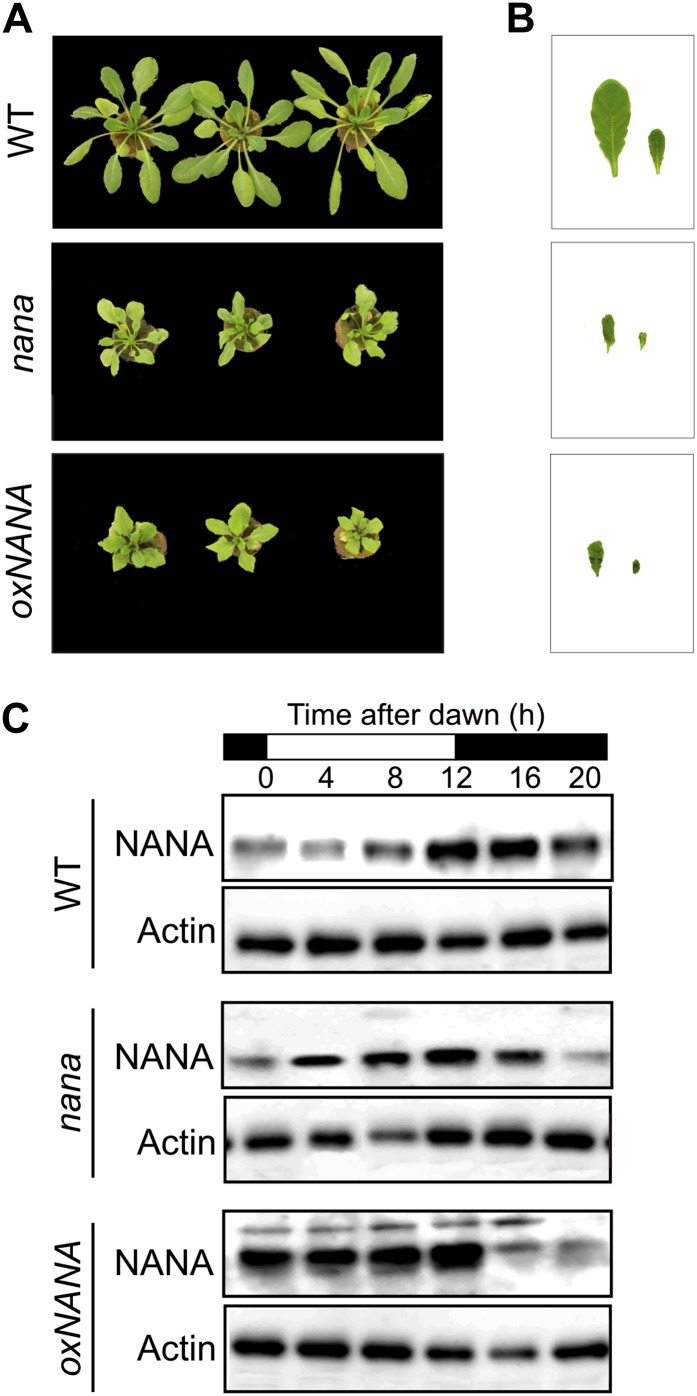
Since At3g12700 is expressed at higher levels in nana, an overexpressor line with the gene under the control of the cauliflower mosaic virus 35S constitutive promoter was produced for comparison. 35S::At3g12700 (oxNANA) plants exhibited a dwarf phenotype that was very similar to that of the nana mutants in terms of reduced rosette diameter (Fig. 3A), leaf morphology (Fig. 3B), and delayed time of flowering (data not shown).
Figure 3.
Effect of the overexpression of NANA on plant growth and levels of the relative protein in the different genotypes. A and B, The overexpressor line (oxNANA) phenocopies the nana mutant in terms of rosette size (A) and leaf morphology (B). C, Immunoblot analysis of the NANA protein in the wild type (WT), nana, and oxNANA during the diurnal cycle. The loading control is Actin11. [See online article for color version of this figure.]
Next, we examined the accumulation of At3g12700 protein (hereafter called NANA) in the wild type, in the nana mutant, and in the oxNANA line during a 24-h time course. Immunoblot analysis of leaves harvested from 12-h-photoperiod-grown wild-type plants at different times of the day showed that NANA was accumulated late during the day and during the early night phase and was partially degraded during the last 4 h of the night (Fig. 3C). In contrast, the nana mutant and the NANA-overproducing line showed a higher accumulation of NANA in the early part of the light period (Fig. 3C). Remarkably, the level of NANA decreased during the night in the oxNANA line, suggesting that a posttranscriptional mechanism operates to prevent excessive NANA accumulation during the night (Fig. 3C). Overall, these results suggested that under normal conditions NANA would be regulated by a day/night rhythm, but the nana mutation alters this regulation.
NANA Encodes a Chloroplast-Located Aspartyl Protease
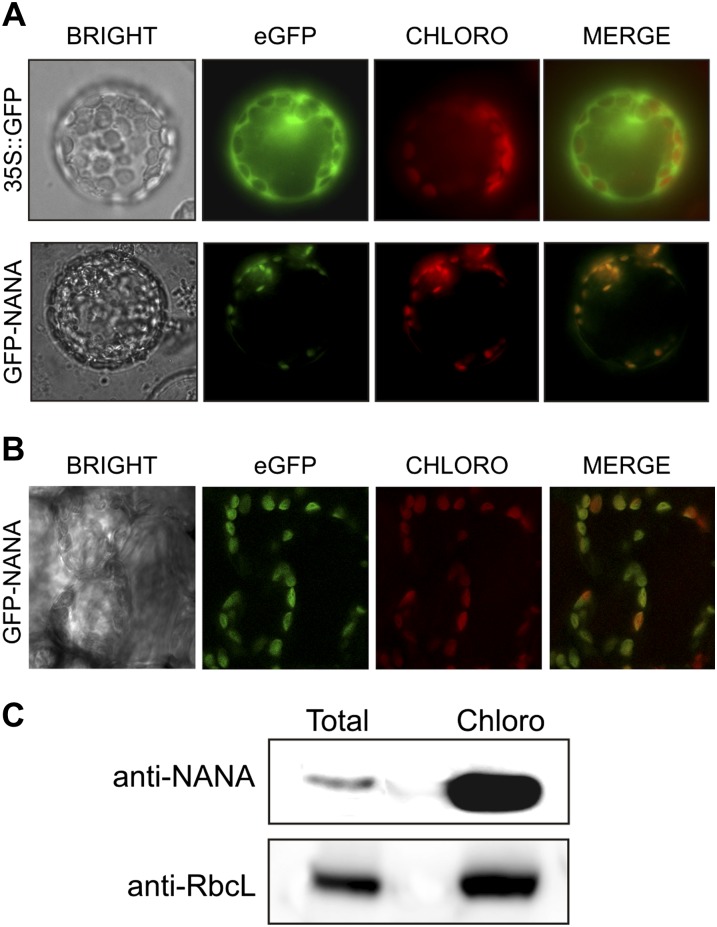
Bioinformatic analysis (http://suba.plantenergy.uwa.edu.au/; Heazlewood et al., 2007) did not predict a predominant subcellular localization for NANA, indicating the lack of a canonical subcellular localization signaling sequence in At3g12700. We examined the NANA localization of an in-frame fusion construct with the enhanced GFP (EGFP). Confocal microscopy showed that GFP fluorescence perfectly overlapped with chlorophyll autofluorescence both in transiently transformed Arabidopsis protoplasts (Fig. 4A) and in stably transformed leaf cells (Fig. 4B). To further confirm the localization of NANA, we performed an immunoblot analysis that showed a clear enrichment of NANA in isolated chloroplasts. The signal arising from the NANA antiserum was strongly enriched in the chloroplast fraction (Fig. 4C). Evidently, even without an identifiable chloroplast transit peptide, NANA was targeted to chloroplasts. Most of the chloroplast proteins do present a recognizable N-terminal transit peptide; however, the chloroplast proteome also contains many other nucleus-encoded proteins not previously predicted as chloroplast localized (Kleffmann et al., 2004; Li and Chiu, 2010). Different and not always completely clarified pathways of sorting of proteins without identifiable plastid transit peptides in chloroplasts seem to take place in plant cells (Asatsuma et al., 2005; Li and Chiu, 2010).
Figure 4.
