Abstract
Stilbenes are a small family of phenylpropanoids produced in a number of unrelated plant species, including grapevine (Vitis vinifera). In addition to their participation in defense mechanisms in plants, stilbenes, such as resveratrol, display important pharmacological properties and are postulated to be involved in the health benefits associated with a moderate consumption of red wine. Stilbene synthases (STSs), which catalyze the biosynthesis of the stilbene backbone, seem to have evolved from chalcone synthases (CHSs) several times independently in stilbene-producing plants. STS genes usually form small families of two to five closely related paralogs. By contrast, the sequence of grapevine reference genome (cv PN40024) has revealed an unusually large STS gene family. Here, we combine molecular evolution and structural and functional analyses to investigate further the high number of STS genes in grapevine. Our reannotation of the STS and CHS gene families yielded 48 STS genes, including at least 32 potentially functional ones. Functional characterization of nine genes representing most of the STS gene family diversity clearly indicated that these genes do encode for proteins with STS activity. Evolutionary analysis of the STS gene family revealed that both STS and CHS evolution are dominated by purifying selection, with no evidence for strong selection for new functions among STS genes. However, we found a few sites under different selection pressures in CHS and STS sequences, whose potential functional consequences are discussed using a structural model of a typical STS from grapevine that we developed.
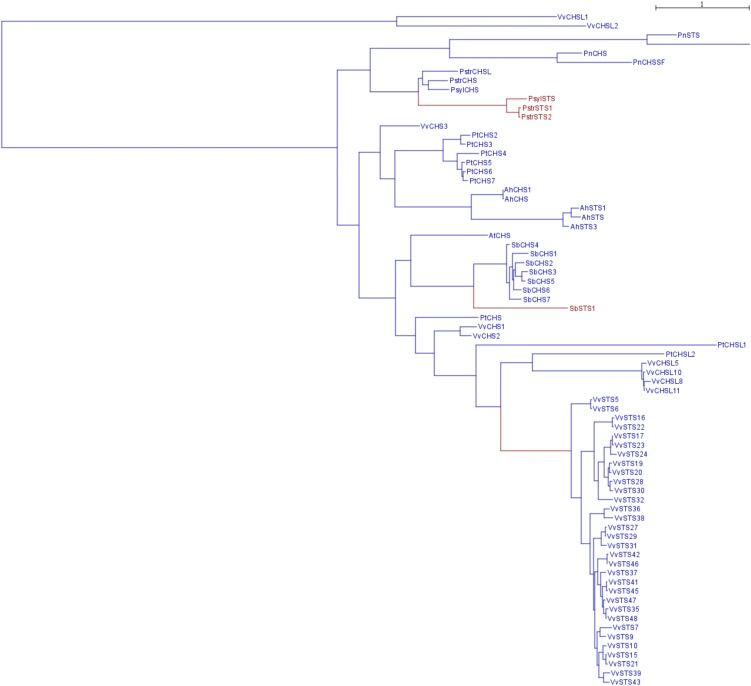
Plants produce a vast array of secondary metabolites, many of them being restricted to specific groups of plant species. This extraordinary chemical diversity is believed to have evolved from a limited number of ubiquitous biosynthetic pathways through gene duplication followed by functional divergence (Pichersky and Gang, 2000). The phenylpropanoid pathway, derived from Phe, illustrates perfectly this phenomenon, as it gives rise to a large diversity of phenolic compounds playing key roles in plants, including participation in structural polymers, defense against herbivores and pathogens, protection from abiotic stress, and important functions in plant-pollinator interactions. Stilbenes are a small family of phenylpropanoids produced in a number of unrelated plant species, including dicotyledon angiosperms such as grapevine (Vitis vinifera), peanut (Arachis hypogaea), and Japanese knotweed (Fallopia japonica, formerly Polygonum cuspidatum), monocotyledons like sorghum (Sorghum bicolor), and gymnosperms such as several Pinus and Picea species. In addition to their participation in both constitutive and inducible defense mechanisms in plants, several stilbenes display important pharmacological properties. Since resveratrol (3,5,4′-trihydroxy-trans-stilbene) was postulated to be involved in the health benefits associated with a moderate consumption of red wine (Renaud and de Lorgeril, 1992), plant stilbenes have received considerable interest. Nowadays, resveratrol ranks among the most extensively studied natural products, and hundreds of studies have shown that it can slow the progression of a wide variety of illnesses, including cancer and cardiovascular disease, as well as extend the life spans of various organisms (Baur and Sinclair, 2006). Stilbene synthases (STSs) are characteristic of stilbene-producing plants and catalyze the biosynthesis of the stilbene backbone from three malonyl-CoA and one CoA-ester of a cinnamic acid derivative. STSs are members of the type III polyketide synthases family, chalcone synthases (CHSs), which catalyze the first step of flavonoid biosynthesis, being the most ubiquitous polyketide synthase in plants. Both CHS and STS use p-coumaroyl-CoA and malonyl-CoA as substrates and synthesize the same linear tetraketide intermediate. However, STS uses a specific cyclization mechanism involving a decarboxylation to form the stilbene backbone. STS proteins share extensive amino acid sequence identity with CHS, and phylogenetic analysis of the STS and CHS gene families has shown that STS genes may have evolved from CHS genes several times independently (Tropf et al., 1994). In most stilbene-producing plants, STS genes form small families of closely related paralogs. For example, two STS cDNAs have been cloned from peanut (Schröder et al., 1988), the genome of Scots pine (Pinus sylvestris) has been shown to contain a small family of four STS genes (Preisig-Müller et al., 1999), and three STS genes have been characterized in Japanese red pine (Pinus densiflora; Kodan et al., 2002). Only one STS gene has been isolated from Japanese knotweed to date (Liu et al., 2011), and the sequencing of sorghum genome has shown that SbSTS1 was the only STS gene in this plant species (Yu et al., 2005; Paterson et al., 2009). Grapevine is a noteworthy exception among stilbene-producing plants, as its genome has been shown to contain a large family of putative STS genes. Early Southern-blot experiments suggested that the grapevine genome contained more than 20 STS genes (Sparvoli et al., 1994). Analyses of the first drafts of the grapevine genome sequence confirmed the large size of this multigene family, with an estimated number of STS genes ranging from 21 to 43 (Jaillon et al., 2007; Velasco et al., 2007). However, these relatively low-coverage sequence drafts did not allow a precise analysis of large families of highly similar genes. The more recently released 12× genome sequence of grapevine inbred Pinot Noir cultivar PN40024 offered an improved sequence quality, allowing an accurate analysis of the STS gene family. In this work, we take advantage of the improved 12× sequence of the grapevine ‘PN40024’ genome to analyze the grapevine STS gene family. Furthermore, we combine molecular evolution to structural and functional analyses to gain more insight into the significance of the remarkable amplification of the STS family in grapevine.
