Abstract
Protein lipid modification of cysteine residues, referred to as S-palmitoylation or S-acylation, is an important secondary and reversible modification that regulates membrane association, trafficking, and function of target proteins. This enzymatic reaction is mediated by protein S-acyl transferases (PATs). Here, the phylogeny, genomic organization, protein topology, expression, and localization pattern of the 24 PAT family members from Arabidopsis (Arabidopsis thaliana) is described. Most PATs are expressed at ubiquitous levels and tissues throughout the development, while few genes are expressed especially during flower development preferentially in pollen and stamen. The proteins display large sequence and structural variations but exhibit a common protein topology that is preserved in PATs from various organisms. Arabidopsis PAT proteins display a complex targeting pattern and were detected at the endoplasmic reticulum, Golgi, endosomal compartments, and the vacuolar membrane. However, most proteins were targeted to the plasma membrane. This large concentration of plant PAT activity to the plasma membrane suggests that the plant cellular S-acylation machinery is functionally different compared with that of yeast (Saccharomyces cerevisiae) and mammalians.
Cellular functions are regulated by various protein modifications. These modifications can occur in a controlled, reversible, and transient manner in response to different signals (Huber and Hardin, 2004; Stulemeijer and Joosten, 2008). Protein phosphorylation is the most prominent modification and is regulated by the balanced activities of protein kinases and protein phosphatases (Hardie, 1999; Luan, 2003). However, other modifications, like ubiquitination and sumoylation (Miura and Hasegawa, 2010) or nitrosylation (Lamattina et al., 2003), also regulate protein functions.
A less well characterized reversible lipid modification is the thioesterification of Cys residues, which is known as S-palmitoylation or more generally referred to as S-acylation (Hemsley and Grierson, 2008; Sorek et al., 2009). S-acyl modifications mainly affect membrane attachment and trafficking of proteins. They are required for the dynamic association of proteins with membrane subdomains (Sorek et al., 2007, 2010) and for the cycling between different cellular membranes (Rocks et al., 2005). In addition, S-acylation can influence the stability of proteins (Valdez-Taubas and Pelham, 2005; Abrami et al., 2006), modulates the functions of proteins (Gubitosi-Klug et al., 2005), or mediates the interaction between different proteins (Charrin et al., 2002; Flannery et al., 2010).
S-acyl modification of proteins is brought about by palmitoyltransferases, correctly referred to as protein S-acyltransferases (PATs; Mitchell et al., 2006). Originally, these enzymes were identified in yeast (Saccharomyces cerevisiae; Lobo et al., 2002; Roth et al., 2002). Sequence analyses of the yeast PAT enzymes revealed that they share a common structure mainly composed of four predicted transmembrane domains (TMDs) and a stretch of Asp-His-His-Cys (DHHC) within a Cys-rich domain (CRD; Pfam accession, PF01529; InterPro entry, IPR001594). Mutational analyses revealed that the Cys residue of the DHHC stretch is necessary for autoacylation and for the modification of target proteins. Therefore, this domain appears to represent the active site of these enzymes (Lobo et al., 2002; Roth et al., 2002). Detailed analyses of the lipid modifications of Rho-of-plants6 (ROP6; Sorek et al., 2007) and of the Calcineurin-B like (CBL) proteins 1 and 2 (Batistič et al., 2008, 2012) revealed that plant PATs either transfer palmitate or stearate to these substrates.
In plants, so far only one PAT was characterized in detail. A screen of Arabidopsis (Arabidopsis thaliana) mutants resulted in the identification of TIP GROWTH DEFECTIVE1 (TIP1), which is affected in root hair formation and growth (Schiefelbein et al., 1993; Ryan et al., 1998). Mapping of the mutant allele revealed that TIP1 encodes a DHHC-CRD-containing protein, and it was shown that this protein is indeed S-acylated (Hemsley et al., 2005, 2008). Moreover, TIP1 contains N-terminal ankyrin repeats, related to the yeast PAT Akr1, and is able to complement the PAT function in the akr1Δ yeast strain, corroborating that TIP1 has a PAT enzyme function (Hemsley et al., 2005). Although our knowledge about the plant S-acylome and S-acylation machinery is still limited (Hemsley, 2009), previous studies on the S-acylation of plant proteins revealed that these modifications could occur at different cellular membranes like the endoplasmic reticulum (ER; Adjobo-Hermans et al., 2006; Batistič et al., 2008), the Golgi (Zeng et al., 2007), or directly at the plasma membrane (PM; Sorek et al., 2007). Furthermore, recent studies on the lipid modification of the tonoplast-associated CBL2 protein from Arabidopsis implicated that S-acylation can also occur at the vacuolar membrane (Batistič et al., 2012). These findings suggested that the PAT enzymes that modify these targets reside at different membranes within the plant cell.
In this report, the complement of the Arabidopsis PAT gene loci was examined to perform a comprehensive analysis of this gene and protein family. These analyses revealed that at least 24 PAT gene loci are encoded in the Arabidopsis genome. All Arabidopsis PATs are expressed. Expression occurs mainly at steady levels throughout development and in most tissues examined. However, some PATs are preferentially expressed at certain developmental stages or cell types especially during flowering and in pollen, respectively. Phylogenetic analysis revealed that the proteins are highly divergent. Nevertheless, most PAT proteins harbor a similar structural topology and preserved sequence motifs that are representative of this class of enzymes. Functional analyses on several proteins by yeast complementation analyses indicate that Arabidopsis PATs represent active enzymes, even if their sequence deviates from the regular standard sequence composition previously reported for functional PATs. Moreover, the protein localizations of all 24 Arabidopsis PATs were examined. This revealed a complex targeting pattern of PATs showing that S-acylation can occur at different cellular membranes within the cell. Remarkably, the plant PATs analyzed are mainly targeted to the PM, indicating that the plant S-acylation machinery has a different organization and function compared with the mammalian and yeast PAT machinery.
RESULTS
The Arabidopsis Genome Encodes 24 DHHC-CRD-Containing PAT Proteins
In a previous study, 23 gene loci that encode DHHC-CRD proteins were identified and preliminary analyzed (Hemsley et al., 2005; Hemsley, 2009). In order to perform a comprehensive and detailed analysis of the DHHC-CRD-containing PAT family from Arabidopsis, the complement of gene loci was first reexamined. The protein sequence of Arabidopsis PAT8 was used in different BLAST searches as a query (see “Material and Methods” for details). This resulted in the identification of overall 24 individual gene loci that could encode for PATs. This included also the At2g14255 locus, which was already described as a gene partially homologous to At5g20350/TIP1 (Hemsley et al., 2005). However, Hemsley and colleagues have classified this gene as a pseudogene and concluded that At5g20350/TIP1 is the only ankyrin-containing PAT in the Arabidopsis genome. Nevertheless, the protein predicted by the databases contains the central DHHC-CRD domain, which is representative for this enzyme class (see below). Therefore, according to the previous naming, the gene product of At2g14255 was designated here as AtPAT23. For a consistent nomenclature (and to exclude a mix-up with the tonoplast intrinsic protein 1) a renaming of TIP1 is proposed here to AtPAT24, while tip1 would still represent a mutant allele of AtPAT24.
The PAT gene loci are distributed throughout the genome and not organized in larger clusters. Most loci are concentrated on chromosome 3 (10 loci), and this includes a single in tandem organized gene pair (AtPATs 1 and 4). Five loci are distributed on the chromosomes 4 and 5, respectively, while three loci reside on chromosome 2. A single PAT gene locus (AtPAT22) was found on chromosome 1 (Supplemental Fig. S1).
To evaluate the mRNA and protein annotations of the Arabidopsis PATs, the open reading frames (ORFs) were PCR amplified from different complementary DNA (cDNA) sources, cloned, and sequenced (Supplemental Materials and Methods). With the exception of AtPAT20, all PAT reading frames were amplified. However, several expressed sequence tags are annotated for the AtPAT20 gene. This implicates that all identified PAT genes are actively transcribed.
Sequences of the amplified ORFs correspond to the mRNA structures annotated in The Arabidopsis Information Resource database (version 1 if alternative versions exist), with the exception of AtPAT9. The 5′ and 3′ end of the AtPAT9 ORF obtained here correlates with At5g50020.1, but the sequence contains 21/7 additional nucleotides/amino acids (Supplemental Fig. S2). These nucleotides/amino acids are also present in At5g50020.2. However, the obtained sequence lacks the N-terminal extension of At5g50020.2 (Supplemental Fig. S2). Therefore, this sequence was designated as AtPAT9-3 and used for all subsequent analyses.
Since At2g14255/AtPAT23 was classified as a pseudogene and not as a PAT yet (Hemsley et al., 2005), its transcript structure was analyzed in detail using cDNAs from different tissues (root, shoot, flower, and pollen specific). An amplification product was obtained by reverse transcription-PCR from all sources (Supplemental Fig. S3A). The DNA obtained from flower tissue was larger than the other PCR products. Sequencing revealed an alternative spliced transcript of AtPAT23, which includes a nonspliced intron and a shortened exon. This transcript contains a premature stop codon, and the protein product would lack the central DHHC site (Supplemental Fig. S3B). The PCR products from root, shoot, and pollen exhibited similar sizes. Sequence analysis revealed that the root cDNA clone contained different alternative splicing events compared with the shoot and pollen cDNA clones. These alternative splicing events result in a premature stop codon, and the resulting protein would lack the DHHC sequence. However, clones obtained from the shoot and pollen cDNA were identical to The Arabidopsis Information Resource annotation and encode for a functional PAT with a DHHC domain (Supplemental Fig. S3B).
Overall, these analyses allow the conclusion that all 24 Arabidopsis genes encode for functional PAT proteins. The Arabidopsis Genome Initiative numbers, nucleotide and protein accessions, nucleotide length of the ORFs, and the predicted protein sizes of all Arabidopsis PATs are summarized in Table I.
Table I. Summary of the Arabidopsis PAT gene family.
