Abstract
Plant resistance to phytopathogenic microorganisms mainly relies on the activation of an innate immune response usually launched after recognition by the plant cells of microbe-associated molecular patterns. The plant hormones, salicylic acid (SA), jasmonic acid, and ethylene have emerged as key players in the signaling networks involved in plant immunity. Rhamnolipids (RLs) are glycolipids produced by bacteria and are involved in surface motility and biofilm development. Here we report that RLs trigger an immune response in Arabidopsis (Arabidopsis thaliana) characterized by signaling molecules accumulation and defense gene activation. This immune response participates to resistance against the hemibiotrophic bacterium Pseudomonas syringae pv tomato, the biotrophic oomycete Hyaloperonospora arabidopsidis, and the necrotrophic fungus Botrytis cinerea. We show that RL-mediated resistance involves different signaling pathways that depend on the type of pathogen. Ethylene is involved in RL-induced resistance to H. arabidopsidis and to P. syringae pv tomato whereas jasmonic acid is essential for the resistance to B. cinerea. SA participates to the restriction of all pathogens. We also show evidence that SA-dependent plant defenses are potentiated by RLs following challenge by B. cinerea or P. syringae pv tomato. These results highlight a central role for SA in RL-mediated resistance. In addition to the activation of plant defense responses, antimicrobial properties of RLs are thought to participate in the protection against the fungus and the oomycete. Our data highlight the intricate mechanisms involved in plant protection triggered by a new type of molecule that can be perceived by plant cells and that can also act directly onto pathogens.
In their environment, plants are challenged by potentially pathogenic microorganisms. In response, they express a set of defense mechanisms including preformed structural and chemical barriers, as well as an innate immune response quickly activated after microorganism perception (Boller and Felix, 2009). Plant innate immunity is triggered after recognition by pattern recognition receptors of conserved pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs, respectively) or by plant endogenous molecules released by pathogen invasion and called danger-associated molecular patterns (Boller and Felix, 2009; Dodds and Rathjen, 2010). This first step of recognition leads to the activation of MAMP-triggered immunity (MTI). Successful pathogens can secrete effectors that interfere or suppress MTI, resulting in effector-triggered susceptibility. A second level of perception involves the direct or indirect recognition by specific receptors of pathogen effectors leading to effector-triggered immunity (ETI; Boller and Felix, 2009; Dodds and Rathjen, 2010). Whereas MTI and ETI are thought to involve common signaling network, ETI is usually quantitatively stronger than MTI and associated with more sustained and robust immune responses (Katagiri and Tsuda, 2010; Tsuda and Katagiri, 2010).
The plant hormones, salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) have emerged as key players in the signaling networks involved in MTI and ETI (Robert-Seilaniantz et al., 2007; Tsuda et al., 2009; Katagiri and Tsuda, 2010; Mersmann et al., 2010; Tsuda and Katagiri, 2010; Robert-Seilaniantz et al., 2011). Interactions between these signal molecules allow the plant to activate and/or modulate an appropriate spectrum of responses, depending on the pathogen lifestyle, necrotroph or biotroph (Glazebrook, 2005; Koornneef and Pieterse, 2008). It is assumed that JA and ET signaling pathways are important for resistance to necrotrophic fungi including Botrytis cinerea and Alternaria brassicicola (Thomma et al., 2001; Ferrari et al., 2003; Glazebrook, 2005). Infection of Arabidopsis (Arabidopsis thaliana) with B. cinerea causes the induction of the JA/ET responsive gene PLANT DEFENSIN1.2 (PDF1.2; Penninckx et al., 1996; Zimmerli et al., 2001). Induction of PDF1.2 by B. cinerea is blocked in ethylene-insensitive2 (ein2) and coronatine-insensitive1 (coi1) mutants that are respectively defective in ET and JA signal transduction pathways. Moreover, ein2 and coi1 plants are highly susceptible to B. cinerea infection (Thomma et al., 1998; Thomma et al., 1999). JA/ET-dependent responses do not seem to be usually induced during resistance to biotrophs, but they can be effective if they are stimulated prior to pathogen challenge (Glazebrook, 2005). Plants