NANA encodes a chloroplast-located aspartyl protease. A and B, Transient (A) and stable (B) expression of EGFP-NANA in Arabidopsis protoplasts and leaves, respectively, show that NANA localizes to chloroplasts. Localization studies were performed using N-terminal GFP fusions with the complete cDNA of the NANA sequence. Chlorophyll autofluorescence (CHLORO) was used as a marker for chloroplasts. C, Immunoblot analysis of protein (30 µg) from total extract and the chloroplast fraction (Chloro) from wild-type plants with antibodies against NANA and RbcL.
The Plant Proteome Database (ppdb.tc.cornell.edu/) classified NANA as a “eukaryotic aspartyl protease family protein.” NANA possesses a highly conserved aspartyl protease domain (PFAM PF00026) located between amino acids 104 and 459 (Fig. 5A; SMART6 bioinformatic tool; Letunic et al., 2009). Phylogenetic analysis based on sequenced plant genomes revealed the presence of homologs with sequence similarity to NANA in chlorophyte algae, mosses, lycophytes, and both monocotyledonous and dicotyledonous flowering plants, including major crop plants (e.g. rice [Oryza sativa], maize [Zea mays], and soybean [Glycine max]), indicating their presence throughout the plant kingdom (RefProt; BLASTP threshold of E < 10−10; Supplemental Fig. S2A). Although the Solanaceae were not represented in the phylogenetic analysis, a putative homolog of NANA was identified in the tomato (Solanum lycopersicum) draft genome (SL2.40ch05:4111801–4114200; Bombarely et al., 2011) with a high sequence similarity (TBLASTN E-value of 8 × 10−50). However, the high sequence similarity between these putative homologs is limited to the aspartyl protease domain. The N-terminal portion of nana (amino acids 1–103) appears to be unique to the Arabidopsis genus and, apart from Arabidopsis, was found only in the Arabidopsis lyrata Alyr-NANA ortholog (protein ARALYDRAFT_478632; XP_002882786). Arabidopsis possesses at least 50 putative paralogs to nana (BLASTP threshold of E < 10−10; Supplemental Fig. S2B), all possessing the aspartyl protease domain. Among these, we identified four genes whose transcripts are regulated in a circadian manner (Covington and Harmer, 2007) and several genes (including nana) whose expression is significantly different in dark- versus light-grown Arabidopsis 7-d-old seedlings (Dohmann et al., 2008; Supplemental Fig. S2B; Supplemental Table S2).
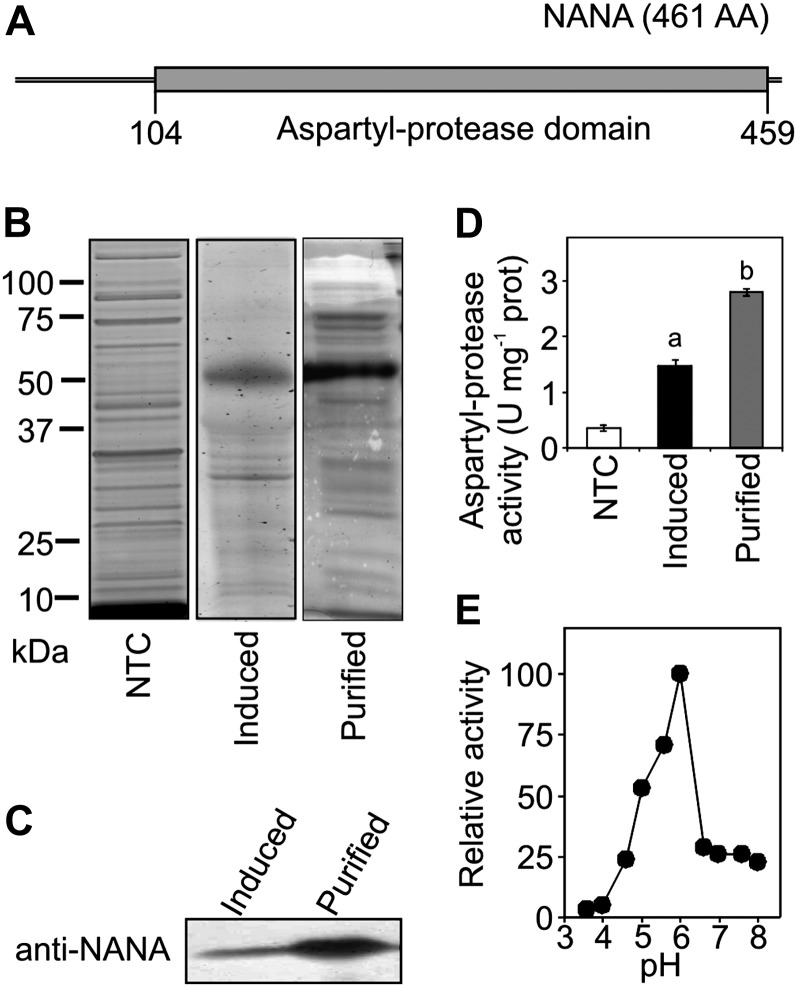
Figure 5.
A, Protein model of Arabidopsis NANA. The gray bar shows the aspartyl-protease domain. AA, Amino acids. B, Purified recombinant NANA protein used in the activity assay. Each fraction was resolved by SDS-PAGE and stained by Coomassie blue. The same volume (20 µL) was loaded for each fraction. C, Immunoblot of induced and purified fractions from E. coli recombinant NANA protein using an anti-NANA antibody. The same volume (20 µL) was loaded for each fraction. D, Aspartyl-proteolytic activity of recombinant NANA (induced and purified fractions) was determined in vitro using the azocasein assay. The non-IPTG-treated fraction (NTC) served as a control. Values are means ± sd (units [U]) from duplicate experiments. The letters indicate significant differences (P < 0.05) by one-way ANOVA. E, The pH optimum of recombinant NANA. The pH optimum was determined using the azocasein assay on the purified protein preparation shown in D. Activity is expressed as a percentage of the maximum value.
To test whether NANA encodes a catalytically active aspartic protease, we expressed the protein as an N-terminal His6-tagged fusion protein in Escherichia coli. The crude lysate from induced E. coli cells contained a prominent 53-kD protein band when analyzed by SDS-PAGE (Fig. 5B) that was recognized by an anti-NANA antibody (Fig. 5C). The 53-kD immunoreactive protein was affinity purified, and enzymatic activity was assayed using azocasein as the substrate, which is specific for endoproteases. Aspartyl protease activity was significantly higher in both the induced cell lysate and the purified protein fraction than in extract from non-isopropylthio-β-d-galactoside (IPTG)-treated E. coli cells (Fig. 5D). The recombinant NANA protein also displayed a pH optimum of 6 (Fig. 5E), a value compatible with the chloroplast environment, likely closer to that of the stroma at night than during the day, when illumination induces a stromal alkalinization (Song et al., 2004). This correlates well with the expression data (Fig. 3C) that indicated accumulation of NANA proteins in the wild type during the night. Interestingly, this pH optimum of NANA appears quite different from that reported for the previously characterized CND41 chloroplast aspartyl protease of tobacco, whose strongest activity was found at an acidic pH (Murakami et al., 2000). Aspartyl protease activity was also measured in crude extracts from rosette leaves. Both nana and oxNANA plants showed significantly higher (2- to 6-fold) activities than the wild type (Supplemental Fig. S3A). All these results confirmed that NANA is an aspartyl protease and that its activity was increased in the nana and oxNANA lines.
nana Affects Endogenous Sugar Levels
Dwarf phenotypes have previously been observed in several mutants with defective chloroplasts or chloroplast function (Nakano et al., 2003; Yabe et al., 2004; Qin et al., 2007; Huang et al., 2009; Myouga et al., 2010). Such observations, together with the chloroplast localization of NANA (Fig. 4, A and B), suggest that higher NANA perturbs chloroplast function in some way. Therefore, we measured several chloroplast-related physiological and biochemical parameters in wild-type and mutant plants. The chlorophyll content of nana leaves was about 50% lower than in the wild type (Fig. 6A), and the electron transport rate (ETR) was similarly decreased, being about 30% lower in nana plants (Fig. 6B). Conversely, CO2 assimilation rate was not significantly affected by the mutation (Fig. 6C). In the oxNANA line, chlorophyll content, ETR, and assimilation rate of CO2 were very similar to those of nana plants (Fig. 6, A–C). In accordance with the unchanged CO2 assimilation rate, Rubisco protein levels seemed to be unaffected by the mutation, with wild-type, nana, and oxNANA plants having similar amounts of RbcL (for Rubisco large subunit) protein to each other, both at the end of the night and at the end of the day (Fig. 6D).
