Abstract
Crop plant development is strongly dependent on the availability of nitrogen (N) in the soil and the efficiency of N utilization for biomass production and yield. However, knowledge about molecular responses to N deprivation derives mainly from the study of model species. In this article, the metabolic adaptation of source leaves to low N was analyzed in maize (Zea mays) seedlings by parallel measurements of transcriptome and metabolome profiling. Inbred lines A188 and B73 were cultivated under sufficient (15 mm) or limiting (0.15 mm) nitrate supply for up to 30 d. Limited availability of N caused strong shifts in the metabolite profile of leaves. The transcriptome was less affected by the N stress but showed strong genotype- and age-dependent patterns. N starvation initiated the selective down-regulation of processes involved in nitrate reduction and amino acid assimilation; ammonium assimilation-related transcripts, on the other hand, were not influenced. Carbon assimilation-related transcripts were characterized by high transcriptional coordination and general down-regulation under low-N conditions. N deprivation caused a slight accumulation of starch but also directed increased amounts of carbohydrates into the cell wall and secondary metabolites. The decrease in N availability also resulted in accumulation of phosphate and strong down-regulation of genes usually involved in phosphate starvation response, underlining the great importance of phosphate homeostasis control under stress conditions.
Maize (Zea mays) represents the most widely cultivated crop plant; it is used for food as well as feedstock and in increasing degree also as a bioenergy crop. The C4 photosynthesis allows very efficient conversion of CO2 into carbohydrates and finally green biomass and yield, especially under conditions of maximal nitrogen (N) supply (for review, see Ghannoum et al., 2011). However, further yield improvements by additional N supply are limited. Excessive use of nitrogenous fertilizers is expensive and leads to increased leakage of N into the environment where it pollutes groundwater reserves and disturbs neighboring ecosystems. Plant breeders are therefore under pressure to provide crop varieties with improved N use efficiency for the future (Hirel et al., 2007). Development of such lines would prevent the need for further increases of N input into industrial agricultural systems and would also allow more stable yields on poorer soils in developing countries.
Nitrate is the main inorganic nitrogenous compound in agricultural soils, and deficiency of nitrate leads to growth retardation and yield losses. However, plants can adapt to limitations in N supply by improving the efficiency of the nitrate uptake system in the root (e.g. an increase in the number of roots and improvement of biochemical properties of the nitrate transporters) and by improving the N utilization efficiency within the plant (Hirel et al., 2001, 2007). Variation in crop performance under N limitation is mainly connected to differences in N utilization efficiency (Hirel et al., 2001). Accumulation of green biomass during vegetative growth is dependent on the source leaves’ capacity for carbon (C) and N assimilation, while remobilization of stored N becomes the dominating pathway during grain filling (Hirel et al., 2001). Improving the efficiency of N use in the source leaf would increase the biomass produced per unit of N (Lawlor, 2002). This strategy would be connected to higher C:N ratios in the plant material, and this could be particularly interesting for the development of specific energy crop lines, where protein content of the final plant material is less important than C content (Torney et al., 2007).
C and N metabolism are highly interconnected (for review, see Nunes-Nesi et al., 2010). N assimilation requires the availability of energy and reducing power as well as C skeletons for incorporation of inorganic N into organic compounds, and in the illuminated leaf, these are provided by photosynthesis. The photosynthetic apparatus, on the other hand, is the main sink for incorporation of N compounds in the leaf. In C3 plants, Rubisco accounts for up to 60% of the leaf protein content. C4 plants, such as maize, can concentrate CO2 at the site of Rubisco activity and need to invest much less N into the production of the Rubisco protein for efficient C assimilation. They have increased proportions of C shuttle proteins and more thylakoids (Ghannoum et al., 2011). Comparison of mRNA sequencing data from source leaves of closely related C3 and C4 species could show that the steady state level of amino acid and protein biosynthesis transcripts is lower in C4 than C3 leaves (Bräutigam et al., 2011). In the end, the C4 shuttle allows higher rates of C assimilation per unit of N. Changes in N supply, however, would also affect the biosynthesis of all other N-containing compounds in the C4 leaf, including amino acids, proteins, polyamines, nucleic acids, lipids, tetrapyrrols, phenylpropanoids, terpenoids, and hormones. In maize plants, N availability affects the chlorophyll content of the leaves; in fact, measurements of the chlorophyll content have reliably been used as estimates for the N content of the leaf (Chapman and Barreto, 1997). Fluorescence measurements showed that the photosystems were generally intact under low-N conditions, but under high illumination, the photosynthetic rate of N-deficient maize was reduced (Lu and Zhang, 2000). This was caused by a feedback inhibition of photosynthesis due to decreased demand for ATP and NADPH in nitrate reduction and other biosynthetic processes (Khamis et al., 1990; Lu and Zhang, 2000). The decrease in chlorophyll is accompanied by considerable reduction in the most abundant proteins of the maize leaf: phosphoenolpyruvate (PEP) carboxylase (PEPC), pyruvate orthophosphate dikinase (PPDK), and Rubisco (Sugiharto et al., 1990). Despite the reduction in C-assimilating enzymes, plants accumulate C under low-N conditions in the form of starch. This can be partly explained by the reduced demand of C skeletons for amino acid assimilation. In maize, starch accumulation is particularly pronounced in the youngest leaves of plants during grain filling (Hirel et al., 2005). The increased biosynthesis of storage carbohydrates is usually connected to a decrease in sink strength in other parts of the plants caused by nutrient stress-related growth retardation of the shoot (Paul and Driscoll, 1997; Hirel et al., 2005). An increase in root growth under N limitation can only partly compensate for the reduction in shoot growth, and under severe N stress, root growth in maize is also inhibited (El-Kereamy et al., 2011). In the end, the reduction in plant sink strength causes feedback reduction of photosynthesis (Paul and Pellny, 2003).
Roots are responsible for active mineral uptake into the plant. Besides being an essential nutrient, nitrate also acts as a signal molecule. This allows roots to react fast to changes in the nitrate availability. Addition or removal of nitrate from roots causes almost instant changes in root transport systems (Wang et al., 2003; Krapp et al., 2011). So far, only two high affinity nitrate transporter have been characterized in maize; ZmNRT2.1 seems responsible for nitrate uptake by the epidermis and ZmNRT2.2 for loading of nitrate into the xylem (Trevisan et al., 2008). Nitrate arrives in the leaves with the xylem flow and is taken up into the leaf cells by another set of nitrate transporters (Dechorgnat et al., 2011). Once in the cytosol, nitrate is then reduced by nitrate reductase to nitrite, which is further transported into the plastid and converted into ammonium by nitrite reductase. The reduction of nitrate requires supply of energy and reducing equivalents; in the nonphotosynthetically active tissue, these come from the oxidative pentose phosphate (OPP) pathway and in the chloroplast from photosynthesis. In the C4 plant maize, nitrate and nitrite reductases could be localized in the mesophyll (Neyra and Hageman, 1978; Becker et al., 2000; Friso et al., 2010), where reducing equivalents are available from primary photosynthetic reactions. Nitrate reduction activity decreases under low N supply but does not seem to represent a limiting step since correlation between nitrate reductase activity and biomass accumulation of different maize lines is negative (Hirel et al., 2001, 2005). Ammonium is incorporated into the first organic compounds by Gln synthetase (GS) and Glu synthase (GOGAT), and in contrast to nitrate reduction, the enzymes for ammonium assimilation seem to be present in both maize leaf cell types (Friso et al., 2010). Especially the activity of the GS has been identified as a key enzyme for N metabolism under low nitrate supply (Hirel et al., 2001). Biosynthesis of most amino acids seems to be concentrated in the mesophyll cell (Friso et al., 2010). In field experiments, total amino acid and protein content of leaves were clearly reduced by N limitation (Hirel et al., 2005).
Our knowledge about metabolic adaptation to N deficiency in crop plants is mainly limited to investigation of photosynthesis and primary N assimilation pathways. However, the central role of the N status requires rearrangements in many further parts of plant metabolism. Large-scale microarray analysis of N status-dependent changes is now available from Arabidopsis (Arabidopsis thaliana; Wang et al., 2000, 2003; Price et al., 2004; Scheible et al., 2004; Bi et al., 2007; Gutiérrez et al., 2007; Krapp et al., 2011), rice (Oryza sativa; Lian et al., 2006), and maize (Yang et al., 2011). These identified numerous N status-responsive transcripts involved in the nitrate transport and assimilation, the OPP pathway, amino acid metabolism, organic acid metabolism, tetrapyrrole biosynthesis, major carbohydrate, and protein, lipid, and secondary metabolism to changes in the N status of the plant (Wang et al., 2000, 2003; Scheible et al., 2004; Bi et al., 2007; Krapp et al., 2011) but also many genes with unknown roles in adaption to low-N stress. Most of the microarray experiments have been done on plant material deriving from a mixture of different plant parts and tissues; this could complicate the identification of specific responses to N deprivation. The different tasks of specialized plant tissues, such as roots, source leaves, young sink leaves, or senescing leaves, would require very different adaptation strategies to low N (Gifford et al., 2008; Krapp et al., 2011).
In this article, we focus on the metabolism of the source leaf during vegetative growth as the site of C and N assimilation for the synthesis of building blocks for biomass production. Since many key enzymes of N and C metabolism are further influenced by posttranslational regulation (Morcuende et al., 2007) and control of activity by the proximate metabolic environment (Paul and Foyer, 2001), the plant material is further analyzed by metabolite profiling. N deficiency causes considerable shifts in the metabolite profiles of plants; amino acids tend to decrease and sugar and secondary metabolites tend to increase under low N supply in leaves from tobacco (Nicotiana tabacum; Fritz et al., 2006a, 2006b), tomato (Solanum lycopersicum; Urbanczyk-Wochniak and Fernie, 2005), and Arabidopsis (Tschoep et al., 2009; Krapp et al., 2011). However, metabolite data are under strong influence of environmental components, and this complicates comparison between different experiments (Massonnet et al., 2010). In order to reduce environmental perturbations to a minimum, plants were cultivated in the highly controlled climate of a growth chamber. With the combined analysis of transcriptional and metabolite responses from the same biological material, it will be possible to assess the influence of N deficiency on different levels of plant metabolism. Correlation analysis of transcript and metabolite data will help to identify the role of genes in a specific metabolic environment. The molecular analysis is further supplemented by phenotypic characterization of the maize seedlings under investigation.
RESULTS AND DISCUSSION
Plant Growth under N Starvation
Limited availability of N fertilizer affects plant growth and metabolism. Therefore, detailed phenotypic characterization preceded sampling of material for transcripts and metabolite analysis. Because we were interested in the general metabolic response pattern to N limitation stress, two inbred lines, A188 and B73, were selected for the study. Both lines have been established as model genotypes in maize research. B73 represents the inbred line used for the sequencing project (Schnable et al., 2009), and A188 is the most common line used in transformation experiments. The two genotypes can be distinguished by differences in their developmental pattern with A188 belonging to the early and B73 belonging to the medium maturation type. However, early development of both lines is comparable. Under control growth conditions (high N), no significant differences could be detected for leaf appearance, leaf growth rate, and shoot biomass accumulation for the time of the experiment (Table I). Seedlings were cultivated in pots with basic peat substrate and watered with Hoagland nutrient solution containing either 15 mm (high-N conditions) or 0.15 mm nitrate (low-N conditions). Leaf growth was monitored continuously by measurements of leaf length. At germination, seedlings still utilize nutrients from the seed, but with increasing plant size, N demand increases. In our experiment, growth rates started to decline in both genotypes under low N about 16 d after germination. Plants were harvested at two different developmental stages. At 20 d after germination, leaf 5 represented the main source leaf and was harvested for analysis of metabolite and transcript profiles. At this stage, plants were mildly affected by the N treatment, and low-N treatment caused a reduction of 20% to 25% in leaf elongation rate and of 25% to 30% in fresh shoot biomass accumulation (Table I). Over the next 10 d, the leaf elongation rate of plants grown under ample N increased only slightly, but their shoot biomass increased about 5-fold (Table I). At the second harvest at 30 d after germination, plants from both treatments had developed leaf 6, a leaf displaying already adult leaf features. At this stage, the shoot biomass in N-stressed plants was 70% lower when compared with plants supplied with 15 mm nitrate in the nutrient solution, indicating severe stress-related growth retardation (Table I). This was mainly due to cessation of growth in young leaves and leaf appearance. Leaf elongation rate was slightly more affected by long-term N deprivation in A188 than in B73. However, for the duration of the experiment, N stress caused very similar biomass reduction in both genotypes. Senescence of the small older leaves had only minor influence on overall biomass reduction.