AGI, Arabidopsis Genome Initiative.
| Gene Name | AGI No. | NCBI Nucleotide Accession | NCBI Protein Accession | Uniprot Protein Accession | Nucleotide Lengtha | Amino Acids | Protein Size in kD |
|---|---|---|---|---|---|---|---|
| AtPAT01 | At3g56920 | NM_115551 | NP_191251 | B3DN87 | 1,017 | 338 | 39 |
| AtPAT02 | At2g40990 | NM_129644 | NP_181632 | O80685 | 1,236 | 411 | 47 |
| AtPAT03 | At5g05070 | NM_120589 | NP_196126 | Q5PNZ1 | 1,242 | 413 | 48 |
| AtPAT04 | At3g56930 | NM_115552 | NP_191252 | Q9M1K5 | 1,434 | 477 | 55 |
| AtPAT05 | At3g48760 | NM_114735 | NP_190445 | Q9M306 | 1,431 | 476 | 54 |
| AtPAT06 | At5g41060 | NM_123471 | NP_198922 | Q9FLM3 | 1,264 | 410 | 46 |
| AtPAT07 | At3g26935 | NM_180307 | NP_850638 | Q0WQK2 | 1,332 | 443 | 50 |
| AtPAT08 | At4g24630 | NM_118596 | NP_194194 | Q9SB58 | 1,244 | 407 | 47 |
| AtPAT09 | At5g50020 | JF792493 | AEF58502 | F6MDM7 | 1,245 | 414 | 48 |
| AtPAT10 | At3g51390 | NM_114998 | NP_566950 | Q7XA86 | 1,023 | 340 | 39 |
| AtPAT11 | At3g18620 | NM_112748 | NP_188492 | Q9LIH7 | 1,038 | 345 | 38 |
| AtPAT12 | At4g00840 | NM_116310 | NP_567193 | Q5M757 | 876 | 291 | 33 |
| AtPAT13 | At4g22750 | NM_118402 | NP_567668 | Q94C49 | 909 | 302 | 34 |
| AtPAT14 | At3g60800 | NM_115944 | NP_191639 | Q8VYP5 | 924 | 307 | 35 |
| AtPAT15 | At5g04270 | NM_120509 | NP_196047 | Q500Z2 | 765 | 254 | 29 |
| AtPAT16 | At3g09320 | NM_111766 | NP_566348 | Q93VV0 | 861 | 286 | 32 |
| AtPAT17 | At3g04970 | NM_111369 | NP_187148 | Q3EBC2 | 1,194 | 397 | 45 |
| AtPAT18 | At4g01730 | NM_116403 | NP_192082 | Q9M115 | 1,499 | 499 | 57 |
| AtPAT19 | At4g15080 | NM_117595 | NP_193244 | Q8L5Y5 | 2,157 | 718 | 79 |
| AtPAT20b | At3g22180 | NM_113115 | NP_188857 | Q9LIE4 | 2,121 | 706 | 77 |
| AtPAT21 | At2g33640 | NM_128924 | NP_180922 | Q6DR03 | 1,698 | 565 | 62 |
| AtPAT22 | At1g69420 | NM_202385 | NP_974114 | Q9C533 | 1,791 | 596 | 66 |
| AtPAT23 | At2g14255 | NM_201724 | NP_973453 | Q3EC11 | 1,697 | 536 | 60 |
| AtPAT24c | At5g20350 | NM_122042 | NP_197535 | Q52T38 | 1,860 | 620 | 68 |
Verified in this study.
PAT20 was not amplified from cDNA, but indirectly obtained by expressing a genomic fragment in N. benthamiana and reamplifying from a cDNA.
A mutant allele was designated as tip1 (Hemsley et al., 2005).
Phylogenetic Analysis of Arabidopsis PATs
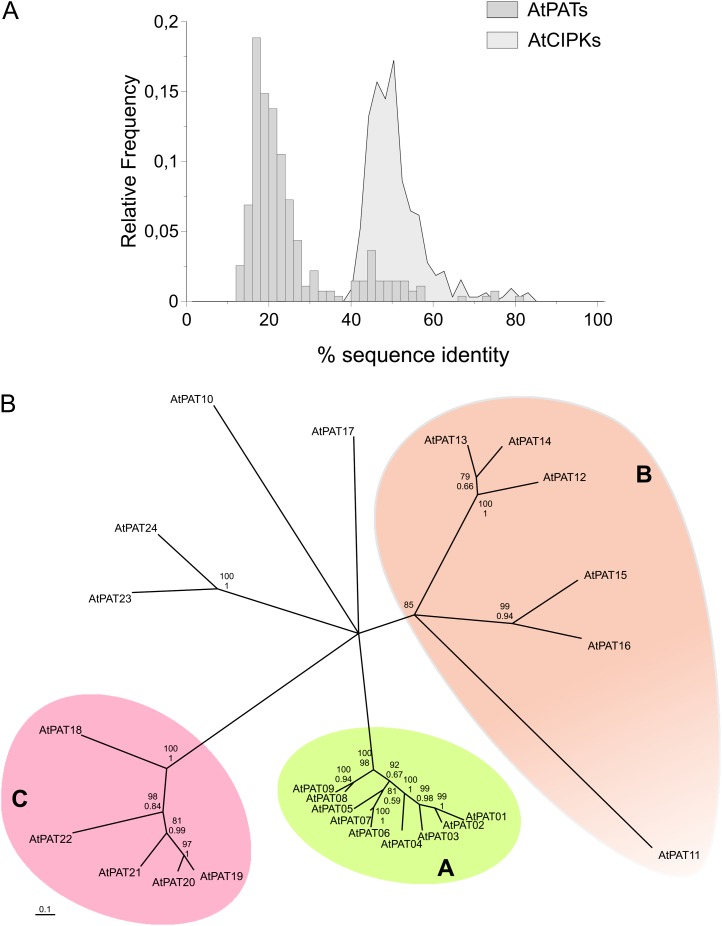
To group AtPAT23 clearly within the protein family, the phylogenetic relationship of all Arabidopsis PATs was analyzed. The degree of sequence conservation (percentage of identical amino acids) between the proteins was first determined based on a combination of alignments generated by different MAFFT algorithms. Interestingly, the overall level of sequence identity is relatively low (Fig. 1A; Supplemental Table S1) compared with other protein families like AtCIPKs (Kolukisaoglu et al., 2004; Fig. 1A). Most of the AtPAT proteins share only approximately 20% identical amino acids. The most divergent proteins AtPAT11 and AtPAT18 display only 12% identity. Overall, AtPAT11 is the least conserved protein compared with all other AtPATs (maximum 24% to AtPAT16; Supplemental Table S1). The AtPATs 6 and 7 (80% identical amino acids), 1 and 2 (73%), 8 and 9 (74%), and the AtPATs 19 and 20 (74%) displayed the highest values of identical amino acids. Less conserved in sequence is the protein pair AtPAT13 and 14 (66%). All residual proteins exhibit <60% identical amino acids. The newly classified AtPAT23 is most closely related to AtPAT24. Both proteins share nearly 50% identical amino acids but only 24% with the next related proteins AtPAT15 and 16.
Figure 1.
Sequence conservation and phylogeny of Arabidopsis PAT proteins. A, Sequence identity values (in percentages) of all Arabidopsis PAT protein pairs were determined, and the distribution (as relative frequency) of the sequence scores was plotted as a histogram (as bars). In comparison with that, the identity values obtained from the Arabidopsis CIPK protein gene family were plotted by the same method (as a filled line graph). Only a few AtPAT proteins show high sequence conservation (>60% identical amino acids), while all others share <60% identical amino acids. The sequence identities of most Arabidopsis PAT protein pairs range from only 10% to 30%. CIPK proteins mainly range from 40% to 60%. B, An NJ tree with collapsed branches (bootstrap values >75%) of Arabidopsis PAT proteins. The proteins are mainly clustered in three groups. Group A consists of the AtPATs 1 to 9, Group B of the AtPATs 11 to 16, and Group C of the AtPATs 18 to 22. The AtPATs 10, 17, 23, and 24 were not clearly positioned. AtPATs 23 and 24 are related proteins as both are on the same branch. Bootstrap values (in percentage values) are given for the respective nodes. The top number (percentage values from 0–100) represents the bootstrap factor obtained from the NJ analysis. The bottom number (percentage values given from 0–1) is the bootstrap factor obtained from a ML analysis. [See online article for color version of this figure.]
A phylogenetic tree was reconstructed by a neighbor joining (NJ) and maximum likelihood (ML) method for comparison (Fig. 1B; Supplemental Fig. S4). The unrooted NJ tree with collapsed branches (node support value >75%) is depicted in Figure 1B. This phylogenetic analysis revealed that most of the Arabidopsis PAT proteins cluster in three main clades (A, AtPATs 1–9; B, 11–16; C, 18–22). The largest and most homogenous cluster (short branch length) is composed of group A proteins. Indeed, proteins within this group preserved a rather high conservation rate (Supplemental Table S1). Moreover, this cluster contains the largest number of highly related protein pairs (AtPAT1-2, 6-7, and 8-9) and the in tandem organized AtPATs 1 and 4.
The overall grouping is mainly well supported by both phylogeny methods except for AtPAT11 within the B group (Fig. 1B; Supplemental Fig. S4). AtPAT11 was resolved on the ML tree with a rather low support value (63% on the ML tree) compared with the NJ tree (84%; Supplemental Fig. S4B). Moreover, the large branch length of AtPAT11 reflects the low sequence conservation compared with the AtPATs 12 to 16 (Supplemental Table S1).
The positions of the residual AtPAT proteins 10, 17, 23, and 24 were not clearly resolved in both phylogenetic analyses. On the NJ tree shown in Supplemental Figure S4A, the AtPATs 10 and 17 were clustered together with the AtPATs 1 to 9 but with only a very low support value. Finally, these analyses confirmed that AtPAT23 is related to AtPAT24 as both were found as a pair on a separate branch.
Taken together, the PAT proteins from Arabidopsis preserved a rather low sequence conservation. Most proteins can be classified in three different phylogenetic groups, while the residual proteins likely represent separate clades.
Identification of Conserved Sequence Motifs within Arabidopsis PATs
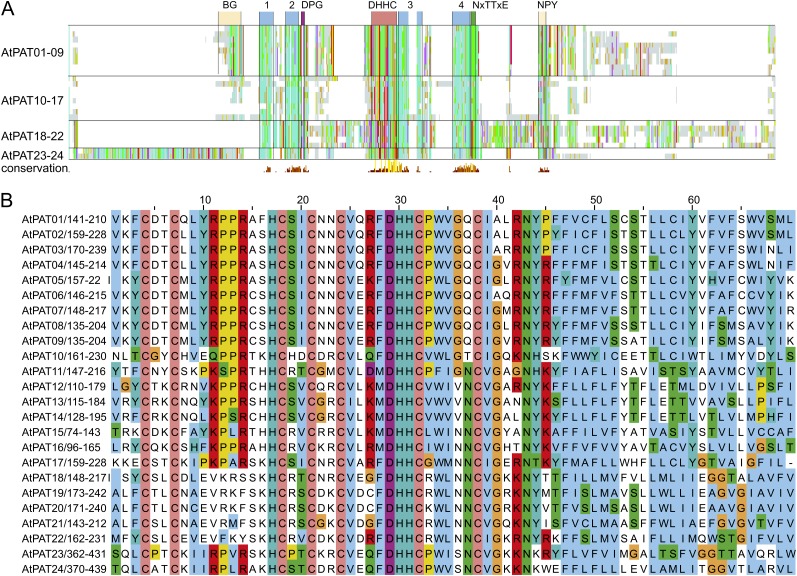
The protein alignment was further inspected visually to identify conserved sequence motifs within the Arabidopsis PATs. An alignment overview is shown in Figure 2A, and the full alignment is shown in Supplemental Figure S5. The DHHC-CRD is present in all PAT proteins (Lobo et al., 2002; Mitchell et al., 2006) and is the only highly conserved domain within all 24 Arabidopsis PAT proteins (Fig. 2A; Hemsley et al., 2005). It is composed of the central DHHC motif (positions 29–32 within the alignment section shown in Fig. 2B) and six invariant Cys residues (amino acids 4, 7, 18, 21, 24, and 38 within the alignment). Asn at position 43 seems to be a further invariant amino acid.
Figure 2.