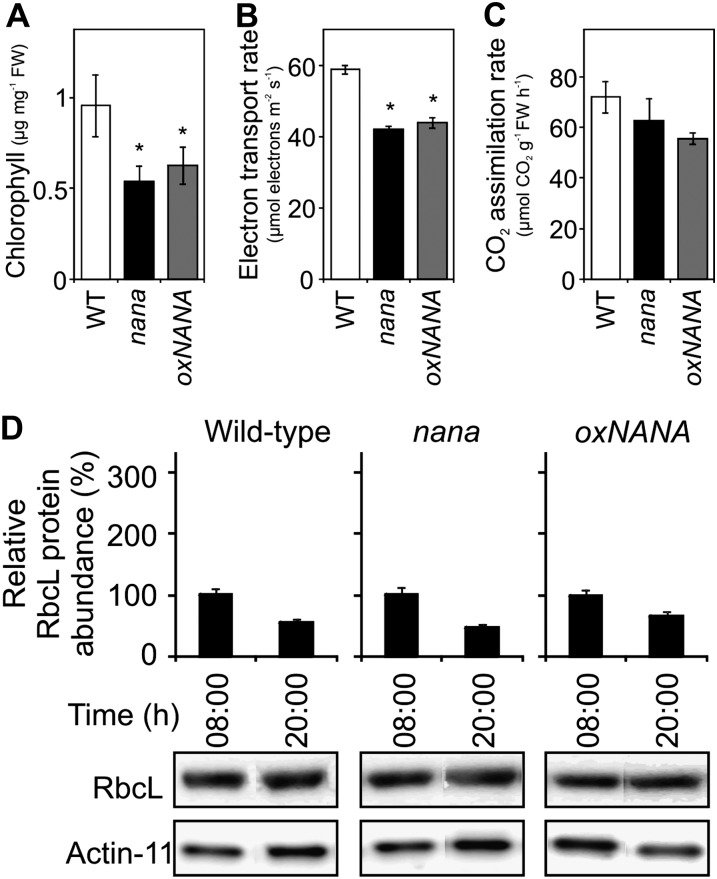
Figure 6.
Impact of the nana mutation on some photosynthetic parameters. A to C, The histograms show the chlorophyll a/b content (A), the photosynthetic ETR under 330 µmol photons m−2 s−1 (B), and the CO2 assimilation rate at 200 µmol photons m−2 s−1 (C) in 4-week-old wild-type (WT; white bars), nana (black bars), and oxNANA (gray bars) plants grown at a light intensity of 80 µmol photons m−2 s−1. In B and C, values shown are the maximum rates. The data represent means ± sd of three independent sets of experiments. Asterisks indicate significant differences (P < 0.05) by one-way ANOVA. FW, Fresh weight. D, Western blot and protein quantification of RbcL (means ± sd; n = 3) at the end of the night (8 am [08.00]) and of the light period (8 pm [20.00]) for each genotype. The data were normalized to the protein quantification of Actin11.
Sugar levels were measured in wild-type and mutant plants. As shown in Figure 7A, the levels of soluble sugars such as Suc, Glc, and Fru were significantly reduced in nana compared with the wild type when the plants were grown at an irradiance of 80 µmol m−2 s−1. Under these conditions, soluble sugar levels in nana leaves were dramatically lower than in the wild type, not only during the day but also at night (Fig. 7A). The oxNANA plants showed the same behavior as the nana mutant (Fig. 7A).
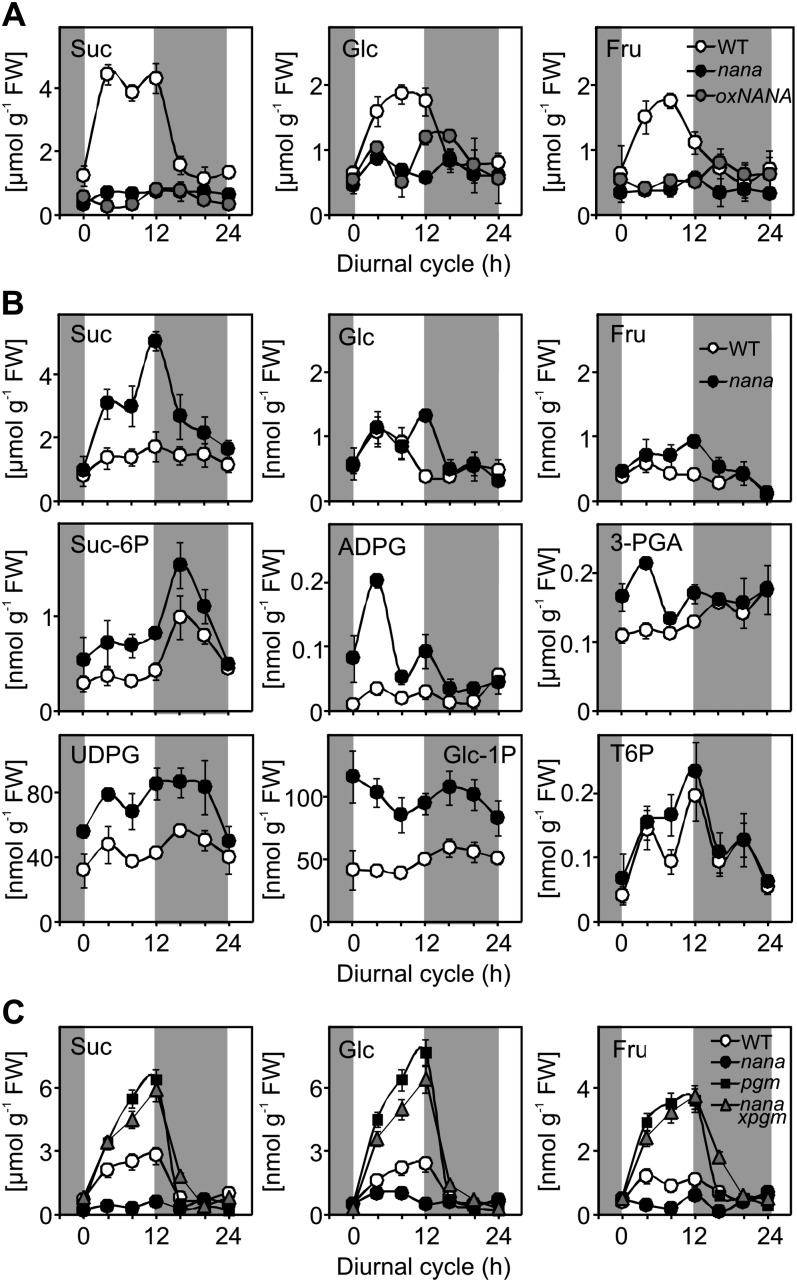
Figure 7.
Changes in the levels of metabolites during the diurnal cycle in wild-type (WT) and nana mutant plants grown in a 12-h-light/12-h-dark cycle. The whole rosette was harvested at the end of the night, 4, 8, and 12 h into the light period, and 4, 8, and 12 h into the dark period. A, Suc, Glc, and Fru content determined for the wild type (white circles), nana (black circles), and oxNANA (gray circles) grown at a light intensity of 80 µmol photons m−2 s−1. B, Suc, Glc, Fru, Suc-6-P, ADP-Glc (ADPG), UDP-Glc (UDPG), Glc-1-P, 3-phosphoglycerate (3-PGA), and T6P measured in the wild type and nana grown at an irradiance of 150 µmol photons m−2 s−1. C, Suc, Glc, and Fru levels determined for the wild type (white circles), nana (black circles), pgm (black squares), and the double mutant nana × pgm (gray triangles) grown at an irradiance of 80 µmol photons m−2 s−1. The results are given as means ± sd (n = 4 independent samples, each consisting of five rosettes). White and dark gray background shading show the light period and the night, respectively. FW, Fresh weight.
In contrast to these observations, growing the plants with a higher irradiance (150 µmol m−2 s−1) led to increased carbohydrate levels in the nana mutant compared with the wild type (Fig. 7B) but did not change the characteristic dwarf phenotype of these plants (data not shown). In addition to the higher Suc content of the nana plants, compared with the wild type, the mutant also contained elevated levels of Suc-6-P, ADP-Glc, UDP-Glc, 3-phosphoglycerate, and Glc-1-P (Fig. 7B). Trehalose-6-phosphate (T6P) has been postulated to act as a signal of Suc status and was found to be higher in the pgm mutant than in wild-type plants (Lunn et al., 2006). Actually, pgm plants, due to a mutation in the gene encoding the plastidial isoform of the phosphoglucomutase enzyme, show a strongly impaired starch synthesis, thus accumulating high levels of Suc during the day (Caspar et al., 1985). Interestingly, the levels of T6P were very similar in the nana and wild-type plants, despite the higher Suc content in the mutant (Fig. 7B). However, within the individual genotypes, the level of T6P showed a similar pattern of diurnal changes to Suc, rising to a peak at the end of the day and then falling at night. This suggests that the molecular mechanism linking T6P to Suc, which is as yet unknown, is still operating in the nana mutant but that the dynamic range of the response is tuned down in the mutant.
