Abstract
procera (pro) is a tall tomato (Solanum lycopersicum) mutant carrying a point mutation in the GRAS region of the gene encoding SlDELLA, a repressor in the gibberellin (GA) signaling pathway. Consistent with the SlDELLA loss of function, pro plants display a GA-constitutive response phenotype, mimicking wild-type plants treated with GA3. The ovaries from both nonemasculated and emasculated pro flowers had very strong parthenocarpic capacity, associated with enhanced growth of preanthesis ovaries due to more and larger cells. pro parthenocarpy is facultative because seeded fruits were obtained by manual pollination. Most pro pistils had exserted stigmas, thus preventing self-pollination, similar to wild-type pistils treated with GA3 or auxins. However, Style2.1, a gene responsible for long styles in noncultivated tomato, may not control the enhanced style elongation of pro pistils, because its expression was not higher in pro styles and did not increase upon GA3 application. Interestingly, a high percentage of pro flowers had meristic alterations, with one additional petal, sepal, stamen, and carpel at each of the four whorls, respectively, thus unveiling a role of SlDELLA in flower organ development. Microarray analysis showed significant changes in the transcriptome of preanthesis pro ovaries compared with the wild type, indicating that the molecular mechanism underlying the parthenocarpic capacity of pro is complex and that it is mainly associated with changes in the expression of genes involved in GA and auxin pathways. Interestingly, it was found that GA activity modulates the expression of cell division and expansion genes and an auxin signaling gene (tomato AUXIN RESPONSE FACTOR7) during fruit-set.
Fruit-set is thought to be under the control of hormones, mainly auxins and GAs, synthesized in the ovary after pollination/fertilization (Pandolfini et al., 2007; Serrani et al., 2007b, de Jong et al., 2009a, Fuentes and Vivian-Smith, 2009). Auxin controls the synthesis of GA during early fruit development (Ozga and Reinecke, 2003; Serrani et al., 2008; Dorcey et al., 2009). In tomato (Solanum lycopersicum), auxin-induced fruit-set is mediated by GA, although auxin may also have a GA-independent effect on fruit growth (Serrani et al., 2008).
Parthenocarpic growth can be induced by the application of different kinds of hormones (Vivian-Smith and Koltunow, 1999; Serrani et al., 2007a; Dorcey et al., 2009). Indeed, there are mutants with parthenocarpic capacity due to the overexpression of genes of auxin biosynthesis (Pandolfini et al., 2007) or the auxin receptor tomato TRANSPORT INHIBITOR RESPONSE1 (SlTIR1; Ren et al., 2011) and to the down-regulation of transcription factors (TFs) involved in the auxin signaling pathway (AUX/IAA [IAA9] and AUXIN RESPONSE FACTOR7 [ARF7] Wang et al., 2005; Goetz et al., 2007; de Jong et al., 2009b). The parthenocarpic capacity of several tomato mutants (pat, pat-2, and pat-3/pat-4) is also associated with higher content of GA and altered expression of GA metabolism genes in the ovary (Fos et al., 2000, 2001; Olimpieri et al., 2007). DELLA proteins, key regulators of the GA-signaling pathway, are destabilized in response to GA through the ubiquitin-26S-proteasome pathway (Sun and Gubler, 2004). Upon GA binding to GA INSENSITIVE DWARF1 (GID1; the GA receptor), the GID1-GA complex interacts with the DELLA proteins through the DELLA/TVHYNP motif, allowing its recognition by the F-box protein (SLEEPY1 in Arabidopsis [Arabidopsis thaliana], GID2 in rice [Oryza sativa]) of the S-phase kinase-associated protein1 (Skp1)/Cullin/F-box E3 ubiquitin ligase complex, which targets the DELLAs for degradation (Harberd et al., 2009; Hirano et al., 2010). Arabidopsis contains five genes encoding DELLA proteins, while rice and tomato, for example, have only one (Martí et al., 2007). The slender la cry DELLA mutant of pea (Pisum sativum) has parthenocarpic capacity (Weston et al., 2008), and antisense-SlDELLA plants of tomato have a tall phenotype and produce parthenocarpic (seedless) fruits (Martí et al., 2007). All these results support the pivotal role played by auxin and GA in fruit-set.
Diverse mutations in the DELLA/TVHYNP domains transform the DELLA protein into a constitutively active, dominant form, resistant to GA-induced degradation (Harberd et al., 2009). By contrast, null and loss-of-function recessive mutations in the DELLA genes of several species provoke a constitutive GA-response phenotype (Sun and Gubler, 2004; Weston et al., 2008; Harberd et al., 2009). This is also the case of the procera (pro) tomato mutant, caused by a point mutation in the VHIID motif of the SlDELLA gene, leading to a single amino acid change (Bassel et al., 2008; Jasinski et al., 2008). pro mutants are taller and have nonserrated leaves (Jones, 1987; Jupe et al., 1988). Interestingly, their GA content is reduced (Jones, 1987; Van Tuinen et al., 1999), indicating that pro is not a GA metabolic mutation. Remarkably, the dwarf phenotype of the dominant Brassica rapa rga1-d mutant is also produced by the change of one amino acid outside the DELLA domain and within the GRAS region (Muangprom et al., 2005). It has been proposed that, in rice, after DELLA binding to GID1 through the DELLA/TVHYNP motif, the GRAS domain helps to stabilize the complex to be recognized by the F-box for degradation (Hirano et al., 2010). This hypothesis agrees with the contention that the higher DELLA level and dwarf phenotype of the dominant B. rapa repressor of GA1 mutant is due to the reduced interaction of DELLA with the F-box needed for degradation (Muangprom et al., 2005; Hirano et al., 2010). By contrast, in the case of the recessive pro mutant, the mutation may lead to lower activity of the repressor protein, for instance causing reduced or no interaction with downstream target TFs, rather than to reduced F-box binding.
In this work, we have further characterized the pro mutant, paying particular attention to the reproductive phenotypes that have been overlooked or poorly considered in previous work with the mutant. The pro mutation induces style elongation (thus preventing self-pollination), meristic changes in the flower (increased number of all floral organs), and strong parthenocarpic capacity. Transcriptome analysis and quantitative PCR (qPCR) of selected genes suggest the involvement of specific TFs in the altered morphology of pro flowers. These results unveil new functions for SlDELLA and that GA activity regulates cell division and expansion and the auxin signaling pathway during fruit-set and development.
RESULTS
Phenotypic Characterization of the pro Mutant
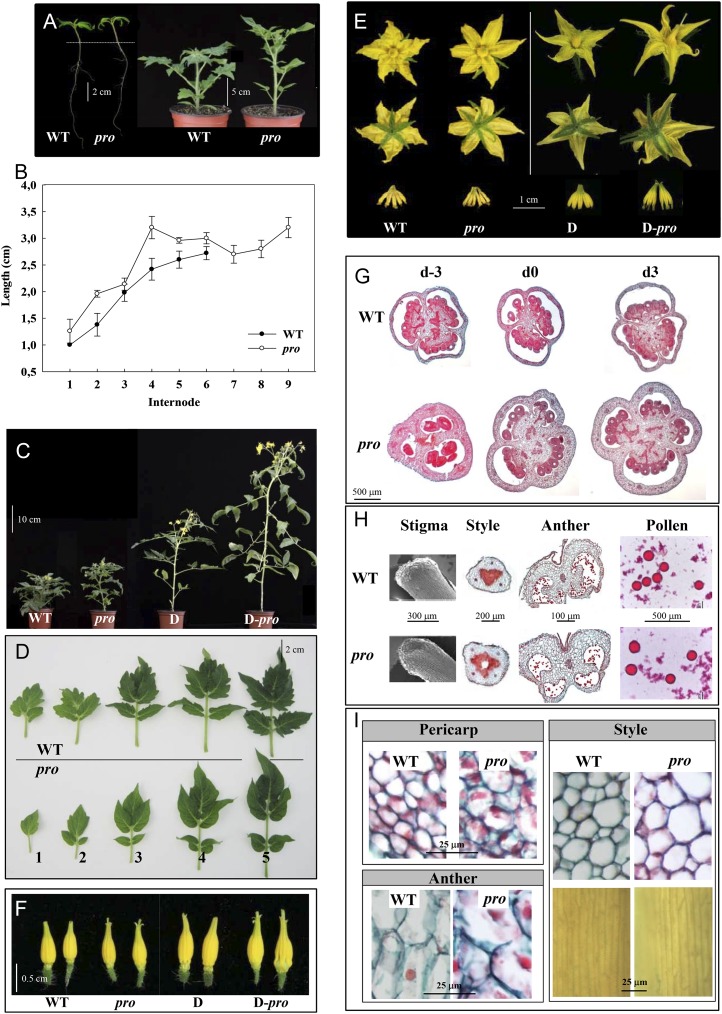
Seedlings of the pro mutant had longer hypocotyls and shorter roots, and the plants were slender, with longer internodes and thinner stems than the controls (Table I; Fig. 1, A and B). pro enhanced growth both in plants with low (wild-type cv Micro-Tom) and high (cv Micro-Tom with introgressed Dwarf [D]) brassinosteroid content (Fig. 1C). In pro plants, the number of leaves before the first inflorescence was higher (eight to nine leaves compared with six to seven leaves in the wild type; Table I), associated with delayed flowering. The leaves had reduced size and, in some cases, fewer leaflets, and they lacked the characteristic serrated borders of the wild type (Fig. 1D; Jasinski et al., 2008).
Table I. Phenotypes of wild-type and pro mutant plants.