Table I. Influence of N supply on biomass accumulation and leaf growth rate.
| Genotype, Plant Age | Shoot Biomass (g) |
Leaf Growth Rate (cm d−1) |
||||
|---|---|---|---|---|---|---|
| High Na | Low Na | %b | High Na | Low Na | %b | |
| A188, 20 d | 12.09 ± 1.840 | 7.66 ± 0.602 | 63.36 | 6.59 ± 0.200 | 4.90 ± 0.144 | 74.36 |
| B73, 20 d | 12.35 ± 1.711 | 7.26 ± 0.726 | 58.79 | 5.86 ± 0.968 | 4.86 ± 0.577 | 82.93 |
| A188, 30 d | 52.89 ± 3.732 | 17.63 ± 2.123 | 33.32 | 7.01 ± 0.333 | 3.86 ± 0.371 | 55.06 |
| B73, 30 d | 59.30 ± 3.854 | 18.34 ± 1.557 | 30.93 | 6.54 ± 1.126 | 4.34 ± 0.718 | 66.36 |
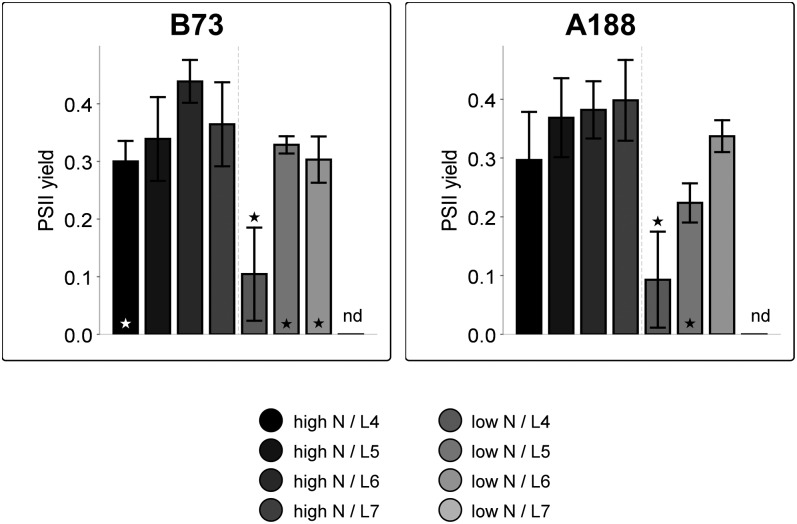
Since we were particularly interested in the metabolic events in source leaves under N limitation, the photosynthetic contribution of different leaves was estimated by measurements of effective PSII quantum yield via a Pulse Amplitude Modulation fluorometer (Mini-PAM; Heinz Walz) 1 d before the harvest at 30 d. At this time point, eight to nine leaves were visible in nonstressed plants, while only the seventh leaf had appeared in the plants grown under N limitation. Effective PSII quantum yield was high in all measured leaves (L4–L7) of high-N-treated plants, indicating that all leaves had intact photosystems and presumably contributed to production of basic molecules for further growth (Fig. 1). In comparison, effective PSII quantum yield was significantly reduced in leaf four of low-N-treated plants, indicating that breakdown of photosynthetic apparatus has already started for remobilization of N-containing compounds. Leaf 7 had not yet expanded in these plants and could not be measured in low-N-treated plants. Growth was therefore dependent on assimilatory power of only two leaves. In comparison with high-N-grown plants, effective PSII quantum yield of leaf 5 was slightly reduced in genotype B73 and even more reduced in genotype A188 (Fig. 1). Results from leaf 6 indicated that this leaf had high photosynthetic capacity even under low-N conditions in both genotypes, and it was harvested for further analysis of source leaf metabolism. Plant material was always harvested in the morning, 2 h after the beginning of the light period, when C and N assimilation are usually high. Phenotypic characterization and molecular analysis were performed on two independent biological experiments, with always two to three individual plants used for microarray and metabolite analysis. The phenotypic response (shoot biomass and growth rate) to N starvation was very similar in both genotypes, and no significant differences could be detected for the tested treatments and growth stages.
Figure 1.
Effective PSII quantum yield of maize leaves at 28 d after germination. Plants were cultivated under control (high-N) or low-N conditions. The asterisk indicates significant differences in comparison with L6 from high-N-grown plants. Fluorescence could not be measured in L7 from low-N-treated plants. nd, Not determined. All data points represent average from four plants ± sd.
Statistical Analysis of Transcript and Metabolite Data
Transcript and metabolite data were analyzed by three statistical methods. Principal component analysis (PCA) revealed the influence of different parameters onto the data set, and differential regulation allowed the identification of features that are statistically different between the two N treatments and their classification into functional groups. Correlation analysis finally organized transcript data into modules with similar expression pattern, and these modules could then be connected to similar pattern in metabolite and phenotype data.
PCA
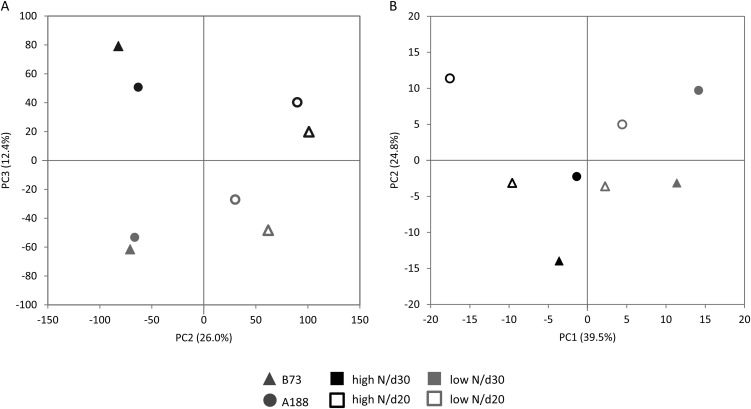
Effects of nitrate starvation on source leaf metabolism were studied in different genotypes and at different developmental stages. The influence of all three parameters could be clearly separated by PCA (Fig. 2A; Supplemental Fig. S1.1). Genotype-specific differences had the greatest influence on the microarray data set (PC1 [for principal component], 46.0%; Supplemental Fig. S1.1), followed by the developmental stage (PC2, 26.0%; Fig. 2A). The strong influence of genotypic background on transcriptome pattern has been described for other species, such as barley (Hordeum vulgare; Kogel et al., 2010). Separation of data by developmental stage can be explained by the different age at time of harvest and the fact that early leaves of maize show differences in morphology, anatomy, cell wall composition, and transcriptome (Strable et al., 2008). The harvested source leaves, numbers 5 and 6, belonged to the intermediate leaves with changing degrees of juvenile and adult leaf characteristics. N treatment represented the third component of PCA analysis (12.4%; Fig. 2A). PCA loading associated with low-N treatment include transcripts for amino acid and peptide transport and for secondary metabolism (e.g. chalcone synthase and isomerase). Under the top 50 of low-N leaf loadings are three transcription factors and three UDP-glucosyl transferases (Supplemental Table S1). High PCA loadings in the high-N sample would indicate that these transcripts are down-regulated under low-N conditions. In the top 50 list are many transcripts associated with inorganic phosphate (Pi) metabolism and regulation (e.g. three different SPX (for SYG1, Pho81, XPR1) domain proteins, four phosphatases, two phosphodiesterases) and six further kinases and proteins for modification of phosphorylated metabolites, such as 3-phosphoglycerate permease, phosphoglycerate mutase, and nucleotide-diphospho-sugar transferase (Supplemental Table S1). The highest loading value was found for the type B monogalactosyldiacetylglycerol synthase, a gene involved in lipid biosynthesis, but also influenced by Pi starvation in Arabidopsis (Kobayashi et al., 2004). Several transcripts involved in N metabolism also showed high-N-specific loadings, e.g. the Arg decarboxylase, which is involved in polyamine biosynthesis, a nitrate reductase, and the Ser-O-acetyltransferase. An MYB- and a lateral organ boundary (LOB)-type transcription factor also possessed strong high-N-specific loadings.
Figure 2.
PCA. A, Calculated from 41,780 transcripts (PC2 versus PC3). B, Calculated from 531 metabolites peaks (PC1 versus PC2).
Compared with the transcript data, metabolite profiles were much more affected by the N supply. The first principal component (39.5%) clearly separated samples from different N treatments; additionally, the component was influenced by the harvest time (Fig. 2B). Metabolites with PC1-specific loadings are shown in Supplemental Table S1. Not surprisingly, N-containing metabolites, such as amino acids, contributed strongly to the separation of this component, and they were associated with high-N conditions. On the other hand, Pi and some sugars and secondary metabolites were specific for low-N-related principal component loadings. Therefore, availability of N has a very immediate effect on the metabolite pattern of plant material, while the transcriptional control exhibits only the first step in a regulatory network and is still strongly influenced by the genotypic background. However, genotypic differences also influenced the metabolite pattern and represent the second principal component (24.8%; Fig. 2B).
Differential Regulation
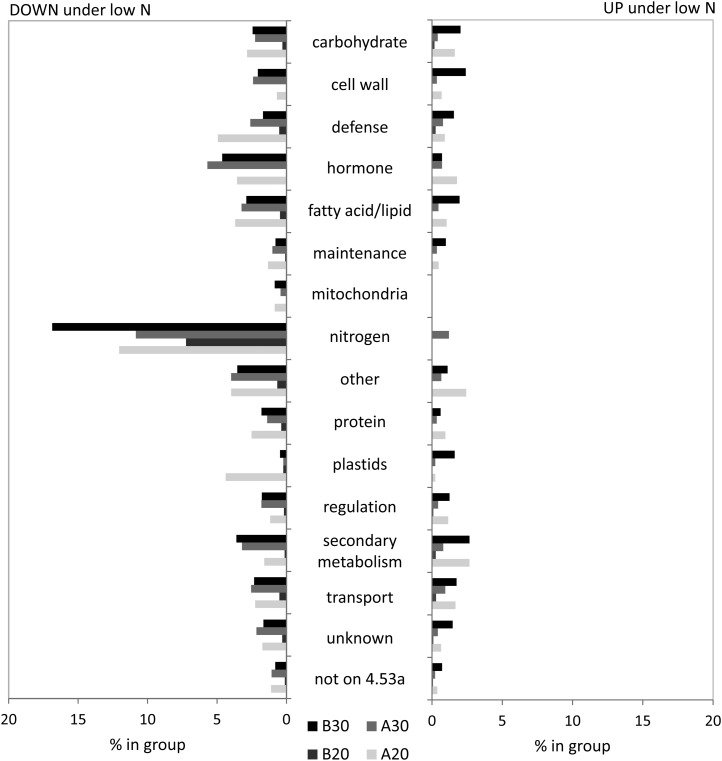
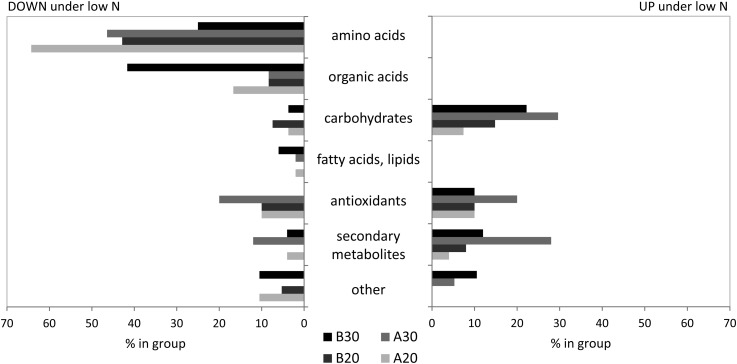
N status-related changes were the main focus of this study, and transcript and metabolite profiles were searched for candidates with significant differences in low- and high-N-treated leaves. In time-course experiments with Arabidopsis, the number of differentially regulated transcripts increased with the duration of the N stress (Krapp et al., 2011). The same pattern was observed in genotype B73, where 144 (109 down/35 up) and 1,208 (684 down/524 up) transcripts were significantly different at early and long-term sampling, respectively (Table II). Only a few of those seemed transiently regulated, and about 60% of sequences affected at 20 d were also significantly different at 30 d. All of these transcripts were regulated into the same direction at both developmental stages (Supplemental Table S2). Genotype A188 showed strong reaction to the early stress, and a similar number of sequences was affected at the two sampling times (748 down/346 up at 20 d; 780 down/169 down at 30 d; Table II). Under long-term N stress conditions, 343 transcripts were down-regulated and 65 transcripts were up-regulated in both genotypes (Table II). This indicated that independent of the genotype background, the transcripts show an N treatment-specific expression pattern. All sequences detected by the microarray chip had been aligned with the corresponding locus on the B73 genome version 4a.53, annotated and classified into functional groups. Figure 3 shows the behavior of all significantly regulated transcripts from these functional groups under low-N conditions compared with high-N conditions in the different genotypes at the analyzed growth stages. Transcripts involved in N metabolism were most affected, and their abundance in the leaves decreased under low-N conditions for both developmental stages and genotypes. Transcripts from defense, hormone, mitochondria, fatty acid, and protein-related metabolism also tended to be down-regulated (Fig. 3). In most other groups, small proportions of up- and down-regulated transcripts could be found. On the metabolite level, N deficiency also caused significant reduction in many amino acids and related compounds (Fig. 4). Organic acids and fatty acids were also reduced under low-N conditions. Under long-term stress, amino acids were thereby more affected in A188, while organic acids showed stronger changes in B73. Carbohydrates and secondary metabolites, on the other hand, accumulated significantly under low-N stress.
Table II. Number of transcripts differentially regulated by low- versus high-N conditions.
Significance threshold was set at fold change of > 2 and false discovery rate of P < 0.05.
| Comparison | A188 Lamina |
B73 Lamina |
A188∩B73 |
|||
|---|---|---|---|---|---|---|
| Down | Up | Down | Up | Down | Up | |
| Low versus high Na | 731 | 175 | 543 | 168 | 366 | 54 |
| D20 low versus high Nb | 748 | 346 | 109 | 35 | 75 | 18 |
| D30 low versus high Nc | 780 | 169 | 684 | 524 | 343 | 65 |
| D20∩30 low versus high Nd | 305 | 38 | 74 | 11 | 45 | 8 |
Data averaged over samples harvested at 20 and 30 d after germination.
Data only from samples harvested at 20 d after germination.
Data only from samples harvested at 30 d after germination.
Intersection of transcripts significantly different at 20 and 30 d after germination.
Figure 3.
Transcripts differentially regulated by low- versus high-N conditions. Transcripts are grouped according to participation in main biological processes.
Figure 4.
Metabolites significantly different in leaves under low- versus high-N conditions. Metabolites are grouped according main compound classes.
Weighted Correlation Network Analysis
Correlation analysis allows the organization of transcript data from individual plants into coexpression modules, and the module eigengenes (MEs) can then be related to metabolite pattern and phenotype characterization. The weighted correlation network analysis (WGCNA) tool was developed specifically for the identification of gene coexpression networks and has so far been applied to transcriptome data from rice and maize (Ficklin et al., 2010; Ficklin and Feltus, 2011). For our experiment, we were interested in identification of modules containing genes with generally high correlation to specific metabolic processes and modules with a strong N-treatment-specific pattern.