In an attempt to understand whether the alteration in carbohydrate metabolism could be a direct consequence of the slightly lower photosynthetic activity (Fig. 6, A–C), nana plants were crossed with the pgm mutant (Caspar et al., 1985). The pgm plants (growth irradiance of 80 µmol m−2 s−1) were smaller than the wild type but still clearly distinguishable from nana plants (Supplemental Fig. S4). The double mutant nana × pgm showed the same dwarf phenotype as nana, in terms of both reduced leaf size and morphology (Supplemental Fig. S4). Interestingly, in nana × pgm, the levels of soluble sugars, such as Glc, Suc, and Fru, perfectly matched the levels found in pgm plants (Fig. 7C), supporting the idea that the metabolic defects in the nana mutant are not directly due to the lower photosynthetic activity but are related rather to differences in the flow of carbon into starch.
Altered Starch Accumulation and Remobilization in the nana Mutant
The impact of NANA on starch metabolism was investigated by staining wild-type, nana, and oxNANA rosettes with Lugol’s solution. A strong staining, indicating the presence of starch, was detected in all three genotypes at the end of the day (Fig. 8A). Interestingly, both nana and oxNANA showed starch staining at the end of the night, whereas no staining was present, as expected, at this time point in the wild-type samples (Fig. 8A). To further confirm these results, the endogenous starch contents were measured in the wild-type, nana, and oxNANA extracts of 4-week-old plants. When grown under 12-h-light/12-h-dark cycles (80 µmol m−2 s−1), starch levels were significantly lower in nana and oxNANA than in the wild type at the end of the day (Fig. 8B). Moreover, the level of starch at the end of the night in both the nana and oxNANA mutants was higher than that of the wild type by a factor of 4.5 and 4.9, respectively (Fig. 8B), in line with the iodine staining results. These results suggested that starch is not properly synthesized and/or degraded in the nana mutant.
Figure 8.
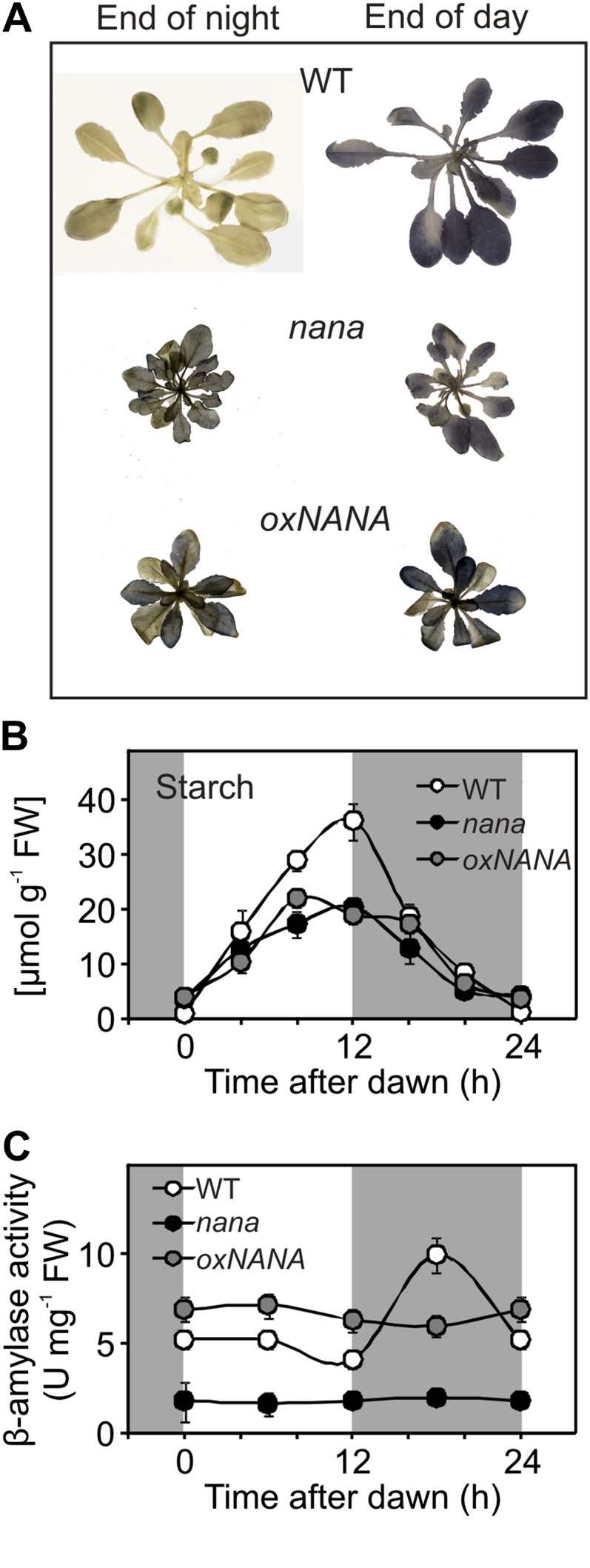
Alterations in starch accumulation and remobilization due to the nana mutation. A, Iodine staining showing the presence of starch in the wild type (WT), nana, and oxNANA mutants at the end of the night and the day. B, Changes in starch content in wild-type and mutant plants during the diurnal cycle at an irradiance of 80 µmol photons m−2 s−1. C, Diurnal changes in β-amylase activity in leaves of the wild type and mutants were determined in vitro using the Betamyl assay. In B and C, leaves of 4-week-old wild-type (white circles), nana (black circles), and oxNANA (gray circles) plants were used. Each point represents the mean ± sd from duplicate experiments. White and dark gray background shading show the light period and the night, respectively. FW, Fresh weight. [See online article for color version of this figure.]
We did not observe any differences in the redox-dependent oligomerization of the small subunit (APS1) of AGPase (for ADP-glucose pyrophosphorylase) in nana when compared with the wild type (Supplemental Fig. S5), indicating that an alteration in the posttranslational activation of this enzyme is not the prime cause of the defects in starch synthesis. The possibility of a defect in starch mobilization was also considered. In Arabidopsis leaves, β-amylases are important enzymes for starch breakdown (Fulton et al., 2008). The time course of the β-amylolytic activities in 4-week-old wild-type, nana, and oxNANA plants was then compared. Whereas the β-amylase activity in the wild-type leaf extracts showed a clear diurnal fluctuation, with a peak during the dark phase, the nana mutant displayed 3- to 6-fold lower levels of β-amylase activity, with no increase in the dark (Fig. 8C). Conversely, the oxNANA leaf extracts showed a constant activity, which was higher than that of the wild type during the light period. Judging from the low β-amylase activity shown by nana plants, we initially inferred that chloroplastic β-amylases could represent a possible target for NANA acting as a protease. We thus studied the diurnal expression patterns of some β-amylases in more detail, both at the gene and protein levels (Supplemental Fig. S6). β-Amylase family members are characterized by an extreme complexity as regards their protein function (Fulton et al., 2008). Among the four chloroplast-targeted β-amylases found in Arabidopsis leaves, only two, BAM1 and BAM3, are considered active in the degradation of starch (Valerio et al., 2011). The BAM1 gene was found to be more expressed in oxNANA compared with the nana mutant and the wild type, while the expression of BAM3 was similar in all three genotypes (Supplemental Fig. S6). Despite the differences in transcript levels, we found that the amounts of BAM1 or BAM3 proteins were fairly similar in the wild type and both the nana mutant and the oxNANA line (Supplemental Fig. S6). Thus, we did not find clues suggesting that NANA could be active on these β-amylases. On the contrary, the strong differences in the β-amylolytic activities (Fig. 8C) could be attributed to another β-amylase, BAM5 (Supplemental Fig. S6), accounting for most of the β-amylase activity in Arabidopsis leaves (Laby et al., 2001). Actually, the high β-amylase activity observed in the oxNANA line (Fig. 8C) correlates well with the strong induction of the BAM5 gene in this genotype (Supplemental Fig. S6). Moreover, the expression levels of BAM5 in nana are clearly lower than in the wild type (Supplemental Fig. S6), in accordance with the lower β-amylolytic activity of nana (Fig. 8C). However, BAM5 encodes an extraplastidic β-amylase (Laby et al., 2001), which thus cannot be a direct target of NANA proteolytic activity.