Values are means ± se. The statistical significance of mean differences was analyzed using a t test: *P < 0.05, **P < 0.01.
| Parameter | Wild Type | pro |
|---|---|---|
| Hypocotyl length (mm)a | 14.3 ± 0.3 | 18.3 ± 0.2** |
| Root length (mm)a | 63.8 ± 0.9 | 58.2 ± 1.4** |
| Height to first inflorescence (cm)b | 10.4 ± 0.4 | 17.2 ± 0.4** |
| Leaves to first inflorescence (n)b | 6.6 ± 0.2 | 8.6 ± 0.2** |
| Stem diameter of fifth internode (mm)b | 6.6 ± 0.1 | 5.0 ± 0.1** |
| Flowers in the two first inflorescences (n)b | 12.5 ± 0.2 | 9.4 ± 1.0* |
| Sepal length (mm)c | 6.8 ± 0.3 | 7.9 ± 0.3** |
| Petal length (mm)c | 11.6 ± 0.3 | 14.0 ± 0.3** |
| Staminal cone length (mm)c | 7.4 ± 0.2 | 8.2 ± 0.2** |
| Style length (mm)c | 5.8 ± 0.1 | 7.7 ± 0.2** |
| Sepals, petals, and stamens per flower (n)d | 5.0 ± 0.1 | 5.7 ± 0.2** |
| Carpels per flower (n)d | 2.9 ± 0.2 | 4.0 ± 0.2** |
| Cell size (µm) | ||
| Style | 1.82 ± 0.08 | 2.43 ± 0.13** |
| Anther wall | 4.53 ± 0.30 | 5.31 ± 0.38* |
| Pericarp | 2.10 ± 0.08 | 2.98 ± 0.16** |
| Pericarp cell layers (n) | 6.9 ± 0.1 | 10.3 ± 0.2** |
| Days to anthesis of first flowere | 38.5 ± 0.1 | 43.7 ± 0.2** |
| Days to breaker (color change) of first mature fruite | 78.0 ± 0.3 | 87.5 ± 0.1** |
| Fruits per plant (n)e | 24.6 ± 2.0 | 42.5 ± 2.1** |
| Fruit weight (g)e | 5.8 ± 0.3 | 2.3 ± 0.2** |
| Fruit production (g per plant)e | 127.6 ± 16.6 | 83.8 ± 5.5* |
| Seeds per fruit (n)e | 34 ± 3 | 0 |
| Brix indexf | 4.7 ± 0.2 | 7.2 ± 0.2* |
Six 7-d-old seedlings. bTwelve fully developed plants. cTwelve random flowers from six plants. dSeventy-five random flowers from two independent batches; in the case of pro flowers, 42% (27 of 63) and 66% (eight of 12) of them, respectively, had six sepals, petals, and stamens and four carpels. eTwelve plants. fBrix index was determined in juice collected from five mature fruits from 12 plants.
Figure 1.
Phenotypes of the tomato pro mutant. A, Seven-day-old seedlings (left) and plants at the time of flowering (right). B, Lengths of different internodes. Values are means from six plants ± se. C, Full developed wild-type cv Micro-Tom (WT), pro, D, and D-pro plants. D, Leaves from different positions in the plant (at nodes 1–5 from cotyledons). E, Entire flowers (top two rows) and stamens (bottom row) from wild-type, pro, D, and D-pro plants. F, Intact staminal cones showing protruding styles in pro and D-pro flowers. G, Transverse sections of day −3, day 0, and day 3 ovaries. H, Stigma and transverse sections of styles and anthers stained with Safranine-Alcian blue and extracted pollen stained with carmine acetate. I, Closeups of transverse sections of pericarp, anther, and style and longitudinal sections of styles at the time of anthesis. [See online article for color version of this figure.]
pro mutant flowers had longer sepals, petals, and stamens (Table I; Fig. 1E), and their stigmas protruded above the staminal cone due to longer styles (Table I; Fig. 1F), also in flowers from D-pro plants (Fig. 1F). Interestingly, the average number of floral organs in all whorls was higher in pro than in the wild type due to a large number of flowers with six sepals, six petals, six stamens, and four carpels, compared with five sepals, petals, and stamens and three carpels in essentially all wild-type flowers (Table I; Fig. 1, E and G). pro pistils had longer cells, rounded stigmas, and the stylar transmitting tissue of those with four carpels was hollow (Fig. 1, H and I), which might affect pollen germination and tube transmission. At late anthesis, when the intersporangial septum of wild-type anthers had degenerated, it still remained intact in pro anthers, probably due to delayed senescence (Fig. 1H). Cells of style, anther wall, and ovary pericarp of pro flowers at the time of anthesis (day 0) were larger than those of the wild type (Table I; Fig. 1, G and I). The number of cell layers in the pericarp was also higher in pro than in wild-type ovaries at anthesis (Table I).
pro had fewer flowers per plant (Table I), although the total number of developed fruits was almost double but smaller (Table I; Fig. 2, A and B). Interestingly, many pro fruits (up to 28% in some batches) grown from spontaneous self-pollinated flowers bore an additional fruit-like structure at the style end (Fig. 2B). Fruit ripening was delayed in the mutant by about 9 d, and the Brix index value of the juice was higher (Table I).
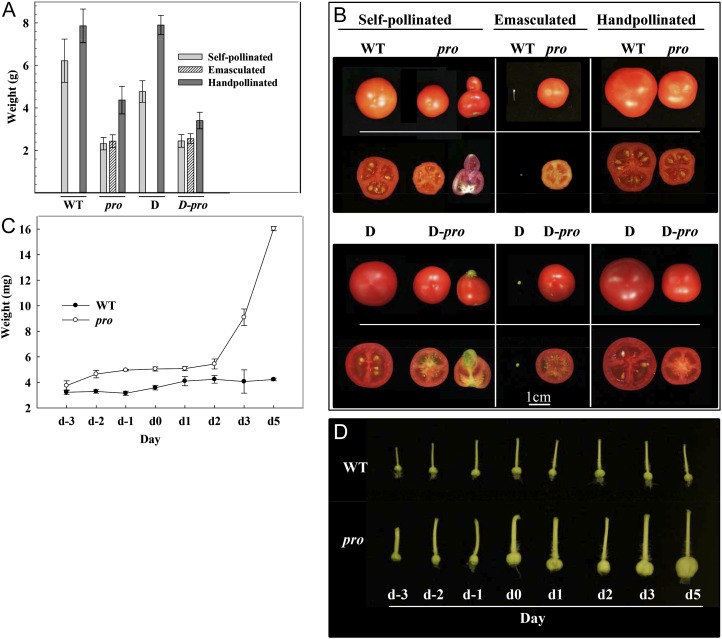
Figure 2.
Growth of wild-type (WT) and pro pistils and fruits. A, Effect of pollination (self-pollination and hand pollination) and emasculation of wild-type, pro, D, and D-pro flowers on final fruit weight. Values are means ± se of 25 (wild-type and pro) and six (D and D-pro) fruits. B, Photographs of representative fruits developed from flowers subjected to different treatments. Fruits with additional fruit-like structures are also presented in the case of self-pollinated pro and D-pro. C, Time course of the weight of unpollinated pistils. Values are means ± se of three replicates, each one consisting of a mixture of 10 pistils. D, Photographs of representative unpollinated pistils from day −3 to day 5. [See online article for color version of this figure.]
The pro Mutant Shows Facultative Parthenocarpy
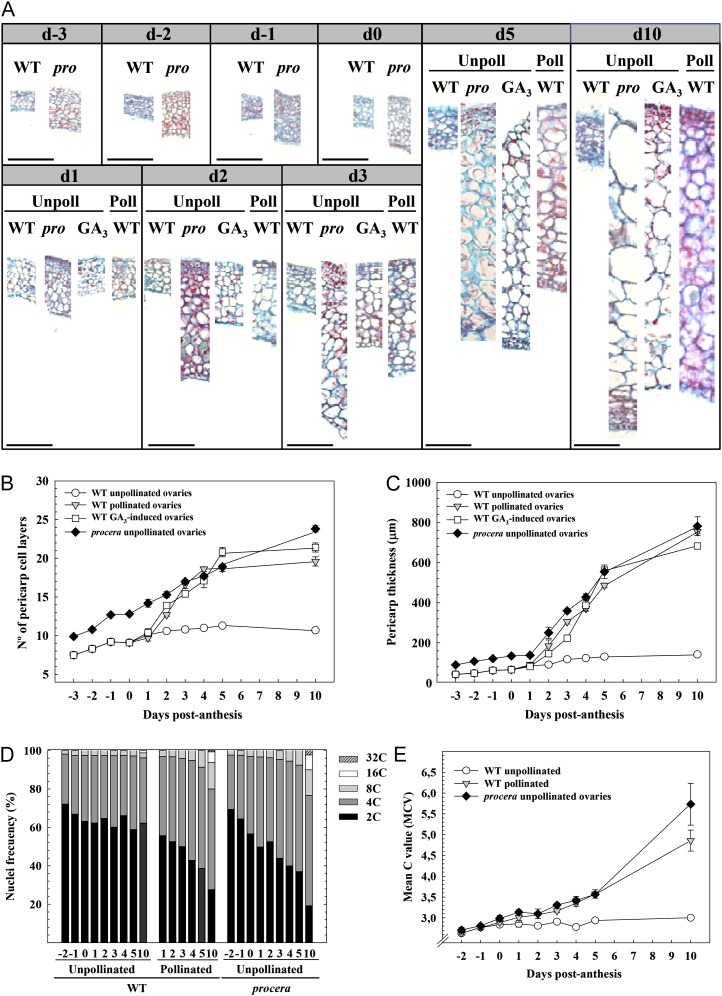
Under our growing conditions, all fruits from self-pollinated flowers of the first three inflorescences of wild-type plants contained seeds. By contrast, in pro plants, more than 95% of fruits that developed from self-pollinated flowers were parthenocarpic (Table II; Fig. 2, A and B). Similar results were obtained with D-pro plants (Fig. 2, A and B). Unpollinated pro ovaries had higher weight (Fig. 2, C and D), with larger cells and more cell layers in the pericarp than the wild type (Fig. 1G). A sudden burst of growth in the mutant unpollinated ovaries occurred at day 3, while unpollinated wild-type ovaries remained essentially unchanged (Fig. 2, C and D).
Table II. Effects of flower emasculation and pollination on fruit-set and growth.
When fruitful, values are means ± se of 36 fruits from six plants, six fruits per plant.
| Flower Treatment | Wild Type |
pro |
||||
|---|---|---|---|---|---|---|
| Fruit-Set | Grams per Fruit | Seeds | Fruit-Set | Grams per Fruit | Seeds | |
| % | % | |||||
| Spontaneous self-pollination | 100 | 6.2 ± 1.0 | Yes | 100 | 2.3 ± 0.3 | No |
| Emasculated | 0 | – | – | 100 | 2.4 ± 0.3 | No |
| Emasculated + manual pollination | 100 | 7.9 ± 0.8 | Yes | 100 | 4.4 ± 0.7 | Yes |
pro ovaries from emasculated flowers developed parthenocarpically and attained the same final weight as parthenocarpic pro fruits developed under spontaneous self-pollination conditions (Table II; Fig. 2A). This shows that flower emasculation did not affect pro parthenocarpic capacity or the fruit developmental program. Manually pollinated ovaries developed into fruits of higher weight, associated with a higher number of seeds (wild type) or with the presence of seeds (pro; Table II). This suggests that in the case of pro, the developing seeds provide additional factor(s) for fruit development.