RESULTS
Characterization and Organization of the Grapevine STS Gene Family
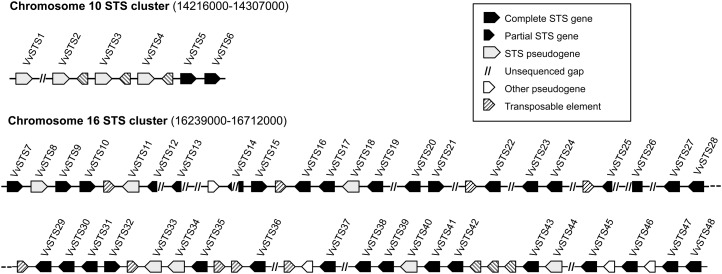
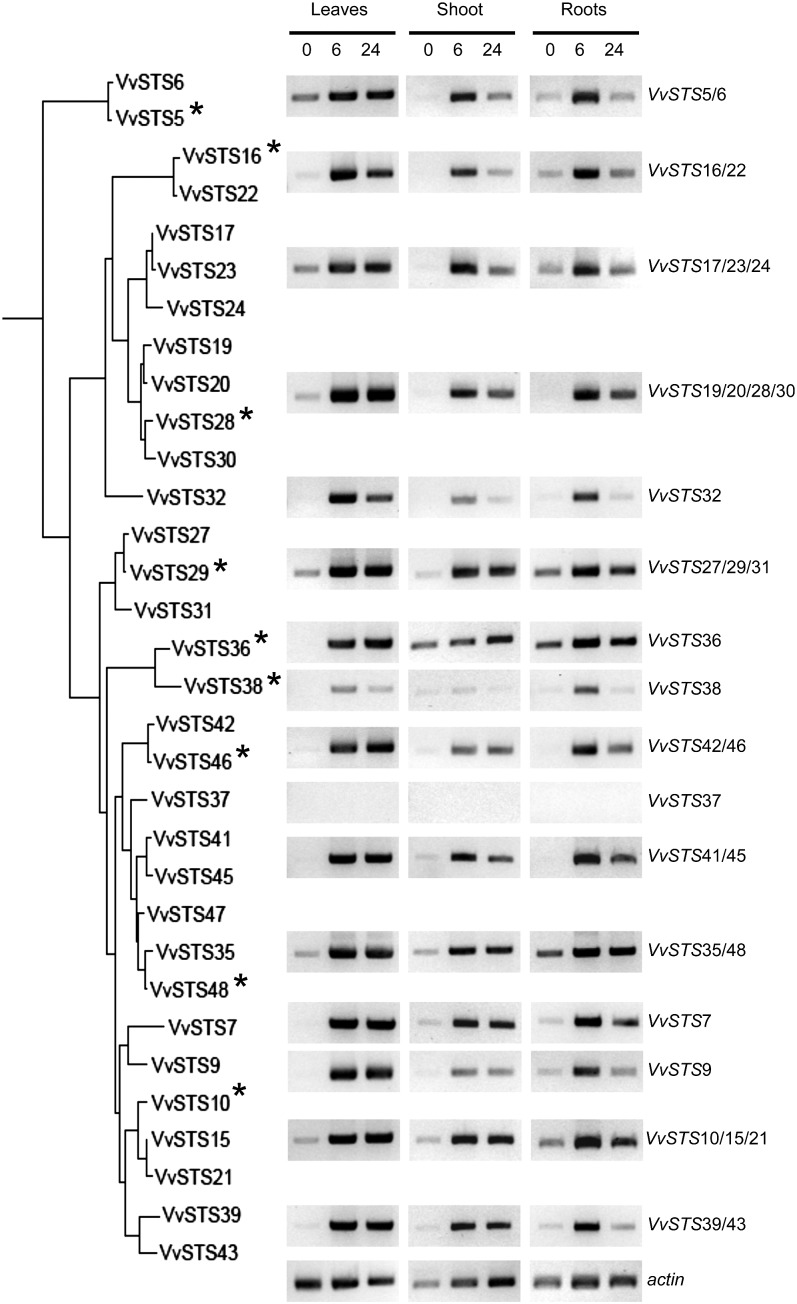
The 12× grapevine ‘PN40024’ genome (National Center for Biotechnology Information Genome ID: 401) has been screened exhaustively thanks to a similarity search approach based on previously characterized STS sequences available in Swiss-Prot (Schneider et al., 2009) and on the HMM profiles PF00195 (chalcone and STS N-ter) and PF02797 (chalcone and STS C-ter) defined in the PFAM database (Finn et al., 2010). Sixty-two locus-exhibiting significant similarities were detected this way. All of them have been manually controlled and reannotated through the ARTEMIS platform (Rutherford et al., 2000), taking into account the predictions of the combiners GAZE (Jaillon et al., 2007) and EuGène (Schiex et al., 2001), specifically trained for the grapevine genome annotation, the results of sequence comparisons (BLAST and HMMer), and the spliced alignments of the available cognate transcript sequences (grapevine EST and cDNA). This curated annotation allowed us to correct and complete the automatic structural annotations and to discriminate between complete genes (i.e. perfect full CDS), partial genes (i.e. suspended by an unsequenced region), and pseudogenes (i.e. disrupted by numerous stop codons, frameshifts, and/or small deletions). The results of this manual annotation are summarized in Supplemental Table S1, and the final CDS structures (Supplemental Data S1) and protein sequences are available in the FLAGdb++ database (Dèrozier et al., 2011). The multiple alignments and the sequence-based classification of all the deduced protein sequences have led to the discrimination between STS-like (48 locus) and CHS-like (14 locus) families. Regarding the CHS family, nine genes out of 14 are complete, including three highly expressed genes (more than 200 cognate ESTs). Phylogenetic analysis associates these three genes with previously characterized plant CHS (from 89% to 94% of identity at the protein level; data not shown). These genes are therefore likely to encode bona fide CHS proteins and have been named VvCHS1 to VvCHS3, while the other genes were considered as putative CHS-like genes, i.e. VvCHSL (Supplemental Table S1). Out of the 48 putative STS genes, 32 are complete, five are partial, and 11 are probable pseudogenes. All the 48 STS genes are distributed on only two tandemly arrayed gene clusters on the chromosomes 10 (six STS over 91 kb) and 16 (42 STS over 473 kb; Fig. 1). The STS family exhibits a highly conserved gene structure since all complete members have two coding exons of 178 and 998 bp, separated by an intron whose length ranges from 136 to 387 bp. The single splicing site is systematically a canonical GT-AG site of type 1. The grapevine STS genes encode 392-amino acid proteins sharing a high level of conservation. Indeed, 307 positions out of 392 are 100% identical in the complete STS proteins. The conservation level inside the family ranges from 90.3% of identity (between VvSTS19 and VvSTS36 proteins) to 99.7% (i.e. only one different residue, between VvSTS15 and VvSTS21 and between VvSTS41 and VvSTS45 proteins). At the transcriptional level, numerous cognate transcripts have been detected in available EST and cDNA libraries (Dèrozier et al., 2011) for all the complete STS genes, except three of them (VvSTS1, VvSTS4, and VvSTS5), as proof of the expression of the nearly whole family. The expression of grapevine STS genes has been shown to be induced under different types of stress conditions (Chong et al., 2009), including pathogen infection and exposure to heavy metals or UV light. In order to get a general view of the stress responsiveness of STS genes, their expression was analyzed in various organs of 3-month-old grapevine plantlets (five leaves stadium) submitted to UV light exposure. Due to the remarkable conservation of STS genes, the design of gene-specific oligonucleotides proved to be very difficult. Several pairs of primers actually matched subsets of two, three, or four STS genes, although they were designed to match the most variable regions of STS genes, including 3′ untranslated regions. Reverse transcription (RT)-PCR analysis of STS gene expression 0, 6, and 24 h after UV exposure confirmed that most STS genes were likely to be expressed, although individual expression rates could not be determined in the case of primers amplifying more than one gene. UV light exposure resulted in a strong induction of the transcription of most STS genes in leaves, shoots, and roots, confirming the stress responsiveness of this gene family (Fig. 2). STS37 was the only genes whose expressions was hardly detectable even after UV exposure. Transcriptional regulation of the STS gene family was not investigated further, as it has been investigated elsewhere (Vannozzi et al., 2012).
Figure 1.
Schematic representation of the grapevine ‘PN40024’ STS gene clusters on chromosomes 10 and 16.
Figure 2.
Stress induction of STS genes expression. STS gene expression was analyzed of by semiquantitative RT-PCR in leaves, shoots, and roots of grapevine plantlets at t = 0 (control), 6, and 24 h after exposure to UV light. The data shown are representative of three independent experiments. Genes selected for functional analysis are indicated with an asterisk.
