Abstract
Chloroplasts and other members of the plastid organelle family contain a small genome of bacterial ancestry. Young chloroplasts contain hundreds of genome copies, but the functional significance of this high genome copy number has been unclear. We describe molecular phenotypes associated with mutations in a nuclear gene in maize (Zea mays), white2 (w2), encoding a predicted organellar DNA polymerase. Weak and strong mutant alleles cause a moderate (approximately 5-fold) and severe (approximately 100-fold) decrease in plastid DNA copy number, respectively, as assayed by quantitative PCR and Southern-blot hybridization of leaf DNA. Both alleles condition a decrease in most chloroplast RNAs, with the magnitude of the RNA deficiencies roughly paralleling that of the DNA deficiency. However, some RNAs are more sensitive to a decrease in genome copy number than others. The rpoB messenger RNA (mRNA) exhibited a unique response, accumulating to dramatically elevated levels in response to a moderate reduction in plastid DNA. Subunits of photosynthetic enzyme complexes were reduced more severely than were plastid mRNAs, possibly because of impaired translation resulting from limiting ribosomal RNA, transfer RNA, and ribosomal protein mRNA. These results indicate that chloroplast genome copy number is a limiting factor for the expression of a subset of chloroplast genes in maize. Whereas in Arabidopsis (Arabidopsis thaliana) a pair of orthologous genes function redundantly to catalyze DNA replication in both mitochondria and chloroplasts, the w2 gene is responsible for virtually all chloroplast DNA replication in maize. Mitochondrial DNA copy number was reduced approximately 2-fold in mutants harboring strong w2 alleles, suggesting that w2 also contributes to mitochondrial DNA replication.
Chloroplasts and mitochondria evolved from bacterial endosymbionts and have retained small genomes encoding components of the organellar gene expression and energy transduction machineries (Timmis et al., 2004). In multicellular plants, chloroplasts belong to a family of organelles, the plastids, that share the same genome but that adopt different functions in different cell types. In most species, plastid genes comprise a circular genetic map that includes a pair of large inverted repeats flanking a “small” and “large” single-copy region (Green, 2011). However, the plastid genome is found primarily in various multimeric, subgenomic, linear, and branched forms in vivo (Bendich, 2004; Day and Madesis, 2007). Developing photosynthetic tissues in angiosperms contain thousands of chloroplast gene copies per cell (for review, see Day and Madesis, 2007). It is generally agreed that the ratio of chloroplast to nuclear gene copy number decreases as leaf cells mature, although the magnitude of this decrease has been a subject of debate (Baumgartner et al., 1989; Oldenburg and Bendich, 2004a; Rowan et al., 2004; Li et al., 2006; Shaver et al., 2006; Zoschke et al., 2007; Rowan et al., 2009).
Two hypotheses have been put forward regarding the functional significance of the high genome copy number in chloroplasts (Bendich, 1987): (1) The high copy number may compensate for stochastic DNA partitioning during plastid division, and (2) the high gene dosage may be necessary to provide an adequate supply of plastid ribosomal RNA (rRNA) to support the initial synthesis of the photosynthetic apparatus at the onset of chloroplast development. In principle, these hypotheses could be addressed by experimentally manipulating plastid DNA levels. Toward this end, a severalfold reduction in plastid DNA content was achieved in the unicellular alga Chlamydomonas reinhardtii by growing cells for up to 2 d in the presence of the DNA replication inhibitor 5-fluorodeoxyuridine (Hosler et al., 1989; Eberhard et al., 2002). Rates of chloroplast transcription roughly paralleled the level of chloroplast DNA, indicating that DNA content limits rates of transcription. However, reduced transcription rates were often not reflected by reduced mRNA levels, translation rates, or the abundance of the protein products. These results indicated that chloroplast mRNAs in C. reinhardtii are synthesized in excess of the amount that is needed to maintain the preassembled photosynthetic apparatus over the short time scale that was probed.
Analogous experiments in vascular plants would be of particular interest because the de novo assembly of the photosynthetic apparatus at the onset of chloroplast differentiation requires a massive synthesis of plastid-encoded proteins. In this study, we explored the quantitative relationship between chloroplast DNA and chloroplast gene products (mRNA, rRNA, transfer RNA [tRNA], and proteins) by taking advantage of maize (Zea mays) mutants with insertions in the nuclear gene white2 (w2), which we show encodes a chloroplast DNA polymerase. We found that a relatively modest decrease in chloroplast DNA content was reflected by a substantial decrease in chloroplast gene expression, with some mRNAs more sensitive to genome copy decrease than others. Our results show further that the division of labor between the pair of nucleus-encoded organelle DNA polymerases in maize and Arabidopsis (Arabidopsis thaliana) differs: in Arabidopsis, both organellar polymerases are dually targeted to chloroplasts and mitochondria and are largely redundant in function (Parent et al., 2011), whereas maize w2 is essential for virtually all chloroplast DNA replication. Finally, the severe loss of plastid DNA revealed by Southern-blot hybridization and real-time quantitative PCR (qPCR) assays of leaf DNA in w2 mutants indicates that the bulk of the signal in normal leaf tissue arises from bona fide chloroplast genomes and not from plastid gene fragments in the nuclear genome, contrasting with conclusions in several prior reports (Kumar and Bendich, 2011; Zheng et al., 2011).
RESULTS
Maize Locus w2 (GRMZM2G480171) Encodes a Chloroplast DNA Polymerase That Is Essential for Chloroplast Development
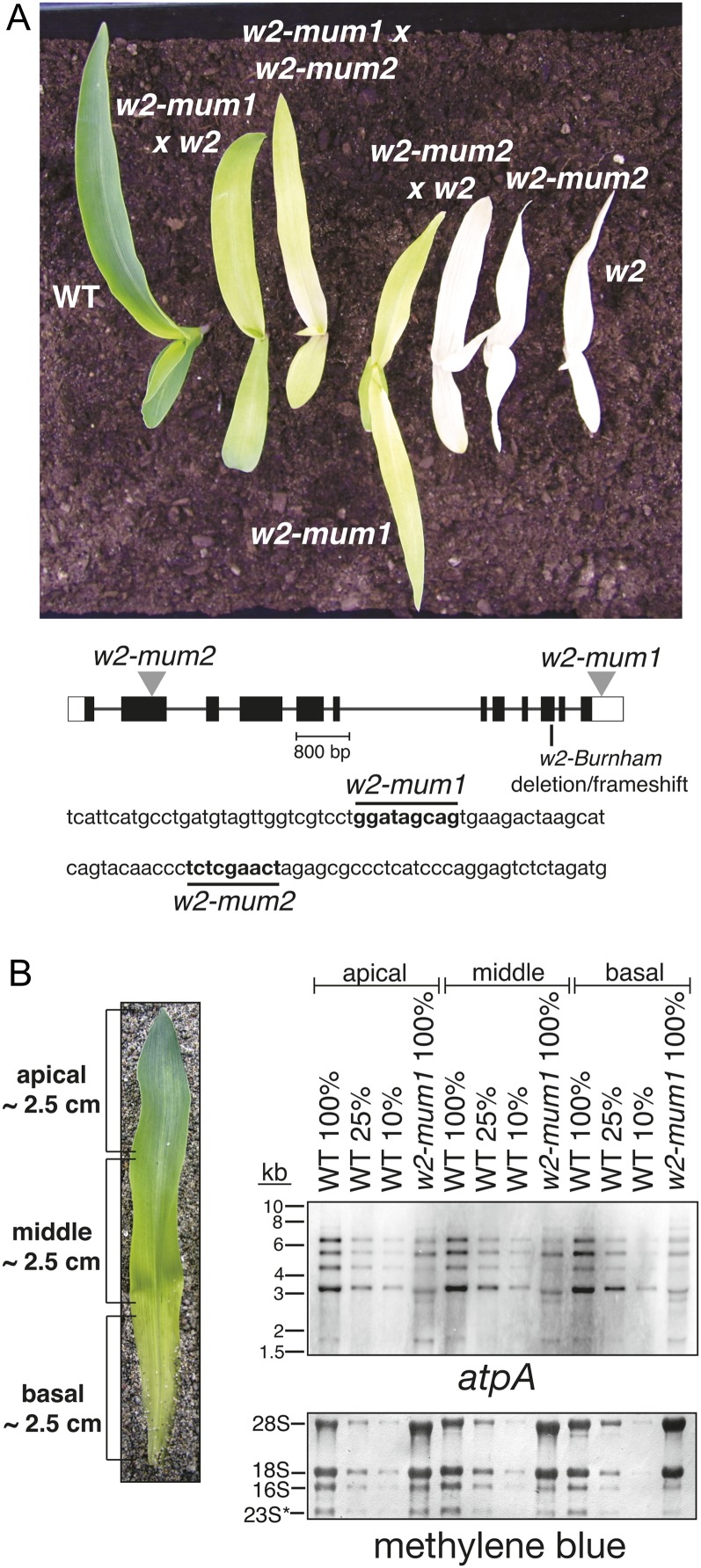
To identify the nuclear gene complement in maize required for chloroplast biogenesis, we developed a large collection of nonphotosynthetic maize mutants selected from lines with active Mu transposons (Stern et al., 2004). As an initial step toward elucidating gene functions and prioritizing mutants for gene identification, the abundance of photosynthetic enzyme complexes and chloroplast mRNAs have been systematically quantified. One mutant with very pale yellow-green leaves (see w2-mum1 in Fig. 1A) was notable in that the abundance of chloroplast atpA mRNA was reduced, but the RNA processing pattern was unaltered (Fig. 1B). Because this phenotype was unique among the many mutants we had analyzed, we selected this mutant for gene identification.