To know whether parthenocarpy induced by pro was facultative or obligatory, we carried out experiments of manual self- and cross-pollination between wild-type and pro ovaries (Table III). Fruit-set in all crosses was 100%. Since 60% of the fruits that developed after pro manual self-pollination had seeds, this means that pro parthenocarpy is facultative and that the absence of seeds under normal culture conditions is probably the result of self-pollination impediment due to the long-style phenotype. The observation that wild-type × wild-type fruits and pro × wild-type fruits (pistil × pollen crosses) had higher numbers of seeds than wild-type × pro fruits and pro × pro fruits (Table III) suggested that the pro mutation reduces male fertility (Fig. 1H). This was confirmed by counting the number of extracted pollen grains per anther (17,300 ± 4,700 in pro versus 55,700 ± 3,400 in the wild type [n = 10]; P < 0.05; Fig. 1H). However, pollen viability in pro was not reduced, according to red carmine diacetate staining and emission of fluorescence after incubation with fluorescein diacetate. In this case, the lower fluorescence found in pro extracts (385 ± 4 versus 586 ± 4 in the wild type [n = 8]; P < 0.01) can be attributed to the reduced number of total pollen grains per extract. Pollen germination capability in the mutant was not reduced either (data not shown). Additionally, the fact that pro × wild-type fruits had fewer seeds than wild-type × wild-type fruits and pro × pro fruits had fewer seeds than wild-type × pro fruits indicates that the pro mutation may also affect ovule viability or pollen tube growth in the pro style and ovary.
Table III. Effects of wild-type and pro crosses on fruit-set and growth.
Successful fruit-set and number of fruits with seeds versus total number of attempts are recorded. The number of seeds per fruit refers only to fruits with seeds.
| Cross | Fruit-Set (Fruits Developed/No. of Attempts) | Seeded Fruits (Fruits with Seeds/No. of Attempts) | Weight | Seeds |
|---|---|---|---|---|
| g per fruit | No. per fruit | |||
| ♀ wild type × ♂ wild typea | 10/10 | 10/10 | 5.9 ± 0.4 | 35.0 ± 3.1 |
| ♀ pro × ♂ proa | 10/10 | 0/10 | 2.5 ± 0.2 | 0 |
| ♀ wild type × ♂ wild type | 10/10 | 10/10 | 5.0 ± 0.8 | 21.1 ± 1.3 |
| ♀ pro × ♂ pro | 10/10 | 6/10 | 4.3 ± 0.5 | 3.2 ± 0.7 |
| ♀ pro × ♂ wild type | 10/10 | 10/10 | 5.5 ± 0.7 | 16.1 ± 1.8 |
| ♀ wild type × ♂ pro | 10/10 | 10/10 | 4.7 ± 0.5 | 8.7 ± 1.3 |
Spontaneous self-pollination.
A time-course analysis showed that both cell size (Fig. 3A) and number of cell layers (Fig. 3, A and B) in the pericarp of pro ovaries was higher than in unpollinated wild-type ovaries from day −3 to day 10, associated with the increase of pericarp thickness (Fig. 3C). This indicated that the mutation induces an enhancement of cell division and expansion before anthesis and in the absence of pollination. The cell size and number of cell layers increased from day 2 following pollination or GA3 application to unpollinated ovaries at day 0, attaining similar values to those of pro from day 5 (Fig. 3).
Figure 3.
Time course between day −3 and day 10 of cell division and expansion in pericarp of pro, wild-type (WT) pollinated, and wild-type unpollinated control and GA3-treated ovaries. A, Photographs of transverse sections from pro and wild-type pollinated ovaries. B, Number of cell layers. C, Pericarp thickness. D, Nuclei frequency. E, Mean C values (MCV). [See online article for color version of this figure.]
Endoreduplication was also monitored to get further insight into the effect of pro on pericarp cytological evolution. The time course of pericarp cell ploidy was determined both as a percentage of nuclei with different C values (Fig. 3D) and as mean C values (sum of the number of nuclei of each ploidy level multiplied by its endoreduplication cycle, divided by the total number of nuclei [Barow and Meister, 2003]; Fig. 3E). The level of endoreduplication was very low and essentially unaltered in wild-type unpollinated ovaries (with mean C values lower than 3), while steady increase of endoreduplication occurred after day 1 in wild-type pollinated and unpollinated pro ovaries. At day 10, mean C values were about 5 in the last two cases, with values up to 32C in a small percentage of nuclei (Fig. 3, D and E).
Response of pro Plants to GA3 and Paclobutrazol Application
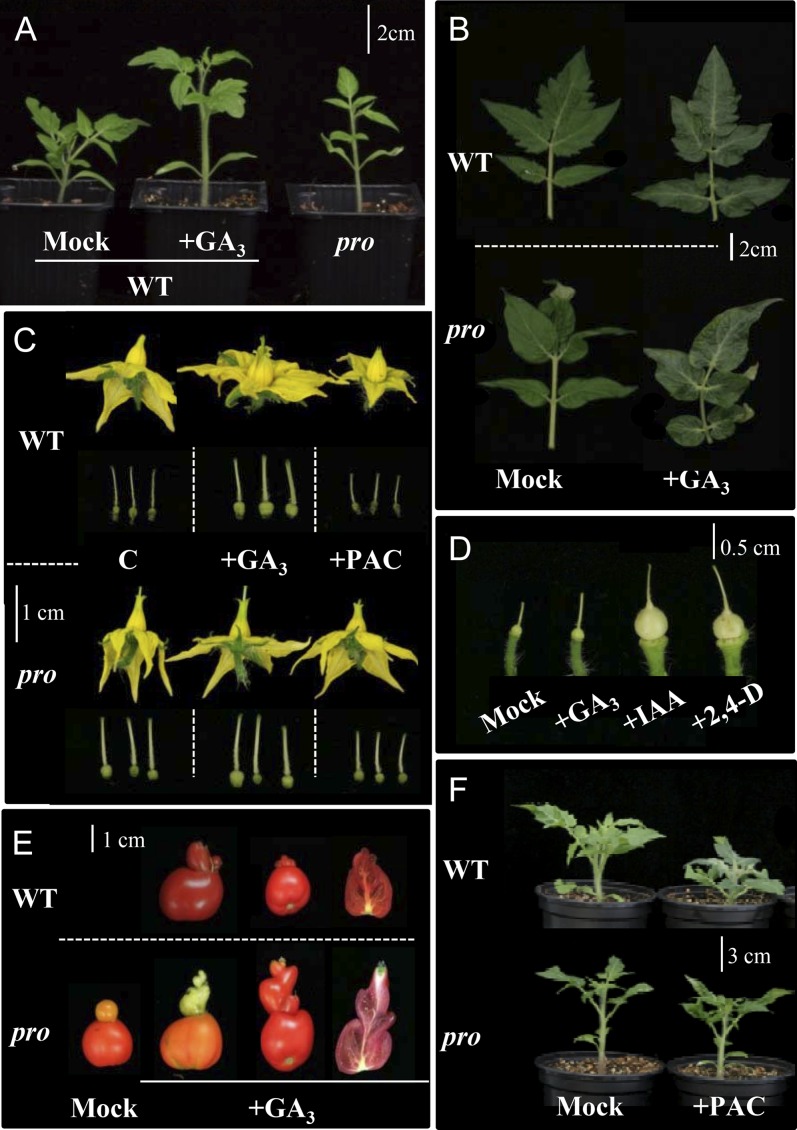
Wild-type tomato plants treated with GA3 display a slender phenotype and have leaves with less serrated borders (Martí et al., 2006), similar to that of pro plants (Fig. 4, A and B). This phenotype was obtained after continuous spray application (every 2 d) of a high GA3 dose (10−4 m). The number of leaves before the first inflorescence was also higher in wild-type plants treated with GA3 (Table IV). Continuous GA3 application to wild-type plants induced the development of flowers with longer styles and protruding stigmas (Fig. 4C) as well as flowers with increased numbers of sepals, petals, stamens, and carpels (sometimes more than four), similar to pro. In these plants, the average fruit weight was similar to pro parthenocarpic fruits, and most of them had no seeds (Table IV), probably due to the prevention of self-pollination by the long-style phenotype. Application of GA3 or auxin (indole-3-acetic acid [IAA] or 2,4-dichlorophenoxyacetic acid [2,4-D]) to pistils of wild-type emasculated flowers also enhanced style elongation, more the latter than the former (Fig. 4D). The Brix index value of the juice was higher in parthenocarpic GA3-induced fruits and similar to that of pro fruits, although GA3 application had no effect on fruit-set and growth on Brix index in pro plants (Table IV). Wild-type plants continuously treated with GA3 showed many fruits with malformations due to the development of one or more additional fruit-like structures at the style end of the fruit, similar to those found in pro (Fig. 4E). When GA3 application was carried out only from flowering onset, that kind of fruit malformation did not occur in wild-type plants but was enhanced in pro plants (data not shown).
Figure 4.
Effects of GA3 and PAC application to wild-type (WT) and pro plants. A, Wild-type control and GA3-treated plants compared with pro plants. B, Morphology of the fifth leaf (from the cotyledons) of nontreated and GA3-treated plants. C, Flower morphology and size of pistils from plants treated with GA3 and PAC. D, Effects of GA3, IAA, and 2,4-D on the style elongation of wild-type pistils. The pistils were treated with hormone solutions on day −5 after removing stamens and petals, and style length was measured on day 0. Final means ± se of style lengths (mm) were as follows: mock, 3.8 ± 0.2; +GA3, 4.7 ± 0.1*; +IAA, 5.3 ± 0.2**; +2,4-D, 6.1 ± 0.2** (n = 7; *P < 0.05, **P < 0.01). E, Effect of continuous GA3 application to plants on the morphology of representative fruits displaying additional fruit-like structures. F, Wild-type and pro plants untreated and treated with PAC. [See online article for color version of this figure.]