Functional Characterization of a Selection of STS Proteins
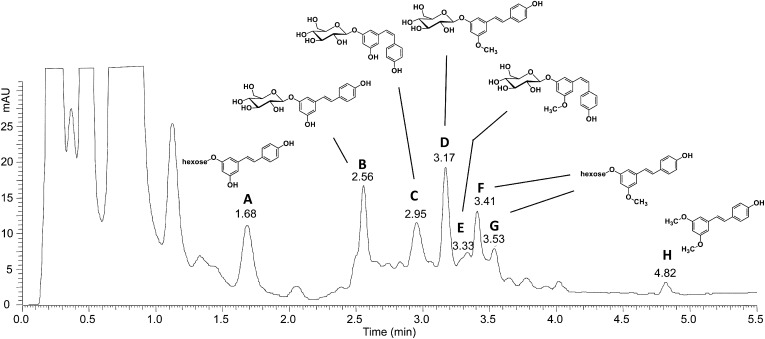
As building blocks for flavonoid biosynthesis, the substrates of STS, 4-coumaroyl-CoA, and malonyl-CoA are ubiquitously present in plants. Indeed, transgenic expression of a STS gene from grapevine in various plant species, including tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), papaya (Carica papaya), and grapevine, has been shown to result in resveratrol and resveratrol derivatives accumulation (Hain et al., 1993; Thomzik et al., 1997; Hipskind and Paiva, 2000; Coutos-Thévenot et al., 2001). In a previous work, we have used Agrobacterium tumefaciens-mediated transient transformation of Nicotiana benthamiana to characterize the activity of a resveratrol O-methyltransferase from grapevine (Schmidlin et al., 2008); we thus chose the same approach here to investigate the activity of a selection of STS proteins in planta. A similar strategy has been used to characterize STS enzymes from peanut (Condori et al., 2009). In order to represent the diversity of the STS family, nine STS full-length genes were selected in the major clades for functional analysis, namely, VvSTS5, VvSTS10, VvSTS16, VvSTS28, VvSTS29, VvSTS36, VvSTS38, VvSTS46, and VvSTS48, based on the fact that they were representative of the diversity of the STS family (Fig. 2). For in planta expression, the selected STS genes were amplified from cv PN40024 genomic DNA, transferred into the Gateway-compatible plant transformation vector pMDC32 (Curtis and Grossniklaus, 2003) to yield the pMDC32-VvSTS constructs and introduced into A. tumefaciens. For each construct, the corresponding A. tumefaciens suspension was infiltrated in two leaves from three different N. benthamiana plants. The resulting six samples were analyzed separately using liquid chromatography-mass spectrometry. N. benthamiana leaves were infiltrated with A. tumefaciens harboring a 35S-GFP construct as a control (Haseloff et al., 1997). No stilbenes were detected in extracts from leaves expressing GFP (data not shown). Conversely, significant amounts of stilbene derivatives accumulated when N. benthamiana leaves were infiltrated with A. tumefaciens harboring pMDC32-VvSTS constructs corresponding to all the selected genes (Fig. 3). Several stilbene derivatives were identified in N. benthamiana leaf extracts, based on comparisons of their retention times and mass spectra with those of authentic standards: trans-piceid (compound B: trans-resveratrol-3-O-glucoside, retention time = 2.56 min), cis-piceid (compound C: cis-resveratrol-3-O-glucoside, retention time = 2.95 min), and trans-pterostilbene (compound H: retention time = 4.82 min). In addition, several previously unreported stilbene derivatives were detected. Based on high-resolution mass data and tandem mass spectrometry (MS) analyses, putative structures proposed for these unknown stilbenes are proposed (Fig. 3). MS data of compounds D and E were consistent with these compounds being methylated derivatives of trans- and cis-piceid, which probably arose through a combination of endogenous glucosyltransferase and O-methyltransferase activities in N. benthamiana leaves. Compounds A, F, and G were tentatively identified as hexose derivatives of resveratrol (A) and resveratrol methyl ether (F and G), respectively. Detailed MS and tandem MS data for stilbene derivatives detected in STS-transformed N. benthamiana leave extracts are presented in Supplemental Table S2. Stilbene quantification following transient expression of the different STS genes in N. benthamiana are presented in Supplemental Table S3. Taken together, these results show that all selected STS genes encode proteins exhibiting STS activity when expressed in N. benthamiana. Considering that the selected STS genes have been chosen to represent the diversity of this gene family, it is very likely that all putative STS genes from grapevine actually encode functional STS enzymes.
Figure 3.
Stilbene accumulation after A. tumefaciens-mediated transient expression of STS genes in N. benthamiana. An N. benthamiana leaf sector (150 mg fresh weight) expressing the VvSTS10 gene (as an example) was excised 72 h after A. tumefaciens-mediated transient transformation. Stilbene content was analyzed using liquid chromatography-mass spectrometry. Peaks corresponding to major resveratrol derivatives are labeled from A to H and their retention times are indicated. Compounds B, C, and H have been identified as trans-piceid, cis-piceid, and trans-pterostilbene, respectively, by comparison with authentic standards. Putative structures of compounds A and D to G are proposed. mAU, Milli-absorbance units.
Phylogenetic dN/dS Analysis of the Grapevine STS Family
The apparent species-specific burst of the number of STS in grapevine raises several interesting questions as to its functional significance and the role of the different paralogs. Why are so many similar genes kept functional (at least 32 genes potentially coding for fully functional STS proteins)? Are there signs of positive selection indicating a recent functional diversification among the duplicates or, on the contrary, did purifying selection tend to restrict sequence divergence, suggesting that high dosage needs may require a large number of functionally equivalent genes? To gain more insight into the origin of the remarkable amplification of the grapevine STS family, we performed a molecular evolution study of this gene family, based on the measure and comparison of the rates of nonsynonymous versus synonymous substitutions (dN/dS = ω) of different members of the CHS/STS family. The dN/dS ratio is classically used to evaluate the selection pressures acting on the sequences: A dN/dS value equal to one indicates neutral evolution, greater than one positive selection, and less than one purifying selection (Yang and Bielawski, 2000).
Two data sets were analyzed independently, with the first set including CHS and STS sequences from different plant species (multispecies set; Supplemental Table S4) and the second set being restricted to the CHS and STS sequences identified in the genome of grapevine ‘PN40024’ (grapevine set). Pseudogenes were excluded from these analyses, as they usually show different evolutionary patterns than functional genes. Several models of the functional evolution of these families were tested, and their adequacy to the CHS/STS sequences was statistically tested (Supplemental Table S5).
Branch Model
The branch model analysis with grapevine CHS and STS sequences showed no significant difference between a model with different dN/dS ratios for the two subfamilies (ω CHS = 0.089 and ω STS = 0.083) and a model with one unique ω for all the sequences (ω global = 0.085, P value of the likelihood ratio test [LRT] = 0.55). This result was confirmed by the branch model analysis of the multispecies sequences set:
-
(1)
The model with two different ω for the grapevine STS sequences (ω grapevine STS = 0.084) and for all the other sequences (ω CHS + other STS = 0.095) showed no significant improvement in the likelihood relative to a model with one unique ω for all the sequences (ω global = 0.093, LRT P value = 0.16).
-
(2)
The model with two different ω for the other species STS sequences (ω other STS = 0.178) and for all the other sequences (ω CHS + grapevine STS = 0.079) does show a significant improvement in the likelihood relatively to a model with one unique ω for all the sequences (LRT P value < 10−308).
-
(3)
Finally, the model with three different ω categories, grapevine STS (ω = 0.084), other species STS (ω = 0.179), and CHS (ω = 0.077), is not significantly better than the one that reassembles grapevine STS and CHS sequences (ω CHS + grapevine STS = 0.079 and ω other STS = 0.178, LRT P value = 0.33).
These results suggest that the STS sequences of some plant species (other than grapevine) present higher dN/dS ratios than the one of the CHS sequences but that this is not the case for grapevine STS. When these other species STS sequences were separated from the CHS, the fit of the model to the data was significantly increased (LRT P values = 3.14 × 10−9 or even smaller, tending to 0, depending on the foreground lineages chosen; Supplemental Table S5).
Clade Model
The clade model analysis (M2a, three site categories where one may vary between defined branches of the tree) showed a majority of sites of the CHS and STS sequences under purifying selection, both in the grapevine and in the multispecies sets of sequences. In the grapevine-specific set of sequences, the free site category (where the dN/dS ratios are allowed to change between branches of the tree) included 53% of the sites, which presented dN/dS ratios of 0.082 (STS) and of 0.148 (CHS). In the multispecies analysis, this site category included 52% of the sites, with ω values of 0.1183 (CHS), 0.1179 (grapevine STS), and a slightly higher value for the STS from other species (ω = 0.2891), which was coherent with the branch model results presented above. Only 5% of the sites were attributed to the neutral evolution category in both cases. The remaining 42% or 43% of the sites exhibited very small ω values (constant for all branches of the tree) in both sets (0.02). These results were statistically significant, giving P values lower than 10−308 when performing LRT with the null model (model with no free site category).