On the whole, all these results appear to indicate a possible involvement of NANA in starch metabolism. On the other hand, whether this involvement can be a direct consequence of NANA proteolytic activity on some enzymes acting in starch synthesis or degradation requires more extensive analyses.
Changes of Gene Expression in nana
As the nana mutant showed alterations in carbohydrate metabolism and different levels of sugars, we investigated whether the expression of sugar-responsive genes was affected in the mutant. In plants grown in 80 µmol m−2 s−1 irradiance, where nana and oxNANA generally have lower levels of soluble sugars, especially during the day (Fig. 7A), we observed differential expression of several sugar-responsive genes in the three genotypes. Two sugar-induced genes, ApL3 (for large subunit ADP-glucose pyrophosphorylase; At4g39210) and a 6-phosphogluconate dehydrogenase family protein-encoding gene, PG (At5g41670; Wingler et al., 2000; Gonzali et al., 2006; Solfanelli et al., 2006; Veyres et al., 2008), were expressed at lower levels in nana and oxNANA leaves than in the wild type (Fig. 9A), consistent with the lower sugar content of the mutants. In contrast, the GPT2 (for glucose6-phosphate/phosphate translocator2; At1g61800) gene, which is also reported to be induced by sugars (Gonzali et al., 2006; Kunz et al., 2010), showed generally higher expression in nana and oxNANA (Fig. 9A).
Figure 9.
Diurnal changes in the transcript levels measured in leaves of 4-week-old plants during a 48-h time course. A, Transcript levels for sugar-modulated genes, ApL3 (At4g39210), PG (At5g41670), and GPT2 (At1g61800). B, Transcript levels for chloroplast-encoded genes linked to photosynthesis, RbcL (AtCG00490), psbA (AtCG00020), and petD (AtCG00730). C, Transcript levels for nucleus-encoded photosynthetic genes, GLK1 (At2g20570), Lhca1 (At3g54890), Lhcb1.3 (At1g29930), Lhcb1.2 (At1g29910), Lhcb1.1 (At1g29920), and RbcS1-A (At1g67090). Wild-type (WT; white circles), nana (black circles), and oxNANA (gray circles) rosettes were used at 80 µmol photons m−2 s−1. Transcript levels are expressed as relative units, with the value of the wild type at the beginning of the day (light input) set to 1. Each value is the mean ± sd of three independent measurements. White and dark gray background shading show the light period and the night, respectively.
Suppression of the close homolog of NANA, CND41 (Supplemental Fig. S7), in tobacco led to marked changes in the expression of several chloroplast-encoded genes (Nakano et al., 1993, 1997). Therefore, we compared the expression level of the RbcL (AtCG00490), psbA (for PSII reaction center protein A; AtCG00020), and petD (for photosynthetic electron transfer D; AtCG00730) genes, as representative of chloroplast-encoded ones, in nana, oxNANA, and wild-type Arabidopsis. Interestingly, the transcript levels of RbcL, encoding the large subunit of Rubisco, and psbA, which encodes a component of the PSII reaction center core, were substantially repressed in both nana and oxNANA (Fig. 9B). In contrast, petD, which encodes the cytochrome b6/f complex subunit IV involved in photosynthetic electron transfer, was similarly expressed in all three genotypes (Fig. 9B), suggesting that in nana and oxNANA plants a process of transcriptional repression of important chloroplast-located genes occurs, although not generally.
Most chloroplast proteins, including many that are essential for photosynthesis, such as the light-harvesting chlorophyll-binding proteins (LHC), are encoded by nuclear genes (Jansson, 1999; Martin et al., 2002). Therefore, we also investigated if the expression of several nuclear genes linked to photosynthesis and other chloroplast functions was affected in the nana and oxNANA mutants. These included GLK1 (At2g20570), a GOLDEN2-LIKE1 transcription factor involved in chloroplast development (Fitter et al., 2002), and Lhca1 (At3g54890), Lhcb1.1 (At1g29920), Lhcb1.2 (At1g29910), and Lhcb1.3 (At1g29930), which encode members of the LHCI and LHCII complexes (Jansson, 1999) and are subject to regulation by plastid-derived signals (Strand et al., 2003; Waters et al., 2009). Interestingly, transcript levels of all four genes were significantly reduced in the nana and oxNANA mutants in comparison with the wild type (Fig. 9C). However, the down-regulation of the transcripts that are directly involved in photosynthetic processes was not a general trend. For example, the RbcS1-A-encoding gene (for ribulose-1,5-bisphosphate carboxylase small chain1A) showed similar expression levels in both the mutants and wild-type plants (Fig. 9C).
Together, these observations indicate that misexpression of NANA results in the down-regulation of different chloroplast-related or chloroplast-located genes. They also suggest that this defect might perturb chloroplast homeostasis, inducing the generation of a retrograde plastid signal that suppresses the expression of chloroplast-related genes in the nucleus.
CONCLUSION
We identified a novel chloroplast-located aspartyl protease (NANA) that not only influences photoassimilate partitioning and transitory starch turnover in a complex manner that is dependent on the growth irradiance but also perturbs both plastid and nuclear gene expression and possibly the cross talk between the chloroplast and nucleus. NANA misexpression causes several severe morphological alterations, such as dwarfism, small curly leaves, and delayed flowering. The major cause of these distinctive traits could be the alteration of carbohydrate metabolism and the consequent modification of the expression of genes involved in chloroplast physiology. Therefore, it is not easy to define the exact function of NANA in the framework of the complexity of chloroplast metabolism. Although the low level of sugars detected in nana and oxNANA grown at 80 µmol m−2 s−1 was suggestive of an impaired photosynthetic activity, the nana × pgm plants contained the same sugar levels as the parental pgm mutant but showed the same dwarf phenotype as nana, suggesting that the slightly lower rates of photosynthetic electron transport are not a major cause of the metabolic defects observed in nana. It appears that the effects of NANA misexpression are highly dependent on growth irradiance, as nana plants grown in higher light (150 µmol m−2 s−1) accumulated more Suc than the wild type and also had higher levels of phosphorylated intermediates involved in CO2 fixation, Suc synthesis, or starch synthesis. Together, these observations suggest that NANA misexpression affects fluxes through all the major pathways of photosynthetic carbon metabolism, affecting the availability of sugars for immediate growth as well as the turnover of transitory starch metabolism that is essential to support respiration, Suc export, and growth at night. The expression of a number of nuclear genes encoding for chloroplast proteins was altered in nana and oxNANA, suggesting the activation of a retrograde signal because of the NANA-related defects in chloroplast functionality. The expression of GLK1, identified as a crucial transcription factor in retrograde signaling and as a positive regulator of photosynthesis-related nuclear gene expression (Kakizaki et al., 2009), was indeed affected by the nana mutation. Thus, it seems that, because of altered NANA expression, a strong alteration in a number of processes involved in chloroplast physiology occurred. This, in turn, might generate a plastid retrograde signal, possibly under the control of GLK1, which consequently results in the down-regulation of a series of nuclear genes, also affecting the chloroplast metabolism and/or functionality, whose result is a strong impairment in plant growth and development.
The strong impact of the nana mutation that leads to a relatively modest alteration in its pattern of expression is suggestive of an important role of this aspartyl protease for chloroplast homeostasis. Our investigation suggested a possible but still unclear involvement of NANA in starch metabolism. Further studies are required to identify the actual protein target of NANA and to elucidate the specific regulatory role of NANA in the context of chloroplast functionality.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accession Columbia glabra (gl1) was used in this study. The nana mutant was identified by screening T-DNA-tagged lines from the Jack T-DNA enhancer trap collection (Campisi et al., 1999), obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk/home.html; Scholl et al., 2000). For the screening experiment, seeds were sterilized with diluted bleach (10-min incubation in 1.7% [v/v] sodium hypochlorite, rinsing, and washing thoroughly in sterile water) and germinated in horizontal petri dishes, prepared using solid growing medium (0.5× Murashige and Skoog solution containing 1% agar and 90 mm Suc).
The nana × pgm double mutant was generated by crossing nana with the pgm mutant that lacks a functional plastidial phosphoglucomutase (Nottingham Arabidopsis Stock Centre identifier N210) and is unable to synthesize starch (Caspar et al., 1985).