Table IV. Effects of PAC and GA3 application to entire plants on vegetative and fruit growth in wild-type and pro plants.
PAC (10−5 m) and GA3 (10−5 m) were applied after the development of the first true leaf, every 2 d until flowering. Values are means of 10 plants and 50 fruits. Brix index was determined in juice collected from five mature fruits from 10 plants.
| Parameter | Wild Type |
pro |
||||
|---|---|---|---|---|---|---|
| Mock | +GA3 | +PAC | Mock | +GA3 | +PAC | |
| Height to first inflorescence (cm) | 10.4 ± 0.3 | 11.7 ± 0.3 | 6.1 ± 0.2 | 17.2 ± 0.2 | 17.6 ± 0.2 | 13.5 ± 0.2 |
| Leaves to first inflorescence (n) | 6.6 ± 0.2 | 7.8 ± 0.3 | 5.6 ± 0.2 | 8.6 ± 0.2 | 8.2 ± 0.3 | 7.8 ± 0.1 |
| Style length (mm) | 5.9 ± 0.4 | 7.0 ± 0.3 | 4.1 ± 0.3 | 7.3 ± 0.3 | 8.8 ± 0.4 | 5.8 ± 0.3 |
| Days to anthesis of first flower (n) | 38.5 ± 0.8 | 36.6 ± 0.1 | 40.4 ± 0.5 | 43.7 ± 0.2 | 42.2 ± 0.2 | 42.2 ± 0.2 |
| Fruits per plant (n) | 25.4 ± 2.0 | 29.6 ± 1.2 | – | 40.5 ± 2.2 | 39.5 ± 1.5 | – |
| Weight (g per fruit) | 5.8 ± 0.5 | 3.0 ± 0.6 | – | 3.0 ± 0.4 | 3.0 ± 0.45 | – |
| Seeds (n per fruit) | 21.8 ± 2.6 | 0.9 ± 1.5 | – | 0 | 0 | – |
| Seedless fruits (%) | 7.7 ± 3.0 | 75.3 ± 1.5 | – | 100 ± 0 | 100 ± 0 | – |
| Brix index | 4.8 ± 0.3 | 6.8 ± 0.4 | – | 7.2 ± 0.2 | 6.5 ± 0.2 | – |
It is known that pro plants have a mutation in the gene encoding the SlDELLA protein, a repressor of GA mode of action (Bassel et al., 2008). This suggests that the observed pro phenotypes are the result of a GA constitutive response induced by this mutation. As expected for a constitutive GA response mutant, pro plants were more resistant than wild-type plants to the reduction of shoot length and flower organ size by paclobutrazol (PAC), an inhibitor of GA biosynthesis (Table IV; Fig. 4, C and F; Supplemental Fig. S1).
Parthenocarpic fruit development can be induced in tomato by the application of GA or auxin (Serrani et al., 2007a, and refs. therein). Fruits developed from unpollinated pro ovaries were of equal size to unpollinated wild-type ovaries induced by GA3 application, and GA3 did not have an additional effect on pro ovaries (Supplemental Table S1), indicating that fruit-set and growth response to GA3 were saturated in the mutant. By contrast, auxin application to unpollinated pro ovaries enhanced fruit growth, although the final weight was lower than that of auxin-treated wild-type ovaries (Supplemental Table S1). Therefore, the additional growth response observed in seedy compared with seedless pro fruits presented in Table III may be due to auxin provided by the developing seeds.
The pro Mutation Causes Substantial Transcriptome Remodeling in the Ovary
Since we wanted to detect early changes in transcript levels that may be involved in parthenocarpy induction, a comparative transcriptome analysis was performed with unpollinated wild-type and pro ovaries 3 d before anthesis, a stage at which they had slightly but significantly different weights (Fig. 2, C and D). Those ovaries were mainly constituted of sporophytic tissues, as they contained nonfertilized ovules (Fig. 1G).
Transcriptome analysis showed significant and remarkable differences in gene expression in pro compared with wild-type ovaries. In pro ovaries, 2,776 unigenes were differentially expressed, 1,346 up-regulated (Supplemental Table S2) and 1,430 down-regulated (Supplemental Table S3). Similarity to Arabidopsis proteins could be assigned to 88.2% of the differentially expressed sequences. For the 617 genes showing at least 2-fold changed expression in pro ovaries that were annotated (325 up-regulated [Supplemental Table S4] and 292 down-regulated [Supplemental Table S5]), their functional roles were examined using the Munich Information Center for Protein Sequences (http://mips.gsf.de) and FunCAt, to search for the most similar corresponding Arabidopsis protein. In most categories, similar proportions of genes were found in both cases (Supplemental Table S6).
To unveil possible key processes that were altered in pro ovaries, we looked for functional enrichment in the differentially expressed set of genes using the AmiGO tool (http://amigo.geneontology.org) based on the Gene Ontology (GO) terms for the most similar Arabidopsis proteins. Biological processes identified as most significantly overrepresented (P < 10−5) among the up-regulated genes (Supplemental Fig. S2A; Supplemental Table S7) were “photosynthesis” (GO:0019684) and “response to stimulus” (GO:0050896), and those among the down-regulated genes were “response to hormone stimulus” (GO:0009725) and “response to endogenous stimulus” (GO:0009719; Supplemental Fig. S2B; Supplemental Table S8). Within cellular components, the GO category corresponding to “chloroplast thylakoid membrane” (GO:0009535) was also overrepresented in the up-regulated set.
In addition to the genes included in the enriched categories described above, some involved in cell organization and in cell wall degradation were up-regulated or down-regulated at least 2-fold in pro ovaries (Supplemental Table S9). Also, the expression of two genes encoding putative peroxidases was altered, one of them up-regulated (homologous to At1g14550) and the other down-regulated (homologous to At5g05340). This is of interest because peroxidase activity has been inversely correlated with cell expansion in pro stem tissues (Jupe and Scott, 1992). In the TF category, 19 genes were significantly up-regulated and 27 down-regulated in pro ovaries, with at least 2-fold differential expression (Supplemental Table S9). In both groups, we identified TFs from the Myb, bHLH, HD-Zip, WRKY, ERF, NAC, and GRAS families. However, MADS box and bZIP-encoding genes (two and one, respectively) were only found within those up-regulated, and C2/H2, Aux/IAA, and ARF genes (two, one, and three, respectively) were only found within those down-regulated.
Quantification of Genes Involved in Cell Division and Expansion
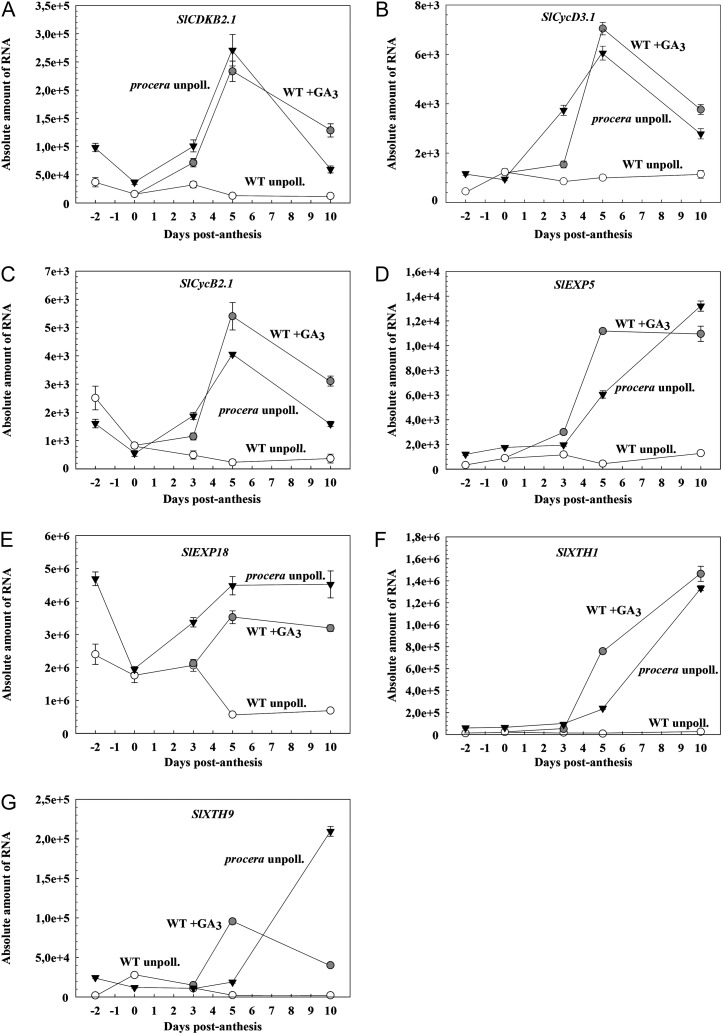
We analyzed by qPCR transcript levels of several genes involved in cell division (tomato CYCLIN DEPENDENT KINASE [SlCDKB2.1], CYCLIN [SlCycB2.1], and SlCycD3.1, encoding a G1 cyclin) and cell expansion (EXPANSINs SlEXP5 and SlEXP18 and XYLOGLUCAN ENDOTRANSGLUCOSYLATE/HYDROLASEs SlXTH1 and SlXTH9), two processes found to be enhanced in pro ovaries even before anthesis (Fig. 3). Those genes belong to large families, were not present in the array, and were selected because they have previously been reported to show early maximum increase after pollination-induced fruit-set (Joubès et al., 2000, 2001; Vriezen et al., 2008). The expression of the three cell division genes was relatively low in unpollinated wild-type and pro ovaries before and at the time equivalent to anthesis (day 0) and started increasing in pro ovaries from day 3 (time of active cell division), with a maximum at day 5, and decreasing afterward (Fig. 5, A–C). Similar results to pro were found in wild-type GA3-treated ovaries (Fig. 5, A–C). Expression of the selected SlEXP and SlXTH cell expansion genes was always low in wild-type unpollinated ovaries, whereas in the case of pro and wild-type GA3-treated ovaries, it increased steadily after day 3 up to at least day 10 (Fig. 5, D–G), associated with the beginning of rapid cell expansion (Fig. 3A). Similar results were found in pollinated and wild-type unpollinated GA3-treated ovaries, except for SlXTH9, which decreased at day 10 (Fig. 5, D–G).