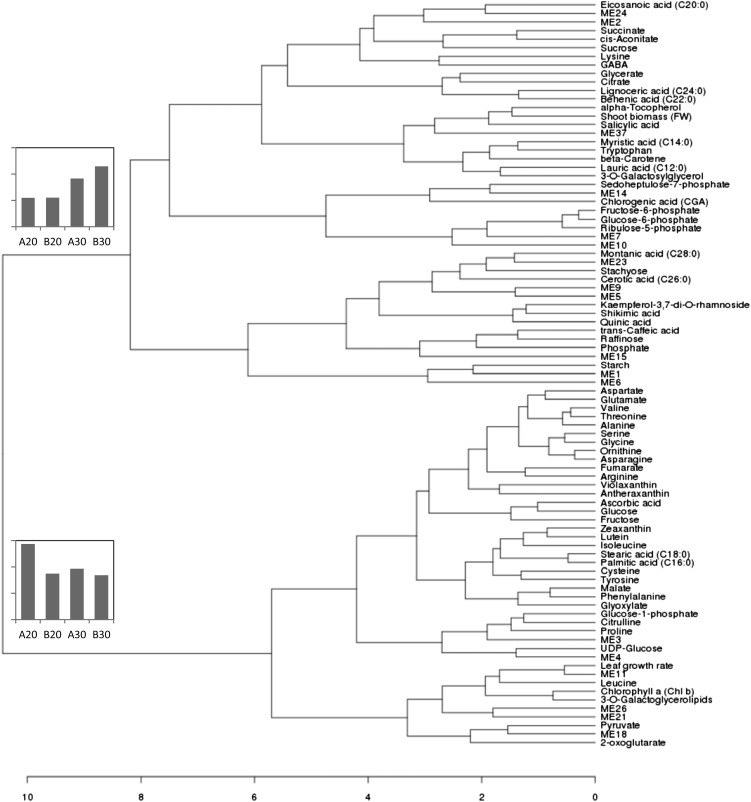
WGCNA of our microarray data set yielded 53 modules that were further analyzed for enrichment of certain gene ontology (GO) categories (AgriGO Web tool; Du et al., 2010) and for association of the module average expression pattern (ME) with the different experimental parameter (genotype, plant age at harvest, or N treatment). Most modules could not be associated with any particular function or experiment-specific pattern. Strongest transcriptional and functional coregulation was found in RNA-related processes (ME2), photosynthesis (ME3), and cell wall metabolism (ME7; Supplemental Table S3). The enrichment of GO terms related to RNA biosynthesis and transcription (ME2) indicates that many of these regulatory processes are strongly connected. The expression pattern of this regulatory module varied greatly between the individual plants, but no common response to N treatment or age at harvest was observed. Modules ME3 and ME4 had similar expression patterns and were enriched for processes regarding chloroplast metabolism, especially the biosynthesis of light-harvesting complex, but also transcripts for key enzymes from Calvin cycle (e.g. large subunit of Rubisco), the C4-specific CO2 enrichment pathway (e.g. PEPC), and tetrapyrrole metabolism (Supplemental Table S4). The high connectivity of photosynthesis-related transcripts also was recently shown along the developmental gradient of the maize leaf (Pick et al., 2011) and in the large maize microarray collection analyzed by Ficklin and Feltus (2011). In our experiment, these modules enriched for chloroplast metabolism were responding mainly to the developmental stage of the maize plants. The same sort of developmental effect also influenced the expression pattern of ME7, a module enriched for transcripts from cell wall metabolism (Supplemental Table S3 and Supplemental Fig. S1.2). Also, this module was influenced by the increasing severity of the N stress. Strongest response to the N treatment was found in the expression pattern of module ME11 (Supplemental Fig. S1.2); the module had a very high overlap with differentially regulated genes. From the significant differentials found in both genotypes under severe N deprivation, 55% belonged to the ME11 module (Supplemental Table S2). ME11 was enriched for GO terms such as carbohydrate metabolism, Pi transport, phosphoric ester hydrolases, and phosphatase activity. Expression pattern ME18 represents 15% of differentially regulated genes (Supplemental Table S2); its pattern responded to N treatment and genotype (Supplemental Fig. S1.2). All main transcript modules with significant functional enrichment (Supplemental Table S3) were related to metabolite pattern and the phenotypic features shoot biomass and leaf growth rate by hierarchical clustering. This produced two main branches (Fig. 5). The first branch contained features affected by low-N treatment at both developmental stages in a similar way; leaf growth rate clustered into this branch together with the majority of amino acids, organic acids, some pigments, and the N-treatment-influenced modules ME11 and ME18. Features in the second main branch were much stronger influenced by the prolonged N treatment; in this branch, shoot biomass could be found together with phosphate and phosphorylated intermediates, many fatty acids, Suc, starch, and secondary metabolites. Trp and Lys were the only amino acids in this branch (Fig. 5).
Figure 5.
Dendrogram of selected transcript modules, metabolites, and phenotypic features calculated by the WGCNA analysis tool. The small graphs show the average pattern from features on the two main branches of the dendrogram.
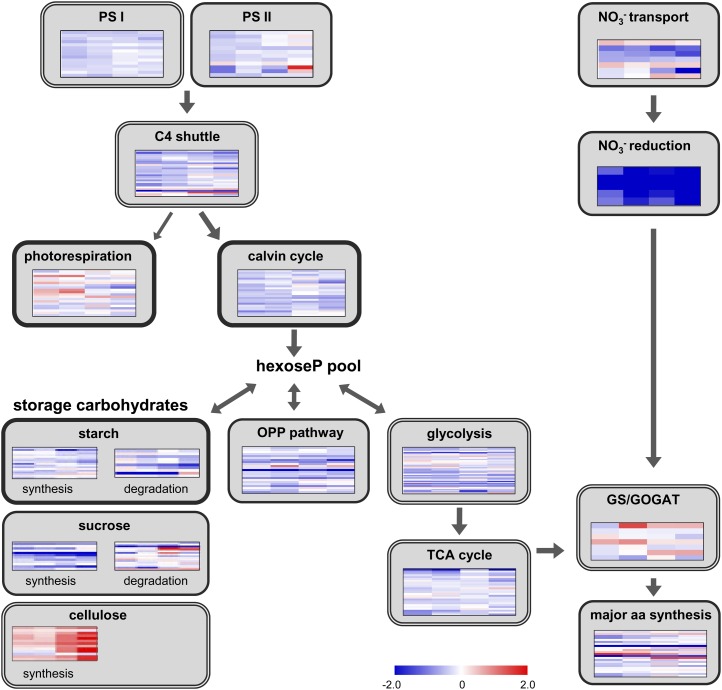
The statistical analysis showed that N starvation strongly affected the metabolome and transcriptome of the maize leaves. As expected, many genes involved in N metabolism are down-regulated by the N treatment. The same response pattern could be found for many transcripts involved in regulation of Pi homeostasis. Compared with N- and Pi-responsive genes, transcripts from primary C metabolism were less affected by N starvation. A detailed discussion of the influence of N starvation on the central metabolism in source leaves is presented below. Figure 6 gives an overview on transcriptional changes in the different pathways of primary N and C metabolism. The data behind the compressed heat maps can be found in Supplemental Table S4, and primary metabolism related transcripts with significant differences in low and high N treatment are listed in Tables III and IV.
Figure 6.
Transcriptional changes of primary C and N metabolism under low N. From left to right, the four columns represent changes measured in A188 at 20 d, in B73 at 20 d, in A188 at 30 d, and in B73 at 30 d. Heat maps show log2 fold changes in low- versus high-N conditions. Thin frame, Pathway preferentially in mesophyll; double frame, pathway in both cell types; thick frame, pathway preferentially in bundle sheath.
Table III. Transcripts with involvement in primary C metabolism.
The table shows transcripts as described in Figure 6 with significant changes (fold change > 2, false discovery rate P < 0.05; marked by asterisks) in at least one genotype/growth stage combination for low- versus high-N conditions. A20, A188 at 20 d; B20, B73 at 20 d; A30, A188 at 30 d; B30, B73 at 30 d.
| Process | MaizeGDB Genome ID | Description | A20 | B20 | A30 | B30 | ME |
|---|---|---|---|---|---|---|---|
| C4 shuttle | GRMZM2G097457 | PPDK | −1.407* | −0.806* | −0.877 | −1.217* | 1 |
| GRMZM2G141289 | Chloroplast NAD-MDH | −0.734 | −0.724 | −0.816 | −1.053* | 5 | |
| GRMZM2G066413 | PEP/P translocator | −0.463 | −0.830 | −0.579 | −1.347* | 15 | |
| GRMZM2G088064 | Ala aminotransferase | −2.590* | −1.568 | −1.349* | −1.571* | 18 | |
| Calvin cycle | GRMZM2G002807 | Triosephosphate isomerase | −1.004* | −0.877 | 0.082 | 0.193 | 3 |
| Photorespiration | GRMZM2G050481 | Ser-glyoxylate aminotransferase | 0.595 | 1.140* | −0.109 | −0.101 | 1 |
| OPP pathway | GRMZM2G177077 | Glc-6-P 1-dehydrogenase | −0.678 | −1.084 | −0.855 | −1.673* | 18 |
| GRMZM2G440208 | 6-Phosphogluconate dehydrogenase | −2.749* | −2.394* | −1.635* | −2.127* | 18 | |
| GRMZM2G035599 | Ribose-5-P isomerase | −0.907 | −0.992 | −1.171* | −0.934 | 11 | |
| GRMZM2G065030 | Ribose-P pyrophosphokinase | −0.556 | −1.071* | −0.975 | −0.778 | 11 | |
| Glycolysis | GRMZM2G059151 | Pyrophosphate-dependent phosphofructokinase | −1.243* | −1.712* | −1.468* | −0.994 | 11 |
| GRMZM2G021605 | Phosphoglycerate mutase family | −1.074* | −1.052 | −0.919 | −1.585* | 11 | |
| GRMZM2G064302 | Enolase | −1.269* | −1.138 | −1.261* | −1.127* | 11 | |
| GRMZM2G446253 | Enolase | −1.432* | −1.142 | −1.257* | −1.202* | 11 | |
| GRMZM2G150098 | Pyruvate kinase | −0.916 | −0.312 | −1.528* | −0.534 | 11 | |
| GRMZM2G008714 | Pyruvate kinase | −0.935 | −0.602 | −1.553* | −0.659 | 11 | |
| TCA cycle | GRMZM2G302832 | Pyruvate dehydrogenase | −1.770* | −2.316* | −0.717 | −2.115* | 15 |
| Suc-related | GRMZM2G098370 | UDP-Glc pyrophosphorylase | −1.063* | −0.781 | −0.993 | 0.161 | 5 |
| AC197705.4_FG011 | UDP-Glc pyrophosphorylase | −2.291* | −2.494 | −2.448* | −2.030* | 11 | |
| GRMZM2G013166 | Suc-P synthase | −0.876 | −1.188 | −1.566* | −1.878* | 11 | |
| GRMZM2G140107 | Suc-P synthase | −0.711 | −1.233 | −1.239* | −1.916* | 11 | |
| GRMZM2G089713 | Suc synthase | −2.771* | −1.421 | −3.169* | −1.922 | 18 | |
| GRMZM2G152908 | Suc synthase | −0.719 | −0.936 | −1.006* | −1.065* | 11 | |
| Starch-related | GRMZM2G082034 | Chloroplast β-amylase | −0.912 | −1.018 | −1.221* | −1.686 | 11 |
| GRMZM2G058310 | β-Amylase | −3.926* | −2.298 | −2.033 | 0.686 | 3 | |
| Cellulose-related | GRMZM2G113137 | Cellulose synthase6 | 0.625 | 0.429 | 0.930 | 1.170* | 14 |
| GRMZM2G113432 | Similar to cellulose synthase | −0.282 | 0.412 | 0.999 | 1.967* | 7 | |
| GRMZM2G164761 | Similar to cellulose synthase | 1.096* | 1.573 | 1.369* | 2.209 | 1 |
Table IV. Transcripts with involvement in primary N processes.