Figure 1.
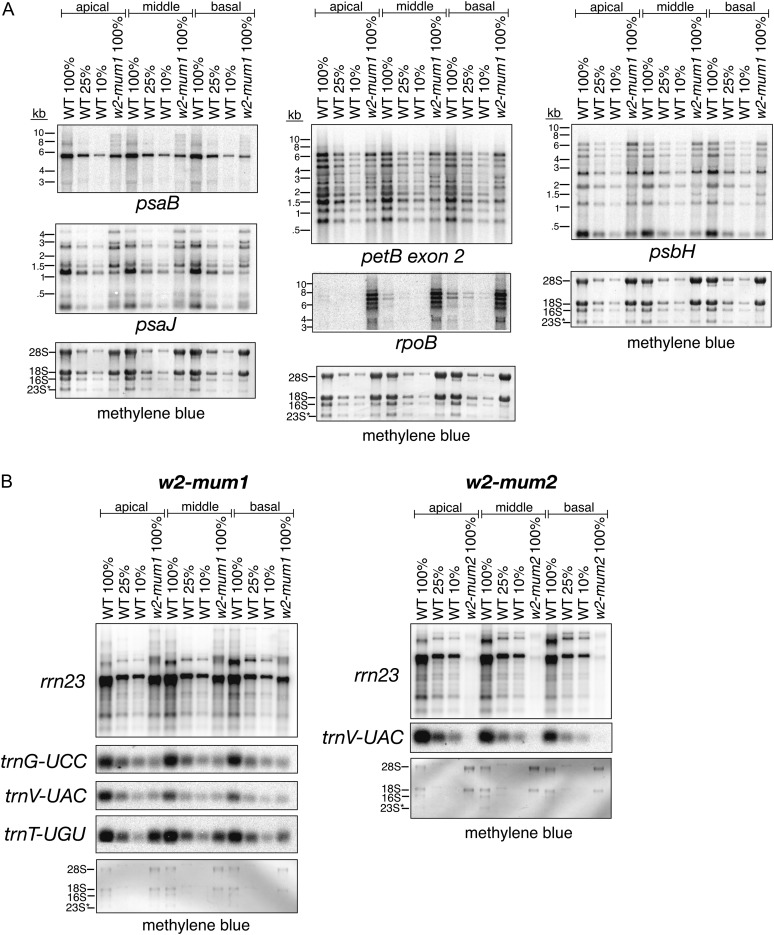
Overview of w2 mutant alleles. A, Seedling phenotype of w2 mutants. Plants of the indicated genotype were grown for 8 d under day/night cycles as described in “Materials and Methods.” Seedlings at this stage, in which the third leaf is emerging from the whorl, were used for all experiments in this study. The w2-Burnham allele is labeled w2 here and throughout. The heteroallelic progeny of complementation crosses are indicated by the two parental alleles. The position of each mutation within the GRMZM2G480171 gene model is diagrammed below. Unshaded rectangles represent transcribed, untranslated regions. The sequences flanking the Mu insertions sites are shown, with the nine base pair target duplication marked in boldface. B, Reduced abundance of chloroplast atpA mRNA in w2-mum1 homozygotes. Four micrograms of RNA purified from each of three sections of the second seedling leaf (left) was analyzed by RNA gel-blot hybridization using an atpA specific probe. A dilution series of the wild type sample was included to aid in quantification. The methylene blue-stained blot below shows the abundance of rRNAs in each sample. 28S and 18S are cytosolic rRNAs; 16S is plastid 16S rRNA, and 23S* is a cleavage product of plastid 23S rRNA.
To determine which of the approximately 100 Mu insertions in this mutant was the cause of the phenotype, we used an Illumina-based method to identify cosegregating insertions (Williams-Carrier et al., 2010; see “Materials and Methods”). An insertion in the 3′-untranslated region of a gene encoding a predicted organellar DNA polymerase (GRMZM2G480171) was identified in this manner; this was an appealing candidate for the causal mutation, as a reduction in plastid DNA copy number could account for the macroscopic and molecular phenotypes observed. To test this premise, a second insertion in the same gene was recovered via a reverse genetic screen of our mutant collection (see “Materials and Methods”); this insertion maps in an exon and cosegregates with an albino small seedling phenotype (Fig. 1A, w2-mum2). Complementation crosses between the two alleles yielded chlorophyll-deficient progeny (Fig. 1A, w2-mum × w2-mum2), confirming that the pigment deficiencies result from disruption of this gene. We suspected these mutants to be allelic to the uncloned maize locus w2 based on concordant map positions and the fact that w2 mutants are albino and have reduced chloroplast DNA and mRNA levels (Han et al., 1993). Complementation crosses confirmed allelism with a w2 allele denoted w2-Burnham (Lambert, 1980; Fig. 1A). Therefore, w2 corresponds to GRMZM2G480171. We named our alleles w2-mum1 and w2-mum2 to respect the original gene nomenclature while also indicating that they are Mu insertion alleles.