Figure 5.
Time course between day −2 and day 10 of transcript levels of cell division and expansion genes in pro and wild-type (WT) unpollinated control and GA3-treated ovaries. A, SlCDKB2.1. B, SlCycD3.1. C, SlCycB2.1. D, SlEXP5. E, SlEXP18. F, SlXTH1. G, SlXTH9. Absolute amount of RNA means molecules of RNA per ng of total RNA.
A 450-bp promoter deletion that results in the down-regulation of SlStyle2.1 (encoding an atypical bHLH protein lacking DNA-binding activity) in developing styles has been reported to reduce style length and favor self-pollination (autogamy) in cultivated tomato (Chen et al., 2007). We have confirmed that cv Micro-Tom also contains this SlStyle2.1 promoter deletion (Supplemental Fig. S3A). SlStyle2.1 transcript levels in pro styles were lower than in the wild type, although they were slightly increased in the ovary (Supplemental Fig. S3B). Moreover, application of 2,4-D but not GA3 to pistils of wild-type emasculated flowers induced a discrete increment on the accumulation of SlStyle2.1 transcripts (Supplemental Fig. S3C). Overall, these results suggest that SlStyle2.1 is not responsible for the long style with elongated cells phenotype of pro flowers.
Quantification of Genes Involved in Hormone Signaling and Metabolism
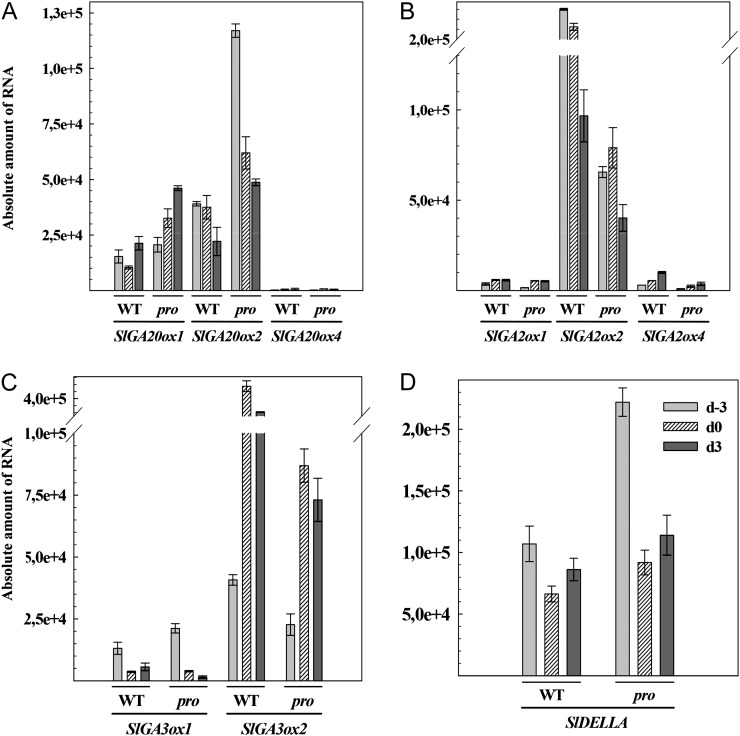
The results of transcriptome analysis showed that the expression of some genes of GA and abscisic acid (ABA) metabolism, as well as several genes involved in the auxin signaling pathway, was altered in day −3 (3 d before anthesis) pro ovaries. This suggests that the pro mutation affects the expression of genes of several hormone pathways and prompted us to further investigate the extent of this effect by first analyzing the expression of the main tomato genes of GA metabolism (Serrani et al., 2007b) and SlDELLA as well as ABA metabolism in day −3, day 0, and day 3 pro and wild-type ovaries by qPCR (Fig. 6).
Figure 6.
Transcript levels of GA metabolism and SlDELLA genes in unpollinated wild-type (WT) and pro ovaries between day −3 and day 3. A, SlGA20ox1, SlGA20ox2, and SlGA20ox4. B, SlGA2ox1, SlGA2ox2, and SlGA2ox4. C, SlGA3ox1 and SlGA3ox2. D, SlDELLA. Absolute amount of RNA means molecules of RNA per ng of total RNA.
Transcript content of SlGA20ox1, encoding the main GA20ox in tomato ovary, and SlGA20ox2 was enhanced (Fig. 6A), while that of SlGA2ox1, -2, and -4 was reduced (Fig. 6B) in pro. This was surprising because the expression of GA20ox genes has been reported to be under negative and that of GA2ox under positive GA feedback regulation, by DELLAs (Yamaguchi, 2008). In the case of GA3ox (also expected to be under GA negative feedback regulation), the expression of SlGA3ox1 was similar in the wild type and pro, while that of SlGA3ox2 was down-regulated in pro ovaries, suggesting that this gene is under constitutive negative feedback regulation in the mutant (Fig. 6C). Transcript levels of SlDELLA were higher in pro ovaries at the three stages investigated (Fig. 6D), indicating that expression of this repressor is up-regulated by the mutation in very young ovaries. Treatment of 7-d-old seedlings with GA3 and PAC confirmed the existence of negative feedback regulation of SlDELLA expression in tomato (Supplemental Fig. S4). Two genes of ABA metabolism encoding 9-cis-epoxy-carotenoid dioxygenase and a putative (+)ABA 8′-hydroxylase, previously shown to regulate ABA content in tomato ovaries (Nitsch et al., 2009), were investigated. However, only in the first case was some reduction of expression in pro ovaries observed (Supplemental Fig. S5).
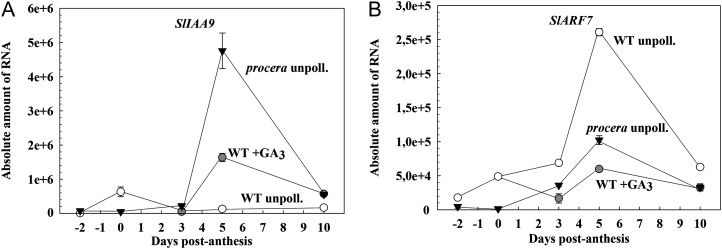
To estimate the effect of the pro mutation on auxin signaling in relation to parthenocarpic growth, we concentrated on two genes previously described to act as repressors of ovary growth not present in the microarray: SlIAA9 (belonging to the Aux/IAA family; Wang et al., 2005) and SlARF7 (de Jong et al., 2009b). The time-course analysis showed that transcript levels of SlIAA9 in unpollinated pro ovaries were very low between day −2 and day 3 but increased afterward, with a maximum at day 5 (Fig. 7). The content of SlIAA9 transcripts in wild-type unpollinated ovaries was always very low, except at day 0, which was higher than in pro. Wild-type unpollinated ovaries treated with GA3 displayed a similar pattern to pro ovaries (Fig. 7). By contrast, expression of SlARF7 was always lower in pro than in wild-type unpollinated ovaries (between day −2 and day 10), and GA3 treatment also dramatically reduced SlARF7 transcript content in wild-type ovaries (Fig. 7). The expression of other genes in the auxin-signaling pathway (SlIAA1, SlIAA3, SlIAA14, and SlARF1), previously shown to display some variation in response to ovary GA3 application (Serrani et al., 2008), was also investigated, but no clear effect of pro on their expression was found (data not shown).
Figure 7.
Time course between day −2 and day 10 of SlIAA9 (A) and SlARF7 (B) transcript content in pro and wild-type (WT) unpollinated control and GA3-treated ovaries. Absolute amount of RNA means molecules of RNA per ng of total RNA.
DISCUSSION
The Longer Style of pro Flowers Is Not Controlled by SlStyle2.1
Wild-type tomato species, like Solanum pennellii, have long styles with stigmas exserted beyond the staminal cone, thus preventing self-pollination. Shorter style length is associated, together with loss of self-incompatibility, with the evolution of autogamy in cultivated tomatoes (Chen et al., 2007). Down-regulation of SlStyle2.1 expression due to a 450-bp promoter deletion (−4,656 to −4,002) has been associated with the transition from cross-pollination to self-pollination. cv Micro-Tom also contains that promoter deletion, although some transcripts are still present, as was previously found by in situ hybridization in styles of flowers from S. lycopersicum ‘M82’ (Chen et al., 2007). However, our results do not support the idea that SlStyle2.1 is responsible for the long-style phenotype of pro flowers, because its transcript levels were not enhanced in the mutant or in the wild type after GA3 application. Nevertheless, since auxin application induces style elongation and the accumulation of SlStyle2.1 transcripts in wild-type pistils, we cannot discard that auxins may play a role in style elongation of pro flowers.
The protein encoded by SlStyle2.1 belongs to the atypical bHLH family lacking DNA-binding activity (Chen et al., 2007). Several proteins from this family have been shown to regulate hormone-induced cell elongation in Arabidopsis and rice. For instance, PACLOBUTRAZOL RESISTANT1 (PRE1) overexpression in the gai-1 genetic background (bearing a GA-insensitive allele of the GAI DELLA protein) suppresses its short-hypocotyl phenotype, suggesting that PRE1 may act as positive regulator of GA signaling, presumably downstream of DELLA proteins (Lee et al., 2006). Transcript accumulation of SPATULA (a DNA-binding bHLH gene), involved in style and stigma growth during gynoecium development in Arabidopsis, is negatively regulated by DELLA (Groszmann et al., 2011). Also, DELLA proteins are known to interact with several phytochrome-interacting factors (members of a subfamily of DNA-binding bHLH proteins) involved in the GA regulation of plant growth and development (Leivar and Quail, 2011). Increase of style length upon SlDELLA depletion in antisense-SlDELLA plants has previously been reported (Martí et al., 2007). Overall, these results suggest a scenario in which the reduction of style cell elongation of tomato flowers may occur by SlDELLA sequestering in an inactive complex a transcriptional regulator related to STYLE2.1, through protein-protein interaction.
pro Flowers Display Meristic Alterations
The high number of pro flowers carrying additional floral organs and the presence of fruits with additional fruit-like structures were surprising. We also found a similar effect in wild-type plants continuously treated with GA3, but not when GA3 was applied only at the time of flowering, probably because this effect of GA3 occurs in the flower meristem at the time of early flower organ formation. Previous reports suggested that the increased number of floral organs in tomato plants grown under low-temperature conditions is mimicked by GA3 application (Sawhney, 1983), associated with alteration in MADS box gene expression (Lozano et al., 1998). The size of the additional fruit-like structures developed in pro fruits was smaller than in wild-type fruits from plants continuously treated with GA3, but it was enhanced in pro plants by GA3 application. This may be because the mutated SlDELLA still displays some repressor activity, thus preventing the development of additional carpels, while continuous GA3 application may be more efficient to induce endogenous SlDELLA degradation in wild-type plants.