impaired in SA signaling are highly susceptible to biotrophic and hemibiotrophic pathogens. Following pathogen infection, SA hydroxylase (NahG), enhanced disease susceptibility5 (eds5), or SA induction-deficient2 (sid2) plants are unable to accumulate high SA levels and they display heightened susceptibility to Pseudomonas syringae pv tomato (Pst), Hyaloperonospora arabidopsidis, or Erysiphe orontii (Delaney et al., 1994; Lawton et al., 1995; Wildermuth et al., 2001; Nawrath et al., 2002; Vlot et al., 2009). Mutants that are insensitive to SA, such as nonexpressor of PATHOGENESIS-RELATED (PR) genes1 (npr1), have enhanced susceptibility to these pathogens (Cao et al., 1994; Glazebrook et al., 1996; Shah et al., 1997; Dong, 2004). According to some reports, plant defense against necrotrophs also involves SA. Arabidopsis plants expressing the nahG gene and infected with B. cinerea show larger lesions compared with wild-type plants (Govrin and Levine, 2002). In tobacco (Nicotiana tabacum), acidic isoforms of PR3 and PR5 gene that are specifically induced by SA (Ménard et al., 2004) are up-regulated after challenge by B. cinerea (El Oirdi et al., 2010). Resistance to some necrotrophs like Fusarium graminearum involves both SA and JA signaling pathways (Makandar et al., 2010). It is assumed that SA and JA signaling can be antagonistic (Bostock, 2005; Koornneef and Pieterse, 2008; Pieterse et al., 2009; Thaler et al., 2012). In Arabidopsis, SA inhibits JA-dependent resistance against A. brassicicola or B. cinerea (Spoel et al., 2007; Koornneef et al., 2008). Recent studies demonstrated that ET modulates the NPR1-mediated antagonism between SA and JA (Leon-Reyes et al., 2009; Leon-Reyes et al., 2010a) and suppression by SA of JA-responsive gene expression is targeted at a position downstream of the JA biosynthesis pathway (Leon-Reyes et al., 2010b). Synergistic effects of SA- and JA-dependent signaling are also well documented (Schenk et al., 2000; van Wees et al., 2000; Mur et al., 2006) and induction of some defense responses after pathogen challenge requires intact JA, ET, and SA signaling pathways (Campbell et al., 2003).
Isolated MAMPs trigger defense responses that also require the activation of SA, JA, and ET signaling pathways (Tsuda et al., 2009; Katagiri and Tsuda, 2010). For instance, treatment with the flagellin peptide flg22 induces many SA-related genes including SID2, EDS5, NPR1, and PR1 (Ferrari et al., 2007; Denoux et al., 2008), causes SA accumulation (Tsuda et al., 2008; Wang et al., 2009), and activates ET signaling (Bethke et al., 2009; Mersmann et al., 2010). Local application of lipopolysaccharides elevates the level of SA (Mishina and Zeier, 2007). The oomycete Pep13 peptide induces defense responses in potato (Solanum tuberosum) that require both SA and JA (Halim et al., 2009). Although signaling networks induced by isolated MAMPs are well documented, the contribution of SA, JA, and ET in MAMP- or PAMP-induced resistance to biotrophs and necrotrophs is poorly understood.
Rhamnolipids (RLs) are glycolipids produced by various bacteria species including some Pseudomonas and Burkholderia species. They are essential for bacterial surface motility and biofilm development (Vatsa et al., 2010; Chrzanowski et al., 2012). RLs are potent stimulators of animal immunity (Vatsa et al., 2010). They have recently been shown to elicit plant defense responses and to induce resistance against B. cinerea in grapevine (Vitis vinifera; Varnier et al., 2009). They also participate to biocontrol activity of the plant beneficial bacteria Pseudomonas aeruginosa PNA1 against oomycetes (Perneel et al., 2008). However, the signaling pathways used by RLs to stimulate plant innate immunity are not known. To gain more insights into RL-induced MTI, we investigated RL-triggered defense responses and resistance to the necrotrophic fungus B. cinerea, the biotroph oomycete H. arabidopsidis, and the hemibiotroph bacterium Pst in Arabidopsis. Our results show that RLs trigger an innate immune response in Arabidopsis that protects the plant against these different lifestyle pathogens. We demonstrate that RL-mediated resistance involves separated signaling sectors that depend on the type of pathogen. In plants challenged by RLs, SA has a central role and participates to the restriction of the three pathogens. ET is fully involved in RL-induced resistance to the biotrophic oomycete and to the hemibiotrophic bacterium whereas JA is essential for the resistance to the necrotrophic fungus.