Branch Site
The branch site analysis with codeml allows ω to vary both among sites in the proteins and between two predefined groups in the tree. The branch site models aim at detecting positive selection affecting specific sites along particular lineages. When ω was freely and independently estimated between the grapevine STS and CHS, only a very small proportion (0.015) of sites exhibited an increase in ω from the background lineage (grapevine CHS ω = 0.059) to the foreground lineage (grapevine STS ω = 1). Note that the foreground ω was not >1 but = 1; this model was thus strictly identical to the null model, where the ω of the eventual positive selection acting on some sites of the foreground lineage is fixed (and equal to one). This means that no positive selection was significantly detected in the STS lineage, which is consistent with the results present above.
Fitmodel
If only very few grapevine STS sites present ω > 1, their influence would not be sufficient to globally affect the dN/dS ratio of the whole grapevine STS lineage. In order to investigate this hypothesis, we used Fitmodel, a more flexible program for detecting sites under positive selection (Guindon et al., 2004). Fitmodel estimates different dN/dS ratios to each sequence site and each branch on the phylogenetic tree. The output of the program indicates, for each site in the alignment and in the different tree branches and leaves, the category of dN/dS values with the biggest posterior probability, among three different ω categories estimated by the program. We performed two different Fitmodel analyses with both data sets: one with three freely estimated ω categories (model M2a, with ω1 < ω2 < ω3) and one with three constrained ω categories (model MX with ω0 ≤ 1; 1 < ω1 ≤ 1.5; ω2 > 1.5). The first global conclusion of the Fitmodel analysis confirmed the conclusion of the codeml analyses: grapevine STS exhibited more sites evolving under purifying selection (389 out of a 405 sites alignment for the M2a model, 383 for the MX model) than sites affected by positive selection (10 out of a 405 sites alignment for the M2a model, 22 for the MX model).
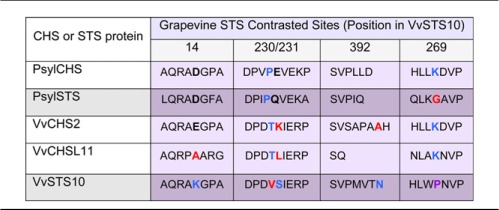
We also looked for specifically contrasted sites in the STS versus the CHS branches, i.e. sites that were classified in extreme ω categories and thus submitted to opposite selection pressures (negative versus positive) specifically in the STS lineages, relative to the background (surrounding CHS branches), and we compared such sites obtained with each of the two sets of data (grapevine-specific and multispecies analysis). Contrasted sites are of interest because they may correspond to sites participating in functional changes between CHS and STS proteins. Remarkable sites are referred to according to their nature and position in VvSTS10, which has been chosen for molecular modeling. Three contrasted sites under purifying selection were detected in grapevine STS: Lys-14, Ser-231, and Asn-392 and only one site (Val-230) specifically subjected to positive selection, in a region of sites affected by negative selection, in all Fitmodel analyses (Supplemental Fig. S1 and Supplemental Table S5). This site was also detected by the codeml branch site analysis, showing a posterior probability of 98.9% of evolving under positive selection.
Some other grapevine STS sites showing probabilities >50% of being under positive selection pressures by the codeml branch site analysis were also detected by the Fitmodel analyses, but either they were not evolving under positive selection in both data sets, were restricted to some (and not all) grapevine STS or were evolving neutrally in the background sequences (Supplemental Table S5).
In the restricted data set (both M2a and MX analyses), one site in grapevine STS (Pro-269) showed a very specific pattern of positive selection on the branch at the base of the STS lineage and purifying selection elsewhere (Fig. 4, Table I). This pattern would be expected for a site whose substitution played a major role in the CHS to STS transition. In the broader data set analysis, evidence for positive selection on this site at the origin of the STS was less clear. However, it still appeared to have an ω (slightly) > 1 both in the branch at the base of the grapevine STS and in all pine STS and sorghum STS (all other branches and sequences are classified into the negative selection category [ω = 0.024]), indicating that this position may be important in different STS lineages.
Figure 4.
Selection patterns of the amino acids corresponding to Pro-269 in VvSTS10. Phylogenetic maximum likelihood tree of the multispecies set of sequences showing the Fitmodel results for a contrasted site evolving under positive selection at the base of the grapevine STS subfamily (Pro-269 in VvSTS10). Different colors indicate higher posterior probabilities of evolving under different selection regimes: red = positive selection; blue = purifying selection; black = neutral evolution. Ah, Peanut; At, Arabidopsis; Sb, sorghum; Psyl, Scots pine; Pstr, Pinus strobus; Pn, Psilotum nudum; Pt, poplar; Vv, grapevine.
Table I. Amino acid sites under contrasted selection pressures in grapevine STS compared with CHS and with STS from Scots pine.
Contrasted amino acid positions are indicated in bold letters. Sites under positive selection are indicated in red, sites evolving neutrally in black, and sites under negative selection pressure are indicated in blue. Pro-269 is colored in purple to indicate positive selection on the basal branch of the grapevine STS. The STS sequence cells are highlighted in darker background. Psyl, Scots pine; Vv, grapevine.
|
Molecular Modeling of a Typical STS Protein from Grapevine
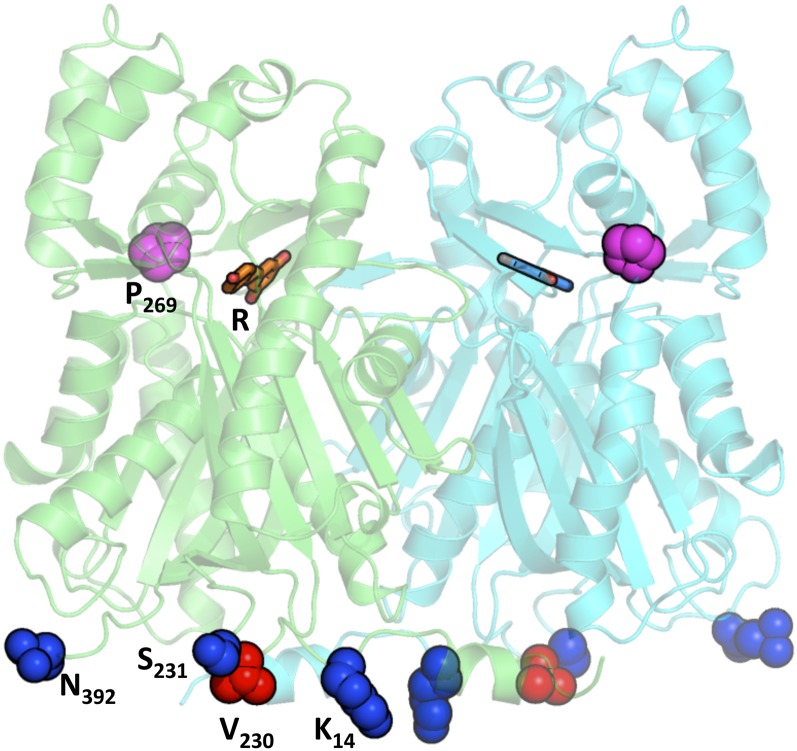
In order to investigate the structural significance of the positive or negative selection affecting specific amino acids, a tridimensional model of a typical STS protein from grapevine was constructed using a homology modeling approach (Fig. 5). VvSTS10 was selected for modeling, as it closely resembles the Vst1 protein from grapevine ‘Optima,’ which has been expressed in various plant species to engineer resveratrol biosynthesis (Hain et al., 1993; Delaunois et al., 2009). The VvSTS10 protein showed a high similarity with other CHSs (1CGZ, 71.1% identity; 1U0W, 69.9% identity) and STSs (1Z1F, 69.4% identity; 1U0U, 64.2% identity). The grapevine STS model displayed few differences from Scots pine STS (the superimposed Cα yield a root mean square deviation of 0.302 Å over 387 Cα positions), the structure of the active site being conserved. The resveratrol ligand adopted a planar conformation as observed in engineered CHS (1U0W) and as it was expected from vibrational spectroscopy studies (Billes et al., 2007). This was slightly different from the resveratrol conformation in the active site of peanut STS (1Z1F; Shomura et al., 2005). Grapevine STS formed the same kind of dimer than other CHSs or STSs, and the buried surface was 7280 Å2 (7169 Å2 for 1U0W and 7860 Å2 for 1Z1F). The three sites under purifying selection (Lys-14, Ser-231, and Asn-392) and the site subjected to positive selection (Val-230) are located in the same region, on the external surface of the STS dimer, suggesting a possible involvement in interactions with other protein partners (Fig. 5). Conversely, Pro-269, whose selection pattern is consistent with a role in the CHS to STS transition, is located nearby the active site of the enzyme.