The visual phenotypes conditioned by w2-mum1 and w2-mum2 suggested that they are weak and strong alleles, respectively. This is consistent with the positions of the Mu insertions (3′-untranslated region versus exon; Fig. 1A) and with the nature of the mRNA produced from each mutant allele (Supplemental Fig. S1). The exon insertion in w2-mum2 results in a missplicing event that joins a known splice donor in the 5′-terminal inverted repeat of the Mu1 element (Ortiz and Strommer, 1990) to the start of the next exon in the w2 gene (Supplemental Fig. S1). The protein produced by this aberrant mRNA lacks 52 amino acids of the 3′→ 5′ exonuclease domain and includes 47 Mu-encoded amino acids in their place. The severe loss of plastid DNA conditioned by this allele (see below) indicates that the aberrant protein encoded by this allele retains little, if any, function. We identified the lesion in the w2-Burnham allele by sequencing PCR products derived from genomic and cDNA. Both methods revealed a deletion of one base pair near the 3′ end of the protein coding region, which results in a frame shift that eliminates 96 amino acids from the highly conserved C terminus of the protein (see Fig. 1A; Supplemental Fig. S1B). This mutation can account for the strong mutant phenotype conditioned by the w2-Burnham allele.
w2 has a closely related paralog (GRMZM2G023422). These genes are predicted co-orthologs of Arabidopsis genes POL1A and POL1B (AT1G50840/AT3G20540), and they encode proteins in the DNA polymerase A superfamily. The Arabidopsis co-orthologs are each dually targeted to mitochondria and chloroplasts and have largely overlapping functions; disruption of each single gene caused no visible phenotype and only a minor reduction in organellar DNA content, whereas the double mutant is embryo lethal (Parent et al., 2011). The strong visible phenotype of w2 mutants contrasts with those findings, and shows that w2 function is essential for chloroplast biogenesis. This is consistent with proteome studies, which have detected W2 but not its paralog in various chloroplast subfractions (Majeran et al., 2012).
w2-mum1 and w2-mum2 Condition a Moderate and Severe Loss of Plastid DNA, Respectively
As an initial test of the effects of these mutations on organelle DNA copy number, we assayed the ratio of organellar to nuclear DNA by Southern-blot hybridization of leaf DNA in w2-mum1 mutants (Fig. 2). Because organelle DNA copy number changes during leaf cell differentiation (Baumgartner et al., 1989; Shaver et al., 2006), we sampled different developmental stages by taking advantage of the developmental gradient in the leaves of maize seedlings (Leech et al., 1973). The second leaf of seedlings at the early three-leaf stage was divided into basal, middle, and apical sections (Fig. 1B), which contain immature (pale green), young, and mature chloroplasts, respectively. The sampled material did not include the least differentiated plastids, which are found in more basal regions. Total DNA was extracted, digested with EcoRI, and used for Southern-blot hybridization using probes for four regions of the plastid genome (Fig. 2). Comparison with equivalent material in normal siblings showed that the ratio of chloroplast to nuclear DNA was reduced approximately 5-fold at all stages, whereas the single mitochondrial gene assayed was unaffected. Southern hybridization of BamHI-digested leaf DNA with a different region of the plastid genome previously showed that the w2-Burnham allele conditions a severe loss of plastid DNA (Han et al., 1993). It has been suggested that Southern analysis with restriction enzymes such as EcoRI and BamHI, which do not distinguish methylated from unmethylated nuclear DNA, cannot accurately quantify organellar DNA because of the confounding signal from plastid gene fragments in the nuclear genome (known as NUPTs; Kumar and Bendich, 2011; Zheng et al., 2011). However, the fact that this Southern blotting approach revealed a consistent reduction in the plastid/nuclear DNA ratio accompanying two mutations affecting a chloroplast DNA polymerase provides strong evidence that the method typically provides valid results.
Figure 2.
Southern-blot hybridization showing reduction in plastid DNA in w2-mum1 leaf tissue. DNA was purified from the apical, middle, and basal sections of the second seedling leaf, digested with EcoRI, and analyzed by Southern-blot hybridization using the indicated probes. Wild-type DNA came from phenotypically normal siblings of the w2-mum1 plants. Mitochondrial DNA was detected with the mt-rps4 probe. The 18S probe detects the highly repetitive nuclear DNA encoding 18S rRNA.
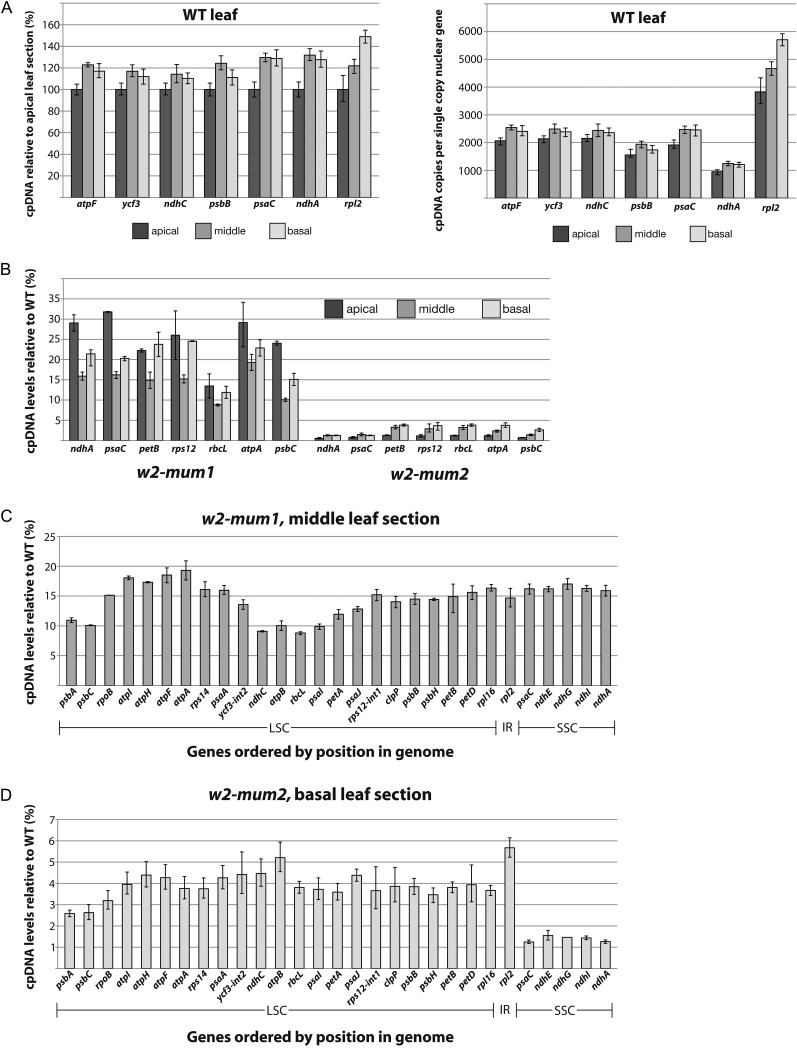
To more thoroughly catalog and quantify the effects of the w2-mum mutations on chloroplast DNA levels, we used qPCR to sample 29 DNA segments distributed throughout the plastid genome. The single copy nuclear gene HMG-1/2 (GRMZM2G024976) was used as the internal control. The ratio of plastid-to-nuclear DNA copy number was approximately 30% lower in the apical section than in the basal section of wild-type leaves (Fig. 3A, left; Supplemental Fig. S2A), in accordance with previous reports that chloroplast DNA copy number decreases modestly in mature chloroplasts, following a maximum early in leaf differentiation (Baumgartner et al., 1989; Zoschke et al., 2007). The ratio of plastid to nuclear gene copy number was estimated for those amplicons whose size and amplification efficiencies were similar to those of the single copy nuclear control (Fig. 3A, right): genes in the plastid single copy regions were approximately 2,000-fold more abundant than the nuclear gene, whereas the rpl2 gene in the plastid inverted repeat was roughly twice that abundant. These numbers varied only slightly among the three developmental stages.