RESULTS
RLs Elicit Defense Responses in Arabidopsis
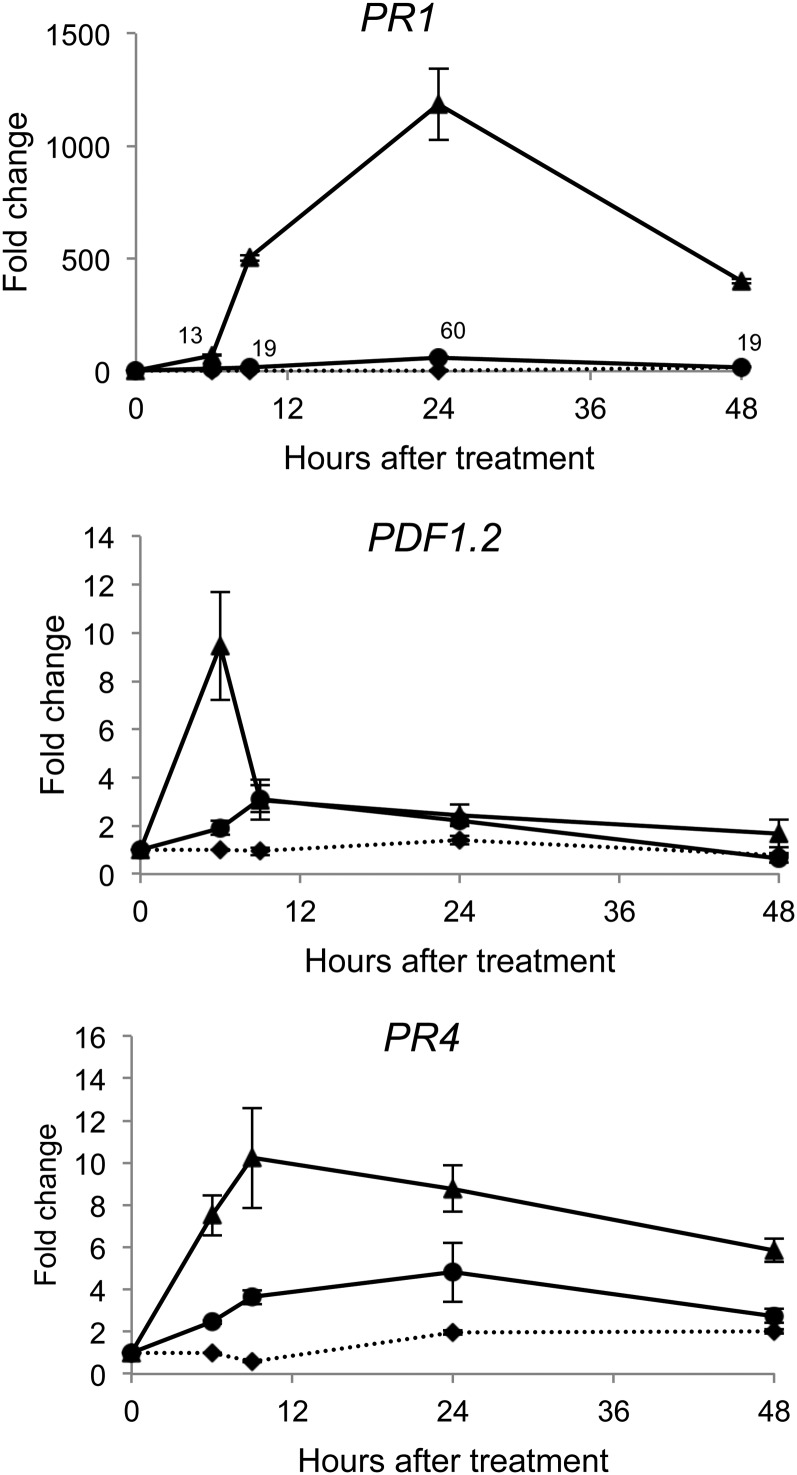
To assess the ability of RLs to induce defense responses in Arabidopsis and the potential links with SA, JA, and ET signaling, we monitored the expression pattern of PR-1, PDF1.2, and PR-4 in plants challenged with RLs. In these experiments and the following, leaves were sprayed with the molecules to monitor local defense responses. PR-1 is well known as a SA-dependent defense gene marker (Lebel et al., 1998; Vlot et al., 2009) whereas PDF1.2 expression is regulated by JA/ET (Penninckx et al., 1996; Penninckx et al., 1998), and PR-4 expression is dependent on ET (Lawton et al., 1994). We used two concentrations of RLs (0.2 and 1 mg mL−1) to compare the responses induced by low and strong RLs stimuli (Varnier et al., 2009). PR-1 expression was induced at 6 h in response to both concentrations of RLs and peaked at 24 h post treatment (hpt; Fig. 1). PDF1.2 expression was strongly and transiently induced in response to the highest dose of RLs, peaking at 6 hpt. A slight increase in PDF1.2 expression was also observed with the lowest concentration of RLs. PR4 expression was stimulated with both concentrations of RLs. Induction of gene expression was stronger with 1 mg mL−1 of RLs. Concomitantly with gene expression, we measured a 2-fold increase in SA level at 24 hpt and a 200-fold increase in JA level at 6 h and 24 hpt with the lowest concentration of RLs (Supplemental Fig. S1). Using the Evans blue test, we did not detect any cell death in Arabidopsis leaves treated with 0.2 mg mL−1 of RLs (Supplemental Fig. S2). At 1 mg mL−1, we observed few microlesions and clear necroses were present when we increased the concentration of RLs to 5 mg mL−1. These results suggest that high concentrations of RLs may trigger a hypersensitive response (HR)-like response as previously described for grapevine (Varnier et al., 2009).