Alignment of Arabidopsis PAT proteins. A, Alignment overview of Arabidopsis PAT proteins. Light-gray lines/bars indicate the respective position of amino acid. Conserved amino acids were decorated using the ClustalX color code. In addition, coloring was performed group-wise (AtPATs 1–9, 10–17, 18–22, and 23–24) to identify conserved blocks within subgroups. Below, the histogram displays the degree of conservation (large yellow bar represents conserved amino acids). Only the central DHHC-CRD is highly conserved in all proteins. Upstream and downstream hydrophobic stretches are indicated as blue boxes (1–4), as well as the position of short, conserved peptide sequences (DPG in purple and NxTTxE in green), which follow the second and fourth hydrophobic stretch. AtPATs 1 to 9 contain a conserved N-terminal region that contains several basic and Gly residues (BG). A less conserved region was discovered in the C-terminal region, which in most proteins consists of the amino acid sequence NPYxxGxxxN. B, Alignment of the central DHHC-CRD domain. Coloring of conserved amino acids was performed according to the ClustalX specifications. Coloring was applied to all proteins as one group. Cys residues (orange) are invariant amino acids, as well as the central DHHC peptide. The DHHC peptide is followed by a hydrophobic stretch of amino acids. Further upstream, a [KR]PPR peptide is nearly conserved in all Arabidopsis PATs but missing in the PAT group 18–22. Number range after the protein name and diagonal slash (e.g. 141–210 in AtPAT01) give the amino acid positions within the protein. Numbers above the alignment give the position of an amino acid within the alignment. [See online article for color version of this figure.]
Moreover, most Arabidopsis PAT proteins contain the [KR]PPR motif within the DHHC-CRD (positions 11–14). At least the last Arg of this motif has been shown to be important for enzyme activity (Mitchell et al., 2010). Remarkably, this sequence of amino acids is missing in the AtPATs 18 to 22. However, here, one or both of the central Pro positions were replaced by basic residues that could thereby substitute the missing Arg on position 14.
Further in-depth analysis revealed less conserved domains and sequence motifs upstream and downstream of the DHHC-CRD. Four stretches of hydrophobic residues were resolved by the alignment, each two flanking the DHHC-CRD (Fig. 2A; Supplemental Fig. S5, blue numbered boxes). Moreover, two short motifs were identified that are conserved within the PAT protein family (Mitchell et al., 2006). The amino acids DPG and Nx[TS]Tx[DE] follow the second and last hydrophobic stretch, respectively, and were preserved in most Arabidopsis PATs. The AtPATs 18 to 22 display a variation of the second motif where the Asn is missing, but they still contain the [TS]Tx[DE] stretch. Interestingly, both conserved sequences are missing in the AtPAT11 protein.
A further less conserved domain, the palmitoyltransferase conserved C terminus (composed of NP[Yx]xxGxxxN), was proposed to be required for enzyme activity (González Montoro et al., 2009). The palmitoyltransferase conserved C terminus domain was identified in most AtPAT 1 to 9 and AtPATs 23 and 24 proteins and is partially preserved in the AtPATs 10 and 12 to 17, which still harbor GxxxN (Supplemental Fig. S5). In AtPAT11 and the AtPAT 18 to 22 protein group, this motif is missing. To exclude that the domain was not recognized in the AtPATs 11 and 18 to 22 due to incorrect alignment, a manual inspection was performed, which corroborated the absence of this domain.
Further group specific motifs were revealed by the alignment. The AtPATs 1 to 9 harbor an N-terminal domain composed of several basic and Gly residues, which precede the first hydrophobic stretch (BG domain; Supplemental Fig. S5). The AtPATs 18 to 22 contain a conserved stretch that consists of the amino acids R[KR]HGWQ and shortly follows the initiator Met. In addition, a sequence overlap was observed within the large N-terminal region between the AtPATs 23 and 24 that precedes the first hydrophobic stretch.
Taken together, despite their low sequence conservation, all Arabidopsis PAT proteins preserved a common general composition and contain a conserved central DHHC-CRD flanked by two hydrophobic stretches. This arrangement is one hallmark of this enzyme class. Moreover, most PAT proteins contain short amino acid motifs that are also present in enzymes from other organisms. The AtPATs 11 and 18 to 22 represent exceptional proteins as they contain several variations or complete substitutions of conserved motifs.
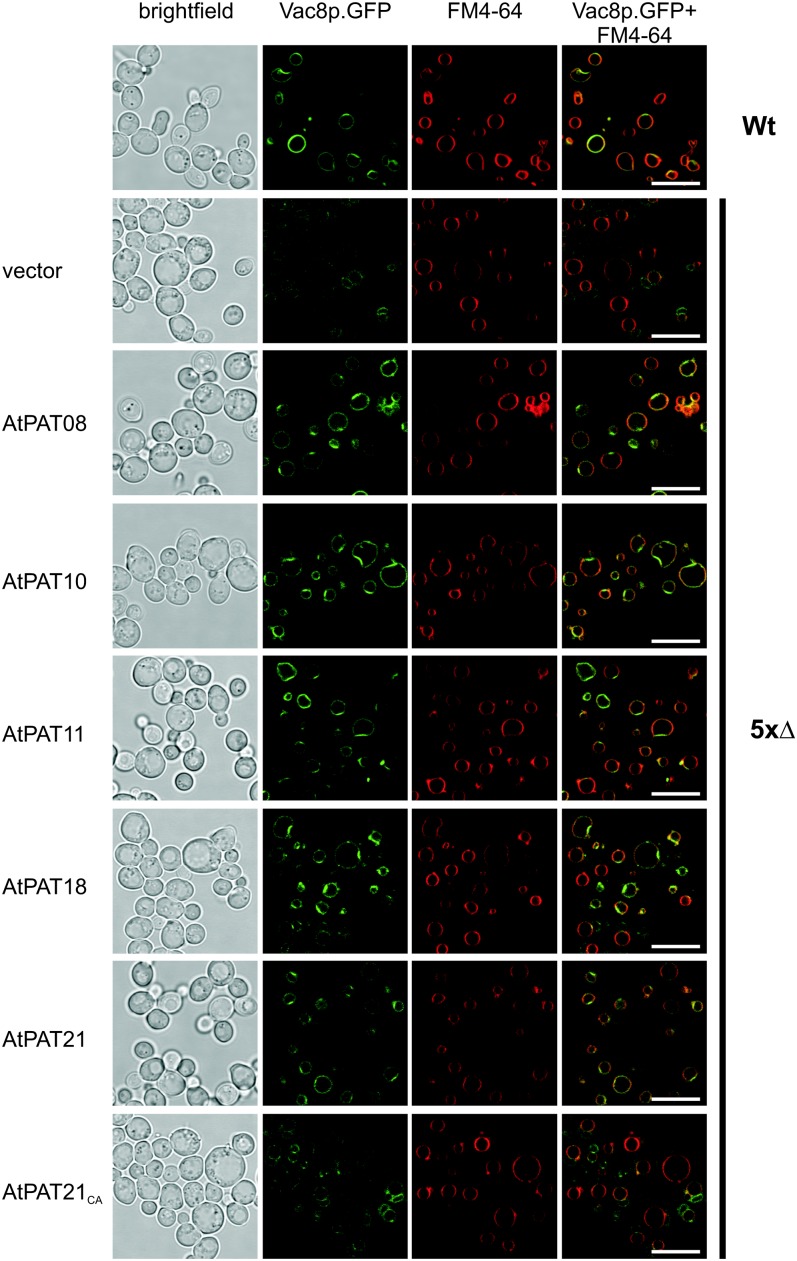
Complementation of the Vac8 Protein Localization to the Vacuolar Membrane in Mutant Yeast Cells by Arabidopsis PATs
To test the functionality of especially the PATs 11 and 18 to 22, which contain unique sequence variations, the enzyme activity of selected proteins was tested using a yeast-based complementation assay (Hou et al., 2009). S-acylation of the yeast vacuole related protein8 (Vac8p) is required for the efficient targeting to the vacuolar membrane (Wang et al., 1998; Subramanian et al., 2006). Localization studies using a quintuple PAT yeast mutant (akr1Δ, akr2Δ, pfa3Δ, pfa4Δ, and pfa5Δ; termed 5xΔ) revealed that Vac8p is not targeted to the vacuolar membrane but to ER structures (Hou et al., 2009; Fig. 3). Moreover, it was shown that overexpression of any of the five knocked-out yeast PATs can efficiently lipid modify Vac8p, which results in retargeting of the protein back to the vacuolar membrane (Hou et al., 2009).
Figure 3.
Targeting of Vac8p to the yeast vacuolar membrane in yeast mutant cells by Arabidopsis PATs. Vac8p.GFP is targeted to the vacuolar membrane in wild-type cells (Wt; indicated on the right). In the 5xΔ strain (indicated on the right) containing the empty plasmid as a control (vector, indicated on the left), Vac8p.GFP associates with ER structures. Expression of AtPATs (indicated on the left) can retarget Vac8p.GFP back to the vacuolar membrane, while the PAT21CA mutant is not able to retarget Vac8p. First row shows a bright-field picture, and the GFP fluorescence is depicted in the second row. FM4-64 staining of the yeast vacuole is shown in the third row, which was merged with the GFP fluorescence picture (shown in the last row). Bars = 10 µm. [See online article for color version of this figure.]
Therefore, the complementation of Vac8p targeting to the vacuolar membrane was tested here by the expression of Arabidopsis PATs in the yeast 5xΔ strain. In the yeast wild-type strain, Vac8p was associated with ring like structures and colocalized with FM4-64 representing the vacuolar membrane (Fig. 3; an enlargement of the GFP fluorescence is shown in Supplemental Fig. S6). In the 5xΔ yeast strain harboring the empty vector (vector 5xΔ), Vac8p was not associated with the vacuolar membrane but instead accumulated with the ER (Hou et al., 2009; Fig. 3). In contrast, expression of the Arabidopsis PATs 8 and 10 (two regular representatives) in the yeast 5xΔ cells resulted in Vac8p localization to the vacuolar membrane colocalizing with the staining of FM4-64. These data indicate that PATs from Arabidopsis can efficiently use the yeast Vac8 proteins as substrate for S-acylation. Most importantly, the AtPATs 11, 18, and 21, which represent PAT proteins with atypical sequence variations, also are able to retarget Vac8p to the vacuolar membrane (Fig. 3; Supplemental Fig. S6). As a control to investigate if this localization depends on the PAT enzyme activity, a mutant version of PAT21 was introduced (termed PAT21CA). This protein harbors a mutation within the active site (DHHC174 → DHHA exchange), which renders PAT proteins inactive (Lobo et al., 2002; Roth et al., 2002). As expected, expression of PAT21CA did not result in retargeting of Vac8p, and fluorescence was observed in structures as in the vector control strain (Fig. 3; Supplemental Fig. S6).
These results indicated that the Arabidopsis PATs 8 and 10 and especially the PATs 11, 18, and 21, which harbor several unusual sequence features, can lipid modify Vac8p and therefore represent active enzymes.