Figure 3.
qPCR analysis of chloroplast DNA levels in wild-type and w2 mutant plants. The average of three technical replicates is plotted, with maxima and minima indicated by error bars. The basal, middle, and apical leaf sections are diagrammed in Figure 1B. A, Chloroplast copy number during the development of wild-type leaves. The signal for the indicated plastid gene relative to the single-copy nuclear control is plotted on the left as a percent of the ratio in the apical leaf section. Results from additional amplicons are provided in Supplemental Figure S2A. The absolute ratio of chloroplast gene copy number relative to the single-copy nuclear control is plotted to the right; amplicons used for this analysis were similar in size and amplification efficiency to the nuclear control. The rpl2 gene is in the inverted-repeat region of the plastid genome, whereas the other genes are in single-copy regions. B, Ratio of plastid-to-nuclear DNA copy number in w2 mutants. Values are expressed as a percentage of the ratio in the corresponding wild-type tissue. Results from additional amplicons are shown in Supplemental Figure S2B. C, Position dependence of plastid-to-nuclear gene copy number in the middle section of w2-mum1 mutant leaf tissue. Amplicons are ordered according to their position in the plastid genome. Values are expressed as a percentage of the ratio in corresponding wild-type tissue. Results with the basal and apical leaf sections were similar (Supplemental Fig. S2B). LSC, Large single-copy region; IR, inverted repeat; SSC, small single-copy region. D, Position dependence of plastid-to-nuclear gene copy number in the basal section of w2-mum2 mutant leaf tissue. Amplicons are ordered according to their position in the plastid genome. Values are expressed as a percentage of the ratio in corresponding wild-type tissue. DNA from the middle leaf section showed a similar but less robust trend (Supplemental Fig. S2B).
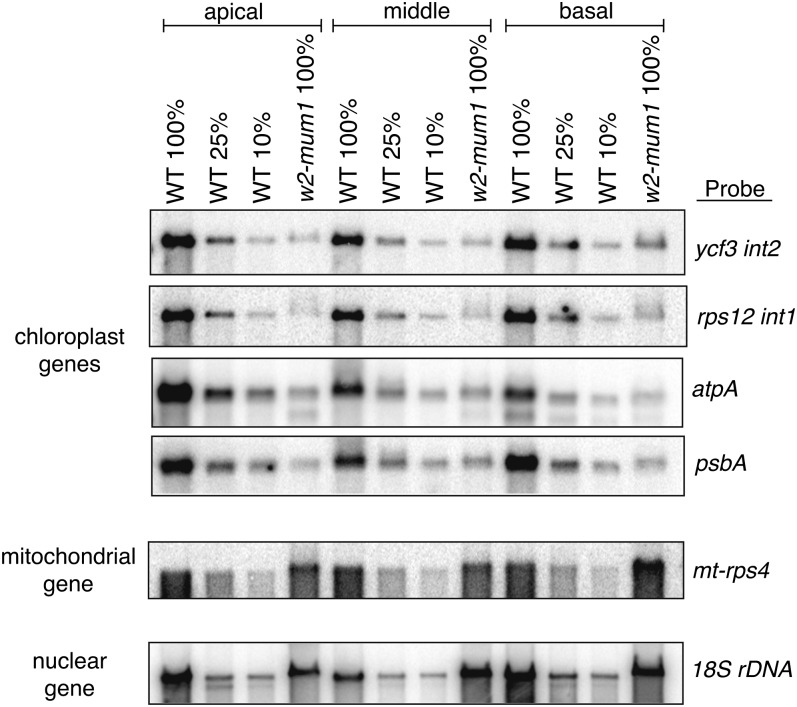
The ratio of chloroplast to nuclear DNA copy number in w2-mum1 and w2-mum2 homozygotes is shown in Figure 3B and Supplemental Figure S2B as a percentage of the value in the corresponding wild-type tissue. The w2-mum1 allele conditions a (roughly) 4- to 6-fold decrease in the ratio of plastid to nuclear DNA, with some variation depending upon the particular plastid gene and developmental stage. The w2-mum2 allele conditions a much more severe loss of chloroplast DNA: values range between 1% and 4% of normal, with the most severe defects consistently observed in the apical tissue. qPCR analysis of the w2-Burnham allele showed that it conditions a severe plastid DNA deficiency as well (Supplemental Fig. S3), consistent with prior Southern-blot data (Han et al., 1993). The approximately 100-fold decrease in qPCR signal for multiple plastid genes observed in w2-Burnham and w2-mum2 mutants places an upper limit of approximately 1% on the fraction of total signal that could arise from NUPTs.
Interestingly, the magnitude of plastid DNA loss depends to some extent on position within the plastid genome. In w2-mum1 mutants, the greatest decrease was observed near the ndhC-rbcL-atpB interval, with hints of a similar trend in the psbA-psbC interval (Fig. 3C; Supplemental Fig. S2B). In the severe w2-mum2 mutants, genes in the small single copy region (psaC through ndhA) were disproportionately reduced (Fig. 3D; Supplemental Fig. S2B). It seems likely that these trends are related to genomic features that influence DNA replication or maintenance (for review, see Day and Madesis, 2007). For example, origins of replication (Kunnimalaiyaan and Nielsen, 1997; Wang et al., 2002; Wang et al., 2003) and ends of linear plastid DNA molecules (Oldenburg and Bendich, 2004b; Scharff and Koop, 2006) map near the junction of the inverted repeat and small single copy region in various angiosperms. The disproportionate loss of DNA from the small single-copy region in w2-mum2 mutants may be related to these features.
Effects of w2 Mutations on Mitochondrial DNA Levels
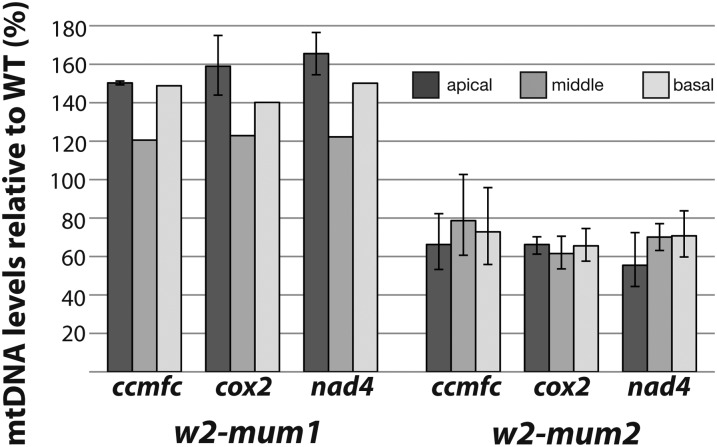
The w2 co-orthologs in Arabidopsis, POL1A and POL1B, are dual targeted to mitochondria and chloroplasts (Christensen et al., 2005). Disruption of each gene causes a small reduction (20%–50%) in mitochondrial and plastid DNA copy number, whereas disruption of both genes is embryo lethal (Parent et al., 2011), implying redundant functions in DNA replication in both organelles. Although the maize genome encodes a paralog of w2, the nearly complete loss of plastid DNA in w2-mum2 mutants indicates that w2 is the primary or possibly sole source of DNA polymerase for DNA replication in the chloroplast. To determine whether w2 also contributes to mitochondrial DNA replication, qPCR was used to quantify mitochondrial DNA in the same samples used for plastid DNA analysis (Fig. 4). The ratio of mitochondrial to nuclear DNA copy number was slightly elevated in w2-mum1 homozygotes, whereas in w2-mum2 homozygotes, mitochondrial DNA copy number was reduced roughly 2-fold. Mitochondrial DNA was reduced to a similar extent in w2-Burnham mutants (Supplemental Fig. S3). Neither mitochondrial nor chloroplast DNA levels are reduced as a secondary effect of an early block in plastid differentiation, as shown by the normal or elevated levels of organellar DNAs observed during a large survey of albino maize mutants in our mutant collection (Supplemental Fig. S3). Therefore, the 2-fold reduction in mitochondrial DNA content conditioned by strong w2 mutant alleles suggests that both w2 and its paralog contribute to mitochondrial DNA replication, similar to Pol1A and Pol1B in Arabidopsis (Parent et al., 2011). In fact, W2 has been detected in proteome analyses of mitochondria purified from ear shoots (S. Briggs, personal communication), supporting the possibility that it functions in both organelles.