Figure 1.
RLs elicit defense gene expression in Arabidopsis. Defense-related gene expression was monitored in control wild-type leaves (diamonds) and after treatment with RLs (0.2 [circles] and 1 mg mL−1 [triangles]). Transcript accumulation of PR1, PDF1.2, and PR4 genes was determined by qRT-PCR. Results are expressed as the fold increase in transcript level compared with time 0 h and are means + /− sd of duplicate data from one representative experiment among three independent repetitions.
RLs that we used are produced by P. aeruginosa and consist of a mix of mono- and di-RLs (Varnier et al., 2009). We previously described the ability of purified mono- and di-RLs to induce plant defense with the same intensity in grapevine cell suspensions (Varnier et al., 2009). To verify that both type of RLs can induce defense in Arabidopsis, we assayed induction of PR1 in a PR1::GUS reporter line using flg22 (1 µM) as positive control (Denoux et al., 2008; Supplemental Fig. S3). GUS expression was observed with similar intensity in Arabidopsis leaves after treatment with mono-RLs, di-RLs, and the mix of mono- and di-RLs at 0.2 mg mL−1. No induction of PR1::GUS was observed after elicitation with a concentration of 0.05 mg mL−1. We also quantified by quantitative reverse transcription-PCR (qRT-PCR) PR1 gene expression at 24 h after flagellin and RLs (mix at 0.2 mg mL−1) treatment. PR1 was induced 217-fold (±3) over the control by flg22 and 160-fold (±6) by RLs (data not shown).
RLs Induce Local Resistance against B. cinerea, Pst, and H. arabidopsidis
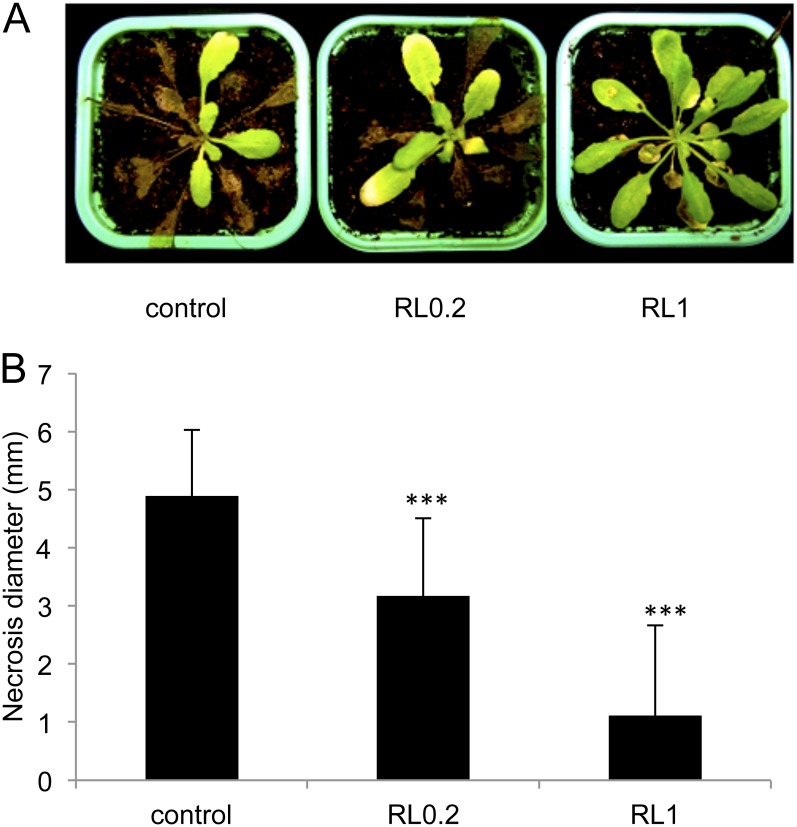
To assess the ability of RLs to enhance Arabidopsis resistance to different lifestyle pathogens, we performed infection experiments with B. cinerea, Pst, and H. arabidopsidis. These three pathosystems have been widely used to decipher disease resistance in Arabidopsis (Glazebrook, 2005; Coates and Beynon, 2010). Arabidopsis plants were sprayed with RLs at low or high concentrations and inoculated with B. cinerea 4 d after pretreatment. Twelve days after infection, most of the leaves from control plants were fully necrotized (Fig. 2A). Some protection effect was observed with 0.2 mg mL−1 of RLs, and most of the leaves treated with 1 mg mL−1 was symptomless or displayed only few necrotic lesions (Fig. 2A). Diameters of lesions on infected leaves were also measured 72 h after B. cinerea challenge (Fig. 2B). Control leaves challenged with B. cinerea displayed very large necrotic lesions (mean of 5 mm). A significant reduction in lesion size was measured in plants treated with RLs at 0.2 mg mL−1, and a strong protective effect of RLs was found at the highest concentration with very small lesions (mean of 1 mm).