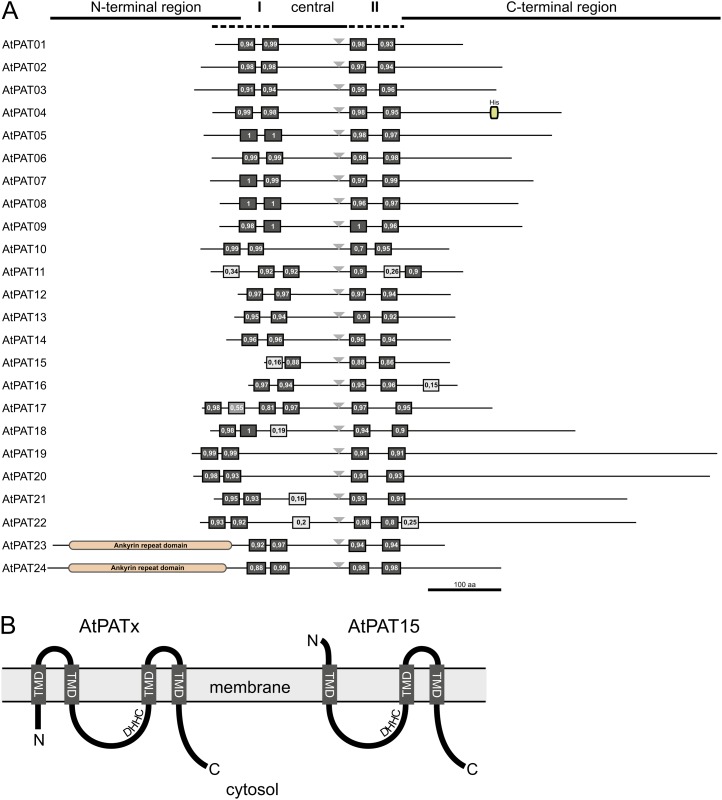
Protein Topology of Arabidopsis PATs
Most of the identified PATs from yeast and mammals contain four predicted TMDs with cytosolic N and C termini (Politis et al., 2005; Ohno et al., 2006). Also here, four hydrophobic stretches were identified in the protein alignment of the Arabidopsis PATs, which could represent TMDs. Therefore, the Aramemnon database was used to further determine the protein topology by bioinformatic analyses. These analyses revealed that most of the proteins indeed harbor four potential TMDs predicted with a high probability score (Fig. 4A). Further hydrophobic domains that were identified by the prediction algorithms in different AtPATs likely do not represent TMDs due to the low probability score (<50%).
Figure 4.
Protein topology of Arabidopsis PATs. A, Overview of the protein topology of Arabidopsis PAT proteins. Given are the relative positions of the potential TMDs and the variable N- and C-terminal regions. The central region contains the DHHC sequence (gray triangle) in the vicinity of the C-terminal TMDs. Black boxes represent predicted TMDs (with support values in percent, given from 0–1). Light-gray boxes represent further hydrophobic domains, which likely do not represent TMDs (support value <0.5). The identified ankyrin domains are shown for the AtPATs 23 and 24 in the N-terminal region. The box within the C-terminal region of AtPAT4 represents a His-rich region. Bar represents 100 amino acids. B, Proposed protein topology of Arabidopsis PATs within the membrane. Most PATs contain two TMDs upstream of the central region (AtPATx). In general, the N terminus, the central DHHC-CRD (DHHC), and the C terminus are directed to the cytosol. AtPAT15 likely contains only one predicted TMD, which is preceding the DHHC-CRD. To direct the active site to the cytosol, the N terminus has to be secreted. [See online article for color version of this figure.]
An exception of the common protein topology is likely represented by AtPAT15, which contains two N-terminal hydrophobic domains. However, only the second hydrophobic domain is highly predicted as a TMD. This potential arrangement of only one N-terminal TMD would require a secretion of the N-terminal end to allow the cytosolic positioning of the DHHC-CRD (Fig. 4B). On the other hand, AtPAT17 contains four predicted TMDs (one weakly predicted with 55%) upstream of the DHHC-CRD (Fig. 4A). However, this would still allow it to direct the active site to the cytosol as proposed for PATs with two N-terminal TMDs. A further variance in the topology was found in the AtPAT11 protein. In AtPAT11, the TMDs behind the DHHC-CRD are separated by a larger spacer, including a further hydrophobic domain.
These topology analyses also revealed the variance of the overall protein structure and within the different N- and C-terminal or central regions. Generally, the protein sizes range from 254 amino acids/29 kD in AtPAT15 to 718 amino acids/79 kD in AtPAT19 (Table I). Larger central (124–157 amino acids) domains are common especially for the AtPATs 18 to 22 and for the AtPAT10 protein (121 amino acids). The central region in the remaining proteins ranges from 68 to 101 amino acids. Despite this variance, the DHHC motif within the central region is always located to the vicinity of the C-terminal TMDs (triangle in Fig. 4A).
With the exception of the AtPATs 23 and 24, which contain extremely long N-terminal regions, the C-terminal region is in general longer than the N-terminal region. The latter region can consist of only a few amino acids (four amino acids in AtPAT17) preceding the first TMD. The size of the C-terminal region can vary from 58 amino acids (AtPAT11) to 428 amino acids (AtPAT19). This extreme C-terminal extension is a common feature of the AtPATs from the B clade, especially of the AtPATs 19 to 22.
To examine the presence of further functional domains, all PAT protein sequences were inspected using bioinformatic tools. These analyses confirmed the presence of the N-terminal ankyrin domain in AtPAT24, as previously reported (Hemsley et al., 2005). Moreover, an N-terminal ankyrin domain is also present in AtPAT23, further confirming the close relationship of AtPATs 23 and 24. For AtPAT4, a His-rich region was identified in the C-terminal region (amino acids 380–390). For the residual PAT proteins, additional domains were not identified by the database scans.
Overall, these analyses revealed that most Arabidopsis PAT proteins exhibit a common but variable protein topology. Larger structural variances are present in the N-terminal, central, and especially in the C-terminal regions of the proteins. Particularly the AtPATs 18 to 22 contain largely extended central and C-terminal domains, while the AtPATs 23 and 24 contain N-terminal extensions composed of ankyrin repeats. Additionally, AtPATs 15 and 17 differ from the residual proteins in their N-terminal topology by the number of predicted TMDs.
Expression of Arabidopsis PAT Genes
Gene expression levels and patterns can provide insights into the physiological function of their products. Often proteins with a general function are expressed at rather constant levels during development and in several tissue types, while a regulated and tissue-specific expression could argue for a more specific function of a protein (Peremyslov et al., 2011). Therefore, it was of interest to address if Arabidopsis PAT enzymes are expressed constantly or in a regulated manner. For this, the expression values were analyzed based on ATH1 chip experiments (from Genevestigator) or tiling arrays (using TileViz; see “Materials and Methods” for details).
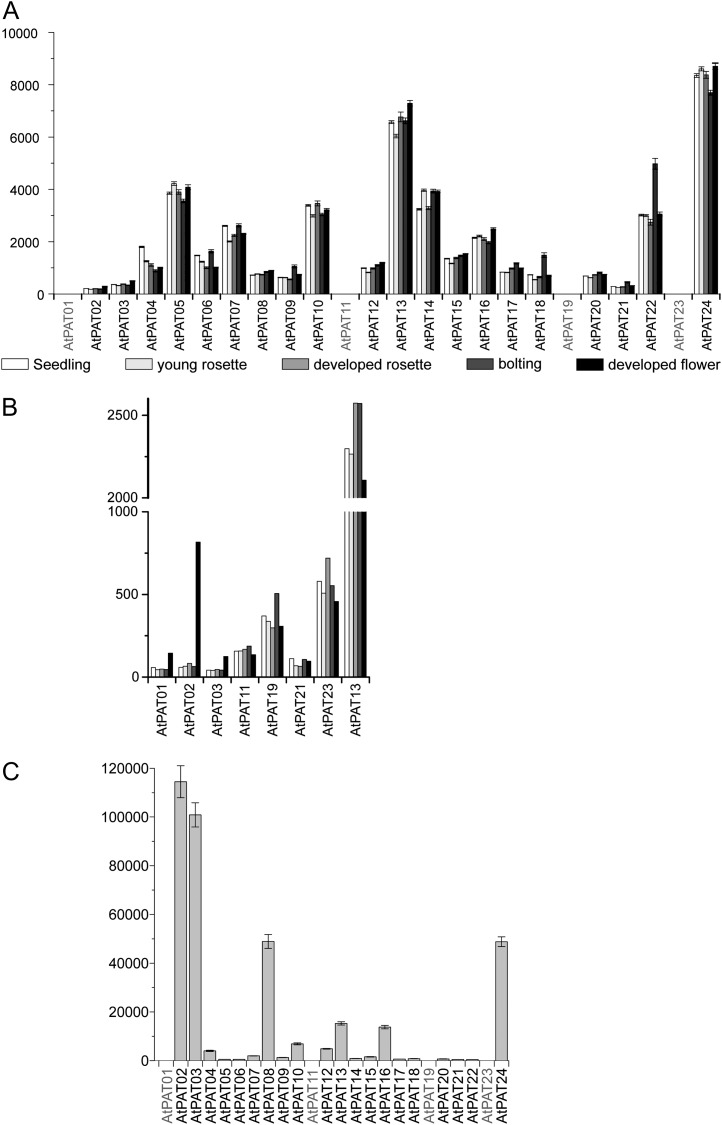
According to the expression values obtained, 19 of the 24 Arabidopsis PATs exhibited a broad and constant expression pattern with transcripts detected at different developmental stages (Fig. 5) and in most tissues (Supplemental Fig. S7). The AtPATs 18 and 22 exhibited a certain degree of regulation as expressions of both genes were stronger in bolting plants.
Figure 5.
Expression of Arabidopsis PATs. A, Global expression values of Arabidopsis PATs obtained from Genevestigator at different developmental stages (seedling, white bar; young rosette, light-gray bar; developed rosette, medium-gray bar; stem, dark-gray bar; flowers, black bar). Note that the AtPATs 1, 11, 19, and 23 are not present on the ATH1 microarray chip. Bars represent mean values. Error bars represent sd. B, Expression of the AtPATs 1, 2, 3, 11, 19, 21, and 23 was compared with the expression of AtPAT13 at different developmental stages (seedling, white bar; young rosette, light-gray bar; developed rosette, medium-gray bar; stem, dark-gray bar; flowers, black bar). Values were obtained from At-TEX using TileViz. C, Expression values of PATs in Arabidopsis pollen obtained from Genevestigator based on ATH1 microarray analysis. Bars represent mean values. Error bars represent sd.
The AtPATs 1 to 3, 11, and 21 were found to exhibit a very low expression level compared with the residual PATs (Fig. 5). In contrast, largely increased transcript levels were found for the AtPATs 1 and 3 and especially for AtPAT2 during flower development (Fig. 5B). This increase is likely due to the particular expression in stamen and pollen (at least AtPATs 2 and 3; Fig. 5C; Supplemental Fig. S6). Rather high transcript levels in pollen were also found for the AtPATs 8 and 24.
On the other hand, the expression of AtPAT21 seems to be constant during development (Fig. 5, A and B) but rather is restricted to a few cell types that would account for the low transcript abundance (Supplemental Fig. S7). Also, AtPAT9 exhibited a certain specialization as transcripts were mainly detected in tissues especially involved in reproduction (Supplemental Fig. S7).
This shows that most Arabidopsis PAT genes are constantly expressed during development and in most tissues. Only few PAT genes exhibited a specific transcription either in certain tissues and/or at a specific developmental stage.