The table shows transcripts described in Figure 6 with significant changes (fold change > 2, false discovery rate P < 0.05; marked by asterisks) in at least one genotype/growth stage combination for low- versus high-N conditions. A20, A188 at 20 d; B20, B73 at 20 d; A30, A188 at 30 d; B30, B73 at 30 d.
| Process | MaizeGDB Genome ID | Description | A20 | B20 | A30 | B30 | ME |
|---|---|---|---|---|---|---|---|
| Nitrate transport | GRMZM2G161459 | NRT1.1, a dual-affinity nitrate transporter | −1.096* | −0.902 | −1.476* | −1.219* | 5 |
| GRMZM2G112154 | NRT1.1, a dual-affinity nitrate transporter | −0.683* | −0.819 | −0.939 | −1.429* | 18 | |
| GRMZM2G455124 | High-affinity nitrate transporter family | −0.247* | −0.105 | −0.809 | −2.454* | 11 | |
| Nitrate reduction | GRMZM2G142386 | Cytosolic isoform of nitrate reductase | −1.215* | −2.046* | −1.944* | −3.109* | 18 |
| GRMZM2G104898 | Cytosolic isoform of nitrate reductase | −2.633* | −2.774* | −2.443* | −4.118* | 18 | |
| GRMZM2G428027 | Cytosolic isoform of nitrate reductase | −2.700* | −2.771* | −2.564* | −4.181* | 18 | |
| GRMZM2G076723 | Nitrate reductase structural gene | −1.206* | −2.076* | −1.828* | −3.013* | 18 | |
| Nitrite reduction | GRMZM2G079381 | Ferredoxin-nitrite reductase. | −1.044* | −1.762 | −1.307 | −2.079* | 11 |
| GRMZM2G000739 | Uroporphyrin III methyltransferase | −1.532* | −2.136* | −1.346* | −3.152* | 11 | |
| Amino acid metabolism | GRMZM2G178415 | Glu dehydrogenase | 0.038 | −0.042 | −0.797 | −1.077* | 41 |
| GRMZM2G078472 | Asn synthetase | −3.466* | −2.733 | −3.800* | −2.155 | 11 | |
| GRMZM2G082032 | Asparaginase | 1.680* | 1.403 | 0.819 | 0.332 | 15 | |
| GRMZM2G088064 | Ala aminotransferase | −2.590* | −1.568 | −1.349* | −1.571* | 18 | |
| GRMZM2G119583 | Acetyl-Orn transaminase | −0.286 | −0.637 | −0.523 | −1.008* | 18 | |
| GRMZM2G028535 | Glu 5-kinase | −1.425* | −2.143 | −0.203 | −1.194* | 1 | |
| GRMZM2G375504 | Glu 5-kinase | −1.007* | −0.662 | 0.309 | 0.410 | 1 | |
| GRMZM2G087103 | Branched-chain amino acid aminotransferase | −1.327* | −0.172 | −2.435* | −1.011 | 7 | |
| GRMZM2G104613 | 3-Isopropylmalate dehydrogenase | −1.017* | −0.559 | 0.017 | 0.081 | 3 | |
| GRMZM2G024686 | Asp kinase | −2.409* | −0.693 | −0.391 | 0.570 | 15 | |
| GRMZM2G104546 | Homo-Ser dehydrogenase | −0.772 | −1.941 | −0.281 | −1.965* | 20 | |
| GRMZM2G050570 | Thr synthase | −1.597* | −1.186 | −1.180* | −1.744* | 5 | |
| GRMZM2G069203 | Ser O-acetyltransferase | −0.726 | −1.296 | −1.058* | −1.231* | 11 | |
| GRMZM2G013430 | Ser O-acetyltransferase | −1.933* | −2.890* | −2.315* | −3.361* | 11 | |
| GRMZM2G396212 | 3-Deoxy-7-phosphoheptulonate synthase | 0.426* | 0.376 | 0.576 | 1.162* | 10 | |
| GRMZM2G070218 | Shikimate kinase | −1.396* | −1.446* | −0.974 | −1.241* | 5 | |
| GRMZM2G161566 | Shikimate kinase | −1.553* | −1.453 | −1.012 | −1.055 | 5 | |
| GRMZM2G106950 | Indole-3-glycerol-P synthase | −2.197* | −2.740 | −3.164* | −1.951* | 11 | |
| GRMZM2G085381 | Trp synthase | −2.503* | −0.411 | −3.421* | 0.415 | 11 | |
| GRMZM2G015892 | Trp synthase | −1.148 | −1.146 | −2.065* | −1.529 | 18 | |
| GRMZM2G046163 | Trp synthase | −1.163 | −1.269 | −2.026* | −1.681* | 18 | |
| GRMZM2G054465 | Trp synthase | −1.045* | −0.358 | −0.283 | −0.104 | 5 | |
| Amino acid transport | GRMZM2G177659 | Amino acid transporter | −1.632* | −1.035 | −0.884 | −0.610 | 10 |
| GRMZM2G010433 | Amino acid transporter | 2.221* | 1.634 | 0.342 | 0.474 | 3 | |
| GRMZM2G101125 | Amino acid transporter | 1.410* | 1.641 | 0.862 | 0.887 | 3 | |
| GRMZM2G108597 | Amino acid transporter | 4.131* | 2.643* | 3.836* | 2.955* | 11 | |
| Peptide transport | GRMZM2G135291 | Oligopeptide transporter OPT superfamily | −1.200 | −1.316 | −0.879 | −1.295* | 6 |
| GRMZM2G026523 | H+/oligopeptide symporter | −0.643 | −0.614 | −0.372 | −1.077* | 4 | |
| GRMZM2G064091 | Oligopeptide transport | −0.642 | −1.037 | −1.879 | −2.104* | 10 | |
| GRMZM2G034389 | Di- and tripeptide transporter | 0.181 | 0.309 | 0.899 | 1.247* | 18 | |
| GRMZM2G470454 | H+/oligopeptide symporter | 1.028* | 0.405 | 0.551 | 1.003 | 4 | |
| GRMZM2G071759 | Oligopeptide transporter | 2.819* | 2.698* | 2.242* | 3.104* | 11 |
Effects of Low N on N Assimilation
Transcripts of genes involved in first steps of nitrate metabolism were mainly down-regulated in maize leaves under N starvation. Compared with the root, the presence of different nitrate transporters is probably less critical for adaptation to N limitation in the leaf. Their specific function is the uptake from the xylem, distribution in the leaf, and remobilization (Fan et al., 2009). In the maize leaf, transcripts for low- and high-affinity transporters could be detected. Only under prolonged stress individual members of the NRT1.1 and the NRT2 group were reduced (Table IV). Transcription of the nitrate-reducing enzymes is usually strongly dependent on the N status of the tissue (Wang et al., 2000, 2003; Scheible et al., 2004), and a considerable decrease was also found for all four nitrate reductase transcripts as well as the nitrite reductase transcript. Transcription patterns of nitrate reductase followed the ME18 and showed also genotype-dependent differences, with raw array signals from B73 being generally higher than from A188 (Table IV). Comparison of transcriptome responses in different maize lines recently also showed that marker genes for N deprivation are not simply characterized by their absolute transcript values at any given time but by their linear response to the analyzed stress situation (Yang et al., 2011). Furthermore, genotype-specific transcript levels would not necessarily translate into different levels of activity because in addition to the transcriptional control, nitrite reductase activity is also regulated on the posttranslational level by phosphorylation and interaction with Mg2+ (Huber et al., 1994). Nitrite reductase activity is dependent on the cofactor siroheme, and a transcript for the branch point of its biosynthesis, Uroporphyrin III methyltransferase, was significantly down-regulated by low-N conditions in the maize leaf (Table IV). The results confirm data from other species describing the direct responsiveness of transcripts for nitrate transporter and primary nitrate assimilation to nitrate status of the tissue (Wang et al., 2000, 2003; Scheible et al., 2004; Krapp et al., 2011).
Incorporation of inorganic ammonium into organic compounds is mediated by the GS/GOGAT cycle. In the maize leaves, transcripts encoding different isoforms of these enzymes were only minorly affected by low-N conditions, confirming the importance of the reaction even under conditions of N limitation (Fig. 6; Supplemental Table S4). Ammonium is not only produced by nitrate assimilation but also an intermediate in protein turnover and photorespiration, and its immediate reassimilation prevents loss of N to the environment. Activity of GS1 is therefore highly regulated and adapted to the specific organ, cell type, or environmental condition (Bernard and Habash, 2009). The increase of ammonium in leaves from plants with reduced GS1 transcription also indicated that reassimilation of ammonium is highly dependent on GS1 presence and might be critical under conditions with N limitation (Bernard and Habash, 2009). Active metabolism in the maize source leaf would also depend on a certain rate of protein turnover. The contribution of photorespiration to ammonium production is reduced in C4 plants such as maize but would still be present. Interruption of photorespiration by repression of the glyoxylate oxidase has shown recently that the presence of photorespiration is also essential for survival of C4 plants (Zelitch et al., 2009). It was proposed that photorespiration is important to avoid accumulation of glycolate to toxic levels, and this protection mechanism would be needed in high- and low-N conditions. Compared with most other pathways of primary leaf metabolism, transcripts for photorespiration were much less affected by the low-N treatment. Therefore, ammonium is constantly released in the active source leaf, and stable transcription of the GS as the main reassimilating enzyme plays an important role in N management under low-N conditions. A second enzyme with the capacity to assimilate ammonium in plants is the Glu dehydrogenase, but in comparison with GS, the enzyme plays only a minor role in ammonium assimilation. Its function is probably connected to protein catabolism under C shortage conditions. In maize source leaves, long-term N stress causes a reduced presence of Glu dehydrogenase transcripts (Fig. 7A).
Figure 7.
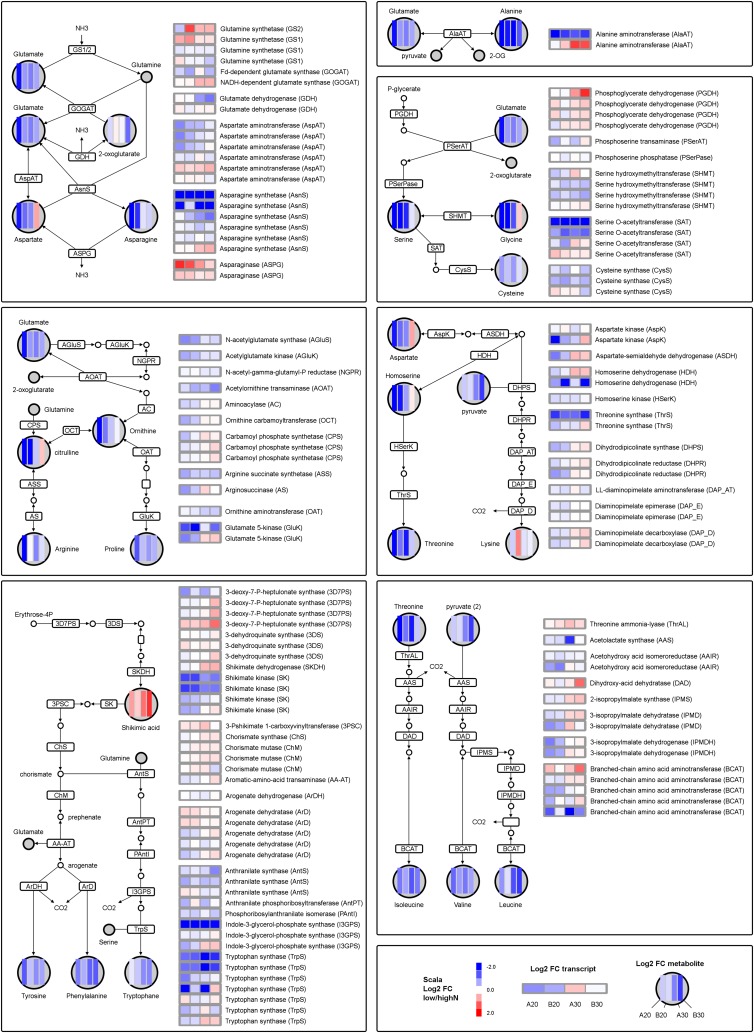
Amino acid metabolism under low-N conditions. In the pathways, changes measured for metabolite concentrations are given as heat maps of log2 fold changes in low- versus high-N-treated maize source leaves, and changes of transcripts of the participating enzymes are given on the right. A, Glu, Gln, Asp, and Asn metabolism. B, Ala metabolism. C, Ser, Gly, and Cys metabolism. D, Arg and Pro metabolism. E, Thr and Lys metabolism. F, Tyr, Phe, and Trp metabolism. G, Val, Leu, and Ile metabolism.
Glu is the primary amino acid produced by the GS/GOGAT cycle; for the synthesis of many other amino acids, Glu serves as donor of the amino group. The amino acid pool of plants is characterized by high flexibility and varies between species, cell types, and physiological situations. The majority of amino acids usually change in coordination; especially high correlation had been described for minor amino acids (Noctor et al., 2002) or amino acids sharing the same biosynthetic pathway (Fritz et al., 2006a). In maize source leaves, Glu was significantly reduced under low-N conditions (Fig. 8; Supplemental Table S2); the effect was similar for early and late N starvation. In the WGCNA dendrogram, this behavior closely correlated to the pattern of the other major amino acids (Asp, Asn, Ala, Ser, and Gly; Fig. 5). With the exception of Trp and Lys, minor amino acids were also down-regulated by low N status in a similar manner (Fig. 5). The general down-regulation of amino acids in N-starved plants is expected and in agreement with many other studies in Arabidopsis (Krapp et al., 2011), wheat (Triticum aestivum; Howarth et al., 2008), tomato (Urbanczyk-Wochniak and Fernie, 2005), and tobacco (Fritz et al., 2006a). However, different amino acids seem to show very individual responses in the different experiments. In contrast to the majority of amino acids, Lys, Arg, and His increased in N-deficient Arabidopsis plants (Krapp et al., 2011). In the study of Tschoep et al. (2009), only Asp, Ala, and Asn decreased under low N, but most other amino acids increased. It was postulated that growth retardation was due to changes in sink demand and allowed accumulation of amino acids under N stress. Strong individual responses to low-N conditions were also found for Lys and Leu in tomato leaves (Urbanczyk-Wochniak and Fernie, 2005), for Gln and Trp in wheat (Howarth et al., 2008), and for Glu, Phe, and Asp in tobacco leaves (Fritz et al., 2006a). It shows that under specific experimental conditions, the response of individual amino acids differs not only quantitatively but also qualitatively from the general trend. Most authors report a very high stability of Glu in changing amino acid profiles, and it has been suggested that Glu homeostasis in plants is regulated by different control mechanisms sensing ATP and redox status as well as 2-oxoglutarate availability (Fritz et al., 2006a; Krapp et al., 2011). However, in our experiment, Glu levels also were affected by N deficiency, and the trend was mirrored in many other amino acids (Figs. 5 and 8). Biosynthesis of many amino acids might therefore also be affected by limited availability of the amino group donor.
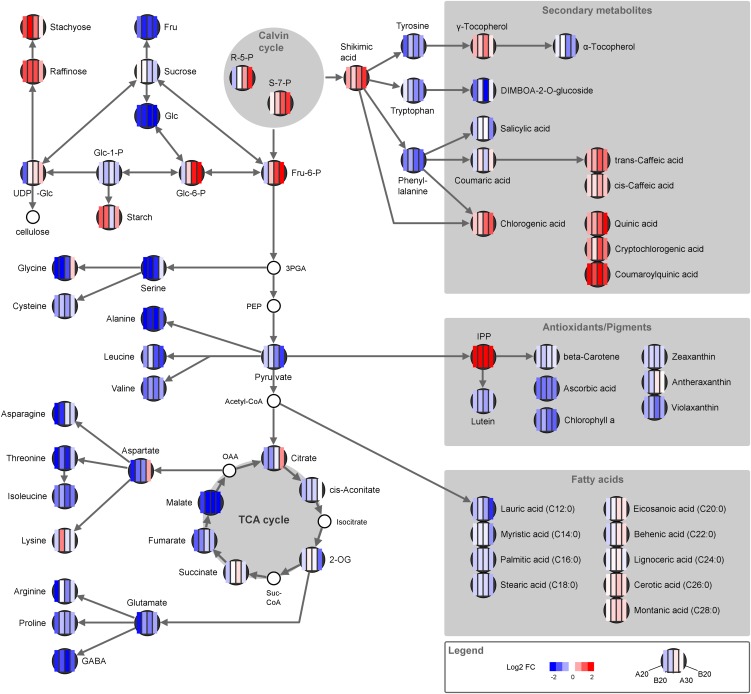
Figure 8.