Figure 2.
RLs induce resistance against B. cinerea. Plants were sprayed with RLs at 0.2 or 1 mg mL−1 or water (control) and, 4 d later, leaves were inoculated with the fungus. A, Symptoms observed 12 d after infection by the fungus. B, Necroses diameter was measured 72 h after infection by the fungus in the control or RL-treated plants. Values shown are means + /− sd (n = 24) from one representative experiment among three independent repetitions. Stars indicate significant differences between the RL-treated sample and the control according to Student’s t test (***P < 0.005). [See online article for color version of this figure.]
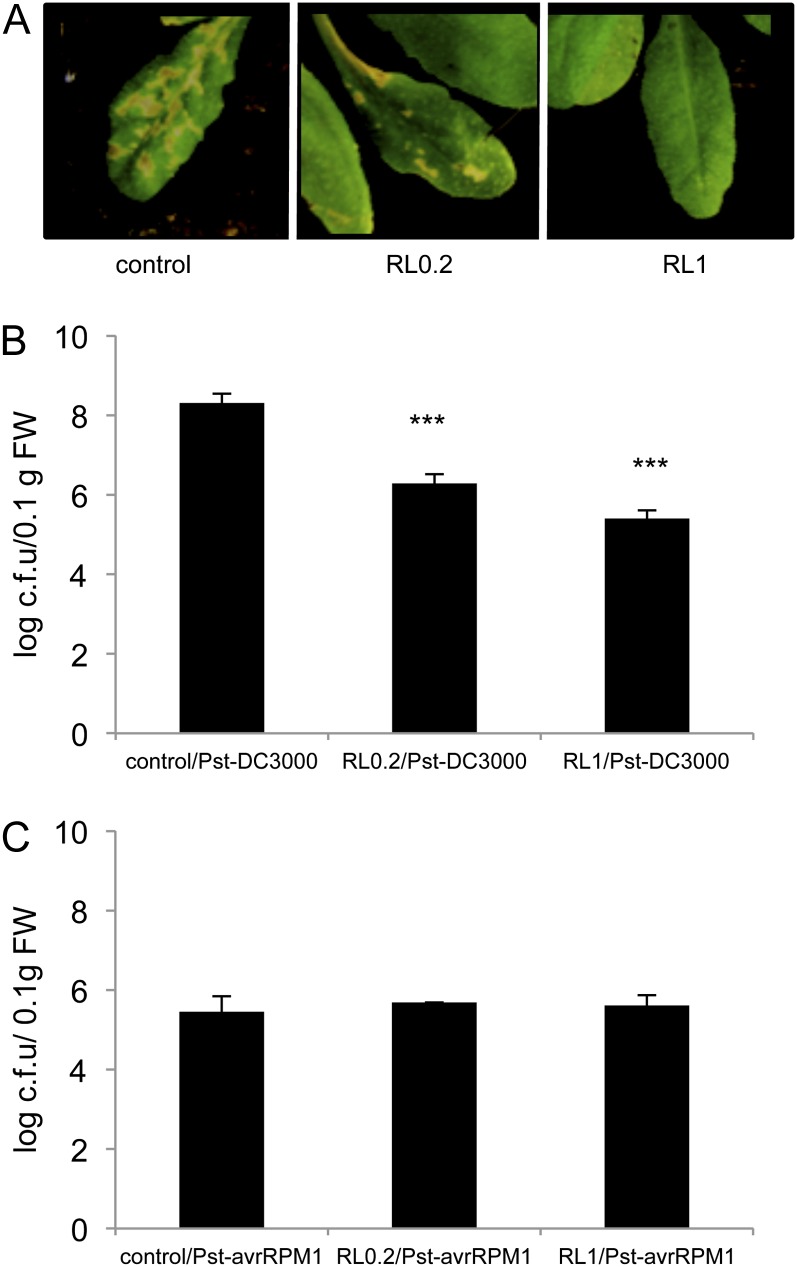
A Pst DC3000/Arabidopsis Columbia-0 (Col-0) pathosystem was used to assess the protective effect of RLs in the context of a compatible interaction. In these experiments, we also monitored the impact of RLs on a typical ETI triggered by the avirulent Pst carrying avrRPM1 (Pst-avrRPM1). Arabidopsis plants dipped with the virulent strain of Pst exhibited typical disease symptoms 7 d post inoculation (Fig. 3A). Symptom development was strongly reduced by pretreatment with 0.2 mg mL−1 of RLs and totally abolished with the highest concentration. We monitored the growth of Pst and Pst-avrRPM1 over a time course in RL-treated and nontreated plants. Pst growth was stopped at 24 h post inoculation (hpi; data not shown) and was still strongly reduced at 72 hpi in RL-treated plants at the highest concentration (Fig. 3B). Bacterial growth was also stopped 24 hpi with 0.2 mg mL−1 of RLs (data not shown) and significantly reduced at 72 hpi (Fig. 3B). No difference was observed between plants treated or not treated with RLs following inoculation with Pst-avrRPM1, so RLs did not interfere with the ETI triggered by the bacteria (Fig. 3C). It is interesting that resistance induced by RLs against the virulent strain of Pst was very similar in intensity with the resistance observed during the typical Pst-avrRPM1-triggered ETI (Fig. 3, B and C).