Subcellular Localization of Arabidopsis PATs
S-acylation of proteins affects membrane binding and targeting of substrates. Therefore, the specific localization pattern of PAT proteins themselves could be important for directing proteins to the right destination (Nadolski and Linder, 2007). To gain insights into the organization of the plant S-acylation machinery, the subcellular localization of all Arabidopsis PATs was analyzed by fusing PAT proteins to GFP and transient expressions in Nicotiana benthamiana leaves.
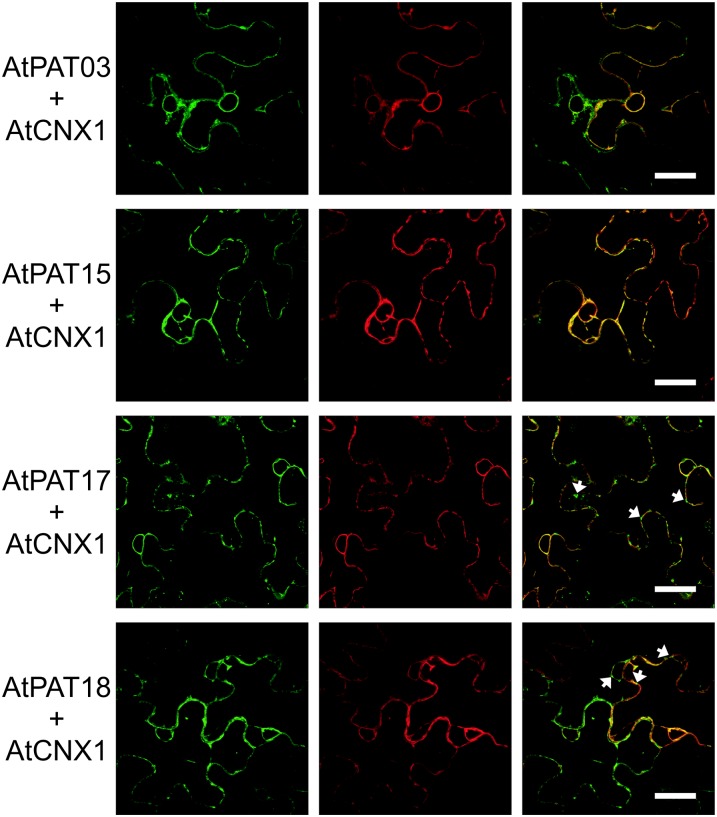
The Arabidopsis PATs 3, 15, 17, and 18 were detected on the ER, as indicated by the colocalization with the ER marker protein mCherry.Calnexin1 (AtCNX1; Irons et al., 2003; Dunkley et al., 2004; Fig. 6). In addition, the fluorescence of the GFP-fused PATs was also observed on vesicular structures that were not observed for mCherry.AtCNX1 (indicated by arrows in Fig. 6). Exemplarily, the localization of AtPATs with the ER and to the vesicles was analyzed in detail observing the fluorescence of AtPAT17.GFP for a longer incubation period (up to 5 d after infiltration). However, a differential targeting of the PAT protein during different time points was not observed corroborating the stable association with these compartments (Supplemental Fig. S8, A and B).
Figure 6.
ER localization of the Arabidopsis PATs 3, 15, 17, and 18. PAT.GFP fusions were transiently expressed in N. benthamiana leaves, and fluorescence is shown at the left. PATs were coexpressed with mCherry.CNX1 shown in the middle. The merged fluorescence is shown at the right. Arrows indicate the vesicle observed for PAT.GFP fusions. Bars, shown in the merged picture, = 20 µm. [See online article for color version of this figure.]
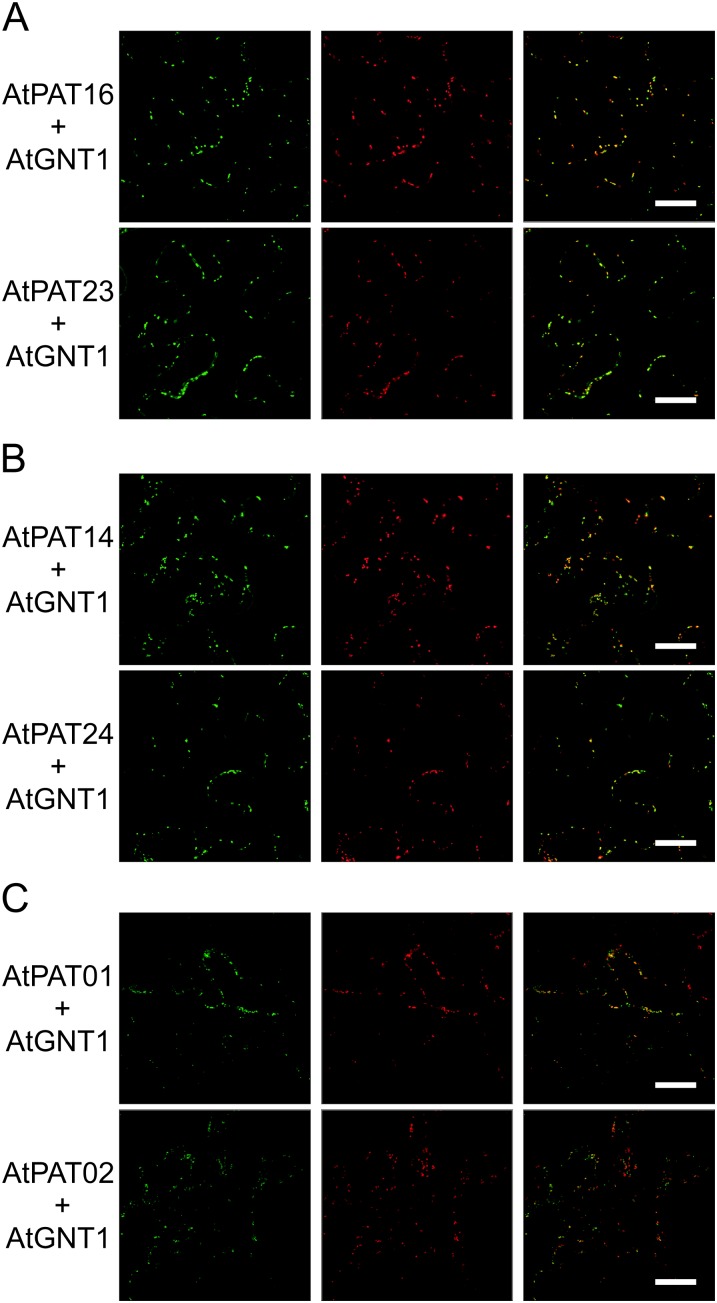
In contrast, AtPATs 16 and 23 exclusively decorated moving vesicles. Colocalization studies performed with the Golgi marker protein N-acetylglucosaminyltransferase1 from Arabidopsis (AtGNT1; Saint-Jore-Dupas et al., 2006; Reichardt et al., 2007) revealed that these vesicles represent the Golgi (Fig. 7A; Supplemental Fig. S9A). This localization was also observed for the AtPATs 14 and 24 (Fig. 7B). Remarkably, at higher magnifications, AtPAT14.GFP and AtGNT1.mCherry appeared to face opposite sites of the Golgi, suggesting that AtPAT14 is rather targeted to the trans-Golgi than to the cis-Golgi face (Supplemental Fig. S9B). Moreover, both AtPAT14 and 24 decorated additional vesicles that were smaller and separated from the AtGNT1 fluorescence (Fig. 7B; Supplemental Fig. S9B). In contrast, the two highly related proteins AtPAT1 and AtPAT2 decorated vesicular structures that colocalized with AtGNT1.mCherry only to some extent (Fig. 7C). Most AtPAT1/2-decorated vesicles were smaller and clearly separated from the Golgi vesicles (Fig. 7C; Supplemental Fig. S10A). Therefore, the smaller vesicles may represent endosomal compartments. This assumption was corroborated by comparing the fluorescence of AtPAT1 and of the coinfiltrated endosomal marker AtRAB5C/RABF1/ARA6.mCherry (Ueda et al., 2001; Supplemental Fig. S10B).
Figure 7.
Localization of the Arabidopsis PATs 16, 23, 14, 24, 1, and 2. AtPAT.GFP fusions were transiently expressed in N. benthamiana leaves (left) and coexpressed with AtGNT1.mCherry (middle). The merged fluorescence is shown at the right. Bars, shown in the merged picture, = 20 µm. [See online article for color version of this figure.]
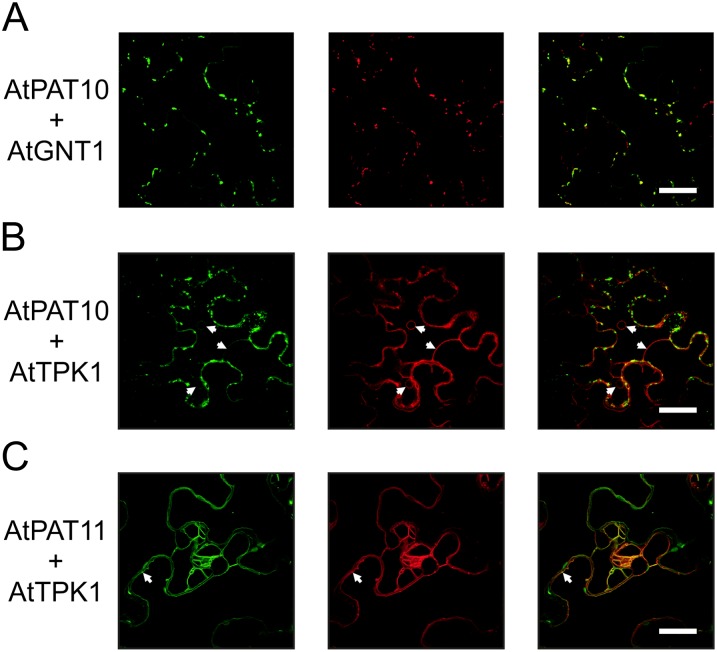
The fluorescence of PAT10.GFP was mainly observed on Golgi vesicles as revealed by colocalization studies (Fig. 8A). However, some additional fluorescence was consistently observed at a large membrane structure after different incubation periods (after 2 d, Fig. 8B; up to 6 d, Supplemental Fig. S11, A–C). Colocalization analysis with the Arabidopsis Two-pore-K+ channel1 (AtTPK1).mCherry revealed that this membrane represents the vacuolar membrane (Fig. 8B; Supplemental Fig. S11A; Batistič et al., 2010). Another protein that localized to the tonoplast was AtPAT11 (Fig. 8C). Additional colocalization analysis of the AtPAT11.GFP protein with CNX1 corroborated that the observed fluorescence was clearly separated from the ER membrane (Supplemental Fig. S11D). However, it should be noted that some of the AtPAT11.GFP fluorescence was detected at small vesicles. These vesicles were also observed for AtTPK1-mCherry and therefore could represent a prevacuolar compartment (Fig. 8C).
Figure 8.
Localization of the Arabidopsis PATs 10 and 11. AtPAT.GFP fusions were transiently expressed in N. benthamiana leaves, and fluorescence is shown at the left. AtPAT10 was coexpressed with GNT1.mCherry (A) and TPK1.mCherry (B). A higher gain was used to enhance green fluorescence in (B) compared with (A) to better display the vacuolar membrane (arrows). AtPAT11 was coexpressed with TPK1.mCherry (C). Both decorated the vacuolar membrane and vesicles (arrows). All coexpressed mCherry fused proteins are shown in the middle. The merged fluorescence is shown at the right. Bars, shown in the merged picture = 20 µm. [See online article for color version of this figure.]