Patterning of the four flower whorls is controlled by the coordinated expression of at least three sets of floral organ identity genes at the initiation of floral organ primordia (Ng and Yanofsky, 2001). However, although the eventual fate of organ primordia is determined by the organ identity genes, the arrangement of the floral meristem in concentric whorls, each one with a particular number of units, is thought to be regulated by auxin (Cheng and Zhao, 2007). The altered expression of genes encoding several Aux/IAA and ARF proteins in pro ovaries indicates that some of them may be involved in the observed flower meristic transformations.
pro Induces Strong Parthenocarpic Capacity Associated with Ovary Transcriptome Remodeling
The pro ovaries have the capacity to develop parthenocarpically. The absence of seeds was due to the long-style phenotype, which prevents self-pollination. The reduced number of pollen grains found in pro flowers is of interest because loss of DELLA repression in rice and barley (Hordeum vulgare) also leads to less pollen production (for review, see Plackett et al., 2011). This means that successful reproductive development needs restriction of the GA response. A long-style phenotype, but not reduced pollen number, was found in tomato antisense-SlDELLA plants (Martí et al., 2007). However, pro parthenocarpy is facultative, because fruits with a certain number of seeds could be obtained by manual self-pollination. pro ovaries from emasculated flowers also developed parthenocarpically, while in antisense-SlDELLA plants, poor parthenocarpic fruit growth occurs in emasculated flowers. This means that the pro mutation activates a pollination-independent developing program in the ovary even under conditions (flower emasculation) where this program is still repressed in antisense-SlDELLA ovaries, probably because the pro mutation has a stronger effect on the reduction of SlDELLA level or activity than the antisense-SlDELLA knockdown mutation. That activation occurs very early in pro, at least 3 d before anthesis, as shown by enhanced weight, larger and more cells, and transcriptome changes in the ovary.
The strong parthenocarpic capacity of pro ovaries described before was found under different growing conditions (in the greenhouse throughout the year, using the conditions described in “Materials and Methods,” and in a growth chamber using artificial lighting only) and is not due to the absence of brassinosteroids, because similar phenotypes (including exserted stigmas) were found in D-pro plants. The high penetrance and expressivity of this phenotype was probably not caused by the presence of the short-pruning mutation in cv Micro-Tom either, because it has been described that many parthenocarpic fruits also develop from nonemasculated flowers in antisense-SlDELLA of UC82 (a determinate cultivar). The possibility that other mutations present in cv Micro-Tom (e.g. miniature) might affect the pro phenotype is uncertain.
Tomato fruit exocarp constitutes a constraint to turgor-driven growth (Andrews et al., 2002). Cell wall-located peroxidases are involved in growth restriction (Passardi et al., 2004) and seem to play an important role in tomato fruit growth regulation (Andrews et al., 2002). Thus, the strong down-regulation of a peroxidase in pro ovaries is of interest in relation to its early parthenocarpic growth. Reduction of peroxidase activity was found in elongating stem tissues of pro and GA3-treated wild-type tomato plants (Jupe and Scott, 1992) as well as in the la cry pea mutant (Jupe and Scott, 1989; Weston et al., 2008). On the other hand, the pro mutation up-regulated CUT1, a gene involved in cuticle biosynthesis, which may help to prevent water loss and facilitate ovary growth. Up-regulation of genes encoding wax and cuticle biosynthesis enzymes occurs in transgenic Carrizo citrange citrus (Citrus sinensis × Poncirus trifoliata hybrid) with elevated GA (Huerta et al., 2008).
Enhanced expression of genes involved in photosynthesis and carbon utilization has been reported after GA application and in citrus overexpressing CcGA20ox1, associated with the increase of net photosynthesis in the leaves (Huerta et al., 2008). In the case of pro ovaries, although net photosynthesis has not been determined, a transcript increase of genes encoding proteins involved in light and dark reactions may be relevant to its parthenocarpic growth capacity.
SlDELLA Modulates Cell Cycle and Expansion Genes during Fruit-Set and Development
Fruit development in tomato, after pollination, is characterized by a cell division phase (up to about 10 d post anthesis) followed by cell expansion (Gillaspy et al., 1993). Unpollinated pro ovaries had more and larger cells than the wild type, indicating that both processes are stimulated by pro mutation in the absence of pollination and fertilization. A higher number of cell layers was found in pro ovaries as early as 3 d before anthesis. The complex cell division cycle is controlled by the ordered action of cyclin-dependent kinases (CDK) and their positive regulatory proteins named cyclins (Cyc). Thus, the higher transcript level of SlCDKB2.1 in pro ovaries from at least 2 d before anthesis, and those of SlCycB2.1 and SlCycD3.1 from day 3, support the hypothesis that the pro positive effect on cell division is mediated, at least partially, by that kinase and cyclins. Increased expression of those genes during the cell division phase also occurs in pollinated ovaries (Joubès et al., 2000, 2001; Czerednik et al., 2012) and in parthenocarpic fruits induced in RNA interference SlARF7 plants (de Jong et al., 2011). By contrast, in antisense-SlDELLA plants, no increase of cell division was found in the fruit (Martí et al., 2007), suggesting that in this case parthenocarpic growth occurs, bypassing the cell division phase. This may mean that the residual SlDELLA present in the antisense plants is still capable of repressing cell divisions in the ovary, an effect that can explain the lower parthenocarpic capacity of antisense-SlDELLA compared with pro plants. On the other hand, EXP and XTH mediate changes in cell wall loosening, and the expression of genes encoding both kind of proteins increases in tomato ovaries after pollination (Vriezen et al., 2008) and in RNA interference SlARF7 parthenocarpic fruits (de Jong et al., 2011). The higher transcript levels of SlEXP5, SlEXP18, SlXTH1, and SlXTH9 in pro ovaries provide a molecular basis for the positive effect of this mutation in cell expansion.
High levels of endopolyploidy occur in tomato during fruit development (Chevalier et al., 2011). We found that increase of mean C levels started at day 1 to 2 in pro and wild-type pollinated ovaries, associated with the first signs of cell expansion, compared with the absence of endoreduplication and cell expansion in wild-type unpollinated ovaries. No difference in the time and degree of endoreduplication between wild-type pollinated and pro ovaries was observed, at least up to day 10. Our results suggest that SlDELLA may act as a repressor of endoreduplication, thus preventing cell expansion. However, since it has been shown that cell and fruit size can be uncoupled from DNA ploidy levels, endoreduplication (both in the wild type and pro) may act just as a limiting factor for cell expansion during fruit growth (Chevalier et al., 2011).
Hormone Response and Metabolism Are Altered in the pro Ovary
The higher SlDELLA transcript content in pro ovaries and seedlings indicates that the gene was subject to negative feedback regulation by its functional protein product in tomato. This agrees with SlDELLA up-regulation in pro leaves (Bassel et al., 2008) and in wild-type ovaries after GA3 application, which is expected to reduce SlDELLA content (Serrani et al., 2008). By contrast, only slight or no feedback regulation of DELLA genes occurs in Arabidopsis, rice, and barley (Sun and Gubler, 2004).
Enhanced expression of SlGA20ox1, which seems to play a pivotal role in tomato fruit-set (Martí et al., 2007; Olimpieri et al., 2007; Serrani et al., 2007b), also occurs in pro ovaries. This was surprising, mainly because feedback regulation of that gene occurs in antisense-SlDELLA (Martí et al., 2007) and in GA3-treated wild-type tomato ovaries (Martí et al., 2010). Other dioxygenases (e.g. SlGA20ox2 and SlGA2ox2) were not subject to feedback regulation in pro ovaries either. In Arabidopsis, high GA content is associated with enhanced expression of AtGA3ox2 during germination (Frigerio et al., 2006) and of AtGA20ox and AtGA3ox in etiolated compared with deetiolated seedlings (Alabadí et al., 2008). This suggests that the feedback regulation of some GA dioxygenases may be uncoupled from GA mode of action in some circumstances to secure a high level of active GAs.