Figure 3.
RLs protect Arabidopsis against Pst DC3000 infection. Plants were sprayed with RLs at 0.2 or 1 mg mL−1 or water (control) and 4 d later leaves were dipped with Pst +/− avrRPM1. A, Symptoms observed 7 d after infection with the virulent strain Pst DC3000. Bacterial growth in Arabidopsis at 72 hpi with Pst DC3000 (B) and Pst-avrRPM1 (C). Asterisks indicate significant differences between the RL-treated sample and the control according to Student’s t test (*** P < 0.005). Values shown are means + /− sd (n = 24) from one representative experiment among three independent repetitions. [See online article for color version of this figure.]
Arabidopsis Col-0 plants pretreated with RLs were also infected by the compatible strain of the biotroph oomycete H. arabidopsidis Noco2. Conidiospores were harvested 7 d after Arabidopsis infection and counted. A strong and significant protection against H. arabidopsidis was observed after treatment with RLs at 0.2 mg mL−1 (Fig. 4). Moreover, at 1 mg mL−1, RLs restricted almost completely pathogen sporulation.
Figure 4.
RLs induce resistance against H. arabidopsidis in Arabidopsis. Plants were treated with RLs (0.2 and 1 mg mL−1) or with water (control) and, 4 d later, leaves were inoculated with the oomycete. Spores were washed from infected leaves (7 d post inoculation) with water and an aliquot of spore suspension was counted under a microscope. Values shown are means + /− sd (n = 15) from one representative experiment among three independent repetitions. Asterisks indicate significant differences between the RL-treated sample and the control according to Student’s t test (***P < 0.005).
RL-Driven Potentiation of Gene Expression in Plants Challenged by B. cinerea, Pst, and H. arabidopsidis
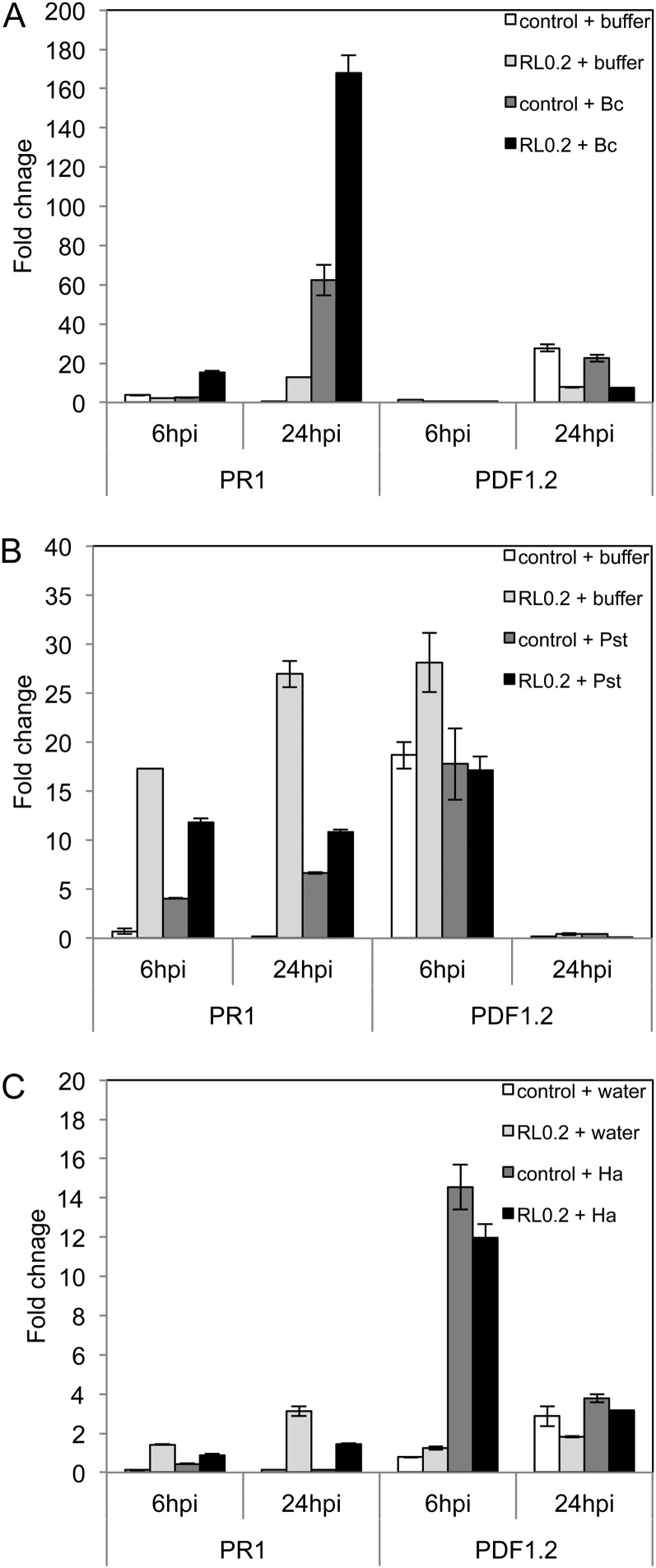
The expression of defense-related genes PR1, PDF1.2, and PR4 was compared at 6 and 24 hpi between pathogen-inoculated or mock-inoculated plants pretreated 4 d before with water control or RLs at 0.2 mg mL−1 (Fig. 5) or at 1 mg mL−1 (Supplemental Fig. S4). Our results indicate that only B. cinerea infection caused a significant induction of PR1 at 24 hpi and that pretreatment with 0.2 mg mL−1 of RLs potentiates PR1 gene expression in response to the fungus as soon as 6 hpi (Fig. 5A). No significant potentiation in PDF1.2 expression (Fig. 5) and PR4 expression (data not shown) was observed for all the conditions tested. Using a higher concentration of RLs (1 mg mL−1), we measured a strong potentiation of PR1 response 24 h after B. cinerea infection or 6 h after Pst challenge and a very small potentiation effect at 24 h after H. arabidopsidis challenge (Supplemental Fig. S4). Again, no significant potentiation of PDF1.2 expression (Supplemental Fig. S4) or PR4 expression (data not shown) was observed in these conditions.