Changes in metabolite profile under low N. From left to right, the four columns represent changes measured in A188 at 20 d, in B73 at 20 d, in A188 at 30 d, and in B73 at 30 d. Heat maps show log2 fold changes in low- versus high-N conditions.
Transcriptional control is supposed to contribute to the regulation of amino acid biosynthesis (Scheible et al., 2004; Howarth et al., 2008). In maize leaves, many enzymes were encoded by multigene families, and the reactions to low-N conditions were not uniform; only few changes in amino acid content could be connected to transcriptional changes, e.g. Asn metabolism and Trp biosynthesis (Fig. 7, A and F). Asn is characterized by a high N:C ratio and usually serves as an N storage metabolite. In our maize leaves, Asn was particularly affected during early N starvation, and the effect leveled off under prolonged N deprivation. Several transcripts for Asn synthetase were down-regulated by the low-N treatment, while transcripts for the degrading enzyme asparaginase were up-regulated (Fig. 7A; Table IV). Significant reduction in transcript abundance was also found for several transcripts involved in biosynthesis of aromatic amino acids (Fig. 7F). Under low-N conditions, the general precursor shikimic acid was increased in maize leaves (Fig. 7F), and the same was observed for related secondary metabolites, such as quinic acid or chlorogenic acid (Fig. 8). Transcripts for shikimate kinase, the enzyme leading from shikimic acid toward the synthesis of Tyr, Phe, and Trp, were down-regulated under low-N conditions. Further down, transcripts specific for Trp biosynthesis, such as indole-3-glycerol-P synthase and Trp synthase, were also reduced under N deficiency. Trp pattern differed from the majority of amino acids in N-starved maize leaves and progressively decreased under long-term stress. Trp metabolism is closely connected to the secondary metabolism and the production of metabolites, such as phytoalexins, glucosinolates, alkaloids, and the growth-promoting auxin (Tzin and Galili, 2010).
N deficiency was generally accompanied by significant changes in different amino acids and oligopeptide transporters (Table IV). Individual transporter showed opposite behavior, suggesting that controlled distribution of amino acids plays an important role in adaptation of source leaf metabolism to N deprivation as well as the export of N resources toward sink organs (Okumoto and Pilot, 2011).
Effects of Low N on Primary C Metabolism
Pathways for C and N assimilation are highly interconnected in source leaves. Chloroplast metabolism belongs to the first processes to be affected by limitation in N supply. Generally, transcription of photosynthesis-related genes were down-regulated in N-deficient maize leaves (Fig. 6; Supplemental Table S4). The concentration of photosynthesis genes in ME3 indicated that the process was regulated in coordination. This is in agreement with similar studies in Arabidopsis (Scheible et al., 2004; Bi et al., 2007; Krapp et al., 2011), rice (Lian et al., 2006), and maize (Yang et al., 2011). In the maize source leaves analyzed in our experiment, transcripts for the antenna proteins of PSI and PSII were down-regulated under low-N conditions (Fig. 6). Metabolite measurements also confirmed a reduction in chlorophyll content of the leaves under low-N conditions (Fig. 8; Supplemental Table S2). However, effective PSII quantum yield in the leaves was only slightly affected by the N status, indicating that the remaining PSII reaction centers were intact (Fig. 1).
In the examined maize leaves, transcription of main enzymes of the C4 carbon shuttle, C4-PEPC, PPDK, NADP malic enzyme, and malate dehydrogenase, and many involved plastidial transporters decreased under N-deficient conditions (Fig. 6; Supplemental Table S4). As described in the introduction, on the protein level, the C4 shuttle proteins PEPC and PPDK were more affected by low-N conditions than Rubisco (Sugiharto et al., 1990). It has recently been suggested that Ala and Asp contribute substantially to the metabolites shuttle between mesophyll and bundle sheath in maize (Pick et al., 2011). Transcripts for the related aminotransferases were therefore also included into the C4 cluster of Figure 6. Only few individual members from the C assimilation pathways were unchanged or up-regulated (Supplemental Table S4). Generally, the overview in Figure 6 indicates that photosynthesis was transcriptionally down-regulated under low-N conditions and that this response was not dominated by regulation of individual enzymes but by a coordinated reduction of all members of the pathways. On the protein level, plants save N by substantial reduction of the most abundant proteins. However, the accumulation of phosphorylated intermediates indicated that under long-term N deficiency, leaf metabolism is more influenced by further processing of C assimilates rather than properties of the photosynthetic machinery.
Hexose-Ps produced in photosynthesis can be further metabolized for production of energy and precursors for different biosynthetic pathways or converted into storage forms. The primary catabolic pathways, such as glycolysis, tricarboxylic acid (TCA) cycle, and OPP pathway, followed the same transcriptional downward trend in response to N deficiency (Fig. 6). In nongreen tissue, the OPP pathway supplies reducing power for N assimilation. Transcription of the OPP pathway is therefore responsive to the N status of the plant (Wang et al., 2000; Scheible et al., 2004). In maize leaves, the first genes of the OPP pathway (plastidial Glc-6-P-1-dehydrogenase and 6-phosphogluconate dehydrogenase) were significantly reduced in low N plants (Table III). This is in agreement with results from Arabidopsis (Bi et al., 2007) and rice leaves (Lian et al., 2006). In contrast to roots, the illuminated leaves receive energy and reducing compounds from photosynthetic electron transport, but the strong transcriptional regulation suggests that the pathway is also active in some leaf cells. Individual genes from the glycolytic pathway were significantly reduced under low-N conditions in maize leaves (Table III), especially the last steps leading to PEP and pyruvate (phosphoglycerate mutase, enolase, and pyruvate kinase). In the opposite situation, when nitrate is added to the plant, transcripts for the Arabidopsis homologs were rapidly induced (Scheible et al., 2004), indicating a strong influence of the C/N regulatory network on transcription of these genes. The following TCA cycle was also characterized by general reduction of transcription under low-N conditions. The organic acids involved in these pathways are C precursors for amino acid biosynthesis, and the influence of N status on organic acid metabolism seems to be a central point in coordination of C and N metabolism (Hodges, 2002).
N limitation caused a general reduction in the organic acid pools of the maize source leaf (Fig. 8); however, the response curves had quite individual patterns. Citrate, aconitate, and fumarate were more affected during early N starvation, while pyruvate, fumarate, and 2-oxoglutarate reacted stronger under prolonged stress (Fig. 8). There was no clear correlation in the patterns of organic acid precursors and the related amino acid, e.g. 2-oxoglutarate and Glu or pyruvate and Ala. Malate concentrations were continuously low under N deficiency, and from the measured organic acids, its signature closely correlated to most of the amino acids (Fig. 5). Besides its involvement in the TCA cycle, malate has a number of different functions in the maize leaf: as shuttle for C and reducing equivalents, as osmoticum, pH regulator, and in stomatal functioning (Fernie and Martinoia, 2009). C and nitrate assimilation declined under low-N conditions; therefore, the demand for malate in the C4-specific shuttle and as counterion balancing the production of hydroxyl ions during nitrate reduction would also decrease. Data from the C3 plant Arabidopsis indicated a strong correlation between malate and nitrate reduction pattern in the diurnal cycle (Tschoep et al., 2009). In the C4 leaf, two major processes with malate participation, namely, the C4 shuttle and the pH regulation during nitrate reduction, would be affected by N limitation and could explain the strong response of the metabolite.
When demand for organic acids decreases, conversion of photosynthates can shift to other end products and starch and Suc biosynthesis are usually induced under nutrient stress (Hermans et al., 2006). In Arabidopsis, starch production is up-regulated under low-N conditions by increased expression of ADP-Glc pyrophosphorylase, the key enzyme in starch biosynthesis (Bi et al., 2007). It was therefore interesting to see that starch metabolism transcripts were actually repressed in maize leaves under low-N conditions, indicating reduced starch turnover in the plants (Fig. 6; Supplemental Table S4). The transcriptional response was very similar in leaves harvested at early or late N stress. The metabolite profile, on the other hand, implied slight differences in starch-related metabolism at early and late N stress. In agreement with many similar studies (Bi et al., 2007; Tschoep et al., 2009; Krapp et al., 2011), starch accumulated in the fifth leaf when plants had grown under low-N conditions (Fig. 8). However, starch accumulation was much less pronounced in the sixth leaf. It is possible that this was connected to the age of the leaf; the harvested sixth leaf was still very young, and even under N limitation the metabolism could be programmed for C export rather than storage. Assimilate export from the sixth leaf may be expected since the photosynthetic capacity of the remaining leaves is strongly reduced under N starvation (Fig. 1). The absence of strong starch accumulation in N-depleted leaves has also been observed in other studies on maize plants in the vegetative stage (Hirel et al., 2005; El-Kereamy et al., 2011). Only during grain filling did the N treatment lead to accumulation of starch in the upper leaves, and this was connected to decreased sink strength of the growing ear under low-N conditions (Hirel et al., 2005).
Microarray analysis revealed further that cellulose synthase genes increased especially under long-term stress, indicating that an increased amount of C was funneled toward cell wall biosynthesis under N deficiency (Fig. 6; Supplemental Table S4). Genes related to Suc metabolism were even more suppressed than starch biosynthesis; and the most abundant transcript for Suc-P synthase in the leaves was significantly reduced by N deficiency (Table III). Suc degradation was less affected, and two isoforms of Suc synthase were up-regulated under prolonged N stress (Fig. 6; Supplemental Table S4). Their expression pattern correlated with the transcripts for cellulose synthases, which could indicate that these isoforms work in close collaboration in cell wall synthesis (Coleman et al., 2009). The amounts of Suc measured in the leaf material were not significantly affected by N deficiency, but the amounts of Glc and Fru decreased considerably (Fig. 8). On the other hand, increased amounts of C were converted into sugars from the raffinose group (Fig. 8), a pathway very common in different abiotic stress situations (Hannah et al., 2006; Krapp et al., 2011). Raffinose serves thereby not only as a C store but can also have protective functions.
Effects of N Deprivation on Lipid and Secondary Metabolism
Galactolipids are the major constituents of the thylakoid membranes. Under N deficiency, the amount of galactoglycerolipids decreased significantly (Fig. 8), indicating a very coordinated reduction of all components of the photosynthetic reaction centers. The same trend had been observed in Arabidopsis. The decrease in galactolipids was accompanied by a shift in the ratio of monogalactosyldiaglycerol to digalactosyldiaglycerol (Gaude et al., 2007). Monogalactosyldiaglycerol is usually tightly bound to the PSI reaction centers (Jordan et al., 2001), and a higher ratio of digalactosyldiaglycerol in comparison to monogalactosyldiaglycerol would imply that the N-deprived changes especially affected the PS reactions. Only few genes of the lipid metabolism seemed to be significantly affected by the N stress; however a monogalactosyldiacylglycerol B synthase was strongly down-regulated in low N-treated leaves (Supplemental Table S1). The fatty acid composition shifted under N deficiency, medium- to long-chain fatty acids (up to C18) were down-regulated by the stress, while very-long-chain fatty acids tended to be up-regulated (Fig. 8). The reduction in shorter chain fatty acids could be an adjustment to the general decrease in chloroplast structures.
Longer chain fatty acids are associated with waxes and surface structures. Especially under long-term N deficiency cell wall biosynthesis was enhanced in the N-deficient plants. Beside the increases in cell wall transcripts, the formation of lignin-related metabolites was also enhanced. Enhanced lignification had already been shown for N-starved tobacco leaves (Fritz et al., 2006b). The precursors for chlorogenic acids are shikimic acid and Phe, and under low-N conditions, both compounds changed in opposite directions: shikimic acid accumulated while Phe decreased (Fig. 8). The leaves also accumulated significant amounts of isopentenyl diphosphate, the precursor of many pigments and protective metabolites (Fig. 8). It was therefore surprising to see that lutein, carotene, ascorbic acid, and the xanthins were reduced under low-N conditions. The decrease in these compounds may be a result of the reduced photosynthetic apparatus of N-deficient leaves, and the precursor might accumulate because of the decreased demand for pigments. Generally, the production of C-rich secondary metabolites is preferred to N-rich compounds under low-N conditions (Fritz et al., 2006b).
Effects of N Deficiency on Hormone Metabolism
Auxin- and cytokinin-dependent regulation is expected to play a role in long-distance N response. Generally, high nitrate concentrations in the root increase cytokinin production, and the movement of the hormone with the xylem stream can transfer the signal to the shoot, where the cytokinin-derived signal modulates N partitioning and influences the developmental program (Sakakibara, 2003). The nitrate response is thereby not limited to the root tissue, and nitrate concentration in shoots can also enhance transcription of cytokinin biosynthesis genes (Miyawaki et al., 2004). N deprivation, on the other hand, is characterized by low cytokinin production, but most cytokinin-related genes in the N-deprived maize leaf were not significantly affected by the treatment. However, one cytokinin oxidase, a cytokinin degrading enzyme, was down-regulated under prolonged N stress and the genes responsible for glycosylation of cytokinin were up-regulated (Supplemental Table S5). This indicates that cytokinin is rather inactivated by glycosylation then degraded. This would have the advantage that under recovery of N supply, a fast release of the cytokinin signal would be possible. Recovery from nitrate stress in maize is characterized by de novo synthesis as well as reactivation of the inactive storage forms (Takei et al., 2002).