Figure 5.
Mapping of evolutionary contrasted amino acid sites on the three-dimensional model of a typical STS protein. VvSTS10 protein was modeled using the structure of STS from Scots pine as a template (Austin et al., 2004). In both STS monomers, remarkable amino acids are highlighted in red or blue, for amino acids subjected to positive or purifying selection, respectively. Pro-269, subjected to early positive selection in the grapevine STS family, is represented in purple. The position of the resveratrol product (R) is indicated.
DISCUSSION
Organization of the Grapevine CHS and STS Families
The grapevine CHS gene family consists of 14 genes, including four putative pseudogenes. Three highly expressed genes are likely to encode bona fide CHS proteins, while the other genes were considered as putative CHS-like genes. The size of this gene family is intermediate between the size of the corresponding families in Arabidopsis (Arabidopsis thaliana) and poplar (Populus trichocarpa), which consist of one CHS and three CHS-like genes and six CHS and seven CHS-like genes, respectively (Tsai et al., 2006). Conversely, the STS gene family has experienced a unique expansion in grapevine compared to other stilbene-producing plants, which usually possess one to five STS genes. Both STS gene clusters are characterized by mixing potentially functional genes and pseudogenes, together with numerous relicts of transposable elements (Fig. 1). These transposable elements probably played a major role in the dynamics of these regions through their impact in the frequency of recombination events (Fiston-Lavier et al., 2007; Xu et al., 2008). It is therefore possible that these STS clusters may be highly polymorph, in terms of gene number, throughout Vitis species and varieties. Such tandemly arrayed gene clusters are known to be refractory to the automatic annotation pipelines. Indeed, out of the 37 complete or partial STS genes, 13 were absent and 17 had erroneous intron-exon structures in the official annotation of the 12× ‘PN40024’ genome sequence (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/). This fact illustrates the important gain of manual and knowledge-driven annotation approach.
The Grapevine STS Gene Family Is Exceptionally Large and Encodes Proteins with Redundant Catalytic Activity
Gene duplication is assumed to be a major driving force in the evolution processes that gave rise to the extraordinary diversity of plant secondary metabolism (Pichersky and Gang, 2000). Duplicated genes may then retain their original function, relaxed functional constraints may lead to pseudogenization, and novel functions may be acquired through neofunctionalization, or subfunctionalization may lead to a partition of the ancestral gene function (Lynch and Conery, 2000). The unusual size of the grapevine STS gene family raises several questions as to the significance of such a large family and the functions of its members. Comparisons of CHS and STS proteins failed to identify STS-specific consensus sequences allowing an unambiguous identification of STS enzymes (Tropf et al., 1994). Therefore, the activity of a selection of STS proteins was assayed to ensure that they indeed possess STS activity. Transient expression of all the selected genes resulted in the accumulation of stilbenes in N. benthamiana, showing that they encode functional STS enzymes. As these genes are representative of the STS family diversity, it is therefore likely that the same is true for all its members.
Early studies have shown that stilbene biosynthesis was induced in response to a wide range of biotic and abiotic stresses, as a result of an increased transcription of STS genes in peanut, grapevine, and pine (Lanz et al., 1990; Fliegmann et al., 1992; Sparvoli et al., 1994). However, due to the lack of genome sequence information, it was difficult to estimate the number of STS genes involved. Nevertheless, RT-PCR analyses indicated that at least 20 different STS genes were expressed in grapevine leaves following infection with downy mildew (Plasmopara viticola; Richter et al., 2006). Our rapid survey of STS genes expression in response to UV light confirmed that the vast majority of STS genes were likely to be functional and stress inducible. Therefore, we conclude that most if not all the 32 complete STS genes present in the genome of grapevine ‘PN40024’ are expressed and are very likely encode functional STS enzymes. Although many genes associated with primary and secondary metabolism exist as multigene families in plant genomes (Xu et al., 2009), the STS family in grapevine represents a unique example of functional redundancy.
The Evolution of the STS Family Is Dominated by Purifying Selection in Grapevine
The exceptional size of grapevine STS family raises the question as to the evolutionary process that gave rise to such a large family of genes encoding proteins with similar catalytic activity. The dN/dS analysis showed that the grapevine CHS/STS family was globally strongly constrained, with almost all ω values lower than one and very close to zero in some cases. No evidence was found for important positive selection pressures acting specifically on grapevine STS proteins, which appeared even more constrained than CHS proteins. Some sites under different selection pressures between the STS and the CHS sequences were identified for all the species present in the phylogeny. These remarkable sites were not the same for all the species, neither were the selection regimes associated to their evolution, which is consistent with an independent, repeated and parallel evolution of the STS subfamilies in different branches of the tree (Tropf et al., 1994; Fig. 4). However, the Fitmodel analysis allowed the identification of one remarkable site that could be related to the evolution of STS activity in both grapevine and other species. Indeed, in grapevine STS, the site P269 (Fig. 4) exhibits a pattern of positive selection on the branch at the base of the lineage and purifying selection elsewhere. This pattern is expected for an amino acid whose substitution is associated with the transition from CHS to STS. Most interestingly, an ω > 1 is detected for this branch, and it is also the case for both pine and sorghum STS (all other branches and sequences presenting ω = 0.024), even if the evidence for positive selection on this specific site is not as strong in the multispecies data set. Pro-269 in grapevine STS corresponds to Gly-272 in pine STS, which belongs to one of the two regions, whose conformation differ substantially between CHS and STS crystal structures (Austin et al., 2004). Gly-272 is located within a small loop on the outer surface of the CoA binding tunnel in pine STS. This region is not predicted to influence the cyclization specificity of STS, but it may rather impact kinetic parameters of this enzyme (Austin et al., 2004). Nevertheless, the specific selection pattern of Pro-269 and the corresponding amino acids in independent lineages of STS enzymes suggests that this site is likely to have played a major role in STS evolution.
Consistent with the apparent similar biochemical activity of STS proteins, no evidence was found for neofunctionalization in the catalytic site of the STS proteins. Only one amino acid site was found under widespread positive selection within the STS family (site Val-230), which is located at the periphery of the enzyme (Fig. 5). Conversely, the Fitmodel analysis allowed the detection of several amino acids subjected to strong purifying selection pressures, namely, Lys-14, Ser-231, and Asn-392 (Table I; Supplemental Fig. S1). Like Val-230, these amino acids are exposed at the outer surface of the STS protein (Fig. 5). Together, they determine a region that could be involved in the interaction with other protein partners. Indeed, studies in different plant species have shown physical interaction and channeling of intermediates between enzymes operating sequentially in the flavonoid pathway (Winkel, 2004). Transgenic expression of STS in various plant species has shown that STS could efficiently compete with CHS for p-coumaroyl-CoA and malonyl-CoA substrates. Constitutive expression of STS in tobacco strongly impacted flavonoid metabolism, leading to changes in flower color from pink to white and to male sterility (Fischer et al., 1997). The same male sterile phenotype was observed in STS-transgenic tomato plants, together with abnormal pollen development (Ingrosso et al., 2011). Male fertility could be restored by application of flavonols on young STS transgenic tobacco plants, indicating that the male sterility was due to the depletion of important flavonoids as a result of the competition for precursors between STS and CHS (Fischer et al., 1997). Substrate channeling has been proposed to occur in the stilbene biosynthetic pathway too (Hammerbacher et al., 2011), and the efficient accumulation of stilbenes at the expense of chalcone-derived flavonoids may partly rely on the insertion of STS into flavonoid biosynthetic multienzyme complexes, allowing the channeling of precursors toward stilbene biosynthesis. One could therefore speculate that the peripheral amino acids subjected to strong purifying selection in STS may be critical for protein-protein interactions within these complexes and for an efficient stilbene biosynthesis.