Figure 5.
RL-mediated potentiation of gene expression after pathogen challenge. Plants were sprayed with 0.2 mg mL−1 of RLs or not treated (control) 4 d before inoculation with pathogen or mock inoculation. Transcript accumulation of PR1 and PDF1.2 genes was determined by qRT-PCR 6 and 24 hpi with B. cinerea (Bc; A), Pst DC3000 (Pst; B), or H. arabidopsidis (Ha; C). Results are expressed as the fold increase in transcript level compared with nontreated leaves just before pathogen inoculation (control 0 hpi). Values shown are means + /− sd of duplicate data from one representative experiment among three independent repetitions.
Effect of RLs on Growth and Swarming Motility of Pst and B. cinerea Spore Germination
RLs are known to inhibit oomycete mycelial growth and to decrease zoospore germination and/or motility in vitro (Stanghellini and Miller, 1997; Yoo et al., 2005). Recently, Varnier et al. (2009) showed that RLs also have a direct effect against the strain T4 of B. cinerea. We performed protection experiments with the strain B05.10 of the fungus, which is widely used for infection tests in Arabidopsis (Williamson et al., 2007). Using in vitro tests, we observed that after incubation with RLs at 0.2 mg mL−1, there was no significant difference in spore germination and hyphae growth of strain B0510 compared with the control (Supplemental Fig. S5). However, we estimated that 1 mg mL−1 of RLs led to around 95% of spore germination inhibition at 24 h. To monitor the potential effect of RLs on Pst and Pst-avrRPM1 growth, bacterial strains were cultivated in King’s B medium supplemented or not with RLs (0.2 and 1 mg mL−1). No effect on bacterial growth was observed in the presence of RLs compared with the control (data no shown). RLs are known to be involved in P. aeruginosa swarming motility (Köhler et al., 2000; Déziel et al., 2003; Caiazza et al., 2005). Swarming motility was assessed by examining and measuring the circular turbid zone formed by the bacterial cells migrating on swarm agar plates supplemented or not with 0.2 and 1 mg mL−1 of RLs. In these conditions, we did not observe any swarming motility effect of RLs on Pst and Pst-avrRPM1 (data not shown).