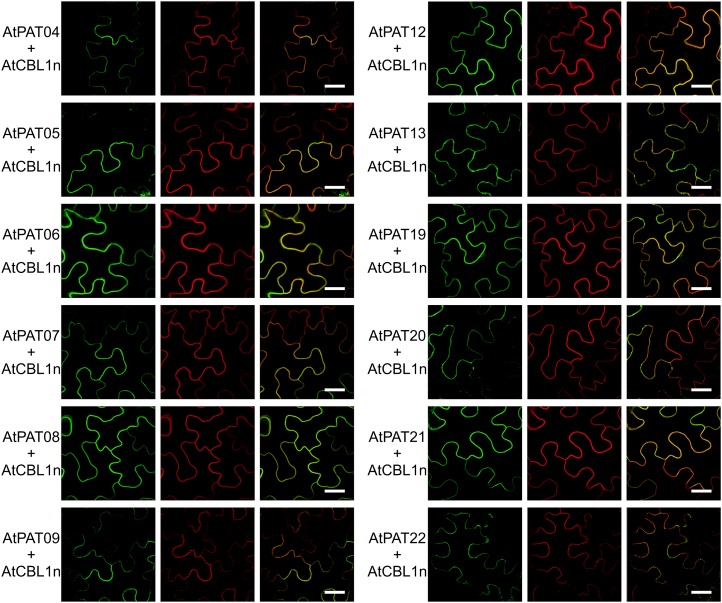
The remaining PATs 4 to 9, 12, 13, and 19 to 22 all decorated the PM membrane, as revealed by colocalization studies using CBL1n.OFP (for orange fluorescent protein; Batistič et al., 2010; Fig. 9). Some additional localization on vesicles was observed for AtPAT13.GFP, which did not colocalize with the Golgi (Supplemental Fig. S12). Similarly, these vesicles were also observed for the AtPATs 20 and 22 (Fig. 9).
Figure 9.
PM localizations of the Arabidopsis PATs 4 to 9, 12-13, and 19 to 22. The Arabidopsis PATs fused to GFP (left) were transiently expressed in N. benthamiana leaves with CBL1n.OFP (middle). The merged fluorescence is shown at the right. Bars, shown in the merged picture, = 20 µm. [See online article for color version of this figure.]
Together, these studies of the subcellular localization of Arabidopsis PATs revealed a complex pattern of the distribution of these proteins at several cellular membranes (summarized in Table II; Supplemental Fig. S13). Most PAT proteins displayed PM localization, which mainly originated from the A and C clades of the protein family. The other proteins were detected at the ER, Golgi, endosomal compartments, and at the vacuolar membrane. Moreover, these studies revealed that proteins within a phylogenetic subgroup are not targeted homogenously and even related proteins, as the AtPATs 1/2 and 3 or the AtPATs 13 and 14 can be associated with different membranes.
Table II. Summary of Arabidopsis PAT localizations.
| Localization | Arabidopsis PAT |
|---|---|
| ER (and vesicles) | AtPAT03 |
| AtPAT15 | |
| AtPAT17 | |
| AtPAT18 | |
| Vesicles (Golgi) | AtPAT10 |
| AtPAT16 | |
| AtPAT23 | |
| Vesicles (Golgi and non-Golgi) | AtPAT14 |
| AtPAT24 | |
| Vesicles (non-Golgi) | AtPAT01 |
| AtPAT02 | |
| PM and vesicles | AtPAT13 |
| AtPAT20 | |
| AtPAT22 | |
| PM | AtPAT04 |
| AtPAT05 | |
| AtPAT06 | |
| AtPAT07 | |
| AtPAT08 | |
| AtPAT09 | |
| AtPAT12 | |
| AtPAT19 | |
| AtPAT21 | |
| Tonoplast | AtPAT10 |
| AtPAT11 |
DISCUSSION
DHHC-CRD-containing proteins mediate the S-acylation of proteins. However, our current knowledge about the plant S-acylation machinery is still very limited. Arabidopsis contains overall 24 expressed gene loci, which is similar to the number of PATs (22) found in humans (Ohno et al., 2006). Phylogenetic analysis revealed that most of the Arabidopsis proteins grouped in three main clusters. The AtPATs 10, 17, and the AtPATs 23/24 were not grouped into one of the main groups and therefore could form independent groups. In addition, although AtPAT11 branched together with the AtPATs 12 to 16, this position is uncertain. Therefore, AtPAT11 could form a further separate group distantly related to AtPATs 12 to 16. These analysis also revealed highly related pairs (AtPATs 1-2, 6-7, and 8-9 within clade A and AtPATs 19-20 within clade C), which likely arose by recent gene duplication events. Moreover, the gene loci of AtPAT1 and AtPAT4 were found to be directly organized in tandem. This suggests that both arose by a tandem duplication event, although sequence conservation between these two proteins is relatively low (53% identity). However, low sequence conservation of tandem duplicated genes also is common in other gene families (Kolukisaoglu et al., 2004), implicating that several tandem duplications arose rather early in evolution.
Remarkably, the sequence conservation between the Arabidopsis PAT proteins is exceptional low. In addition, the three main clusters display different rates of conservation. This indicates an overall early and differential evolution of the three main groups. Of these, clade A represents the largest and most homogenous one. It is likely that genes within the A clade evolved later in evolution, which would explain the larger degree of sequence conservation. Moreover, the high number of duplicated gene pairs within this clade further reflects a selection process that somehow favored the evolution of clade A genes. Such a process could be the evolution of reproductive organs like flowers in higher plants. In accordance with this, several clade A genes were found to be preferentially expressed in flower organs.
Although protein sequences can largely vary within this enzyme class, the overall topology and general domain structure of PAT proteins is preserved throughout different organisms (Ohno et al., 2006). This was also predicted here for the Arabidopsis PATs by bioinformatic analysis. Most proteins contain four hydrophobic domains that likely represent TMDs. They are flanked by N-terminal and C-terminal regions and separated by the central conserved DHHC-CRD, which represents the active site of these enzymes (Figs. 2 and 3). In general, the N- and C-terminal and central domains are thereby arranged to the cytosol (Politis et al., 2005). Major topological variations were observed for the AtPATs 15 and 17. AtPAT17 contains four predicted N-terminal TMDs, while AtPAT15 likely harbors only a single predicted N-terminal TMD. AtPAT15 would therefore require a secreted N terminus to arrange the DHHC-CRD domain back into the cytosol (Fig. 4B). On the other, it could be possible that all N-terminal hydrophobic domains in AtPAT15 represent TMDs similar to the arrangement found for the other PATs. Hence, the topology of the PATs 15 and 17 should be analyzed experimentally in more detail in the future.
While the overall topology of the Arabidopsis PAT proteins was mainly preserved, the different cytosolic domains vary in size and amino acid composition. Variations were found especially in the subgroup that includes the Arabidopsis PATs 18 to 22. These proteins contain largely extended C-terminal regions as well as central domains. Moreover, the active site of the AtPATs 18 to 22 contains a unique sequence variation that was not found in other Arabidopsis PATs. A further exceptional protein represents AtPAT11, which lacks several sequence motifs that are otherwise preserved in this protein family, like the DPG and NxTTxE sequence. However, functional analysis of these proteins in yeast PAT mutant cells revealed a complementation of the Vac8 protein targeting to the vacuolar membrane, indicating that PAT11 and proteins from the 18-22 group represent active enzymes. This implicates that the conserved domains that are missing in PAT11 or the sequence variations found in PATs 18 to 22 are not necessarily required for function. Therefore, these variations could point to a so far not recognized variability in the enzymatic mechanism within this protein family or to a yet unknown mechanistic specialization specific to these enzymes. Essentially, how target specificity and activity of PAT proteins is regulated within a cell is currently not well known. The C-terminal regions could be required for correct targeting of the proteins, as shown for human enzymes (Greaves et al., 2011). Moreover, it was assumed that the C-terminal regions could act as scaffolding domains for substrates or for accessory proteins that regulate enzyme function (Yang et al., 2010). Interestingly, the scaffolding function could be independent of enzyme function as recently reported. The PAT DHHC5 from mouse contains a largely extended C-terminal region. This region is sufficient for interaction with the postsynaptic density protein-95 (PSD-95). However, DHHC5 seems to be not responsible for lipid modification of PSD-95, implicating that DHHC5 is mediating a scaffolding function for PSD-95 and thereby modulating synaptic functions (Li et al., 2010). This would extend the role of PATs beyond the known lipid modification of proteins. Therefore, it is tempting to speculate if plant PATs also mediate such scaffolding functions via their variable C-terminal regions to regulate targeting of proteins, besides or in addition to their enzymatic activity. Moreover, the large domain variations of the PAT proteins allow the assumption that they mediate substrate specificity, the specific regulation by putative accessory proteins, and/or the targeting of the enzymes.
To address potential roles of the variable domains, different protein motif identification algorithms were applied. However, these analyses revealed classical domains in only three proteins of the family. AtPAT4 contains a His-rich region within the C-terminal domain. Such regions can bind metals and can be required for enzyme activity (Kasher et al., 1993). AtPAT23 and AtPAT24 both contain ankyrin repeats within the large N-terminal region. Ankyrin domains are known to mediate protein-protein interactions (Li et al., 2006), as shown for the yeast ankyrin repeat containing PAT Akr1 with the γ-subunit of the yeast heterotrimeric G protein (STE18). However, this interaction does not result in an enzymatic modification of STE18 by Akr1 (Hemsley and Grierson, 2011). On the other hand, the enzymatic modification of the Akr1 substrate yeast caseine kinase2 does not require the presence of the ankyrin repeats. Therefore, the N-terminal ankyrin repeats in Akr1, and in AtPAT23/24, may fulfill a scaffolding function similar to the C terminus of the mouse DHHC5 protein (Hemsley and Grierson, 2011). The related AtPAT23 and AtPAT24 may modify similar targets and fulfill similar functions. However, the partial differential localization pattern of these related PATs observed in this study are suggestive for certain trend to specialization between the two PATs. Taken together, these analyses established the close structural and phylogenetic relationships between AtPAT23 and AtPAT24 and corroborated that Arabidopsis contains two functional ankyrin repeat PATs as also observed in mammalians and yeasts.
Protein function can be further specified by a timely and spatially regulated expression. The expression values of most Arabidopsis PATs appear rather constant and occur in most tissues analyzed. This may reflect the general requirement of PATs and S-acylation for cellular function and suggests that a large number of targets exist that require lipid modifications. However, certain Arabidopsis PATs exhibited an exceptional high expression level especially in pollen. This included AtPAT24, which is known to be required for pollen development and growth (Hemsley et al., 2005). Therefore, a similar role could be proposed for AtPATs that displayed high transcript abundance during flower development and in pollen, respectively. This implicates that certain substrates that are required for pollen function are lipid modified by PATs.