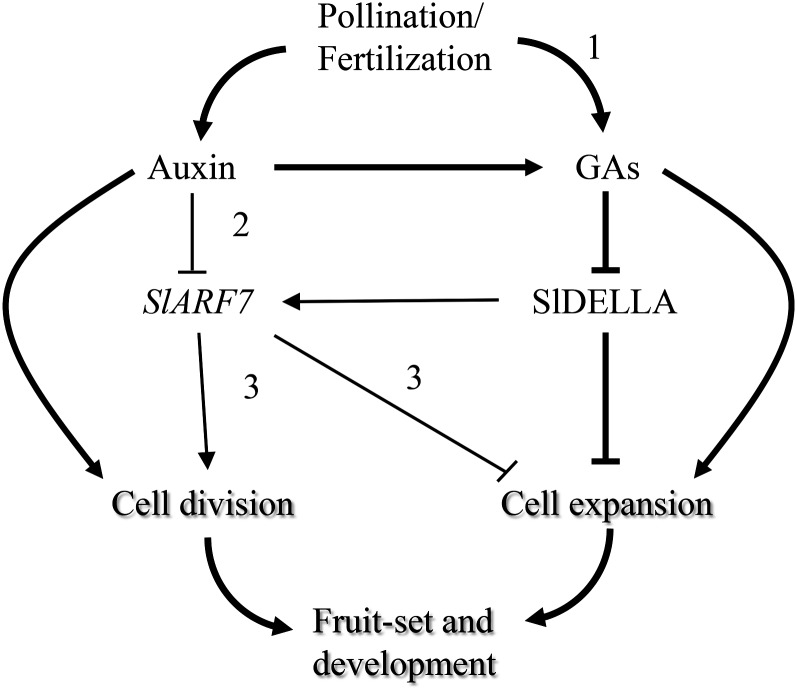
Induction of parthenocarpic growth in pea, Arabidopsis, and tomato by auxin is mediated in part by GAs through the regulation of GA metabolism (Ozga and Reinecke, 2003; Serrani et al., 2008; Dorcey et al., 2009). We provide here new information on the auxin-GA cross talk during fruit-set by showing that GA activity modulates the expression of a gene in the auxin signaling pathway (SlARF7), previously reported to act as a repressor of ovary growth. The pro mutation produced down-regulation of SlARF7 expression in the ovaries from before anthesis up to day 10, and a similar effect was observed upon GA3 application to wild-type unpollinated ovaries. Lower transcript content of SlARF7 in the cell wall of GA3-treated unpollinated ovaries during the 3 d after treatment, as well as in pollinated ovaries, has been found before (de Jong et al., 2009b). All these results support the conclusion that SlDELLA up-regulates the transcription of SlARF7, which encodes a repressor of cell expansion (de Jong et al., 2011). By contrast, the transcript content of SlIAA9 was low in unpollinated ovaries, showing a small transient maximum peak in wild-type ovaries at day 0. Scarce information on SlIAA9 expression in relation to fruit-set has been reported before. It is of interest that in situ hybridization analysis has shown that pollination triggers the initiation of fruit development associated with the release of a tissue-specific gradient of SlIAA9 expression established during flower development (although no quantitative data on SlIAA9 transcript content were reported; Wang et al., 2009). A proposed model showing the control of fruit-set and development by auxin and GAs, mediated by SlDELLA and the auxin signaling factor SlARF7, based on the results presented in this work and previously by other authors, is given in Figure 8.
Figure 8.
Model for auxin and GA interaction through SlDELLA during tomato fruit-set and development. Thick lines correspond to previously well-established processes and thin lines to proposed relationships based on the results of this work or from other authors. Path 1, possible auxin-independent regulation of GA metabolism; path 2, from de Jong et al. (2009b) and O. Ruiz-Rivero and J.L. García-Martínez (unpublished data); path 3, from de Jong et al. (2011). In addition to down-regulation of SlARF7 transcription by auxin, this hormone would be expected to promote the capacity of SlARF7 to regulate gene expression by virtue of auxin-induced degradation of Aux/IAA proteins.
In summary, our results with pro unveiled new roles of SlDELLA in the control of flower morphology and ovary cell division and expansion. They also suggest that a transcriptional regulator similar to SlStyle2.1, encoding an atypical bHLH, is a possible target of that GA repressor. These results showed a new kind of GA-auxin cross talk during fruit-set, where auxin and GA activities regulate the expression of SlARF7 encoding a repressor of ovary growth, and support the idea that fruit-set and development are controlled by a complex regulatory gene network.
MATERIALS AND METHODS
Plant Material
Near-isogenic lines of tomato (Solanum lycopersicum ‘Micro-Tom’) wild type, pro, D, and introgressed D-pro, as well as Solanum pennellii (accession LA0716; Tomato Genomics Research Center, University of California at Davis), were used in the experiments. The pro allele was introgressed in cv Micro-Tom (BC6F2) from LA0565 cv Condine Red, and D (a wild-type gene encoding 6-deoxocatasterone oxidase) from cv Manyel, as described by Carvalho et al. (2011). The plants were grown in a greenhouse under 24°C (day)/20°C (night) conditions, in 0.6-L pots with peat:vermiculite (1:1), and irrigated with Hoagland solution. Natural light was supplemented with Osram lamps (Powerstar HQI-BT; 400 W) to get a 16-h-light photoperiod. Two to three flowers per truss and the first two to three trusses were normally used for the experiments. Flower emasculation was carried out 2 d before anthesis (day −2) to prevent self-pollination. Cross-pollination was carried out at day 0 on previously emasculated flowers. All nonselected flowers were removed.
Application of Plant Growth Substances
GA3 (10−5 and 10−4 m) was sprayed on the entire plants as aqueous 0.1% Tween 20 solution. PAC (10−5 and 10−4 m) was sprayed or applied to the roots in the nutrient solution. GA3 (2,000 ng), IAA (2,000 ng), and 2,4-D (200 ng) were also applied to unpollinated ovaries from previously emasculated flowers in 10 µL of 5% ethanol, 0.1% Tween 20 solution. All plant growth substances were from Duchefa. Control plants and ovaries were treated with the same volumes of solvent solutions.
Histology
Tissue sections of day −2, day 0, day 3, day 5, and day 10 ovaries were fixed in 4% paraformaldehyde, 0.1 m sodium phosphate buffer, pH 7.2. After dehydration in ethanol, the samples were embedded in paraffin (Paraplast Plus; Sigma-Aldrich). 8-µm-thick sections were stained with Safranine-Alcian blue solution (a mixture of 2 mL of 0.1% Safranine in 50% ethanol and 5 mL of 0.1% Alcian blue in 50% ethanol, diluted in 200 mL of 0.1 m acetate buffer, pH 5.0), viewed with a microscope, and photographed with a spot digital camera (DMX1200F; Nikon).
Cell size was estimated by measuring the longest diameter of 10 cells in transverse sections from at least four fruits, anthers, and styles (one section per organ). In the case of pericarp, only cells from internal mesocarp were considered. The number of cell layers was determined by counting the number of cells along a line across the pericarp perpendicular to the epidermis and endocarp (avoiding vascular vessels).
Ploidy Determination
Ploidy of day −2 to day 10 wild-type ovaries was determined as described before (Serrani et al., 2007a). In the case of wild-type pollinated and pro day 3 to day 10 developing fruits, only the pericarp was used. Briefly, the material was sliced with a razor blade into 0.4 mL of nuclei isolation buffer (High Resolution DNA Kit, Solution A: Nuclei Isolation; Partec), mixed with 1 mL of staining buffer (High Resolution DNA Kit, Solution B: DAPI Staining; Partec), shaken for 1 min, and filtered through 100-µm nylon mesh (Nyblot). The filtrates (more than 5,000 nuclei per extract) were analyzed using a Partec PA-II flow cytometer. Peak areas corresponding to nuclei with different ploidy, taking young C values as reference, were used to calculate nuclei percentage distribution.
Pollen Quantification and Viability
Anthers were extracted with a vortex in Eppendorf vials with 500 μL of 0.5 m Suc solution. The pollen was pelleted and resuspended in 200 μL, deposited on a microscope slide, and the number of pollen grains was counted in 20 randomly selected field trials per slide.
Pollen viability was determined using two staining methods. First, by looking at the number of pollen grains stained in red with carmine acetate solution. Second, by incubating extracted pollen in 2.4 × 10−5 m fluorescein diacetate solution for 10 min and measuring fluorescence intensity (λex = 494 nm, λem = 521 nm) in a Perkin-Elmer LS50B spectrofluorimeter (Pinillos and Cuevas, 2008).
Pollen germination tests were performed on glass slides coated with germination medium [0.292 m Suc, 1.27 mm Ca(NO3)2, 1.62 mm H3BO3, 1 mm KH2PO4, and 0.5% agarose]. The number of germinated pollen grains was counted with a microscope after 2 h of incubation at 25°C in the dark.
Transcriptome Analysis
Transcript profiles corresponding to wild-type and pro d −3 ovaries were compared using the tomato 70-mer oligo microarray (EU-TOM2) containing 11,857 unigenes and the updated version TomatoCombine#5 (SGN-U5xxxxx) of the tomato unigene database (Bombarely et al., 2011; http://solgenomics.net/). According to the prepublished ITAG2.3 release of the tomato genome annotation, the EU-TOM2 oligo microarray represents 7,575 different gene models.
Total RNA was extracted using the RNAqueous and Turbo DNA-free kits (Ambion, Applied Biosystems). Labeled cDNA was prepared using 1 µg of total RNA and the Amino Allyl MessageAmpII aRNA amplification kit (Ambion). Cy3/Cy5-labeled cDNAs corresponding to three wild-type and pro independent biological samples with a dye swap were hybridized to four independent array slides. Images were acquired using the Genepix 6.0 image acquisition program. Statistical analysis of data was performed with the GeneSpring GX version 9.0.5 software (Agilent Technologies). Only genes showing at least 2-fold differential expression between samples, with P < 0.05 in one-way ANOVA (parametric test without the assumption of equal variances), were used for further analyses.
Quantitative PCR
One microgram of total RNA (RNAqueous-4PCR kit and Plant RNA Isolation Aid; Ambion, Applied Biosystems) was used to synthesize first-strand cDNA (TaqMan reverse transcription kit; Ambion). Transcript levels were determined by absolute qPCR according to the methodology described in detail elsewhere (Serrani et al., 2008) using specific primers (Supplemental Table S10; Serrani et al., 2008). Amounts of mRNA in samples were quantified using three biological replicates.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Style2.1, EU161281; SlCycB2.1, AJ243455; SlCycD3.1, AJ245415; SlCDKB2.1, AJ297917; SlXTH1, D16456; SlXTH9, AY497479; SlEXP5, AF059489; SlEXP18, AJ004997; SlDELLA, AY269087; SlIAA9, AJ937282; SlARF7, EF121545; SlCYP707A1, EU183406; and SlNCDE1, AJ439079.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of PAC and GA3 on wild-type and pro plants.
Supplemental Figure S2. Hierarchical view of GO biological process.
Supplemental Figure S3. Transcript level analysis of SlStyle2.1.
Supplemental Figure S4. Effect of GA3 and PAC on SlDELLA expression in wild-type seedlings.
Supplemental Figure S5. Transcript levels of ABA metabolism genes in wild-type and pro ovaries.
Supplemental Table S1. Effect of GA3 and 2,4-D on unpollinated wild-type and pro ovaries.
Supplemental Table S2. Tomato gene annotations of significantly up-regulated genes in pro ovaries.
Supplemental Table S3. Tomato gene annotations of significantly down-regulated genes in pro ovaries.
Supplemental Table S4. Tomato gene annotations of up-regulated genes in pro ovaries with at least 2-fold change.
Supplemental Table S5. Tomato gene annotations of down-regulated genes in pro ovaries with at least 2-fold change.