In most experiments, seeds were sown in moist soil or in a hydroponic system, as described by Gibeaut et al. (1997), stratified at 4°C in the dark for 48 h, and germinated at 22°C day/18°C night with a 12-h-light/12-h-dark photoperiod at 80 µmol photons m−2 s−1 irradiance, unless otherwise specified. Plants were harvested by transferring rosettes into liquid nitrogen under ambient irradiance.
Identification of the nana Mutation
For the screening of the insertional mutants, 630 different T-DNA-tagged lines from the Jack T-DNA enhancer trap collection (Campisi et al., 1999) were used. The screening was carried out in petri dishes containing 0.5× Murashige and Skoog solution, 1% agar, and 90 mm Suc, and putative Suc-hypersensitive mutants were collected as described previously (Gonzali et al., 2005). The nana mutant described here was initially selected for its Suc-hypersensitive phenotype.
For the molecular characterization of nana, DNA extraction was performed with the GenElute Plant Genomic DNA Miniprep Kit (Sigma-Aldrich) using approximately 100 mg of rosette leaves from 4-week-old nana plants. All PCR amplifications were carried out with 20 ng of genomic DNA and a PCR Master Mix (Promega). For the amplification of the T-DNA/plant genome junctions, thermal asymmetric interlaced-PCR was used, as described by Liu et al. (1995). Primers used were AD1, AD2, and AD3 and different TR1, TR2, and TR3 oligonucleotides for T-DNA left border or right border. All primer sequences are reported in Supplemental Table S3.
Cloning
The NANA coding sequence was amplified from a cDNA template using Phusion High Fidelity DNA polymerase (New England Biolabs) and cloned into the pENTR-D/TOPO Gateway vector (Invitrogen) to obtain pENTR-NANA. The resulting entry vector was recombined into destination vectors pK7WG2 and p7WGF2 (Karimi et al., 2002) using the LR Reaction Mix II (Invitrogen) to obtain the expression vectors 35S::NANA and 35S::GFP-NANA, respectively. The GFP-NANA in-frame fusion construct has the EGFP at the N terminus of NANA. EGFP at the C terminus of NANA resulted in the absence of fluorescence, and this is the reason for using EGFP at the N terminus. The cauliflower mosaic virus 35S promoter drove expression of the chimeric gene in both the constructs. Primers used for cloning in the pENTR-D/TOPO plasmid are reported in Supplemental Table S4.
Plant Transformation
Stable transgenic plants were obtained using the floral dip method (Clough and Bent, 1998; Zhang et al., 2006). T0 seeds were screened for kanamycin resistance, and single-insertion lines were identified by real-time PCR. Homozygous T3 or subsequent generations were used in the following experiments. Arabidopsis mesophyll protoplasts were prepared and transiently transformed with 10 µg of plasmid DNA according to Yoo et al. (2007).
Microscopy
For GFP imaging, fluorescence was observed with a Nikon ViCo videoconfocal microscope (http://www.nikon.com/) using a GFP filter. To observe epidermal cell size, leaves of wild-type and nana plants were plunged in liquid N2 and analyzed in a frozen-hydrated state by cryo-scanning electron microscopy, as described by Di Baccio et al. (2009).
Total RNA Extraction and Quantitative PCR
Total RNA was extracted from Arabidopsis rosettes as described previously (Perata et al., 1997) with a minor modification (omission of aurintricarboxylic acid) to make the protocol compatible with the subsequent PCR procedures. Electrophoresis using a 1% agarose gel was performed for all RNA samples to check for RNA integrity, followed by spectrophotometric quantification. Contaminating DNA was removed using a TURBO DNA-free kit (Ambion). RNA was then reverse transcribed using an iScript cDNA Synthesis kit (Bio-Rad Laboratories). Gene expression analysis was performed by real-time PCR using an ABI Prism 7300 sequence detection system (Applied Biosystems). Quantitative PCR was performed using 40 ng of cDNA (nuclear genes) or 0.005 ng of cDNA (chloroplastic genes) and iQSYBRGreen Supermix (Bio-Rad Laboratories), according to the manufacturer’s instructions. Transcripts of GAPDH and 40SrRNA for nuclear genes and of 16SrRNA and accD for plastid genes were used as reference genes. Relative expression levels were calculated using Genorm (http://medgen.ugent.be/∼jvdesomp/genorm). For a list of the primers used, see Supplemental Table S5.
SDS-PAGE and Western Blots
Plant samples (about 500 mg) were ground in liquid nitrogen. The extraction buffer, as described by Siddique et al. (2008), was added at a 1:2 ratio (plant tissue:buffer). Total protein content was quantified using the BCA Protein Assay Reagent (Pierce). SDS-PAGE was performed on a 10% Criterion polyacrylamide gel (Bio-Rad Laboratories). Blotting on an Amersham Hybond-P polyvinylidene difluoride membrane was performed with a Novablot electrophoretic transfer system (Amersham Pharmacia Biotech). After blocking and challenge with appropriate antibodies, immunoreactive proteins were detected using an Immun-Star HRP Chemiluminescent Detection Kit (Bio-Rad Laboratories). An antibody against NANA was produced by Agrisera (www.agrisera.com) using a synthetic peptide (CLFWKQNPTGDKKNQ) based on amino acid residues 6 to 19 (underlined) from the deduced sequence of the NANA protein. An antibody against the large subunit of Rubisco (AS03037) was purchased from Agrisera. The antibody against Actin11 (AS10702; Agrisera) was used to confirm even loading and transfer. Antisera specific for the BAM1 and BAM3 proteins were kindly provided by Prof. Samuel C. Zeeman. Densitometric analysis of the protein signals on the western blots was performed with the software package UVP VisionWork LS (Ultra-Violet Products). Normalization was carried out using Actin11 signal and setting to 100 the relative protein signal value for each “end of the day” time point. Extraction of AGPase, separation by nonreducing SDS-PAGE, and electroblotting onto nitrocellulose membrane were performed as described by Hendriks et al. (2003). After blocking, the membrane was incubated with anti-potato AGPB antiserum (Tiessen et al., 2002) for 1 h at 20°C. After washing, the membrane was incubated with IRDye800-conjugated anti-rabbit IgG F(c) antibody from goat (Biomol; http://www.biomol.de/). The fluorescent infrared signal from the dye-labeled secondary antibody was detected using an Odyssey Infrared Imaging System (LI-COR; http://www.licor.com/).
Chloroplast Isolation and Protein Extraction
Rosette leaves of Arabidopsis (20 g fresh weight), previously kept in the dark for 14 to 16 h, were homogenized in 400 mL of cold grinding buffer (330 mm sorbitol, 50 mm HEPES/KOH, pH 8.0, 2 mm EDTA, 1 mm MnCl2, and 1 mm MgCl2) using a Waring blender. The homogenate was filtered through four layers of Miracloth. The filtered samples were centrifuged at 6,000g for 30 min. The precipitate was gently resuspended in 5 mL of grinding buffer and separated by a step gradient with 80% and 40% Percoll. After centrifugation at 3,600 rpm for 30 min, intact chloroplasts were collected, resuspended in 2 volumes of grinding buffer, and centrifuged at 10,000g for 1 min. The supernatant was removed, and the pellet was vigorously resuspended in 1 mL of a hypotonic buffer containing 10 mm HEPES/KOH, pH 8.0, 5 mm dithiothreitol (DTT), and 1 mm EDTA. All procedures were carried out at 4°C.
Chloroplast proteins were extracted according to Yang et al. (2007), with some modifications. Chloroplasts were ground in liquid nitrogen and homogenized with 1 mL of extraction buffer (5 m urea, 2 m thiourea, 40 mm Tris-HCl, 2% CHAPS, and 50 mm DTT). The homogenate was centrifuged at 15,000g for 15 min. Supernatant was precipitated using TCA (15%, v/v) containing 0.007% β-mercaptoethanol in acetone at −20°C for 2 h and then at 4°C for a minimum of 2 h. Samples were centrifuged at 4°C at 14,000g for 15 min, supernatant was discarded, and pellets were washed twice with ice-cold acetone containing 0.007% β-mercaptoethanol. Pellets were dissolved in a rehydration buffer (5 m urea, 2 m thiourea, 2% CHAPS, and 50 mm DTT). The immunoblot analysis was performed as described above, loading 30 µg of protein for each fraction.