The most intriguing feature of the Arabidopsis S-acylation machinery is its diverse cellular organization when compared with yeast and mammalian cells. In yeast, three of the seven PAT proteins are localized to the ER, two to the Golgi, and one PAT to the PM and the vacuolar membrane. In Drosophila melanogaster and mammalian cells, almost all of the PAT proteins reside at the ER and the Golgi (Ohno et al., 2006; Bannan et al., 2008). This was corroborated by in situ assays to analyze the cellular activity of the S-acylation machinery directly in mammalian cells. These analyses revealed that most of the PAT activity resides at the Golgi and implicated that the targeting of proteins to the PM or to other membranes is mainly mediated by Golgi PATs (Rocks et al., 2010). Here and in contrast to yeast and mammalians, most of the Arabidopsis PAT proteins identified reside at the PM (overall 12 proteins) and only seven at the Golgi and/or endosomal compartments (Table II). The large number of PM-targeted PATs implicates that the plant S-acylation machinery is largely modifying substrates at the PM either after the targeting of proteins to this destination and/or by a direct trapping from the cytoplasm. By this, several lipid modified proteins could be directly bound to the PM and would bypass vesicular pathways to the PM. This would be in contrast to the S-acylation machinery in animal cells where the lipid modification of proteins mainly occurs at the Golgi for the subsequent vesicle-mediated targeting to the PM or to other compartments (Rocks et al., 2010). Moreover, it is tempting to speculate that the PM PATs are further involved in several dynamic lipid modification processes that influence protein-protein interactions or protein functions (Gubitosi-Klug et al., 2005). These dynamic modifications could regulate the localization of signaling proteins into subdomains of the PM as described for the AtROP6 protein (Sorek et al., 2007, 2010). Therefore, it can be expected that there are many more proteins in plants that are rather dynamically regulated by PAT enzymes at the PM.
Four of the Arabidopsis PATs were observed to localize at the ER. All four proteins belong to different phylogenetic subclasses. This may indicate a certain diverse function of the ER-targeted enzymes that is further supported by the differential expression patterns observed (e.g. AtPAT3 in pollen). Besides being targeted to the ER, all these proteins were detected additionally at vesicular structures. This could either reflect a concentration of the PAT proteins to ER export sites or to Golgi vesicles, implicating that these proteins could have a certain function in the regulated exit of substrates from the ER.
The AtPAT proteins 10 and 11 were identified to reside at the vacuole. AtPAT10 was previously identified as a vacuolar protein by a mass spectrometric-based proteomic approach corroborating the observed localization (Jaquinod et al., 2007). The association of PAT enzymes with the vacuolar membrane allows direct trapping of target proteins to the tonoplast by lipid modifications. Such a mechanism was recently proposed for tonoplast-associated CBL proteins (Batistič et al., 2012). In addition, these proteins could also function in targeting of proteins into potential membrane microdomains of the tonoplast similar to the role of the vacuolar membrane-targeted PAT Pfa3 in yeast cells (Hou et al., 2005; Peng et al., 2006). However, it should be noted that AtPAT10 was additionally detected at Golgi vesicles, suggesting a dual function of this enzyme.
Dual localizations were observed for several proteins (Table II). Similarly, several mammalian PATs exhibited a dual ER/Golgi or Golgi/PM localization (Ohno et al., 2006; Bannan et al., 2008). Such a dual localization could simply reflect the continuous cycling of proteins between different membranes. On the other hand, the dual localization of PAT proteins could be required for the correct function. In fact, target proteins, like the mammalian Ras proteins and the Man 6-P receptor, which cycle between membranes, are known to be lipid modified by enzymes that target to different membranes (Stöckli and Rohrer, 2004; Rocks et al., 2005; Baekkeskov and Kanaani, 2009). Such function appears to be conceivable for the plant PAT proteins that therefore require the observed dual localizations.
Collectively, the results of this study provide insights into the variable structural composition, phylogenetic relationship, and functionality of the Arabidopsis PATs. The expression of most proteins is not regulated developmentally and occurs in most plant tissues. This supports the conclusion that PATs are required for general cellular functions. Nevertheless, few PATs seem to fulfill a particular role according to their specific expression profiles. Finally, the PAT proteins were found at different cellular membranes. Remarkably, most enzymes were targeted to the PM. This suggests that the cellular S-acylation machinery in plants is functionally different compared with other organisms. In summary, the insights from this report provide an initial framework for further evolutionary and functional studies of this protein family.
MATERIAL AND METHODS
BLASTP Searches, Protein Alignments, Phylogenetic and Protein Topology Analysis
BLASTP searches were performed using the Arabidopsis (Arabidopsis thaliana) PAT8 protein sequence as a query (Altschul et al., 1990). This allowed direct identification of the central DHHC-CRD in the obtained sequences and to immediately exclude non-PAT proteins from further analysis. BLASTP searches were performed using the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) and Phytozome databases (http://www.phytozome.net/) by restricting the search to Arabidopsis.
Protein alignments were performed using a combination of two MAFFT algorithms. First alignment was performed using the L-INS-i setting (Katoh and Toh, 2008), which is recommended for proteins with one conserved domain and large gaps. The second alignment was performed using the G-INS-i setting, which is recommended for proteins with global homology. Both alignments were combined using the COMBINE program provided by T-COFFEE (Poirot et al., 2003). The resulting protein alignment was used for subsequent analysis using Jalview (Waterhouse et al., 2009) and for phylogenetic analyses. A protein identity matrix was calculated from the alignment using the PFAAT program (Caffrey et al., 2007).
Phylogenetic tree analysis were performed using the NJ method from the MEGA program package version 5.01 (Kumar et al., 2008) using a JTT substitution matrix with a γ substitution rate of 4. Tree robustness was tested using a bootstrapping method with 1,000 replicates. To generate a tree by ML, phylogenetic analysis was performed using the PALM pipeline (http://palm.iis.sinica.edu.tw/), which automatically tests different settings to provide the best ML tree (Chen et al., 2009). Tree robustness was further tested using a bootstrap method with 100 replicates (automatically restricted by the pipeline). The best tree provided by PALM was used in this analysis, and the tree was visualized using the MEGA program.
Protein topology was determined using the Aramemnon database, which provides a consensus transmembrane topology generated from 10 different TMD prediction algorithms (Schwacke et al., 2003). PrositeScan (http://expasy.org/tools/scanprosite/) and InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/) were used to identify further known protein domains (McDowall and Hunter, 2011).
Gene Expression and Protein Localization Analysis in Plants and Yeast Cells
All PAT ORFs were amplified using either Pwo- (Peqlab) or Pfu-Polymerase (Agilent Technologies) and inserted in front of the GFP reading frame behind the Mannopine Synthase gene promoter within a pGPTVII backbone plasmid (Walter et al., 2004). Schematic representation of the expression cassette, primers used for PCR amplification, and template resources are given in Supplemental Materials and Methods S1 online. The expression values of the PAT genes from seedlings, young and old rosette leaves, bolting plants, and flowers were obtained and analyzed using Genevestigator (https://www.genevestigator.com) based on ATH1 chip analysis. Expression values for the PAT genes 1, 2, 3, 11, 13, 19, and 23 were additionally obtained from Arabidopsis Tiling Array Express (Laubinger et al., 2008) using TileViz (http://jsp.weigelworld.org/tileviz/tileviz.jsp). A tissue expression heat map was obtained from Genevestigator.
Nicotiana benthamiana plants for infiltration experiments were cultivated under a 12-h-light/12-h-dark cycle with 60% atmospheric humidity at 20°C in a greenhouse. Transient in planta protein expression was performed in N. benthamiana leaves that were infiltrated with the Agrobacterium tumefaciens strain GV31010 pMP90 in combination with the 19K helper strain (Waadt and Kudla, 2008). Agrobacteria were transformed with the respective plasmids by electroporation. For leaf infiltration A. tumefaciens were prepared as described previously (Waadt and Kudla, 2008). Leaf samples were analyzed 2 to 3 d (maximum 6 d) after infiltration.
For yeast (Saccharomyces cerevisiae) complementation analysis of the Vac8 protein targeting, a strain was used that lacks five of the seven PAT enzymes (Roth et al., 2006; Hou et al., 2009). Within this strain (5xΔ), and in the respective wild-type strain, Vac8p.GFP expression is driven from the genomic locus (Hou et al., 2009). To overexpress Arabidopsis PATs, the 5xΔ strain was transformed with plasmids, containing the respective PAT reading frames, and selected on synthetic dropout media lacking the amino acid Leu. Expression of Arabidopsis PATs is driven by the constitutive ADH promoter from a 2µ-containing plasmid, which is a derivative of the pGAD.GH plasmid. The fragment of the activation domain from the pGAD.GH plasmid was excised and replaced by the multiple cloning site from the pGPTVII plasmids (Walter et al., 2004) to enable insertion of the PAT reading frames. For microscopy analysis, yeast cells were grown in a shaker overnight in respective media at 28°C (yeast peptone dextrose for wild-type strains and selective synthetic dropout-Leu media for transformed 5xΔ cells). Cells were harvested for 1 min at 4,000g at room temperature and resuspended in fresh media supplemented with 20 µM FM4-64 (from a 20 mm stock solution dissolved in water). Yeast cells were incubated under shaking for 20 min for pulse-chase uptake of FM4-64 (Molecular Probes Life Technologies), again harvested and resuspended in fresh media lacking FM4-64. For vacuolar staining, the cells were analyzed after an incubation period of 1 h.
Fluorescence microscopy of plant epidermal cells was performed with an inverted microscope (Leica DMIRE2 and Leica DMI6000) equipped with the Leica TCS SP2 or SP5 confocal laser scanning device, respectively (Leica Microsystems). Detection of fluorescence was performed as follows: GFP, excitation at 488 nm (Ar/Kr laser) and scanning at 500 to 535 nm; OFP, excitation at 543 nm (He/Ne laser; SP2 system) or 561 nm (SP5 system) and scanning at 565 to 595 nm. All images were acquired using a Leica 63×/1.20 water immersion objective (HCX PL Apo CS). Yeast cells were microscopically analyzed using the following laser settings: GFP, excitation at 488 nm (Ar/Kr laser) and scanning at 500 to 560 nm; FM4-64 excitation at 488 to 561 nm and scanning at 590 to 650 nm. Images were acquired using a Leica 100×/1.44 oil immersion objective (HCX PLAPO).
All laser scanning confocal micrographs presented in this study are single optical sections and were processed using either the Leica Imaging software or using the Corel Photo Paint program.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of Arabidopsis PAT gene loci.
Supplemental Figure S2. Protein sequence alignment of AtPAT9 isoforms.
Supplemental Figure S3. PCR on cDNAs and transcript structure of AtPAT23.
Supplemental Figure S4. Comparison of the Arabidopsis PAT phylogeny by a neighbor joining and maximum likelihood approach.
Supplemental Figure S5. Alignment of Arabidopsis PAT proteins.
Supplemental Figure S6. Targeting of Vac8p in yeast mutant cells by AtPATs.
Supplemental Figure S7. Expression heat map of Arabidopsis PATs.
Supplemental Figure S8. Colocalization studies of Arabidopsis PAT17.
Supplemental Figure S9. Colocalization studies of selected PATs.
Supplemental Figure S10. Colocalization studies of selected PATs.
Supplemental Figure S11. Colocalization studies of selected PATs.