Hypotheses for High STS Gene Number in Grapevine
The hypotheses classically proposed to explain gene family expansion are subfunctionalization, neofunctionalization, and selection for increasing dosage (Conant and Wolfe, 2008). Subfunctionalization or duplication-degeneration-complementation (DDC; Lynch and Conery, 2000) is a mere neutral process of duplicate genes diversification, which does not account well for the sudden expansion of single gene families, such as the one of the STS in grapevine, and is probably not very likely here. Moreover, purifying selection is expected to be weaker within duplicates evolving under DDC compared to related genes with few or no duplicates, and this is not what we found comparing grapevine STS with CHS or with STS from other species in our dN/dS analysis. Selection for increased dosage may have led to the amplification of the STS family. Increased dosage can be obtained through the evolution of enhancers that will increase expression levels also by simply duplicating a gene over and over. In this case, no or very little functional diversification and similar expression patterns would be expected among STS genes. This hypothesis predicts that STS dosage should be unusually elevated in grapevine compared to other stilbene-producing plants, which should result in a very high stilbene biosynthetic capacity. Roots and woody parts of grapevine constitutively accumulate stilbenes, which can account for up to 0.5% of the dry weight (Vergara et al., 2012). However, roots of Japanese knotweed contain up to 1.6% dry weight piceid (Benová et al., 2008) and in pine heartwood, the natural accumulation of pinosylvin derivatives ranges from 0.1% to 4% dry weight (Hart and Shrimpton, 1979). Although the grapevine genome contains an unparalleled number of STS genes, the amounts of stilbenes accumulated in grapevine do not seem particularly larger than in other stilbene-producing plants. Alternatively, functional diversification among STS copies could explain why the family has become so large. In this case, STS copies would be expected to show evidence for positive selection at the protein level and/or diversified expression patterns, suggesting the evolution of new functions among STS genes. Our dN/dS analyses do not support an important functional diversification at the protein level. STS and CHS genes show very similar global dN/dS and very few amino acid sites have distinct evolution in STS and CHS (see above). Only one amino acid site was found under widespread positive selection within the STS gene family (site 230), this site corresponding to a branched-chain amino acid (Val, Leu, or Ile) or to a Ser or Thr, depending on STS proteins. Due to its peripheral localization, this site is unlikely to affect STS catalytic activity, but it may rather be involved in interactions with other protein partners. The large size of the STS family may also be linked to a diversification of expression patterns among paralogs. Stilbenes have been shown to accumulate either constitutively or in a developmentally regulated way or following stresses in various organs of grapevine. Stilbenes accumulate constitutively in roots and in woody parts of grapevine (Vergara et al., 2012), and developmentally regulated stilbene synthesis occurs in the skin of healthy grapes during the ripening process, from véraison to maturity (Gatto et al., 2008). Stress regulation of stilbene biosynthesis is well documented in grapevine, where expression of STS genes and synthesis of stilbenes are induced upon both abiotic and biotic stresses, including infection with different fungal pathogens, such as powdery mildew (Erysiphe necator), downy mildew, or gray mold (Botrytis cinerea; Chong et al., 2009). Fine-tuning of stilbene biosynthesis in such diverse situations may thus require multiple regulation pathways operating on specific subsets of STS genes. Although the high similarity of STS genes makes it difficult to accurately assess individual transcript levels, STS gene subfamilies have been shown to exhibit different expression profiles. Microarray and RNA-seq analysis of STS gene expression in different physiological conditions showed that the STS genes located on chromosome 10 were likely to be involved in constitutive and developmentally regulated stilbene biosynthesis and stress-induced stilbene synthesis depending rather on the gene cluster located on chromosome 16 (Vannozzi et al., 2012). However, why STS expansion may have been advantageous in grapevine remains unclear at this stage. Comparative studies of STS and other gene families (such as terpene synthases) that underwent similar grapevine-specific expansion may help address this question. An interesting possibility is that such expansion events may be linked to the domestication of grapevine. Sequencing and comparative genomics of domesticated and wild grapevine will help testing this idea.
CONCLUSION
The availability of the grapevine ‘PN40024’ complete genome sequence has shed a new light on grapevine metabolism. Indeed, analysis of the grapevine genome has shown a remarkable expansion of several gene families linked to secondary metabolism compared to other plants (Jaillon et al., 2007). A first example is the terpene synthase family that generates aromatic volatile molecules contributing to grape and wine flavors. Indeed, grapevine exhibits the largest terpene synthase family of all plant species for which genome sequences are available (Martin et al., 2010). Another striking example of gene family expansion is the STS family, which is nearly 10 times as large as the STS families characterized to date in other stilbene-producing plants. Phylogenetic dN/dS analysis of the STS family revealed that STS evolution was dominated by purifying selection in grapevine, with no evidence for strong selection for new function among STS copies. Moreover, subsets of STS genes have been shown to have different expression patterns, suggesting that the evolution of this unusually large gene family may allow a fine spatial and temporal regulation of stilbene biosynthesis under both normal and stress conditions. There is currently considerable interest in breeding new grapevine varieties resistant to diseases of major economical importance in viticulture. To this aim, resistant Vitis species from North America have been extensively crossed with various grapevine varieties to introgress resistance into the grapevine background (Peressotti et al., 2010). Recent work has shown that resistance to downy mildew in a grapevine segregating population is associated with stilbene accumulation (Malacarne et al., 2011). Due to the presence of transposable elements in the STS gene clusters, they may be highly polymorph throughout Vitis species and varieties. It will be of interest to investigate STS gene families among wild Vitis species to take full advantage of natural defense mechanism in current and future breeding programs.
MATERIALS AND METHODS
Plant Material
Grapevine plantlets (Vitis vinifera ‘PN40024’) were obtained from seeds and grown on potting soil in a greenhouse at a temperature of 22°C and 19°C (day and night, respectively), with a photoperiod of 16 h of light (supplemental light provided by sodium lamp illumination), until they developed five to six fully expanded leaves. For UV treatment, plantlets were dug out, spread on a wet filter paper (leaves with the abaxial face up), and exposed for 6 min to UV light (90 µW cm−2) from a UV-C tube (Osram; 30 W, 254 nm).
Chemicals
Trans-resveratrol and trans-pterostilbene were from Sigma-Aldrich. Trans-piceid and cis-piceid standards were kindly provided by R. Pezet (Changins, Switzerland). MS-grade solvents (acetonitrile and methanol) were from Merck and used in combination with sterilized water from Aguettant. All other chemicals and reagents were from Sigma-Aldrich.
Cloning of STS Genes
The selected STS gene coding regions were amplified by PCR using the primers described in Supplemental Table S6. PCR amplification was carried out for 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C to 60°C for 30 s, and extension at 72°C for 1 min with a final extension of 5 min, in a GeneAmp PCR system 9700 cycler (Perkin-Elmer), using Phusion DNA polymerase (Thermo Fisher Scientific). Amplified DNA fragments were cloned into pGEM T-easy (Promega) and the inserts sequenced. STS genes were then amplified by PCR using primers containing att recombination sites and transferred into the pDONR207 Gateway-compatible vector using the Clonase II cloning kit (Invitrogen). STS genes cloned into the pDONR207 were sequenced to verify that no mutation had been introduced.
Transient Expression in Nicotiana benthamiana
For Agrobacterium tumefaciens-mediated transient expression, STS genes were transferred into the Gateway-compatible binary vector pMDC32 (Curtis and Grossniklaus, 2003) and m-GFP4 was used as a control (Haseloff et al., 1997). All constructs were introduced into A. tumefaciens strain GV3101 (Koncz and Schell, 1986) by electroporation. N. benthamiana leaves were infiltrated with A. tumefaciens cultures (optical density at 600 nm = 0.3 to 0.5) according to Batoko et al. (2000). Disks were punched from N. benthamiana leaves 72 h after A. tumefaciens infiltration and analyzed for stilbene content.