Figure 4.
qPCR analysis of mitochondrial DNA abundance in w2 mutants. The ratio of signal for the indicated mitochondrial gene relative to the single-copy nuclear control is plotted as a percentage of the ratio in the corresponding wild-type tissue. The basal, middle, and apical leaf sections are diagrammed in Figure 1B. The average of three technical replicates is plotted, with maxima and minima indicated by error bars. The w2-Burnham allele conditions an approximately 2-fold decrease in leaf mitochondrial DNA content (Supplemental Fig. S3). Mitochondrial DNA content was decreased approximately 2-fold in seedling roots in w2-mum2 and w2-Burham mutants as well (data not shown).
Plastid Genome Copy Number Limits the Accumulation of Many Chloroplast RNAs
The findings above imply that the atpA RNA deficiency that originally drew our attention to w2 mutants is caused by a reduction in chloroplast DNA template. To survey the relationship between plastid genome copy number and RNA abundance, RNA was extracted from the same w2-mum1 tissue samples that had been used for DNA analysis. RNA gel-blot hybridizations demonstrated a reduction of approximately 4-fold for both atpA and psaB transcripts in all three leaf sections of w2-mum1 mutants (Figs. 1B and 5A). The magnitude of these RNA deficiencies is similar to that of the DNA deficiency, implying that the quantity of DNA template limits RNA accumulation from these genes. All transcript isoforms from the petB gene were reduced, but the effect was weaker (Fig. 5A), indicating that the DNA template is not the sole limiting factor in this case. Transcripts from the psbH and psaJ genes exhibited a different response, in that the monocistronic transcript isoforms were more strongly affected than were the precursors (Fig. 5A). This raises the possibility that chloroplast DNA deficiency impacts the expression of nuclear genes such as HCF107, HCF152, and PPR10, which are required for the stabilization of processed psbH or psaJ RNA (Felder et al., 2001; Meierhoff et al., 2003; Pfalz et al., 2009; Hammani et al., 2012; Zoschke et al., 2012). Yet another response was observed for the plastid rpoB gene, whose mRNA accumulated to greatly elevated levels in w2-mum1 tissue (Fig. 5A).
Figure 5.
RNA gel-blot hybridization assays of plastid RNAs in w2-mum1 mutants. Total RNA from the same tissue samples used for DNA analysis was analyzed by hybridization to the indicated probes. The methylene blue-stained blots shown below illustrate loading of the rRNAs. 28S and 18S are cytosolic rRNAs. 16S and 23S* are plastid-encoded rRNAs. A, RNA from protein-coding genes. Lanes contained 4 µg of total RNA (100%) or dilutions as indicated. B, RNA from rRNA and tRNA genes. Lanes contained 0.5 µg of RNA (100%) or dilutions as indicated.
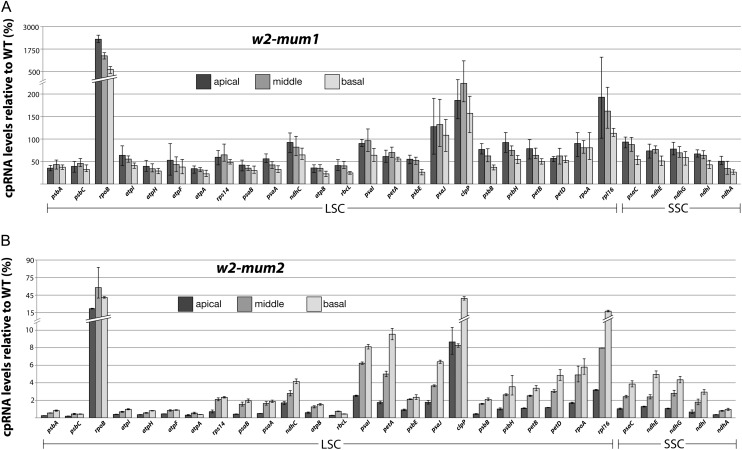
To gain a more global perspective, quantitative reverse transcription (qRT)-PCR was used to quantify the abundance of RNA segments from many plastid genes in each leaf section of w2-mum1 mutants (Fig. 6A). Amplicons from intron-containing genes mapped within exons, so that both spliced and unspliced RNAs contribute to the signal. All of the RNAs that were assayed by RNA gel-blot hybridization were also assayed by qRT-PCR; the results were generally in agreement, with the exception of psaJ, for which the qRT-PCR assay showed an unusual degree of experimental variation. The qRT-PCR data show that w2-mum1 conditions a moderate loss of most plastid mRNAs, with the strongest effects in the basal leaf section. For example, atpB, rbcL, psbE, and ndhA RNAs were reduced roughly 4-fold in the basal leaf section but only approximately 2-fold in the apical leaf section. Thus, DNA template abundance appears to limit the accumulation of these mRNAs during the initial buildup of the photosynthetic apparatus, but transcription or RNA stabilization factors may become limiting as development proceeds. Another set of mRNAs was less sensitive to the loss of plastid DNA; for example, psaC and psbB mRNAs exhibited only a 2-fold reduction in the basal leaf section and increased in the apical section to near normal levels. As predicted by the RNA gel blot data, rpoB exhibited a strikingly different response: rpoB RNA was elevated 25-fold in the apical section of w2-mum1 mutant leaves, despite the approximately 5-fold decrease in plastid DNA content. RNA from the clpP and rpl16 genes was also elevated with respect to DNA template concentration, although much less dramatically.
Figure 6.
qRT-PCR analysis of plastid RNAs in w2-mum1 and w2-mum2 mutants. Total RNA from the basal, middle, or apical leaf section was analyzed by qRT-PCR, using primers specific for exon sequences in the indicated plastid genes. The average of three technical replicates and two biological replicates is plotted, with maxima and minima indicated by error bars. Signals from each plastid gene were normalized to the cytosolic mRNA encoding HMG-1/2. Values are presented as the percentage of the ratio in the corresponding wild-type tissue.
qRT-PCR data for homozygous w2-mum2 mutants is shown in Figure 6B. Most RNAs are reduced roughly 100-fold, in accordance with the severe plastid DNA deficiency in this material. The fold decrease for the rpoB, clpP, and rpl16 RNAs is minor in comparison with the other RNAs examined; most striking is the rpoB mRNA that accumulates to roughly one-half of its normal level in young w2-mum2 leaf tissue, despite the more than 50-fold decrease in rpoB DNA. The rpoB, clpP, and rpl16 genes are transcribed primarily by the nuclear encoded polymerase (NEP), a single subunit phage-like polymerase, whereas the other genes examined are transcribed primarily by the bacterial type plastid encoded polymerase (PEP; for review, see Liere and Börner, 2007). The overrepresentation of NEP transcripts in w2 mutants is consistent with reports that NEP-mediated transcription increases in response to the loss of plastid ribosomes (Emanuel et al., 2004) or mutations in PEP genes (Legen et al., 2002). However, the very different degrees to which different NEP transcripts are overrepresented in response to the loss of plastid DNA indicates that differential RNA stability or transcription contribute to this phenomenon.
Several of the stable RNAs of the plastid gene expression machinery (rRNAs and tRNAs) were examined by RNA gel-blot hybridization in w2-mum1 and w2-mum2 mutants (Fig. 5B). These were reduced roughly 4-fold in the basal tissue, implying a limitation by plastid DNA template during the initial buildup of the photosynthetic apparatus. However, as chloroplast development proceeds, 23S rRNA (rrn23) and trnT-UGU build up to near normal levels whereas trnV-UAC and trnG-UCC do not. The trnV-UAC and trnG-UCC genes contain a group II intron, whereas the trnT-UGU gene lacks introns. Thus, the presence of an intron correlates with the severity of the tRNA deficiency in w2 mutants, suggesting that tRNA splicing may be compromised in w2 mutants (presumably as a secondary effect). 23S rRNA and trnV-UCC are severely depleted in w2-mum2 mutants (Fig. 5B), as is to be expected from the very low plastid DNA levels conditioned by this allele.