Supplemental Table S6. Protein Sequences functional analysis of differentially expressed genes in pro ovaries.
Supplemental Table S7. GO biological process categories overrepresented in the up-regulated set.
Supplemental Table S8. GO biological process categories overrepresented in the down-regulated set.
Supplemental Table S9. Genes encoding TFs and cell division and cell wall biosynthesis.
Supplemental Table S10. Primers used for qPCR.
Supplementary Material
Acknowledgments
We thank Dr. A. Bombarely (Sol Genomics Networks database) for help with tomato gene annotation, Dr. C. Gisbert (Instituto Universitario para la Conservación y Mejora de la Agrodiversidad Valenciana) for providing seedlings of S. pennellii, Mrs. T. Sabater (Instituto de Biología Molecular y Celular de Plantas) for care of plants in the greenhouse, and Drs. D. Alabadí and M.A. Pérez-Amador (Instituto de Biologia Molecular y Celular de Plantas) for critical reading of the manuscript.
Glossary
- TF
transcription factor
- qPCR
quantitative PCR
- IAA
indole-3-acetic acid
- 2,4-D
2,4-dichlorophenoxyacetic acid
- PAC
paclobutrazol
- GO
Gene Ontology
- ABA
abscisic acid
References
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Andrews J, Adams SR, Burton KS, Edmondson RN. (2002) Partial purification of tomato fruit peroxidase and its effect on the mechanical properties of tomato fruit skin. J Exp Bot 53: 2393–2399 [DOI] [PubMed] [Google Scholar]
- Barow M, Meister A. (2003) Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant Cell Environ 26: 571–584 [Google Scholar]
- Bassel GW, Mullen RT, Bewley JD. (2008) procera is a putative DELLA mutant in tomato (Solanum lycopersicum): effects on the seed and vegetative plant. J Exp Bot 59: 585–593 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho RF, Campos ML, Pino LE, Crestana SL, Zsögön A, Lima JE, Benedito VA, Peres LEP. (2011) Convergence of developmental mutants into a single tomato model system: ‘Micro-Tom’ as an effective toolkit for plant development research. Plant Methods 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K-Y, Cong B, Wing R, Vrebalov J, Tanksley SD. (2007) Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318: 643–645 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zhao Y. (2007) A role for auxin in flower development. J Integr Plant Biol 49: 99–104 [Google Scholar]
- Chevalier C, Nafati M, Mathieu-Rivet E, Bourdon M, Frangne N, Cheniclet C, Renaudin JP, Gévaudant F, Hernould M. (2011) Elucidating the functional role of endoreduplication in tomato fruit development. Ann Bot 107: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerednik A, Busscher M, Bielen BAM, Wolters-Arts M, de Maagd RA, Angenent GC. (2012) Regulation of tomato fruit pericarp development by an interplay between CDKB and CDKA1 cell cycle genes. J Exp Bot 63: 2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M, Mariani C, Vriezen WH. (2009a) The role of auxin and gibberellin in tomato fruit set. J Exp Bot 60: 1523–1532 [DOI] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH. (2009b) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57: 160–170 [DOI] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, García-Martínez JL, Mariani C, Vriezen WH. (2011) The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J Exp Bot 62: 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorcey E, Urbez C, Blázquez MA, Carbonell J, Pérez-Amador MA. (2009) Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J 58: 318–332 [DOI] [PubMed] [Google Scholar]
- Fos M, Nuez F, García-Martínez JL. (2000) The gene pat-2, which induces natural parthenocarpy, alters the gibberellin content in unpollinated tomato ovaries. Plant Physiol 122: 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fos M, Proaño K, Nuez F, García-Martínez JL. (2001) Role of gibberellins on parthenocarpic fruit-set in tomato induced by the genetic system pat-3/pat-4. Physiol Plant 111: 545–550 [DOI] [PubMed] [Google Scholar]
- Frigerio M, Alabadí D, Pérez-Gómez J, García-Cárcel L, Phillips AL, Hedden P, Blázquez MA. (2006) Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S, Vivian-Smith A. (2009) Fertilization and fruit initiation. Annu Plant Reviews 38: 107–171 [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. (1993) Fruits: a developmental perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Hooper LC, Johnson SD, Rodrigues JC, Vivian-Smith A, Koltunow AM. (2007) Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Paicu T, Alvarez JP, Swain SM, Smyth DR. (2011) SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J 68: 816–829 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. (2010) Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22: 2680–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta L, Forment J, Gadea J, Fagoaga C, Peña L, Pérez-Amador MA, García-Martínez JL. (2008) Gene expression analysis in citrus reveals the role of gibberellins on photosynthesis and stress. Plant Cell Environ 31: 1620–1633 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Tattersall A, Piazza P, Hay A, Martínez-Garcia JF, Schmitz G, Theres K, McCormick S, Tsiantis M. (2008) PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J 56: 603–612 [DOI] [PubMed] [Google Scholar]
- Jones MG. (1987) Gibberellins and the procera mutant of tomato. Planta 172: 280–284 [DOI] [PubMed] [Google Scholar]
- Joubès J, Lemaire-Chamley M, Delmas F, Walter J, Hernould M, Mouras A, Raymond P, Chevalier C. (2001) A new C-type cyclin-dependent kinase from tomato expressed in dividing tissues does not interact with mitotic and G1 cyclins. Plant Physiol 126: 1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Walsh D, Raymond P, Chevalier C. (2000) Molecular characterization of the expression of distinct classes of cyclins during the early development of tomato fruit. Planta 211: 430–439 [DOI] [PubMed] [Google Scholar]
- Jupe SC, Causton DR, Scott IM. (1988) Cellular basis of the effects of gibberellin and the pro gene on stem growth in tomato. Planta 174: 106–111 [DOI] [PubMed] [Google Scholar]
- Jupe SC, Scott IM. (1989) The slender phenotype of pea: stem growth, peroxidase levels and ethylene responses. Physiol Plant 77: 59–66 [Google Scholar]
- Jupe SC, Scott IM. (1992) Gibberellin and the pro gene suppress peroxidase activity in elongating tomato (Lycopersicon esculentum Mill.) stem tissues. Ann Bot (Lond) 69: 33–37 [Google Scholar]
- Lee S, Lee S, Yang K-Y, Kim Y-M, Park S-Y, Kim SY, Soh M-S. (2006) Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol 47: 591–600 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Angosto T, Gómez P, Payán C, Capel J, Huijser P, Salinas J, Martínez-Zapater JM. (1998) Tomato flower abnormalities induced by low temperatures are associated with changes of expression of MADS-box genes. Plant Physiol 117: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí C, Orzáez D, Ellul P, Moreno V, Carbonell J, Granell A. (2007) Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J 52: 865–876 [DOI] [PubMed] [Google Scholar]
- Martí E, Carrera E, Ruiz-Rivero O, García-Martínez JL. (2010) Hormonal regulation of tomato gibberellin 20-oxidase1 expressed in Arabidopsis. J Plant Physiol 167: 1188–1196 [DOI] [PubMed] [Google Scholar]
- Martí E, Gisbert C, Bishop GJ, Dixon MS, García-Martínez JL. (2006) Genetic and physiological characterization of tomato cv. Micro-Tom. J Exp Bot 57: 2037–2047 [DOI] [PubMed] [Google Scholar]
- Muangprom A, Thomas SG, Sun T-P, Osborn TC. (2005) A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiol 137: 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2: 186–195 [DOI] [PubMed] [Google Scholar]
- Nitsch LMC, Oplaat C, Feron R, Ma Q, Wolters-Arts M, Hedden P, Mariani C, Vriezen WH. (2009) Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1. Planta 229: 1335–1346 [DOI] [PubMed] [Google Scholar]
- Olimpieri I, Siligato F, Caccia R, Mariotti L, Ceccarelli N, Soressi GP, Mazzucato A. (2007) Tomato fruit set driven by pollination or by the parthenocarpic fruit allele are mediated by transcriptionally regulated gibberellin biosynthesis. Planta 226: 877–888 [DOI] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM. (2003) Hormonal interactions in fruit development. J Plant Growth Regul 22: 73–81 [Google Scholar]
- Pandolfini T, Molesini B, Spena A. (2007) Molecular dissection of the role of auxin in fruit initiation. Trends Plant Sci 12: 327–329 [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Pinillos V, Cuevas J. (2008) Standardization of the fluorochromatic reaction test to assess pollen viability. Biotech Histochem 83: 15–21 [DOI] [PubMed] [Google Scholar]
- Plackett ARG, Thomas SG, Wilson ZA, Hedden P. (2011) Gibberellin control of stamen development: a fertile field. Trends Plant Sci 16: 568–578 [DOI] [PubMed] [Google Scholar]
- Ren Z, Li Z, Miao Q, Yang Y, Deng W, Hao Y. (2011) The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. J Exp Bot 62: 2815–2826 [DOI] [PubMed] [Google Scholar]
- Sawhney VK. (1983) The role of temperature and its relationship with gibberellic acid in the development of floral organs of tomato (Lycopersicum esculentum). Can J Bot 61: 1258–1265 [Google Scholar]
- Serrani JC, Fos M, Atarés A, García-Martínez JL. (2007a) Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv Micro-Tom of tomato. J Plant Growth Regul 26: 211–221 [Google Scholar]
- Serrani JC, Ruiz-Rivero O, Fos M, García-Martínez JL. (2008) Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J 56: 922–934 [DOI] [PubMed] [Google Scholar]
- Serrani JC, Sanjuán R, Ruiz-Rivero O, Fos M, García-Martínez JL. (2007b) Gibberellin regulation of fruit set and growth in tomato. Plant Physiol 145: 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-P, Gubler F. (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Van Tuinen A, Peters AHLJ, Kendrick RE, Zeevaart JAD, Koornneef M. (1999) Characterisation of the procera mutant of tomato and the interaction of gibberellins with end-of-day far-red light treatments. Physiol Plant 106: 121–128 [Google Scholar]
- Vivian-Smith A, Koltunow AM. (1999) Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol 121: 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C. (2008) Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 177: 60–76 [DOI] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech JC, Bouzayen M. (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech JC, Fernie AR, Bouzayen M. (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston DE, Elliott RC, Lester DR, Rameau C, Reid JB, Murfet IC, Ross JJ. (2008) The pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiol 147: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.