Auxin is mainly transported basipetaly and has been a candidate for communication of nitrate status from the shoot to the root. The transfer of Arabidopsis from high-N to low-N medium causes an increase of auxin in the phloem sap, which is followed by lateral root outgrowth (Tian et al., 2008). The biosynthesis of auxins can be mediated by different Trp-dependent and -independent pathways. As mentioned above, the Trp content of maize leaves under N deprivation decreased and the development was intensified with increasing severity of the stress. The main auxin-synthesizing transcripts YUC and TAA (Zhao, 2012) were not significantly affected by the stress (Supplemental Table S5).
Effects of N Deficiency on Transcription Factors
Transcription factors can synchronize the expression of different genes involved in specific physiological responses. Identification of transcription factors regulating the response to stress could provide a tool for the coordinated manipulation of many adaptive processes. In Arabidopsis, several transcription factors have been identified with nitrate starvation-related changes in expression (for review, see Kant et al., 2011a). However, the knowledge transfer concerning transcriptional regulators from only distantly related species might be problematic, and research on transcriptional regulators need to be carried out in the important crop species directly (Calderon-Vazquez et al., 2008). For instance, the maize chip used in our experiment did not contain clear orthologs of the Arabidopsis transcription factors ANR1 and Dof1, which are supposed to be involved in regulation of the nitrate response (Kant et al., 2011a).
Nevertheless, 60 transcription factors could be identified with strong N deprivation response patterns (Supplemental Table S5). All these transcription factors were significantly different in at least two analyzed genotype/growth stage comparisons or significant in one comparison and present in one of the N-specific modules ME11 and ME18. The genotype-independent, N-dependent response would thereby indicate a general response in N stress adaptation. The majority of the transcription factors (47) were down-regulated by N starvation, and only 13 of them were continuously up-regulated under N stress (Supplemental Table S5). An MYB transcription factor (GRMZM2G016370) and a LOB transcription factor (GRMZM2G044902) were among the most highly down-regulated transcripts (Supplemental Table S1). Another MYB transcription factor (GRMZM2G159119) had recently been identified for its strong correlation to the expression pattern of C4-specific PEPC (Pick et al., 2011). In our experiment, this transcription factor was strongly down-regulated (Supplemental Table S5). It would be interesting to further investigate the specific regulatory role of this transcription factor and the possible involvement in control of C4-specific gene expression. Under N deprivation, the transcription factor could be involved in down-regulation of the PEPC and possibly other C4-specific transcripts. In the same article (Pick et al., 2011), a C2H2 zinc finger had been correlated to the expression of the C4-specific NADP malic enzyme. In our experiment, the same transcript (GRMZM2G426125) was also down-regulated especially under prolonged N stress (Supplemental Table S5).
The abundance of five members from the LOB class of transcription factors also decreased significantly under N deprivation. Individual members of this family have been identified for their involvement in N-specific responses. Three LOB transcription factors from Arabidopsis that were induced by nitrate and reduced under nitrate limitation had been characterized by their involvement in N-dependent regulation of anthocyanin biosynthesis and N-dependent gene expression (Rubin et al., 2009). N limitation also caused strong induction of few transcription factors, especially members of the bHLH (basic Helix-Loop-Helix) and HD (homeodomain) family. Another member of the MYB transcription factor family was strongly induced only in plants at the later growth stage suffering from prolonged stress and severe growth retardation.
Effects of N Deficiency on Pi Metabolism
Phosphate (inorganic and from organic phosphate) was the metabolite peak with the strongest increase under low-N conditions (Supplemental Table S1). Availability of Pi is essential for survival of all living systems as part of many organic compounds and universal energy transfer system. It is further involved in many regulatory processes. The subcellular distribution of Pi influences the metabolic activity of the different compartments and the metabolite exchange between them (Flügge, 1999). Metabolic balance between different pathways is therefore dependent on Pi homeostasis. Under natural conditions, plants are more likely to suffer from Pi limitation than excess. In contrast to nitrate, high Pi levels can damage the leaf. Pi toxicity syndromes were described from Arabidopsis mutants with increased Pi concentration in the shoot (Delhaize and Randall, 1995; Zhou et al., 2008). However, it is not clear if the damage is caused directly by the high Pi concentration or by the knockout of a regulatory gene. The leaves in our experiment showed strong increase in phosphate under N deprivation, but none of the toxicity syndromes could be observed. For the continued functioning of photosynthesis, excess Pi would need to be stored away from active metabolism in the vacuole (Smyth and Chevalier, 1984). Accumulation of Pi in the cytoplasm would initiate increased exchange of triose-P with the chloroplast and deplete the reductive C pathway of intermediates (Edwards et al., 1978), finally causing inhibition of photosynthesis. Additionally, Pi accumulation in the chloroplast would reduce starch synthesis by inhibition of the AGPase reaction (Preiss, 1982). However, feeding Pi to isolated wheat protoplasts showed that intact plant cells have considerable capacity for uptake and storage of Pi without any damage to photosynthesis and starch synthesis (Smyth and Chevalier, 1984). In C4 leaves, the Calvin cycle is mainly localized to the bundle sheath, but for the reduction phase, 3-phosohoglycerate (3PGA) and triose-Ps are moved to the mesophyll where they are transported into the chloroplast via exchange with Pi. Uneven distribution of Pi would impact the metabolite exchange differently in the different cells. Two transcripts of the triose-P transporter responsible for the exchange between chloroplast and cytosol were only slightly affected by the N deficiency and consequent Pi accumulation (Supplemental Table S4).
The presence of Pi in the different cell compartments would further influence the activity of some key regulators of C metabolism, such as PEPC, Suc-P synthase, cytosolic FBPase, and AGPase (Iglesias et al., 1993). Protein phosphorylation plays an important regulatory role in signaling pathways and control of enzyme activity under changing N conditions (Engelsberger and Schulze, 2012), and in our experiment, N limitation caused significant changes in several kinases and phosphatases (Supplemental Table S5). Among them was a kinase regulating the activity of PEPC (PEPK4) that showed strong down-regulation under low-N conditions (Supplemental Table S5). However, this identified isoform is not involved in regulation of the C4-specific PEPC enzyme (Shenton et al., 2006). A transcript for Fru-6-P 2-kinase/Fru-2,6-bisphosphatase, an enzyme with an important role in control of glycolysis was also down-regulated by N limitation (Supplemental Table S5). Another kinase with predicted regulatory function in carbohydrate metabolism was the SnRK1 subunit, which was strongly down-regulated by nitrate deprivation (Supplemental Table S5), although the differences did not classify as significant due to high variation between the replicates. The strong link between phosphate- and sugar-dependent regulation was recently shown for plants inhibited in the activity of a Suc transporter (Lei et al., 2011). The inactivation of the transporter severely impaired the plant’s ability to adapt to Pi starvation stress. Since N starvation impacts on central C metabolism as well as phosphate levels, the regulatory networks of the plant cells are challenged to readjust the system.
The increases in phosphate were accompanied by changes in expression of many genes involved in phosphate-related metabolism underlining the importance of Pi balance regulation under changing conditions. Calderón-Vázquez et al. (2011) recently published a list of Pi regulators in maize, and the majority of those regulators are strongly down-regulated in the N-deficient maize leaves (Table V). This could indicate that a large part of transcripts down-regulated in the experiment were secondary responses reacting toward the increased phosphate level. The reduction in phosphatases under low-N conditions would contribute to an increased binding of phosphate in organic compounds. Our metabolite profile supports this and most phosphorylated metabolites increased under low-N conditions. Decreased expression of transporters might limit the movement of Pi in the cells. Many regulatory elements identified as responsive to Pi starvation, such as the SPX domain proteins, were down-regulated under N deprivation. Generally, the expression of many genes observed in our experiment was just opposite to the situation in plant suffering from Pi starvation (Morcuende et al., 2007).
Table V. Transcripts with involvement in Pi homeostasis.
Response to N deprivation in maize leaves. Asterisks mark significant differences. A20, A188 at 20 d; B20, B73 at 20 d; A30, A188 at 30 d; B30, B73 at 30 d.
| MaizeGDB Genome ID | Description | A20 | B20 | A30 | B30 | ME |
|---|---|---|---|---|---|---|
| Pi transporter | ||||||
| AC147602.5_FGT005 | Pi transporter | −0.016 | 0.088 | 0.757 | 0.508 | 1 |
| AC235543.1_FGT002 | Pi transporter | 0.296 | 0.416 | 0.217 | −0.176 | 12 |
| GRMZM2G009779 | Pi transporter | −1.098* | −0.902 | −1.816* | −1.349* | 11 |
| GRMZM2G017047 | Pi transporter | 0.129 | 0.204 | 0.245 | 0.331 | 7 |
| GRMZM2G046681 | Pi transporter | −0.376 | −0.123 | 0.438 | 0.346 | 7 |
| GRMZM2G075870 | Pi transporter | −1.177* | −0.785 | −1.809* | −1.408* | 11 |
| GRMZM2G088196 | Pi transporter | −0.300 | −0.131 | 0.470 | 0.354 | 7 |
| GRMZM2G092793 | Pi transporter | 0.043 | 0.015 | 0.007 | 0.307 | 2 |
| GRMZM2G139639 | Pi transporter | −1.364* | −1.134 | −2.161* | −1.671* | 11 |
| GRMZM2G146240 | Pi transporter | 0.193 | −0.063 | −0.085 | −0.012 | 4 |
| GRMZM2G310175 | Pi transporter | −0.043 | 0.348 | 0.159 | 0.332 | 1 |
| GRMZM2G045473 | PHT1-like high-affinity Pi transporter | −1.233* | −1.255 | −1.906* | −1.360* | 11a |
| GRMZM2G154090 | PHT1-like high-affinity Pi transporter | −1.098* | −0.902 | −1.816* | −1.349* | 11a |
| GRMZM2G092780 | PHT12-like low-affinity Pi transporter | −0.407 | −0.383 | −0.574 | −0.761 | 15a |
| GRMZM2G158489 | Protein transport protein SEC12p | −1.310* | −1.555 | −1.362* | −1.154* | 11a |
| SPX domain proteins | ||||||
| GRMZM2G335989 | PHO1 family, Pi transport and homeostasis | −1.033 | −1.038 | −2.287* | −1.770* | 11 |
| GRMZM2G466545 | PHO1 family, Pi transport and homeostasis | −0.243 | −0.134 | −0.656 | −0.782 | 20a |
| GRMZM2G083655 | SPX domain-containing protein1-like | −1.789* | −2.317 | −3.013* | −2.472* | 11 |
| GRMZM2G035579 | SPX domain-containing protein1-like | −3.868* | −3.555 | −3.896* | −3.527* | 11 |
| GRMZM2G171423 | SPX domain-containing protein1-like | −1.789* | −2.317 | −3.013* | −2.472* | 11a |
| GRMZM2G122108 | SPX domain-containing protein4-like | −1.109* | −1.350 | −2.090* | −1.228* | 11 |
| GRMZM2G370780 | SPX domain-containing protein5-like | −2.783* | −2.915 | −4.292* | −2.884* | 11a |
| GRMZM2G065989 | SPX domain-containing protein6-like | −4.573* | −4.197 | −6.061* | −4.368* | 11 |
| GRMZM2G086430 | SPX-MFS protein, SPX domain membrane protein | −1.854* | −2.408* | −2.691* | −2.058 | 11 |
| GRMZM2G050329 | SPX-MFS protein, SPX domain membrane protein | 1.358* | 1.662 | 1.456* | 3.520* | 1 |
| Transcription factors involved in Pi regulation | ||||||
| GRMZM2G024530 | PFT1-like bHLH transcription factor | 0.190 | 0.148 | 0.539 | 0.219 | 1a |
| GRMZM2G006477 | PHR1-like transcription factor | 0.093 | −0.082 | 0.006 | −0.402 | 14a |
| GRMZM2G162409 | PHR1-like transcription factor | −0.028 | −0.306 | −0.796 | −1.472 | 1a |
| GRMZM2G096358 | Transcription factor, Myb superfamily | −0.259 | −1.903 | −1.173* | −1.161 | 46a |
| Other Pi regulators | ||||||
| GRMZM2G436295 | Noncoding RNA involved in Pi allocation | −2.767 | −3.731 | −3.826* | −3.054* | 1a |
| GRMZM2G026391 | Oligopeptide transporter OPT superfamily | −1.200 | −1.316 | −0.879 | −1.295* | 6 |
| GRMZM2G464572 | PHO2-like ubiquitin E2 conjugase | −1.086* | −1.793 | −1.705* | −1.215* | 11a |
| GRMZM2G100652 | SQD2-like sulfolipid synthesis protein | −1.795* | −1.717 | −2.391* | −1.759* | 11a |
| GRMZM2G150496 | IPK1-like inositol-pentakisphosphate 2-kinase | −0.643 | −0.301 | −0.527 | −0.685 | 14a |
| GRMZM2G054050 | LPR1-like multicopper oxidase | −0.101 | −0.393 | −0.695 | −0.365 | 1a |
| GRMZM2G086727 | LPR1-like multicopper oxidase | −0.140 | −0.356 | −0.636 | −0.467 | 1a |
| GRMZM2G004414 | CAX1-like K+-dependent Na:Ca2+ antiporter | 0.337 | 0.294 | −0.535 | −0.576 | 28a |
| GRMZM2G011592 | CAX3-like Ca2+/H+ antiporter | −0.669 | −0.838 | −1.589 | −1.291 | 1a |
| GRMZM2G002999 | SIZ1-like SUMO E3 ligase | −0.036 | −0.070 | 0.193 | −0.105 | 1a |
| GRMZM2G155123 | SIZ1-like SUMO E3 ligase | 0.323 | 0.258 | 0.103 | 0.114 | 5a |
| GRMZM2G060824 | PDR2-like P-type ATPase cation pumps | 0.083 | 0.184 | 0.221 | −0.013 | 9a |
| GRMZM2G088487 | ARP6-like component in chromatin complex | 0.074 | 0.186 | 0.233 | 0.314 | 23a |
Transcripts that have been described by Calderón-Vázquez et al. (2011).