Decreased Abundance of Photosynthetic Complexes in w2-mum1 Mutants
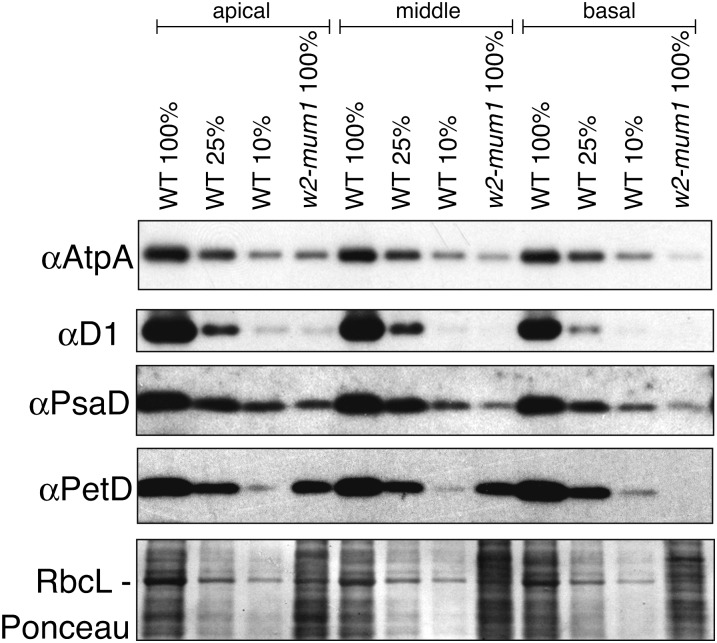
The chloroplast genome encodes at least one core subunit of each of the major photosynthetic enzyme complexes (Rubisco, PSII, cytochrome b6f, PSI, and the ATP synthase). To assess the effects of DNA template deficiency on the accumulation of the photosynthetic apparatus, one subunit of each complex was examined by immunoblotting (Fig. 7). As for the mRNAs, the protein deficiencies were most severe in the basal leaf section. AtpA, D1, and PsaD (subunits of the ATP synthase, PSII, and PSI, respectively) accumulated to less than 10% of normal levels in the basal section, and to approximately 10% of normal levels in the apical section. PetD, representing the cytochrome b6f complex, was likewise reduced more than 10-fold in the basal section, but accumulated to approximately 25% of normal levels in the apical section. RbcL, the large subunit of Rubisco, was reduced approximately 10-fold in the apical material, based on the intensity of its band on the Ponceau-stained filter. The magnitude of the protein losses exceeded those of corresponding plastid mRNAs, suggesting that the protein defects result from the combined effects of reduced mRNA, reduced tRNA, and reduced ribosome abundance (as reflected by reduced rRNA) early in chloroplast development.
Figure 7.
Reduced accumulation of photosynthetic enzyme complexes in w2-mum1 mutants. Immunoblots of leaf extract (5 µg of protein or the indicated dilutions) were probed with antibodies to core subunits of photosynthetic enzyme complexes: AtpA (ATP synthase), D1 (PSII), PsaD (PSI), and PetD (cytochrome b6f complex). The same blot stained with Ponceau S is shown below to illustrate sample loading and the abundance of RbcL.
DISCUSSION
Results presented here show that w2 encodes a DNA polymerase that is essential for chloroplast DNA replication, affirming the speculation made two decades ago that w2 is involved in plastid DNA replication (Han et al., 1993). Our results suggest that W2 also contributes to mitochondrial DNA replication and that another DNA polymerase (likely the paralog GRMZM2G023422) makes an equal contribution. The division of labor between organelle DNA polymerases differs in Arabidopsis, where a pair of paralogs function redundantly to support DNA replication in both organelles (Parent et al., 2011). W2 has been detected in proteome analyses of purified chloroplasts (Majeran et al., 2012), but the W2 paralog has not been detected in either chloroplasts or mitochondria. Our results are consistent with the possibility that W2 is the only source of DNA polymerase in chloroplasts, as the residual chloroplast DNA signal in the most severely affected samples (approximately 1% of normal in the apical section of w2-mum2 leaves) might arise from small revertant sectors. Furthermore, w2-mum2 may not be a null allele, as the aberrantly spliced mRNA resulting from this insertion maintains the normal open reading frame, albeit resulting in a deletion of 52 amino acids from the 3′→5′ exonuclease domain. The w2-Burham allele is unlikely to be null, as its phenotype is slightly milder than that of w2-mum2.
We used the weak w2-mum1 allele to assess the correlation between chloroplast DNA copy number and the abundance of chloroplasts RNAs. In immature chloroplasts, the abundance of many chloroplast RNAs, including mRNA, tRNA, and rRNA, decrease in proportion to the decrease in genome copy number. It has been proposed that high plastid gene dosage is necessary for adequate rRNA production during the initial buildup of the photosynthetic apparatus, when massive plastid protein synthesis is required for the biogenesis of the abundant photosynthetic enzyme complexes (Bendich, 1987). Our results are consistent with that view, but suggest that enhanced tRNA and mRNA accumulation are likewise promoted by high gene dosage and may also be important for optimal gene expression during the biogenesis phase. Chloroplast RNA deficiencies were generally less severe in the apical leaf section of w2-mum1 mutants despite the similar loss of plastid DNA at the early and late developmental stages. The apical leaf section, with its fully developed photosynthetic apparatus that requires relatively little gene expression for maintenance, is comparable with the situation in the constitutively green alga C. reinhardtii, where related questions were addressed. Growth of C. reinhardtii for 2 d in the presence of an inhibitor of DNA replication reduced chloroplast DNA and transcription rates severalfold but the levels of most of the RNAs assayed did not change appreciably (Eberhard et al., 2002). The authors proposed that proteins that stabilize chloroplast mRNAs are in limiting concentrations such that only a fraction of newly synthesized transcripts escape rapid degradation. Several nucleus-encoded proteins that stabilize specific chloroplast RNAs have been discovered in maize and Arabidopsis (Barkan et al., 1994; Felder et al., 2001; Meierhoff et al., 2003; Lezhneva and Meurer, 2004; Yamazaki et al., 2004; Beick et al., 2008; Pfalz et al., 2009; Johnson et al., 2010; Stoppel et al., 2011; Hammani et al., 2012), and there is evidence that the majority of chloroplast mRNAs in angiosperms are stabilized by proteins that block exoribonucleases (Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012a). Our results imply that such proteins may typically be in limiting concentrations in mature chloroplasts in angiosperms, as in C. reinhardtii chloroplasts.
The rpoB and, to a lesser extent, the clpP and rpl16 transcripts responded differently to the loss of plastid DNA than did the other genes assayed: instead of decreasing in rough proportion to the loss of plastid DNA, these transcripts accumulated to increased levels in w2-mum1 mutants and were only modestly reduced in the severe w2-mum2 mutants. These genes are transcribed primarily by NEP, whose mRNA increases 8-fold in barley (Hordeum vulgare) mutants lacking PEP due to a plastid ribosome deficiency (Emanuel et al., 2004). However, the particularly dramatic overrepresentation of rpoB mRNA when plastid DNA content is reduced suggests the existence of a dedicated regulatory mechanism affecting the synthesis or stability of this transcript, which encodes catalytic subunits of the PEP enzyme.