Expression of Recombinant Protein in Escherichia coli
The NANA coding sequence was recombined from pENTR-NANA into the pDEST17 plasmid (Invitrogen) and transformed into the E. coli Rosetta2 strain (Novagen). The expression of the His6-tagged NANA fusion protein was induced by the addition of IPTG (final concentration, 0.5 mm) to early log-phase cultures and incubation overnight at 20°C. The overexpressed protein was purified using TALON immobilized metal ion affinity chromatography resin (Clontech). The purified protein was eluted in a buffer containing 50 mm HEPES, 300 mm NaCl, 150 mm imidazole, and 5 mm MgCl2, pH 7.0, and the identity of the protein was confirmed by SDS-PAGE and immunoblot analysis with anti-NANA antibody.
Detection of Enzymatic Activity
An azocasein assay was performed for the determination of the aspartic proteolytic activity of NANA. Protease activity was determined by measuring the release of acid-soluble material from azocasein (Sigma-Aldrich). Soluble protein were extracted from leaves by homogenization in 100 mm sodium acetate (pH 5.0) buffer and incubated for 30 min at 4°C. The reactions were performed in 50 mm sodium acetate (pH 5.0) and 0.8% (w/v) azocasein reaction buffer, with or without 100 µM pepstatin A (Promega), as aspartyl protease inhibitor. The mixture was incubated for 3 h at 30°C, and the reaction was stopped by the addition of TCA to a final concentration of 10%. After incubation on ice for 15 min, the reaction mixture was centrifuged (5,000g, 5 min) to remove precipitated protein and unreacted azocasein. Absorbance of the azopeptide-containing supernatant was measured at 330 nm. Aspartyl-protease activity units were calculated as change in optical density at 330 nm h−1 mg−1 protein. Aspartyl protease activity units were calculated as the difference between the total protease activities in the presence and absence of pepstatin A. The activity of the recombinant NANA protein was assayed as above using a 50 mm sodium acetate (pH 6.0) buffer. The pH optimum for both the recombinant protein and the crude extract was determined within a pH range of 3.6 to 8.0, using 50 mm sodium acetate (pH 3.6–5.6) and 100 mm sodium phosphate (pH 6.0–8.0) buffers.
Activities of β-amylase were measured using the Betamyl assay kit from Megazyme, according to Zeeman et al. (1998).
Extraction and Measurement of Metabolites
Analyses of Suc, Fru, and Glc were carried out as described previously (Guglielminetti et al., 1995). Samples (0.05–0.30 g fresh weight) were rapidly frozen in liquid nitrogen and ground to a powder. Samples were then extracted as described by Tobias et al. (1992). After centrifugation, the supernatant was used for analysis of Suc, Fru, and Glc. The starch-containing pellet was extracted with 10% KOH, centrifuged, and neutralized with 18% (v/v) acetic acid. The resulting suspension was treated with 2.5 units of amyloglucosidase (from Rhizopus niger) for 3 h to release Glc. Samples were assayed by coupled enzymatic assay methods, measuring the increase in A340. Starch was quantified on the basis of the Glc units released after the amyloglucosidase treatment. For qualitative assay of starch, leaves were stained using an iodine solution after first decolorizing them in 80% (v/v) ethanol. T6P and phosphorylated intermediates were extracted and measured by anion-exchange liquid chromatography coupled to tandem mass spectrometry as described by Lunn et al. (2006).
Photosynthesis Measurements
Fluorescence emission was measured in vivo using a PAM fluorometer (Walz) on 5-week-old plants maintained at fixed irradiance (250 and 700 μmol photons m−2 s−1) for 30 min prior to measurement of chlorophyll fluorescence yield and relative ETR at 330 μmol photons m−2 s−1, which were calculated using the WinControl software package (Walz). Chlorophyll quantification was performed as described by Lichtenthaler (1987) and Porra (2002). Gas-exchange measurements were performed in a special custom-designed open system at 200 μmol photons m−2 s−1 irradiance (Lytovchenko et al., 2002).
Sequence and Phylogenetic Gene Analysis
BLASTP 2.2.26+ (Altschul et al., 1990) was used to identify protein sequences similar to NANA (Supplemental Fig. S2A), using the BLOSUM62 scoring matrix and applying an E-value threshold of 10−10, against the RefProt database (National Center for Biotechnology Information; release November 1, 2012). The tomato (Solanum lycopersicum) homologous sequences were obtained from the Sol Genomics Network draft genome (Bombarely et al., 2011). Arabidopsis putative paralogous protein sequences were taken within the same threshold (E < 10−10) and aligned using MUSCLE (Edgar, 2004). The subsequent maximum likelihood phylogenetic tree (Supplemental Fig. S2B) was built using MEGA5 (Tamura et al., 2011) with default parameters and 100 bootstraps for branch point validation (branch points with less than 50 bootstrap concordance are collapsed). In the tree in Supplemental Figure S2B, gene expression alterations are indicated in a data set from 7-d-old Arabidopsis wild-type Columbia dark-grown seedlings compared with light-grown seedlings (Dohmann et al., 2008; Gene Expression Omnibus entry GSE9728); the P value significance threshold for differential expression was set to 0.05 using the Genevestigator software (Hruz et al., 2008). “Clock-regulated genes” in the phylogenetic tree (with respect to the transcript quantity oscillation) are indicated according to Covington and Harmer (2007). A list of all these genes is given in Supplemental Table S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. DNA ploidy levels in the wild type and the nana mutant determined by flow cytometric analysis.
Supplemental Figure S2. List of NANA putative homologs grouped by organism, selected by BLASTP (E-value threshold of 10−10) from the RefProt database (National Center for Biotechnology Information), and consensus phylogenetic tree (maximum likelihood) of putative NANA in paralogs in Arabidopsis.
Supplemental Figure S3. Aspartyl proteolytic activities in wild-type, nana, and oxNANA rosette extracts using the azocasein assay.
Supplemental Figure S4. Phenotype of the double mutant nana × pgm, in comparison with the wild type and the nana and pgm parental single mutants.
Supplemental Figure S5. Dimerization of the APS1 subunit of ADPglucose pyrophosphorylase wild-type and nana plants, resolved by nonreducing SDS-PAGE and detected by immunoblotting.
Supplemental Figure S6. Diurnal changes in transcript and protein levels of genes encoding β-amylase enzymes in leaves of 4-week-old plants during a time course of 48 h.
Supplemental Figure S7. Clustal alignment of NANA and tobacco CND41 (Nakano et al., 2003) polypeptide sequences.
Supplemental Table S1. Hypocotyl and primary root length in wild-type and nana seedlings, grown in the presence or absence of 90 mm Suc either in light or dark conditions.
Supplemental Table S2. List of putative Arabidopsis paralogs of the NANA (At3g12700) gene.
Supplemental Table S3. List of primers used in thermal asymmetric interlaced-PCR and to test T-DNA insertion.
Supplemental Table S4. List of primers used for cloning in the pENTR-D/TOPO plasmid.
Supplemental Table S5. List of primers used for gene expression analysis using real-time quantitative reverse transcription-PCR.
Supplementary Material
Acknowledgments
We thank Prof. Samuel C. Zeeman (Eidgenössisch Technische Hochschule) for kindly providing us with the BAM antibodies. We also thank Dr. Antonio Minnocci (Scuola Superiore Sant’Anna) for cryo-scanning electron microscopy analysis, Dr. Paola Collecchi (University of Pisa) for help with the endoreduplication experiment, and Dr. Valentina Lucarotti (University of Pisa) for chloroplast isolation. We furthermore are particularly grateful to Dr. Joachim Fisahn and Dr. Alexander Ivakov (both Max Plank Institute) for helping in the measurement of photosynthetic parameters.