The extreme increase in phosphate concentration in leaves under low-N conditions had not been described in comparable studies with other plant species. In Arabidopsis, increases in phosphate concentration were limited to the root or much less dramatic (Tschoep et al., 2009; Krapp et al., 2011). In tomato shoots, phosphate accumulation only started after longer N stress (Urbanczyk-Wochniak and Fernie, 2005). The high phosphate concentration in the maize leaves could be caused by comparably high Pi supply in the nutrient solution and the length of the experiment. However, comparison of our results with the transcriptional response of maize leaves to low-N conditions in the experiments by Yang et al. (2011) shows that our results are in agreement with other studies. Nitrate and Pi deficiencies are both very common in nature. Both types of stresses have very different effects on cellular biochemistry and expression pattern but cause very similar responses on the whole-plant level, such as decrease in photosynthesis and growth retardation (Paul and Stitt, 1993; Morcuende et al., 2007). However, not much is known about the interaction of N- and Pi-responsive systems. A recently identified E3 ubiquitin ligase (N limitation adaptation) from Arabidopsis had regulatory function in nitrate response but could also modulate Pi homeostasis. In the proposed system, Pi uptake is enhanced by low nitrate conditions (Peng et al., 2007; Kant et al., 2011b). If a similar interaction is present in maize, the high phosphate concentration of the leaves could not just be a result of constant uptake during retarded growth but would have been further enhanced by increased uptake by the root.
Particularly interesting was the strong effect of nitrate deficiency on proteins containing the SPX domain (Table V). The proteins are represented in plants by a multigene family named after the SYG1 domain from yeast (Saccharomyces cerevisiae), which is involved in pheromone signal transduction; the Pho81 domain also from yeast, which is involved in inhibition of a cyclin-dependent kinase; and the XRP1 domain from humans (Homo sapiens), which works as a cell surface receptor. The characterization so far also suggests a regulatory function for the SPX domain, but the specific mechanism could not be elucidated so far (Duan et al., 2008). Many proteins carry the SPX domain alongside with other functional domains (including the above mentioned N limitation adaptation protein), but some proteins are just characterized by the SPX. Particularly prominent seemed the role of SPX domain proteins in regulation of Pi homeostasis (for review, see Secco et al., 2012). However, differential regulation of SPX family members had also been found for Fe stress (Nakanishi et al., 1993), cold stress and osmotic stress (Zhao et al., 2009), N stress (Peng et al., 2007), and light stress (Kang and Ni, 2006). N-deficient maize leaves were characterized by high Pi concentration and significant down-regulation of six SPX domain-containing proteins and one significantly up-regulated member of the family. Such a differential regulation had also been observed in Arabidopsis where three of the studied SPX genes were induced and one reduced by Pi starvation (Duan et al., 2008). One of the down-regulated SPX domain proteins in the N-deficient maize leaves belonged to the PHO1 like subfamily, and the proteins are supposed to be involved in Pi loading into the xylem, Pi sensing, and sorting of proteins to the endomembranes (Wang et al., 2004). Interestingly, by down-regulation of the PHO1 gene in Arabidopsis, it was recently possible to partly uncouple the Pi deficiency signal from the growth response, and plant shoots showed normal biomass accumulation despite a significant decrease in Pi supply (Rouached et al., 2011). Transgenic manipulation of Pi regulators had also impact onto the cold adaptation of plants (Hurry et al., 2000; Zhao et al., 2009).
CONCLUSION
Maize leaf samples were taken from two inbred lines at two different growth stages and from two different N treatments. Parallel transcript and metabolite profiling of the same biological material allowed the multilevel analysis of cellular responses to N deprivation. The limitation in N availability caused direct effects on the metabolite profile of the leaves, while transcript patterns were additionally influenced by the genotype and the growth stage of the maize plant. Genotype-specific differences occurred mainly in the quantity and in the temporal pattern of the response to low-N stress. Under early stress condition, A188 showed stronger disturbances in the transcript and metabolite profiles than B73. PSII efficiency measurements also indicated that A188 leaves might senesce faster under N deprivation. However, the final shoot biomass was affected by the stress to a very similar degree in A188 and B73, and additional experiments with a higher number of mature plants would be required for establishment of genotype-specific differences in sensitivity to N stress.
The adaptation to N deprivation was dominated in both genotypes by a readjustment of the primary N and C metabolism. Changes in the primary N assimilation pathways of maize leaves confirmed the picture obtained from other species, such as Arabidopsis, under comparable growth condition (Bi et al., 2007; Krapp et al., 2011). Regulation of nitrate reduction and ammonium assimilation seem to be controlled by independent mechanism. Nitrate reduction transcripts were significantly down-regulated by the stress, but ammonium assimilation into amino acids was not affected. This underlines the importance of the GS/GOGAT not just in primary assimilation but also for protein and amino acid turnover in the metabolically active leaf (Bernard and Habash, 2009). The amino acid pool decreased under N deprivation and the major amino acids reacted in coordination. Glu had previously been predicted to be stable under a variety of environmental stimuli (Krapp et al., 2011), but in the maize leaves, the pattern for this primary amino acid followed (or led) the general trend. Comparison with data from other species revealed that different amino acids are often individually regulated under N deprivation. In maize leaves, Trp and Lys deviated from the general pattern. Individual members of the Trp biosynthesis pathway were thereby down-regulated on transcriptional level.
The decrease in the N-containing amino acids was accompanied by a decrease in the organic acid pool, demonstrating that the biosynthesis of these C skeleton precursors was regulated according to their demand by amino acid synthesis in the maize source leaf. Reduction in transcript abundance could be detected for the later steps of glycolysis, e.g. pyrophosphate-dependent phosphofructokinase, phosphoglycerate mutase, enolase, and pyruvate kinase. A similar picture was observed for the OPP pathway, which is also connected to nitrate assimilation in nonphotosynthetic tissue. Other major C pathways, including synthesis of photosystems, the C4 shuttle, and the Calvin cycle, were also down-regulated. The close correlation in the behavior of genes from chloroplast metabolism was confirmed by correlation analysis and the enrichment of the transcripts in the same module (ME3). In contrast to the results from other species, N deprivation did not cause major accumulation of starch, especially under prolonged stress. Instead, parts of the assimilated C were directed into the synthesis of raffinose and related sugars, into the cell wall and some secondary products. Detailed knowledge about the composition of maize biomass under low N supply will be important for the development of specific lines for cultivation of energy crops. A significant difference to previous studies is the C4-specific division of primary N and C assimilation into mesophyll- and bundle sheath-specific processes. It will be interesting to analyze the relevance of this metabolic separation for the adaptation to N deficiency in more detail.
In the experimental setup, N deprivation caused a strong accumulation of Pi in the leaves. Since Pi concentration and compartmentation strongly influence the activity of many primary metabolic pathways, its homeostasis is tightly regulated. The increase in Pi was therefore accompanied by the down-regulation of almost all genes previously described for their involvement in the opposite situation during Pi starvation. To our surprise, the readjustment of Pi homeostasis actually dominated the transcriptional changes recorded under N deprivation. This demonstrates the importance of the Pi homeostasis for cellular metabolism, but so far the control of Pi balance in stress adaptation has not received much attention. Especially in crop systems under sufficient Pi supply, stress-related growth retardation could lead to a similar Pi accumulation in the plants. The importance of the Pi control mechanisms has been already been shown for cold adaptation in Arabidopsis (Hurry et al., 2000).
MATERIALS AND METHODS
Plant Growth
Maize (Zea mays) seeds were obtained from the University of Regensburg. After incubation on wet filter paper for 3 d, uniform seedlings were transferred into pots of 1.5-L volume containing nutrient-poor peat soil (Basissubstrat 2; Klasmann-Deilmann). Fertilization started at day 7 of the experiment with a modified Hoagland solution (5 mm CaCl2, 2 mm MgSO4, 2 mg/L Fe, 0.5 mm KH2PO4, 50 µm H3BO4, 10 µm MnCl2, 1 µm ZnSO4, 0.3 µm CuSO4, and 0.5 µm Na2MoO4). Depending on the N treatment scheme, the nutrient solution contained 15 mm for high-N treatment or 0.15 mm KNO3 for low-N treatment. The differences in potassium supply were balanced with KCl. Plants received 100 mL of nutrient solution every third day, between the fertilization supply, and plants were watered with distilled water depending on water status in the pot. The last watering with nutrient solution always happened 2 d before the harvest. The fertilization schema causes some fluctuation in nutrient supply of both treatments; therefore, vitality of the plants was monitored by daily measurements of leaf length from ligule of the most previous developed leaf until tip. Measurements started with appearance of leaf 3, and first differences in growth of high- and low-N-treated plants were recorded in leaf 4 around 16 d after germination. From that time point on, low-N-treated plants displayed lower leaf elongation rates. For the data in Table I, measurements from at least four consecutive days were used for calculation of leaf growth rate in cm d−1. Growth conditions in the plant climate chamber (PlantMaster PGR 3045; CLF Plant Climatics) were set at diurnal rhythm of 14 h of light (approximately 200 µmol m−2 s−1 at level of the soil surface and approximately 650 µmol m−2 s−1 just under the light source, which was adjusted according to plant height to give maximal possible irradiance) at 28°C and 80% humidity, and 10 h of night at 20°C and 50% humidity. Effective PSII quantum yield was measured with a Mini-PAM fluorometer (Heinz Walz). Harvest of plant material always started 2 h after start of the light period and was always completed before midday. The whole plant was cut at the soil surface and immediately weighed for shoot fresh weight analysis. The main leaf lamina (L5 at 20 d and L6 at 30 d after the start of the experiment) was harvested 2 cm above the appearance point and the top 6 cm from the tip were removed. The remaining middle part of the leaf was immediately frozen in liquid N and stored at −80°C. The whole leaf was homogenized with cooled mortar and pistil and aliquoted under liquid N for transcriptome and metabolome analysis. A subsample of leaf and stem material was dried at 80°C for 3 d for determination of plant dry weight. Since there was no significant difference in fresh/dry weight ratio of low- and high-N plants, biomass of plant is usually given as fresh weight. The study was conducted in two independent biological experiments, and data presented per genotype, growth stage, and treatment always combined samples from both experiments.
Microarray Analysis
RNA was extracted from four biological replicates per genotype, growth stage, and treatment after the method described by Logemann et al. (1987) using 100 mg of frozen leaf material. The isolated RNA was purified with the RNeasy purification kit according to the manufacturer’s instructions (Qiagen). The quality was checked with the Agilent 2100 Bioanalyzer using the RNA 6000 Nano kit. The copy DNA and following antisense copy RNA synthesis was performed according the one-color microarray-based gene expression analysis protocol (Agilent Technologies). An aliquot of 1.65 mg of this RNA was loaded on one-color microarrays with custom-designed oligonucleotide probes (Agilent 025271). Validation of data for the described microarray method by quantitative reverse transcription-PCR was demonstrated by Pick et al. (2011). Transcripts were normalized to the 75th percentile followed by baseline correction using the Agilent Gene spring GX11.0 software. Annotation and classification followed the characterization of the Agilent 025271 array chip by Pick et al. (2011). Transcripts from the primary C and N pathways presented in the article were manually characterized in more detail by another round of blast searches in the National Center for Biotechnology Information database. In the summarized data, only the data from one representative signal output are shown when multiple identifiers were present for the same transcript (alignment according to the 4a.53 B73 genome assembly). Arrays can be accessed under accession number GSE40678 in the National Center for Biotechnology Information Gene Expression Omnibus database.
Metabolites
Lyophilized tissue equivalent to 200 mg of fresh weight was used for metabolite profiling. Metabolites were extracted with the use of accelerated solvent extraction with polar (methanol + water, 80 + 20 by volume) and nonpolar (methanol + dichloromethane, 40 + 60 by volume) solvents. Subsequent analyses of metabolites by gas chromatography-mass spectrometry were performed as described elsewhere (Roessner et al., 2000; Walk, 2007). Suc and starch levels were determined in 100 mg of homogenized leaf material extracted with 80% (v/v) ethanol and 20 mm HEPES, pH 7.5, as described by Stitt et al. (1989), but adapted for determination in a plate reader by direct downscaling the assay to a volume of 200 µL. Five replicate samples were measured per genotype, growth stage, and N treatment. Significant differences were calculated by Student’s t test and cutoff level set at P > 0.05.
Data Analysis
Statistical analysis of microarray data was performed using the Agilent GeneSpring GX11.0 program. MarkerView software 1.2 (Applied Biosystems) was used for PCA of transcript and metabolite data using the Pareto algorithm. In the top 50 lists, double readings for the same genome loci were reduced to one entry. Correlation network analysis was performed in the R statistical environment (version 2.13.1) with the package WGCNA (Langfelder and Horvath, 2008). WGCNA is an integrated pipeline that automatically performs several steps for network construction. First, the Pearson correlation over all samples of the gene expression profiles is calculated. The absolute values of the pairwise correlation coefficients are raised to the power b (default b = 6) and are then transformed into a topological overlap matrix that stores connectivity information between expression profiles. Genes are partitioned into coexpression modules by a hierarchical clustering of the topological overlap matrix. Finally, each module is assigned a representative expression profile, its ME. Hub genes (i.e. those genes highly connected to many other genes) have been shown to be strongly correlated to the ME of the module they are contained in (Horvath and Dong, 2008). We applied the WGCNA pipeline with default parameters to the complete set of normalized expression profiles. The WGCNA network screening function was then called to find genes that are both hub genes and highly correlated to fresh weight, growth rate, or metabolite profiles. Pairwise correlation coefficients between selected MEs as well as phenotypic and metabolic measurements were clustered using the standard R function hclust and visualized in a dendrogram. GO term enrichment was performed using the AgriGO Web tool (Du et al., 2010). VANTED (Junker et al., 2006) and the Multiexperiment Viewer (Saeed et al., 2003) were used for visualization of data.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1.1. PCA organization of transcript data (41,780 features) after PC1 (46%) and PC2 (26%).