A previous study of the w2-Burnham allele revealed reduced levels of chloroplast DNA and a reduction in several chloroplast RNAs, with little change in the relative abundance of various transcript isoforms (Han et al., 1993). These authors contrasted the w2 RNA phenotype with those of two other albino mutants (w1 and iojap) that had normal levels of chloroplast DNA but aberrant transcript populations. In hindsight, it is clear that the w1 and iojap RNA and DNA content are typical of mutants in the grasses that lack plastid ribosomes: the ribosome deficiency results in stereotypical changes in transcript patterns presumably caused by the loss of PEP and enhanced transcription by NEP (Williams and Barkan, 2003; Prikryl et al., 2008; Zhelyazkova et al., 2012b). The fact that the transcript populations in w2-Burnham mutants more closely resemble those in wild-type plants than those in mutants lacking plastid ribosomes is interesting, because plastid ribosome abundance is drastically reduced in ω2-Burnham mutants. This reveals an unanticipated complexity in the signaling mechanisms through which defects in plastid gene expression trigger changes in chloroplast transcription and posttranscriptional processes.
Our findings bear on controversies surrounding changes in chloroplast DNA content during leaf development, and the degree to which NUPTs contribute to the plastid DNA signal in various types of assays (Oldenburg and Bendich, 2004a; Li et al., 2006; Zoschke et al., 2007; Kumar and Bendich, 2011). It has been proposed that signals from NUPTs mask signals from authentic plastid DNA in qPCR and Southern-blot assays of leaf DNA, unless the two DNA classes are distinguished by digestion with methylation sensitive restriction enzymes (Kumar and Bendich, 2011; Zheng et al., 2011). Our results are not consistent with that view. qPCR of undigested leaf DNA with many different primer pairs revealed a consistent degree of DNA loss across the plastid genome in each of two w2 mutant alleles (Fig. 3; Supplemental Fig. S2); Southern-blot analysis using restriction enzymes that are not sensitive to nuclear DNA methylation gave similar results (Fig. 2; Han et al., 1993). The approximately 100-fold loss of plastid DNA signal in w2-mum2 and w2-Burnham leaf tissue implies that no more than approximately 1% of the qPCR signal can arise from NUPTs in the corresponding normal tissue. Therefore, signal from NUPTs does not generally mask the signal from authentic plastid DNA in standard qPCR and Southern-blot assays of total leaf DNA. We can thus conclude with confidence that the ratio of chloroplast to nuclear gene copy number decreases only modestly during the development of maize seedling leaves, and that even the apical leaf tissue retains roughly 1,000 copies of the plastid genome per nuclear genome complement (Fig. 3A).
MATERIALS AND METHODS
Plant Material
A recessive mutation causing a very pale yellow green (vpyg) seedling phenotype arose in our mutant collection (http://pml.uoregon.edu/photosyntheticml.html), and was assigned the temporary name vpyg72. vpyg72 was shown to condition a reduction in chloroplast atpA mRNAs during a large-scale screen for plastid RNA defects in this mutant collection. The vpyg72 mutation was propagated by crossing heterozygotes to the A632 or A188 inbred line, followed by self-pollination to recover ears segregating homozygous mutants. DNA was isolated from mutant individuals from each of four F2 ears and analyzed by Mu-Illumina to map the positions of all of the Mu insertions in each plant, as described previously (Williams-Carrier et al., 2010). An insertion in GRMZM2G480171 was found in all four mutant individuals, and subsequent gene-specific PCR showed this insertion to be absent from closely related +/+ ears. Complementation crosses between vpyg72 heterozygotes and plants harboring the w2-Burnham allele (obtained from the Maize Genetics Cooperative) segregated approximately 25% chlorotic seedlings, demonstrating allelism; vpyg72 was therefore renamed w2-mum1. Another Mu-induced allele, w2-mum2, was obtained via a reverse-genetic effort in which we are cataloging all of the approximately 100 Mu insertion sites in each mutant in the PML mutant collection with the Mu-Illumina method. Phenotypically normal siblings of each mutant allele were used for the wild-type sample in each experiment.
Extraction of DNA, RNA, and Protein
Plants used for DNA, RNA, and protein analysis were grown in a growth chamber under 16-h-light (28°C)/8-h-dark (26°C) cycles. Tissue was harvested when the third leaf was beginning to emerge from the whorl (approximately 8 d after planting). The second leaf was divided into basal, middle, and apical sections (see Fig. 1B). Material from three to five individuals of the same genotype was pooled, frozen in liquid nitrogen, ground to a fine powder with a mortar and pestle while frozen, and divided into three aliquots for DNA, RNA, or protein extraction. DNA was extracted with Plant DNAzol Reagent (Invitrogen) according to the manufacturer’s protocol. The DNA pellets were resuspended in 100 µL of 10 mm Tris-HCl, 1 mm EDTA, pH 8, extracted with phenol/chloroform/isoamyl alcohol (25:24:1), and ethanol precipitated. The final pellet was resuspended in 100 µL deionized water. RNA was extracted with Tri Reagent (Molecular Research Center, Inc.) according to the manufacturer’s protocol. RNA was resuspended in 10 mm Tris-HCl, 0.1 mm EDTA, pH 8. Ten micrograms of each RNA sample was treated with TURBO DNase (Ambion) according to the manufacturer’s protocol. End-point PCR reactions of the DNase-treated RNA samples were performed to ensure that residual DNA did not contribute to signals in qRT-PCR assays.
RNA, DNA, and Protein-Gel Blots
Southern (DNA), northern (RNA), and western (protein) blots were performed using the methods, probes, and antibodies described previously (Prikryl et al., 2008).
qPCR and qRT-PCR
cDNA for qRT-PCR was generated as follows. Two micrograms of DNase-treated RNA was combined with 2 µg of random hexamers, heated to 70°C for 5 min and chilled on ice. Reverse transcription was performed with Avian Myeloblastosis Virus reverse transcriptase (Promega) in a volume of 50 µL at 37°C for 60 min, using the buffer recommended by the manufacturer. After the addition of 50 µL water and 3 µL of 3 m NaCl, samples were extracted with phenol/chloroform/isoamyl alcohol, and ethanol precipitated. The cDNA was resuspended in 100 µL of water.
qPCR and qRT-PCR were performed in a StepOnePlus real-time PCR machine (Applied Biosystems). Reactions contained 1× KAPA SYBR FAST Master Mix with primers at 200 nm in a volume of 10 µL. Primer sequences are given in Supplemental Table S1. Primers were designed using Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/). The amplicon size was designed to be 150 to 200 bp, and the melting temperature (Tm) was set to 60°C (minimum 57°C and maximum 63°C) with a maximum Tm difference of 3°C. Primer specificity was checked against the complete maize (Zea mays) genome (taxid: 4577) to reduce the risk of mispriming. If no primers could be designed using those conditions, the maximum and minimum Tm limits were changed (the maximum Tm difference was kept at 3°C). Primers were checked for efficiency using the standard curve method. All primers were checked for specificity at 60°C using end-point PCR before being used for qPCR. Specificity was also checked using a melt-curve analysis after all qPCR runs. Many primers had already been designed and validated by other researchers (see Supplemental Table S1); these primers were also validated for use at 60°C using end-point PCR, and their specificity was always checked using melt-curve analysis after all qPCR thermocycling runs. The single-copy nuclear gene HMG1/2 (GRMZM2G024976) was used as the reference gene for the qPCR experiments and the mRNA from this gene was used as the reference for qRT-PCR experiments. Early experiments used an actin internal control, which gave similar results in the DNA assays, but showed more variation in the RNA assays. The thermocycling program used the following parameters: 20 s at 95°C, 40 cycles of 3 s at 95°C and 30 s at 60°C, and a final incubation at 95°C for 15 s. After the final 95°C step, a melt-curve analysis was used to confirm that only one product had been amplified: 60°C for 1 min, increased by 0.5°C every 30 s and 95°C for 15 s. The comparative cycle threshold method was used to calculate relative gene expression levels using StepOnePlus Software, version 2.2.2. All qPCR experiments included three technical replicates. The w2-mum1 qRT-PCR analyses included, in addition, two biological replicates.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effects of mutations on w2 mRNA and protein.