Glossary
- AP
aspartic proteinase
- EGFP
enhanced GFP
- IPTG
isopropylthio-β-d-galactoside
- ETR
electron transport rate
- T6P
trehalose-6-phosphate
- DTT
dithiothreitol
References
- Adam Z. (1996) Protein stability and degradation in chloroplasts. Plant Mol Biol 32: 773–783 [DOI] [PubMed] [Google Scholar]
- Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K, et al. (2001) Chloroplast and mitochondrial proteases in Arabidopsis: a proposed nomenclature. Plant Physiol 125: 1912–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajjawi I, Lu Y, Savage LJ, Bell SM, Last RL. (2010) Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project. Plant Physiol 152: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andersson B, Aro E-M. (1997) Proteolytic activities and proteases of plant chloroplasts. Physiol Plant 100: 780–793 [Google Scholar]
- Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T. (2005) Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol 46: 858–869 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T. (1999) Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J 17: 699–707 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C. (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Bush DR. (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol 125: 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Harmer SL. (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Lemaître T, Christ A, Azzopardi M, Kato Y, Sato F, Morot-Gaudry JF, Le Dily F, Masclaux-Daubresse C. (2008) Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol 147: 1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Baccio D, Tognetti R, Minnocci A, Sebastiani L. (2009) Responses of the Populus × euramericana clone I-214 to excess zinc: carbon assimilation, structural modifications, metal distribution and cellular localization. Environ Exp Bot 67: 153–163 [Google Scholar]
- Dohmann EM, Levesque MP, De Veylder L, Reichardt I, Jürgens G, Schmid M, Schwechheimer C. (2008) The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135: 2013–2022 [DOI] [PubMed] [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ. (2004) Ancient invasions: from endosymbionts to organelles. Science 304: 253–257 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G, et al. (2008) β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR. (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios-Rojas N, Pankovic D, Hendriks JH, Fisahn J, Höhne M, Günther M, Stitt M. (2004) Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Gibson SI. (2000) Plant sugar-response pathways: part of a complex regulatory web. Plant Physiol 124: 1532–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. (2004) Sugar and phytohormone response pathways: navigating a signalling network. J Exp Bot 55: 253–264 [DOI] [PubMed] [Google Scholar]
- Gibson SI. (2005) Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol 8: 93–102 [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Solfanelli C, Novi G, Alpi A, Perata P. (2006) Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J Plant Res 119: 115–123 [DOI] [PubMed] [Google Scholar]
- Gonzali S, Novi G, Loreti E, Paolicchi F, Poggi A, Alpi A, Perata P. (2005) A turanose-insensitive mutant suggests a role for WOX5 in auxin homeostasis in Arabidopsis thaliana. Plant J 44: 633–645 [DOI] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM. (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plant at night. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Perata P, Alpi A. (1995) Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol 108: 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis subcellular database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P. (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang X, Yang S. (2009) A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res 19: 1205–1216 [DOI] [PubMed] [Google Scholar]
- Inaba T. (2010) Bilateral communication between plastid and the nucleus: plastid protein import and plastid-to-nucleus retrograde signaling. Biosci Biotechnol Biochem 74: 471–476 [DOI] [PubMed] [Google Scholar]
- Jang JC, Sheen J. (1997) Sugar sensing in higher plants. Trends Plant Sci 2: 208–214 [Google Scholar]
- Jansson S. (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che F-S, Terauchi R, Inaba T. (2009) Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol 151: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kato Y, Murakami S, Yamamoto Y, Chatani H, Kondo Y, Nakano T, Yokota A, Sato F. (2004) The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta 220: 97–104 [DOI] [PubMed] [Google Scholar]
- Kato Y, Saito N, Yamamoto Y, Sato F. (2005a) Regulation of senescence by aspartic protease: CND41 in tobacco and CND41 homologues in Arabidopsis. In A van der Est, D Bruce, eds, Proceedings of the Photosynthesis Congress 2004, Photosynthesis: Fundamental Aspects to Global Perspectives. Alliance Communications Group, Lawrence, KS, pp 821–823 [Google Scholar]
- Kato Y, Yamamoto Y, Murakami S, Sato F. (2005b) Post-translational regulation of CND41 protease activity in senescent tobacco leaves. Planta 222: 643–651 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjölander K, Gruissem W, Baginsky S. (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Koch KE. (1996) Carbohydrate modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Koch KE. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Kunz HH, Häusler RE, Fettke J, Herbst K, Niewiadomski P, Gierth M, Bell K, Steup M, Flügge UI, Schneider A. (2010) The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol (Stuttg) (Suppl 1) 12: 115–128 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kim D, Gibson SI. (2001) The ram1 mutant of Arabidopsis exhibits severely decreased β-amylase activity. Plant Physiol 127: 1798–1807 [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37: D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HM, Chiu C-C. (2010) Protein transport into chloroplasts. Annu Rev Plant Biol 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M. (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytovchenko A, Sweetlove LJ, Pauly M, Fernie AR. (2002) The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 215: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Yohn CB, Cohen A, Danon A. (1995) Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol 46: 147–166 [Google Scholar]
- Milisavljevic MD, Timotijevic GS, Radovic SR, Konstantinovic MM, Maksimovic VR. (2008) Two types of aspartic proteinases from buckwheat seed: gene structure and expression analysis. J Plant Physiol 165: 983–990 [DOI] [PubMed] [Google Scholar]
- Murakami S, Kondo Y, Nakano T, Sato F. (2000) Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Lett 468: 15–18 [DOI] [PubMed] [Google Scholar]
- Mutlu A, Gal S. (1999) Plant aspartic proteinases: enzymes on the way to a function. Physiol Plant 105: 569–576 [Google Scholar]
- Myouga F, Akiyama K, Motohashi R, Kuromori T, Ito T, Iizumi H, Ryusui R, Sakurai T, Shinozaki K. (2010) The Chloroplast Function Database: a large-scale collection of Arabidopsis Ds/Spm- or T-DNA-tagged homozygous lines for nuclear-encoded chloroplast proteins, and their systematic phenotype analysis. Plant J 61: 529–542 [DOI] [PubMed] [Google Scholar]
- Nair JS, Ramaswamy NK. (2004) Chloroplast proteases. Biol Plant 48: 321–326 [Google Scholar]
- Nakano T, Murakami S, Shoji T, Yoshida S, Yamada Y, Sato F. (1997) A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell 9: 1673–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Nagata N, Kimur T, Sekimoto M, Kawaide H, Murakami S, Kaneko Y, Matsushima H, Kamiya Y, Sato F, et al. (2003) CND41, a chloroplast nucleoid protein that regulates plastid development, causes reduced gibberellin content and dwarfism in tobacco. Physiol Plant 117: 130–136 [Google Scholar]
- Nakano T, Sato F, Yamada Y. (1993) Analysis of nucleoid-proteins in tobacco chloroplasts. Plant Cell Physiol 34: 873–880 [Google Scholar]
- Neff MM, Fankhauser C, Chory J. (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Perata P, Matsukura C, Vernieri P, Yamaguchi J. (1997) Sugar repression of a gibberellin-dependent signaling pathway in barley embryos. Plant Cell 9: 2197–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ. (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ. (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17: 471–482 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26: 421–433 [DOI] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf KD. (2008) The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões I, Faro C. (2004) Structure and function of plant aspartic proteinases. Eur J Biochem 271: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C-P, Guo Y, Qiu Q, Lambert G, Galbraith DW, Jagendorf A, Zhu J-K. (2004) A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 10211–10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Lunn J, Usadel B. (2010) Arabidopsis and primary photosynthetic metabolism: more than the icing on the cake. Plant J 61: 1067–1091 [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farré EM, Geigenberger P. (2002) Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timotijević GS, Milisavljević MD, Radović SR, Konstantinović MM, Maksimović VR. (2010) Ubiquitous aspartic proteinase as an actor in the stress response in buckwheat. J Plant Physiol 167: 61–68 [DOI] [PubMed] [Google Scholar]
- Tobias RB, Boyer CD, Shannon JC. (1992) Alterations in carbohydrate intermediates in the endosperm of starch-deficient maize (Zea mays L) genotypes. Plant Physiol 99: 146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio C, Costa A, Marri L, Issakidis-Bourguet E, Pupillo P, Trost P, Sparla F. (2011) Thioredoxin-regulated beta-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J Exp Bot 62: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyres N, Danon A, Aono M, Galliot S, Karibasappa YB, Diet A, Grandmottet F, Tamaoki M, Lesur D, Pilard S, et al. (2008) The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence. Plant J 55: 665–686 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Fritzius T, Wiemken A, Boller T, Aeschbacher RA. (2000) Trehalose induces the ADP-glucose pyrophosphorylase gene, ApL3, and starch synthesis in Arabidopsis. Plant Physiol 124: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J. (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, Morimoto K, Kikuchi S, Nishio K, Terashima I, Nakai M. (2004) The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16: 993–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Wang Y, Zhang J, Shi W, Qian C, Peng X. (2007) Identification of aluminum-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics 7: 737–749 [DOI] [PubMed] [Google Scholar]
- Yazdanbakhsh N, Fisahn J. (2011) Mutations in leaf starch metabolism modulate the diurnal root growth profiles of Arabidopsis thaliana. Plant Signal Behav 6: 995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, Rees T. (1998) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J 15: 357–365 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.