Supplemental Figure S1.2. Overview of expression pattern in ME.
Supplemental Table S1. Top 50 transcripts associated with high-N and low-N treatment (PC3 loadings from PCA; see Figure 2A), and top 20 of known metabolites associated with high- and low-N treatment (PC1 from PCA in Figure 2B).
Supplemental Table S2. Metabolites significantly different in source leaf lamina from low- and high-N-treated maize seedlings, and transcripts significantly different in source leaf lamina from low- and high-N-treated maize seedlings.
Supplemental Table S3. Organization of transcripts according into modules by WGCNA and association of the modules with GO terms and experimental parameters.
Supplemental Table S4. Overview of N-treatment-dependent changes in transcripts of primary C and N metabolism.
Supplemental Table S5. Overview of significant N-treatment-dependent changes in transcripts associated with regulatory processes (cytokinin and auxin metabolism, transcription factors, kinases, and phosphatases).
Supplementary Material
Acknowledgments
We thank Dr. Sophia Sonnewald and Stephen Reid for setup and realization of the microarray experiments and all other members of the OPTIMAS project for useful discussions.
Glossary
- PEPC
phosphoenolpyruvate carboxylase
- PEP
phosphoenolpyruvate
- PPDK
pyruvate orthophosphate dikinase
- GS
Gln synthetase
- GOGAT
Glu synthase
- PCA
principal component analysis
- WGCNA
weighted correlation network analysis
- GO
gene ontology
- TCA
tricarboxylic acid
- OPP
oxidative pentose phosphate
- ME
module eigengene
- C
carbon
- N
nitrogen
- P
phosphate
- Pi
inorganic phosphate
References
- Becker TW, Carrayol E, Hirel B. (2000) Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: localization, relative proportion and their role in ammonium assimilation or nitrogen transport. Planta 211: 800–806 [DOI] [PubMed] [Google Scholar]
- Bernard SM, Habash DZ. (2009) The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol 182: 608–620 [DOI] [PubMed] [Google Scholar]
- Bi YM, Wang RL, Zhu T, Rothstein SJ. (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics 8: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, Sommer M, Gagneul D, Weber KL, Carr KM, Gowik U, Mass J, Lercher MJ, et al. (2011) An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol 155: 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L. (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59: 2479–2497 [DOI] [PubMed] [Google Scholar]
- Calderón-Vázquez C, Sawers RJ, Herrera-Estrella L. (2011) Phosphate deprivation in maize: genetics and genomics. Plant Physiol 156: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Barreto H. (1997) Using a chlorophyll meter to estimate specific leaf nitrogen of tropical maize during vegetative growth. Agron J 89: 557–562 [Google Scholar]
- Coleman HD, Yan J, Mansfield SD. (2009) Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci USA 106: 13118–13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechorgnat J, Nguyen CT, Armengaud P, Jossier M, Diatloff E, Filleur S, Daniel-Vedele F. (2011) From the soil to the seeds: the long journey of nitrate in plants. J Exp Bot 62: 1349–1359 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Edwards GE, Robinson SP, Tyler NJ, Walker DA. (1978) Photosynthesis by isolated protoplasts, protoplast extracts, and chloroplasts of wheat: influence of orthophosphate, pyrophosphate, and adenylates. Plant Physiol 62: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kereamy A, Guevara D, Bi Y-M, Chen X, Rothstein SJ. (2011) Exploring the molecular and metabolic factors contributing to the adaptation of maize seedlings to nitrate limitation. Front Plant Sci 2: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX. (2012) Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J 69: 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S-C, Lin C-S, Hsu P-K, Lin S-H, Tsay Y-F. (2009) The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Martinoia E. (2009) Malate. Jack of all trades or master of a few? Phytochemistry 70: 828–832 [DOI] [PubMed] [Google Scholar]
- Ficklin SP, Feltus FA. (2011) Gene coexpression network alignment and conservation of gene modules between two grass species: maize and rice. Plant Physiol 156: 1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficklin SP, Luo F, Feltus FA. (2010) The association of multiple interacting genes with specific phenotypes in rice using gene coexpression networks. Plant Physiol 154: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge U-I. (1999) Phosphate translocators in plastids. Annu Rev Plant Physiol Plant Mol Biol 50: 27–45 [DOI] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. (2010) Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz C, Mueller C, Matt P, Feil R, Stitt M. (2006a) Impact of the C-N status on the amino acid profile in tobacco source leaves. Plant Cell Environ 29: 2055–2076 [DOI] [PubMed] [Google Scholar]
- Fritz C, Palacios-Rojas N, Feil R, Stitt M. (2006b) Regulation of secondary metabolism by the carbon-nitrogen status in tobacco: nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J 46: 533–548 [DOI] [PubMed] [Google Scholar]
- Gaude N, Bréhélin C, Tischendorf G, Kessler F, Dörmann P. (2007) Nitrogen deficiency in Arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. Plant J 49: 729–739 [DOI] [PubMed] [Google Scholar]
- Ghannoum O, Evans JR, Caemmerer SV. (2011) Nitrogen and water use efficiency of C4 plants. In AS Raghavendra, RF Sage, eds, C4 Photosynthesis and Related CO2 Concentration Mechanisms, Springer Science and Business Media, Dordrecht, The Netherlands, pp 129–146 [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah MA, Zuther E, Buchel K, Heyer AG. (2006) Transport and metabolism of raffinose family oligosaccharides in transgenic potato. J Exp Bot 57: 3801–3811 [DOI] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N. (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Hirel B, Bertin P, Quilleré I, Bourdoncle W, Attagnant C, Dellay C, Gouy A, Cadiou S, Retailliau C, Falque M, et al. (2001) Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol 125: 1258–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Le Gouis J, Ney B, Gallais A. (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58: 2369–2387 [DOI] [PubMed] [Google Scholar]
- Hirel B, Martin A, Terce-Laforgue T, Gonzalez-Moro M-B, Estavillo J-M. (2005) Physiology of maize I: A comprehensive and integrated view of nitrogen metabolism in a C4 plant. Physiol Plant 124: 167–177 [Google Scholar]
- Hodges M. (2002) Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J Exp Bot 53: 905–916 [DOI] [PubMed] [Google Scholar]
- Horvath S, Dong J. (2008) Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol 4: e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth JR, Parmar S, Jones J, Shepherd CE, Corol D-I, Galster AM, Hawkins ND, Miller SJ, Baker JM, Verrier PJ, et al. (2008) Co-ordinated expression of amino acid metabolism in response to N and S deficiency during wheat grain filling. J Exp Bot 59: 3675–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JL, Redinbaugh MG, Huber SC, Campbell WH. (1994) Regulation of maize leaf nitrate reductase activity involves both gene expression and protein phosphorylation. Plant Physiol 106: 1667–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurry V, Strand A, Furbank R, Stitt M. (2000) The role of inorganic phosphate in the development of freezing tolerance and the acclimatization of photosynthesis to low temperature is revealed by the pho mutants of Arabidopsis thaliana. Plant J 24: 383–396 [DOI] [PubMed] [Google Scholar]
- Iglesias AA, Plaxton WC, Podesta FE. (1993) The role of inorganic phosphate in the regulation of C4 photosynthesis. Photosynth Res 35: 205–211 [DOI] [PubMed] [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N. (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Junker BH, Klukas C, Schreiber F. (2006) VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Ni M. (2006) Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi Y-M, Rothstein SJ. (2011a) Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J Exp Bot 62: 1499–1509 [DOI] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ. (2011b) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamis S, Lamaze T, Lemoine Y, Foyer C. (1990) Adaptation of the photosynthetic apparatus in maize leaves as a result of nitrogen limitation: relationships between electron transport and carbon assimilation. Plant Physiol 94: 1436–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Takamiya K, Ohta H. (2004) Arabidopsis type B monogalactosyldiacylglycerol synthase genes are expressed during pollen tube growth and induced by phosphate starvation. Plant Physiol 134: 640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogel KH, Voll LM, Schäfer P, Jansen C, Wu Y, Langen G, Imani J, Hofmann J, Schmiedl A, Sonnewald S, et al. (2010) Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific variances. Proc Natl Acad Sci USA 107: 6198–6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou J-P, Daniel-Vedele F. (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DW. (2002) Carbon and nitrogen assimilation in relation to yield: mechanisms are the key to understanding production systems. J Exp Bot 53: 773–787 [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. (2011) Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol 156: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Wang S, Zhang J, Feng Q, Zhang L, Fan D, Li X, Yuan D, Han B, Zhang Q. (2006) Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol Biol 60: 617–631 [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Lu C, Zhang J. (2000) Photosynthetic CO2 assimilation, chlorophyll fluorescence and photoinhibition as affected by nitrogen deficiency in maize plants. Plant Sci 151: 135–143 [DOI] [PubMed] [Google Scholar]
- Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GT, Meyer RC, Messerli G, Gronlund JT, et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152: 2142–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, Scheible WR. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Okumura N, Umehara Y, Nishizawa NK, Chino M, Mori S. (1993) Expression of a gene specific for iron deficiency (Ids3) in the roots of Hordeum vulgare. Plant Cell Physiol 34: 401–410 [PubMed] [Google Scholar]
- Neyra CA, Hageman RH. (1978) Pathway for nitrate assimilation in corn (Zea mays L.) leaves: cellular distribution of enzymes and energy sources for nitrate reduction. Plant Physiol 62: 618–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Novitskaya L, Lea PJ, Foyer CH. (2002) Co-ordination of leaf minor amino acid contents in crop species: significance and interpretation. J Exp Bot 53: 939–945 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Okumoto S, Pilot G. (2011) Amino acid export in plants: a missing link in nitrogen cycling. Mol Plant 4: 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Driscoll SP. (1997) Sugar repression of photosynthesis: the role of carbohydrates in signalling nitrogen deficiency through source:sink imbalance. Plant Cell Environ 20: 110–116 [Google Scholar]
- Paul MJ, Foyer CH. (2001) Sink regulation of photosynthesis. J Exp Bot 52: 1383–1400 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Pellny TK. (2003) Carbon metabolite feedback regulation of leaf photosynthesis and development. J Exp Bot 54: 539–547 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Stitt M. (1993) Effects of nitrogen and phosphorus deficiency on levels of carbohydrates, respiratory enzymes and metabolites in seedlings of tobacco and their response to exogenous sucrose. Plant Cell Environ 16: 1047–1057 [Google Scholar]
- Peng M, Bi YM, Zhu T, Rothstein SJ. (2007) Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol 65: 775–797 [DOI] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schlüter U, Denton AK, Colmsee C, Scholz U, Fahnenstich H, Pieruschka R, Rascher U, Sonnewald U, et al. (2011) Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23: 4208–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J. (1982) Regulation of the biosynthesis and degradation of starch. Annu Rev Plant Physiol 33: 431–454 [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC. (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Rouached H, Stefanovic A, Secco D, Bulak Arpat A, Gout E, Bligny R, Poirier Y. (2011) Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J 65: 557–570 [DOI] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2003) Nitrate-specific and cytokinin-mediated nitrogen signaling pathways in plants. J Plant Res 116: 253–257 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J. (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Shenton M, Fontaine V, Hartwell J, Marsh JT, Jenkins GI, Nimmo HG. (2006) Distinct patterns of control and expression amongst members of the PEP carboxylase kinase gene family in C4 plants. Plant J 48: 45–53 [DOI] [PubMed] [Google Scholar]
- Smyth DA, Chevalier P. (1984) Phosphate accumulation in leaves of wheat seedlings measured in leaf cell protoplasts isolated after root or foliar treatment with orthophosphate. Physiol Plant 61: 135–140 [Google Scholar]
- Stitt M, Lilley R, Gerhardt R, Heldt H. (1989) Determination of metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol 174: 518–552 [Google Scholar]
- Strable J, Borsuk L, Nettleton D, Schnable PS, Irish EE. (2008) Microarray analysis of vegetative phase change in maize. Plant J 56: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Sugiharto B, Miyata K, Nakamoto H, Sasakawa H, Sugiyama T. (1990) Regulation of expression of carbon-assimilating enzymes by nitrogen in maize leaf. Plant Physiol 92: 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Takahashi T, Sugiyama T, Yamaya T, Sakakibara H. (2002) Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J Exp Bot 53: 971–977 [DOI] [PubMed] [Google Scholar]
- Tian Q, Chen F, Liu J, Zhang F, Mi G. (2008) Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J Plant Physiol 165: 942–951 [DOI] [PubMed] [Google Scholar]
- Torney F, Moeller L, Scarpa A, Wang K. (2007) Genetic engineering approaches to improve bioethanol production from maize. Curr Opin Biotechnol 18: 193–199 [DOI] [PubMed] [Google Scholar]
- Trevisan S, Borsa P, Botton A, Varotto S, Malagoli M, Ruperti B, Quaggiotti S. (2008) Expression of two maize putative nitrate transporters in response to nitrate and sugar availability. Plant Biol (Stuttg) 10: 462–475 [DOI] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M. (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32: 300–318 [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G. (2010) New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant 3: 956–972 [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Fernie AR. (2005) Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. J Exp Bot 56: 309–321 [DOI] [PubMed] [Google Scholar]
- Walk T, inventor. (February 1, 2007) Means and methods for analyzing a sample by means of chromatography-mass spectrometry. International Patent WO 2007/012643
- Wang R, Guegler K, LaBrie ST, Crawford NM. (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y. (2004) Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol 135: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XS, Wu J, Ziegler TE, Yang X, Zayed A, Rajani MS, Zhou D, Basra AS, Schachtman DP, Peng M, et al. (2011) Gene expression biomarkers provide sensitive indicators of in planta nitrogen status in maize. Plant Physiol 157: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelitch I, Schultes NP, Peterson RB, Brown P, Brutnell TP. (2009) High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol 149: 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Liu F, Xu W, Di C, Zhou S, Xue Y, Yu J, Su Z. (2009) Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnol J 7: 550–561 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.