Stilbene Analyses
Stilbene extractions were performed as described previously (Poutaraud et al., 2007), and stilbenes were analyzed by HPLC-diode array detector (DAD)/electrospray ionization-MS. Separations were performed using a Dionex Ultimate 3000 ultra-HPLC-DAD system, on a Nucleodur C18 HTec column (50 × 2 mm i.d., 1.8-µm particle size; Macherey-Nagel), operated at 20°C. Mobile phase consisted of water/formic acid (0.1%, v/v; eluant A) and acetonitrile/formic acid (0.1%, v/v; eluant B). Flow rate was 0.4 mL/min. The elution program was as follows: isocratic for 1 min with 15% B, 15% to 95% B (5 min), isocratic with 95% B (1 min). The sample volume injected was 2 µL. The liquid chromatography system was coupled to an Exactive Orbitrap mass spectrometer (Thermo Fischer Scientific) equipped with an electrospray ionization source operating in negative mode. Parameters were set at 275°C for ion transfer capillary temperature and −2,500 V for needle voltage. Nebulization with nitrogen sheath gas and auxiliary gas were maintained at 40 and 6 arbitrary units, respectively. The desolvating temperature was 275°C. The spectra were acquired within the mass-to-charge ratio range of 100 to 1,000 atomic mass units, using a resolution of 50,000 (full width at half maximum). The instrument was operated using the ExactiveTune software, and data were processed using the XcaliburQual software. The system was calibrated externally using the Thermo Fischer calibration mixture in the range of mass-to-charge ratio 100 to 2,000 a.m.u., giving a mass accuracy lower than 2 nL L−1. Stilbenes were identified according to their mass spectra, UV absorption spectra, and retention time, compared to those of authentic stilbene standards. Stilbene quantifications were based on calibration curves obtained with authentic standards.
RNA Isolation and Semiquantitative RT-PCR
Total RNAs were isolated using the RNeasy plant mini kit (Qiagen) according to the manufacturer’s instructions and quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific). Residual genomic DNA was removed by performing on-column DNase I digestion with the RNase-Free DNase set (Qiagen). One microgram of total RNA was used as template for RT, using RevertAid M-MuLV reverse transcriptase (Fermentas), with 0.5 µg of oligo(dT)18, for 1 h at 42°C. PCR amplifications were performed on 5 µL of the 10× diluted cDNA solution using Taq DNA Polymerase from Promega, with 25 cycles of 94°C for 15 s, 50°C to 60°C for 30 s, and 72°C for 30 s. Primers are described in Supplemental Table S6. All PCR products were separated on 1% agarose gels stained with ethidium bromide, and image processing was carried out with a Bio-Rad GelDoc apparatus (Bio-Rad).
Phylogenetic dN/dS Analysis
CHS and STS coding sequences from other species than grapevine were retrieved from the EMBL database (http://www.ebi.ac.uk/embl/). Accession numbers of the coding sequence and protein sequences are shown in Supplemental Table S4. Coding sequences were aligned with MUSCLE (Edgar, 2004), and phylogenies were built using PhyML (Guindon and Gascuel, 2003) using the GTR model with γ distribution (four rate classes of sites with optimized alfa) via Seaview software (http://pbil.univ-lyon1.fr/; Gouy et al., 2010). PAML (codeml; http://abacus.gene.ucl.ac.uk/software/paml.html; Yang, 2007) and Fitmodel (Guindon et al., 2004) were applied to the coding sequence alignments and phylogenetic trees to perform a multispecies and a grapevine-specific dN/dS analysis of the CHS/STS family. Seventy-four CHS/STS sequences from eight different species were used for the multispecies analysis (peanut [Arachis hypogaea], Arabidopsis [Arabidopsis thaliana], sorghum [Sorghum bicolor], Scots pine [Pinus sylvestris], Pinus strobus, Psilotum nudum, Populus trichocarpa, and grapevine). We ran branch model, site model, clade model, and branch site analyses following codeml standard procedures. We compared nested models using the LRT framework to test statistically differences in dN/dS. We ran Fitmodel M2a+S1 (switch between selection regimes allowed) as an alternative branch site analysis and M2a (no switch between selection regimes allowed) as a null model to perform LRT (Guindon et al., 2004). Fitmodel results were analyzed site by site, and we counted the number of sites showing patterns consistent with STS-specific positive or purifying selection.
Homology Modeling of a STS Protein
A dimer of the grapevine VvSTS10 protein was modeled in resveratrol bound state by homology using MODELER (Sali and Blundell, 1993). Structural alignment was performed with the following known structures: alfalfa (Medicago sativa) CHSs (PDB 1CGZ) and engineered bound to resveratrol (PDB 1U0W; Austin et al., 2004), peanut STS bound to resveratrol (PDB 1Z1F; Shomura et al., 2005), and Scots pine STS ligand free (1U0U; Austin et al., 2004). Resveratrol structure and topology was generated with PRODRG (Schüttelkopf and van Aalten, 2004) and was introduced in the model at the beginning of homology modeling. Since several loops carrying enzymatic specificity have been identified between CHS and STS synthases (Austin et al., 2004), multiple cycles of loop refinement were performed in these regions (sequence EIITAE 96-101, sequence TTSGVEM 131-137, sequence VMLYHQ 157-162, and sequence WPNVPT 268-273). The model with the lowest DOPE score among 100 generated models was selected for energy minimization and molecular dynamics using the GROMACS package (Van Der Spoel et al., 2005). Simulation was performed in cubic boxes filled with SPC216 water molecules and GROMOS43a1 as force field. Before molecular dynamics, the protein was subjected to energy minimization and positional restraints cycles. The simulation was carried out with periodic boundary conditions by adding sodium ions to have a value of zero as net electrostatic charge of the system. The bond lengths were constrained by the all atoms LINCS algorithm. The particle mesh ewald algorithm was used for the electrostatic interactions with a cutoff of 0.9 nm. The simulations were performed at neutral pH with runs of 10 ns at 300K coupling the system to an external bath. GROMACS routines were used to check the trajectories and the quality of the simulations. The structure of the final model was checked using MOLPROBITY (Chen et al., 2010).
Supplemental Data
The following materials are available in the online version of this article.
-
Phylogenetic trees showing Fitmodel results.
-
Detailed annotation of grapevine STS and CHS genes.
-
Characterization of stilbenes produced following transient STS expression.
-
Quantification of stilbenes produced following transient STS expression.
-
Accession numbers of the sequences used in this work.
-
Main results of the dN/dS analyses.
-
Primers used for PCR amplifications.
-
Grapevine STS and CHS coding sequences.
Supplementary Material
Acknowledgments
We thank Pascale Coste, Bernard Delnatte, Vincent Dumas, Denise Hartmann, Charlotte Knichel, Jacky Misbach, and Christian Vivant (Institut National de la Recherche Agronomique, Colmar) for assistance with plant material. We thank Sophie Meyer (Institut National de la Recherche Agronomique, Colmar) for excellent technical assistance. We thank Stéphane Guindon for help with Fitmodel.