Supplemental Figure S2. Additional qPCR assays of plastid DNA content in the wild type and w2 basal, middle, and apical leaf sections.
Supplemental Figure S3. qPCR survey of chloroplast and mitochondrial DNA levels in independently arising albino mutants in the PML mutant collection.
Supplemental Table S1. Probes and primers used in this study.
Supplementary Material
Acknowledgments
We are grateful to the members of David Stern’s laboratory (Boyce Thompson Institute) who performed an RNA gel blot survey of several hundred mutants in the PML collection, including w2-mum1. We also wish to thank Reimo Zoschke (University of Oregon) for helpful comments on the manuscript, and Doug Turnbull and Nick Stiffler of the University of Oregon Genomics Facility for the Illumina sequencing and data analysis.
Glossary
- rRNA
ribosomal RNA
- tRNA
transfer RNA
- qRT
quantitative reverse transcription
- qPCR
quantitative PCR
- NUPT
to be defined
- NEP
nuclear encoded polymerase
- PEP
plastid encoded polymerase
- PML
to be defined
- Tm
to be defined
References
- Barkan A, Walker M, Nolasco M, Johnson D. (1994) A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J 13: 3170–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE. (1989) Plastid transcription activity and DNA copy number increase early in barley chloroplast development. Plant Physiol 89: 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich AJ. (1987) Why do chloroplasts and mitochondria contain so many copies of their genome? Bioessays 6: 279–282 [DOI] [PubMed] [Google Scholar]
- Bendich AJ. (2004) Circular chloroplast chromosomes: the grand illusion. Plant Cell 16: 1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA. (2005) Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17: 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A, Madesis P. (2007) DNA replication, recombination, and repair in plastids. Bock R, , Cell and Molecular Biology of Plastids. Springer-Verlag, Berlin, pp 65–119 [Google Scholar]
- Eberhard S, Drapier D, Wollman FA. (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31: 149–160 [DOI] [PubMed] [Google Scholar]
- Emanuel C, Weihe A, Graner A, Hess WR, Börner T. (2004) Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J 38: 460–472 [DOI] [PubMed] [Google Scholar]
- Felder S, Meierhoff K, Sane AP, Meurer J, Driemel C, Plücken H, Klaff P, Stein B, Bechtold N, Westhoff P. (2001) The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13: 2127–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BR. (2011) Chloroplast genomes of photosynthetic eukaryotes. Plant J 66: 34–44 [DOI] [PubMed] [Google Scholar]
- Hammani K, Cook WB, Barkan A. (2012) RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc Natl Acad Sci USA 109: 5651–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C-D, Patrie W, Polacco M, Coe EH. (1993) Aberrations in plastid transcripts and deficiency of plastid DNA in striped and albino mutants in maize. Planta 191: 552–563 [Google Scholar]
- Hosler JP, Wurtz EA, Harris EH, Gillham NW, Boynton JE. (1989) Relationship between gene dosage and gene expression in the chloroplast of Chlamydomonas reinhardtii. Plant Physiol 91: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. (2010) MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, Bendich AJ. (2011) Distinguishing authentic mitochondrial and plastid DNAs from similar DNA sequences in the nucleus using the polymerase chain reaction. Curr Genet 57: 287–295 [DOI] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL. (1997) Fine mapping of replication origins (ori A and ori B) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res 25: 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert RJ. (1980) Report of maize genetics cooperation stock center. Maize Genetics Cooperation Newsletter 54: 131–135
- Leech RM, Rumsby MG, Thomson WW. (1973) Plastid differentiation, acyl lipid, and fatty acid changes in developing green maize leaves. Plant Physiol 52: 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM. (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Lezhneva L, Meurer J. (2004) The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J 38: 740–753 [DOI] [PubMed] [Google Scholar]
- Li W, Ruf S, Bock R. (2006) Constancy of organellar genome copy numbers during leaf development and senescence in higher plants. Mol Genet Genomics 275: 185–192 [DOI] [PubMed] [Google Scholar]
- Liere K, Börner T. (2007) Transcription and transcriptional regulation in chloroplasts. Bock R, , Cell and Molecular Biology of Plastids. Springer-Verlag, Berlin, pp 121–174 [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ. (2012) Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. (2004a) Changes in the structure of DNA molecules and the amount of DNA per plastid during chloroplast development in maize. J Mol Biol 344: 1311–1330 [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. (2004b) Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J Mol Biol 335: 953–970 [DOI] [PubMed] [Google Scholar]
- Ortiz DF, Strommer JN. (1990) The Mu1 maize transposable element induces tissue-specific aberrant splicing and polyadenylation in two Adh1 mutants. Mol Cell Biol 10: 2090–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent JS, Lepage E, Brisson N. (2011) Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol 156: 254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan BA, Oldenburg DJ, Bendich AJ. (2004) The demise of chloroplast DNA in Arabidopsis. Curr Genet 46: 176–181 [DOI] [PubMed] [Google Scholar]
- Rowan BA, Oldenburg DJ, Bendich AJ. (2009) A multiple-method approach reveals a declining amount of chloroplast DNA during development in Arabidopsis. BMC Plant Biol 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H, Schmitz-Linneweber C. (2012) Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res 40: 3106–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Koop HU. (2006) Linear molecules of tobacco ptDNA end at known replication origins and additional loci. Plant Mol Biol 62: 611–621 [DOI] [PubMed] [Google Scholar]
- Shaver JM, Oldenburg DJ, Bendich AJ. (2006) Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224: 72–82 [DOI] [PubMed] [Google Scholar]
- Stern DB, Hanson MR, Barkan A. (2004) Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci 9: 293–301 [DOI] [PubMed] [Google Scholar]
- Stoppel R, Lezhneva L, Schwenkert S, Torabi S, Felder S, Meierhoff K, Westhoff P, Meurer J. (2011) Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell 23: 2680–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Wang Y, Saitoh Y, Sato T, Hidaka S, Tsutsumi K. (2003) Comparison of plastid DNA replication in different cells and tissues of the rice plant. Plant Mol Biol 52: 905–913 [DOI] [PubMed] [Google Scholar]
- Wang Y, Tamura K, Saitoh Y, Sato T, Hidaka S, Tsutsumi K. (2002) Mapping major replication origins on the rice plastid DNA. Plant Biotechnol 19: 27–35 [Google Scholar]
- Williams PM, Barkan A. (2003) A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J 36: 675–686 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stern DB, Monde RA, Coalter R, Barkan A. (2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy mutator lines of maize. Plant J 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tasaka M, Shikanai T. (2004) PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J 38: 152–163 [DOI] [PubMed] [Google Scholar]
- Zhelyazkova P, Hammani K, Rojas M, Voelker R, Vargas-Suárez M, Börner T, Barkan A. (2012a) Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res 40: 3092–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T. (2012b) The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Oldenburg DJ, Bendich AJ. (2011) Independent effects of leaf growth and light on the development of the plastid and its DNA content in Zea species. J Exp Bot 62: 2715–2730 [DOI] [PubMed] [Google Scholar]
- Zoschke R, Kroeger T, Belcher S, Schöttler MA, Barkan A, Schmitz-Linneweber C. (2012) The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J (in press) [DOI] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T. (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50: 710–722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.