Supplemental Figure S12. Colocalization studies of AtPAT13 and AtGNT1.
Supplemental Figure S13. Overlay of the phylogenetic relation and localization of Arabidopsis PATs.
Supplemental Table S1. Protein identity matrix of Arabidopsis PATs.
Supplementary Material
Acknowledgments
I thank Dr. Dirk Becker (University of Würzburg) for providing pollen-specific cDNA, Christian Eckert and Anette Mähs (University of Münster) for root and shoot cDNA, The Arabidopsis Stock Centre and RIKEN for providing cDNA clones, and Drs. Piers Hemsley and Clair Grierson (University of Bristol) for providing the TIP1 plasmid. The yeast strains (Vac8.GFP in the wild type and 5xΔ) were kindly provided by Thomas Arun and Dr. Christian Ungermann (University of Osnabrück). I thank Christoph Baumann and Lukas Wallrad (University of Münster) for experimental assistance and Dr. Jörg Kudla (University of Münster) for support, helpful discussions, and for critical reading of the article.
Glossary
- PAT
protein S-acyltransferase
- DHHC
Asp-His-His-Cys
- CRD
Cys-rich domain
- ER
endoplasmic reticulum
- PM
plasma membrane
- ORF
open reading frame
- NJ
neighbor joining
- ML
maximum likelihood
- TMD
transmembrane domain
- NCBI
National Center for Biotechnology Information
- cDNA
complementary DNA
References
- Abrami L, Leppla SH, van der Goot FG. (2006) Receptor palmitoylation and ubiquitination regulate anthrax toxin endocytosis. J Cell Biol 172: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjobo-Hermans MJ, Goedhart J, Gadella TW., Jr (2006) Plant G protein heterotrimers require dual lipidation motifs of Gα and Gγ and do not dissociate upon activation. J Cell Sci 119: 5087–5097 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Baekkeskov S, Kanaani J. (2009) Palmitoylation cycles and regulation of protein function (Review). Mol Membr Biol 26: 42–54 [DOI] [PubMed] [Google Scholar]
- Bannan BA, Van Etten J, Kohler JA, Tsoi Y, Hansen NM, Sigmon S, Fowler E, Buff H, Williams TS, Ault JG, et al. (2008) The Drosophila protein palmitoylome: characterizing palmitoyl-thioesterases and DHHC palmitoyl-transferases. Fly (Austin) 2: 198–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Rehers M, Akerman A, Schlücking K, Steinhorst L, Yalovsky S, Kudla J. (2012) S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res 22: 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Sorek N, Schültke S, Yalovsky S, Kudla J. (2008) Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20: 1346–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Waadt R, Steinhorst L, Held K, Kudla J. (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Caffrey DR, Dana PH, Mathur V, Ocano M, Hong EJ, Wang YE, Somaroo S, Caffrey BE, Potluri S, Huang ES. (2007) PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. BMC Bioinformatics 8: 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S, Manié S, Oualid M, Billard M, Boucheix C, Rubinstein E. (2002) Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett 516: 139–144 [DOI] [PubMed] [Google Scholar]
- Chen SH, Su SY, Lo CZ, Chen KH, Huang TJ, Kuo BH, Lin CY. (2009) PALM: a paralleled and integrated framework for phylogenetic inference with automatic likelihood model selectors. PLoS ONE 4: e8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley TP, Watson R, Griffin JL, Dupree P, Lilley KS. (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol Cell Proteomics 3: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Flannery AR, Czibener C, Andrews NW. (2010) Palmitoylation-dependent association with CD63 targets the Ca2+ sensor synaptotagmin VII to lysosomes. J Cell Biol 191: 599–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Montoro A, Quiroga R, Maccioni HJ, Valdez Taubas J. (2009) A novel motif at the C-terminus of palmitoyltransferases is essential for Swf1 and Pfa3 function in vivo. Biochem J 419: 301–308 [DOI] [PubMed] [Google Scholar]
- Greaves J, Carmichael JA, Chamberlain LH. (2011) The palmitoyl transferase DHHC2 targets a dynamic membrane cycling pathway: regulation by a C-terminal domain. Mol Biol Cell 22: 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitosi-Klug RA, Mancuso DJ, Gross RW. (2005) The human Kv1.1 channel is palmitoylated, modulating voltage sensing: Identification of a palmitoylation consensus sequence. Proc Natl Acad Sci USA 102: 5964–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. (1999) Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol 50: 97–131 [DOI] [PubMed] [Google Scholar]
- Hemsley PA. (2009) Protein S-acylation in plants (Review). Mol Membr Biol 26: 114–125 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Grierson CS. (2008) Multiple roles for protein palmitoylation in plants. Trends Plant Sci 13: 295–302 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Grierson CS. (2011) The ankyrin repeats and DHHC S-acyl transferase domain of AKR1 act independently to regulate switching from vegetative to mating states in yeast. PLoS ONE 6: e28799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Kemp AC, Grierson CS. (2005) The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 17: 2554–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Taylor L, Grierson CS. (2008) Assaying protein palmitoylation in plants. Plant Methods 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, John Peter AT, Meiringer C, Subramanian K, Ungermann C. (2009) Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism. Traffic 10: 1061–1073 [DOI] [PubMed] [Google Scholar]
- Hou H, Subramanian K, LaGrassa TJ, Markgraf D, Dietrich LE, Urban J, Decker N, Ungermann C. (2005) The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8. Proc Natl Acad Sci USA 102: 17366–17371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Hardin SC. (2004) Numerous posttranslational modifications provide opportunities for the intricate regulation of metabolic enzymes at multiple levels. Curr Opin Plant Biol 7: 318–322 [DOI] [PubMed] [Google Scholar]
- Irons SL, Evans DE, Brandizzi F. (2003) The first 238 amino acids of the human lamin B receptor are targeted to the nuclear envelope in plants. J Exp Bot 54: 943–950 [DOI] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J. (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics 6: 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasher MS, Wakulchik M, Cook JA, Smith MC. (1993) One-step purification of recombinant human papillomavirus type 16 E7 oncoprotein and its binding to the retinoblastoma gene product. Biotechniques 14: 630–641 [PubMed] [Google Scholar]
- Katoh K, Toh H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistič O, Kudla J. (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L, García-Mata C, Graziano M, Pagnussat G. (2003) Nitric oxide: the versatility of an extensive signal molecule. Annu Rev Plant Biol 54: 109–136 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Zeller G, Henz SR, Sachsenberg T, Widmer CK, Naouar N, Vuylsteke M, Schölkopf B, Rätsch G, Weigel D. (2008) At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol 9: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mahajan A, Tsai MD. (2006) Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45: 15168–15178 [DOI] [PubMed] [Google Scholar]
- Li Y, Hu J, Höfer K, Wong AM, Cooper JD, Birnbaum SG, Hammer RE, Hofmann SL. (2010) DHHC5 interacts with PDZ domain 3 of post-synaptic density-95 (PSD-95) protein and plays a role in learning and memory. J Biol Chem 285: 13022–13031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo S, Greentree WK, Linder ME, Deschenes RJ. (2002) Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem 277: 41268–41273 [DOI] [PubMed] [Google Scholar]
- Luan S. (2003) Protein phosphatases in plants. Annu Rev Plant Biol 54: 63–92 [DOI] [PubMed] [Google Scholar]
- McDowall J, Hunter S. (2011) InterPro protein classification. Methods Mol Biol 694: 37–47 [DOI] [PubMed] [Google Scholar]
- Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. (2010) Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem 285: 38104–38114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. (2006) Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res 47: 1118–1127 [DOI] [PubMed] [Google Scholar]
- Miura K, Hasegawa PM. (2010) Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol 20: 223–232 [DOI] [PubMed] [Google Scholar]
- Nadolski MJ, Linder ME. (2007) Protein lipidation. FEBS J 274: 5202–5210 [DOI] [PubMed] [Google Scholar]
- Ohno Y, Kihara A, Sano T, Igarashi Y. (2006) Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta 1761: 474–483 [DOI] [PubMed] [Google Scholar]
- Peng Y, Tang F, Weisman LS. (2006) Palmitoylation plays a role in targeting Vac8p to specific membrane subdomains. Traffic 7: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Peremyslov VV, Mockler TC, Filichkin SA, Fox SE, Jaiswal P, Makarova KS, Koonin EV, Dolja VV. (2011) Expression, splicing, and evolution of the myosin gene family in plants. Plant Physiol 155: 1191–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot O, O’Toole E, Notredame C. (2003) Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res 31: 3503–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis EG, Roth AF, Davis NG. (2005) Transmembrane topology of the protein palmitoyl transferase Akr1. J Biol Chem 280: 10156–10163 [DOI] [PubMed] [Google Scholar]
- Reichardt I, Stierhof YD, Mayer U, Richter S, Schwarz H, Schumacher K, Jürgens G. (2007) Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr Biol 17: 2047–2053 [DOI] [PubMed] [Google Scholar]
- Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, et al. (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141: 458–471 [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307: 1746–1752 [DOI] [PubMed] [Google Scholar]
- Roth AF, Feng Y, Chen L, Davis NG. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol 159: 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, III, Davis NG. (2006) Global analysis of protein palmitoylation in yeast. Cell 125: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan E, Grierson CS, Cavell A, Steer M, Dolan L. (1998) TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol 138: 49–58 [Google Scholar]
- Saint-Jore-Dupas C, Nebenführ A, Boulaflous A, Follet-Gueye ML, Plasson C, Hawes C, Driouich A, Faye L, Gomord V. (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18: 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J, Galway M, Masucci J, Ford S. (1993) Pollen tube and root-hair tip growth is disrupted in a mutant of Arabidopsis thaliana. Plant Physiol 103: 979–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N, Bloch D, Yalovsky S. (2009) Protein lipid modifications in signaling and subcellular targeting. Curr Opin Plant Biol 12: 714–720 [DOI] [PubMed] [Google Scholar]
- Sorek N, Poraty L, Sternberg H, Bar E, Lewinsohn E, Yalovsky S. (2007) Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol Cell Biol 27: 2144–2154 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sorek N, Segev O, Gutman O, Bar E, Richter S, Poraty L, Hirsch JA, Henis YI, Lewinsohn E, Jürgens G, et al. (2010) An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr Biol 20: 914–920 [DOI] [PubMed] [Google Scholar]
- Stöckli J, Rohrer J. (2004) The palmitoyltransferase of the cation-dependent mannose 6-phosphate receptor cycles between the plasma membrane and endosomes. Mol Biol Cell 15: 2617–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulemeijer IJ, Joosten MH. (2008) Post-translational modification of host proteins in pathogen-triggered defence signalling in plants. Mol Plant Pathol 9: 545–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K, Dietrich LE, Hou H, LaGrassa TJ, Meiringer CT, Ungermann C. (2006) Palmitoylation determines the function of Vac8 at the yeast vacuole. J Cell Sci 119: 2477–2485 [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A. (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas J, Pelham H. (2005) Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J 24: 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Kudla J. (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc 2008: pdb.prot4995 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistič O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang YX, Catlett NL, Weisman LS. (1998) Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol 140: 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. (2010) Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics 9: 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Wang X, Running MP. (2007) Dual lipid modification of Arabidopsis Gγ-subunits is required for efficient plasma membrane targeting. Plant Physiol 143: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.