Glossary
- dN/dS
to be defined
- STS
stilbene synthases
- CHS
chalcone synthase
- MS
mass spectrometry
- LRT
likelihood ratio test
- RT
reverse transcription
References
- Austin MB, Bowman ME, Ferrer J-L, Schröder J, Noel JP. (2004) An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol 11: 1179–1194 [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I. (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. (2006) Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov 5: 493–506 [DOI] [PubMed] [Google Scholar]
- Benová B, Adam M, Onderková K, Královský J, Krajícek M. (2008) Analysis of selected stilbenes in Polygonum cuspidatum by HPLC coupled with CoulArray detection. J Sep Sci 31: 2404–2409 [DOI] [PubMed] [Google Scholar]
- Billes F, Mohammed-Ziegler I, Mikosch H, Tyihák E. (2007) Vibrational spectroscopy of resveratrol. Spectrochim Acta A Mol Biomol Spectrosc 68: 669–679 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Poutaraud A, Hugueney P. (2009) Metabolism and roles of stilbenes in plants. Plant Sci 177: 143–155 [Google Scholar]
- Conant GC, Wolfe KH. (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Condori J, Medrano G, Sivakumar G, Nair V, Cramer C, Medina-Bolivar F. (2009) Functional characterization of a stilbene synthase gene using a transient expression system in planta. Plant Cell Rep 28: 589–599 [DOI] [PubMed] [Google Scholar]
- Coutos-Thévenot P, Poinssot B, Bonomelli A, Yean H, Breda C, Buffard D, Esnault R, Hain R, Boulay M. (2001) In vitro tolerance to Botrytis cinerea of grapevine 41B rootstock in transgenic plants expressing the stilbene synthase Vst1 gene under the control of a pathogen-inducible PR 10 promoter. J Exp Bot 52: 901–910 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunois B, Cordelier S, Conreux A, Clément C, Jeandet P. (2009) Molecular engineering of resveratrol in plants. Plant Biotechnol J 7: 2–12 [DOI] [PubMed] [Google Scholar]
- Dèrozier S, Samson F, Tamby JP, Guichard C, Brunaud V, Grevet P, Gagnot S, Label P, Leplé J-C, Lecharny A, et al. (2011) Exploration of plant genomes in the FLAGdb++ environment. Plant Methods 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Budde I, Hain R. (1997) Stilbene synthase gene expression causes changes in flower colour and male sterility in tobacco. Plant J 11: 489–498 [Google Scholar]
- Fiston-Lavier A-S, Anxolabehere D, Quesneville H. (2007) A model of segmental duplication formation in Drosophila melanogaster. Genome Res 17: 1458–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann J, Schröder G, Schanz S, Britsch L, Schröder J. (1992) Molecular analysis of chalcone and dihydropinosylvin synthase from Scots pine (Pinus sylvestris), and differential regulation of these and related enzyme activities in stressed plants. Plant Mol Biol 18: 489–503 [DOI] [PubMed] [Google Scholar]
- Gatto P, Vrhovsek U, Muth J, Segala C, Romualdi C, Fontana P, Pruefer D, Stefanini M, Moser C, Mattivi F, Velasco R. (2008) Ripening and genotype control stilbene accumulation in healthy grapes. J Agric Food Chem 56: 11773–11785 [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2010) SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Guindon S, Rodrigo AG, Dyer KA, Huelsenbeck JP. (2004) Modeling the site-specific variation of selection patterns along lineages. Proc Natl Acad Sci USA 101: 12957–12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain R, Reif HJ, Krause E, Langebartels R, Kindl H, Vornam B, Wiese W, Schmelzer E, et al. (1993) Disease resistance results from foreign phytoalexin expression in a novel plant. Nature 361: 153–156 [DOI] [PubMed] [Google Scholar]
- Hammerbacher A, Ralph SG, Bohlmann J, Fenning TM, Gershenzon J, Schmidt A. (2011) Biosynthesis of the major tetrahydroxystilbenes in spruce, astringin and isorhapontin, proceeds via resveratrol and is enhanced by fungal infection. Plant Physiol 157: 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart JH, Shrimpton D. (1979) Role of stilbenes in resistance of wood to decay. Phytopathology 69: 1138–1143 [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipskind JD, Paiva NL. (2000) Constitutive accumulation of a resveratrol-glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol Plant Microbe Interact 13: 551–562 [DOI] [PubMed] [Google Scholar]
- Ingrosso I, Bonsegna S, De Domenico S, Laddomada B, Blando F, Santino A, Giovinazzo G. (2011) Over-expression of a grape stilbene synthase gene in tomato induces parthenocarpy and causes abnormal pollen development. Plant Physiol Biochem 49: 1092–1099 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. French-Italian Public Consortium for Grapevine Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Kodan A, Kuroda H, Sakai F. (2002) A stilbene synthase from Japanese red pine (Pinus densiflora): implications for phytoalexin accumulation and down-regulation of flavonoid biosynthesis. Proc Natl Acad Sci USA 99: 3335–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of T L-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lanz T, Schröder G, Schröder J. (1990) Differential regulation of genes for resveratrol synthase in cell cultures of Arachis hypogaea L. Planta 181: 169–175 [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhuang C, Sheng S, Shao L, Zhao W, Zhao S. (2011) Overexpression of a resveratrol synthase gene (PcRS) from Polygonum cuspidatum in transgenic Arabidopsis causes the accumulation of trans-piceid with antifungal activity. Plant Cell Rep 30: 2027–2036 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Malacarne G, Vrhovsek U, Zulini L, Cestaro A, Stefanini M, Mattivi F, Delledonne M, Velasco R, Moser C. (2011) Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol 11: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J. (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Peressotti E, Wiedemann-Merdinoglu S, Delmotte F, Bellin D, Di Gaspero G, Testolin R, Merdinoglu D, Mestre P. (2010) Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol 10: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Gang DR. (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Poutaraud A, Latouche G, Martins S, Meyer S, Merdinoglu D, Cerovic ZG. (2007) Fast and local assessment of stilbene content in grapevine leaf by in vivo fluorometry. J Agric Food Chem 55: 4913–4920 [DOI] [PubMed] [Google Scholar]
- Preisig-Müller R, Schwekendiek A, Brehm I, Reif HJ, Kindl H. (1999) Characterization of a pine multigene family containing elicitor-responsive stilbene synthase genes. Plant Mol Biol 39: 221–229 [DOI] [PubMed] [Google Scholar]
- Renaud S, de Lorgeril M. (1992) Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet 339: 1523–1526 [DOI] [PubMed] [Google Scholar]
- Richter H, Pezet R, Viret O, Gindro K. (2006) Characterization of 3 new partial stilbene synthase genes out of over 20 expressed in Vitis vinifera during the interaction with Plasmopara viticola. Physiol Mol Plant Pathol 67: 248–260 [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. (2000) Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945 [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Schiex T, Moisan A, Rouzé P. (2001) EuGene: an eukaryotic gene finder that combines several sources of evidence. In O Gascuel, MF Sagot, eds, Lecture Notes in Computer Science, Computational Biology, Vol 2066. Springer-Verlag, Berlin, pp 111–125
- Schmidlin L, Poutaraud A, Claudel P, Mestre P, Prado E, Santos-Rosa M, Wiedemann-Merdinoglu S, Karst F, Merdinoglu D, Hugueney P. (2008) A stress-inducible resveratrol O-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol 148: 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Lane L, Boutet E, Lieberherr D, Tognolli M, Bougueleret L, Bairoch A. (2009) The UniProtKB/Swiss-Prot knowledgebase and its Plant Proteome Annotation Program. J Proteomics 72: 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder G, Brown JW, Schröder J. (1988) Molecular analysis of resveratrol synthase. cDNA, genomic clones and relationship with chalcone synthase. Eur J Biochem 172: 161–169 [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten DMF. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60: 1355–1363 [DOI] [PubMed] [Google Scholar]
- Shomura Y, Torayama I, Suh D-Y, Xiang T, Kita A, Sankawa U, Miki K. (2005) Crystal structure of stilbene synthase from Arachis hypogaea. Proteins 60: 803–806 [DOI] [PubMed] [Google Scholar]
- Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C. (1994) Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol Biol 24: 743–755 [DOI] [PubMed] [Google Scholar]
- Thomzik J, Stenzel K, Stöcker R, Schreier P, Gunata Z, Stahl D. (1997) Synthesis of a grapevine phytoalexin in transgenic tomatoes (Lycopersicon esculentum Mill.) conditions resistance against Phytophthora infestans. Physiol Mol Plant Pathol 51: 265–278 [Google Scholar]
- Tropf S, Lanz T, Rensing SA, Schröder J, Schröder G. (1994) Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J Mol Evol 38: 610–618 [DOI] [PubMed] [Google Scholar]
- Tsai C-J, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y. (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172: 47–62 [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. (2005) GROMACS: fast, flexible, and free. J Comput Chem 26: 1701–1718 [DOI] [PubMed] [Google Scholar]
- Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M. (2012) Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol 12: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J, et al. (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara C, von Baer D, Mardones C, Wilkens A, Wernekinck K, Damm A, Macke S, Gorena T, Winterhalter P. (2012) Stilbene levels in grape cane of different cultivars in southern Chile: determination by HPLC-DAD-MS/MS method. J Agric Food Chem 60: 929–933 [DOI] [PubMed] [Google Scholar]
- Winkel BSJ. (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Xu S, Clark T, Zheng H, Vang S, Li R, Wong GK, Wang J, Zheng X. (2008) Gene conversion in the rice genome. BMC Genomics 9: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zhang D, Hu J, Zhou X, Ye X, Reichel KL, Stewart NR, Syrenne RD, Yang X, Gao P, et al. (2009) Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 10: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP. (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15: 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CKY, Springob K, Schmidt J, Nicholson RL, Chu IK, Yip WK, Lo C. (2005) A stilbene synthase gene (SbSTS1) is involved in host and nonhost defense responses in sorghum. Plant Physiol 138: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.