Changes in Gene Expression after Perception of RLs in Arabidopsis Mutants Affected in SA, JA, and ET Signaling Pathways
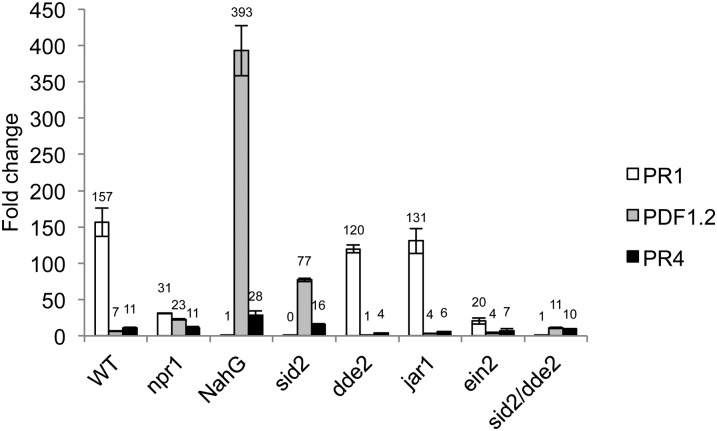
To assess the role of SA, JA, and ET in RL-mediated resistance to pathogens, we used Arabidopsis mutants impaired in their ability to accumulate or perceive these signal molecules. In these experiments, we chose NahG plants that totally degrade SA to be sure that no remaining traces of SA would be detected after RLs treatment and before protection assays (Heck et al., 2003; Ménard et al., 2004; Ferrari et al., 2007). We also used sid2 plants that are impaired in SA synthesis (Nawrath and Métraux, 1999; Wildermuth et al., 2001), npr1 that is insensitive to SA (Cao et al., 1994), and ein2 mutants that are insensitive to ET (Guzmán and Ecker, 1990). jasmonate-resistant1 (jar1) plants insensitive to JA (Staswick et al., 2002), delayed-dehiscence2 (dde2) mutants affected in JA biosynthesis (von Malek et al., 2002), and the double mutant sid2/dde2 affected in both SA and JA pathways (Tsuda et al., 2009) were also used in the following experiments. We first monitored signaling-specific marker gene expression in these mutants and in wild-type plants after RL treatments. In these experiments and the following, we used the lowest concentration of RLs to be in conditions where there is no direct effect of the molecules on the pathogens. PR1 expression was totally abolished in NahG plants, sid2, and sid2/dde2 mutants and strongly reduced in npr1 and ein2 mutants (Fig. 6). This unexpected result for ein2 mutant can be explained by a high basal level of the mock-treated control (9×) compared with the basal level in the wild type. Otherwise, the PR1 expression in the treated plants is similar in ein2 mutant and wild-type plants (data not illustrated). The expression of PR1 was not significantly affected in jar1 and dde2 mutants. PDF1.2 was slightly overexpressed in npr1 mutants but was strongly overexpressed in sid2 and NahG plants. A 2-fold reduction in PDF1.2 expression was observed in jar1 or ein2 mutants compared with wild-type plants, and its expression was totally abolished in the dde2 plants. PR4 expression was similar in npr1, sid2/dde2, and wild-type plants, whereas slightly overexpressed in NahG and sid2 mutants. As for PDF1.2, PR4 expression was weaker in jar1, dde2, and ein2 mutants compared with wild-type plants.
Figure 6.
Defense-related gene expression in leaves of wild-type, npr1, NahG, sid2, dde2, jar1, ein2, and sid2/dde2 plants 24 h after treatment with RLs at 0.2 mg mL−1. Transcript accumulation of PR1, PDF1.2, and PR4 genes was determined by qRT-PCR. Results are expressed as the fold increase in transcript level compared with mock-treated leaves and are means + /− sd of duplicate data from one representative experiment among three independent repetitions.
Role of SA, JA, and ET in RL-Mediated Resistance to B. cinerea, Pst, and H. arabidopsidis
To further elucidate the mechanisms responsible for the resistance triggered by RLs, wild-type plants, and plants affected in the different signaling pathways were analyzed in protection experiments after pretreatment with RLs and challenge with pathogens. As expected, control ein2, dde2, sid2/dde2, and NahG plants were more susceptible to B. cinerea compared with wild-type plants (Fig. 7A; Thomma et al., 1999; Ferrari et al., 2003; Raacke et al., 2006; Ferrari et al., 2007). Surprisingly, we did not observe any differences in susceptibility to the fungus between jar1 and wild-type controls. Resistance induced by RLs was not compromised in ein2 mutants, suggesting that the ET pathway is not involved in the process. No protection was observed in NahG, sid2, jar1, dde2, and sid2/dde2 RL-treated plants (and to a lesser extent in npr1 mutants), suggesting that SA and JA participate in the induced resistance (Fig. 7A).
Figure 7.
RLs induce disease resistance that requires different signaling sectors depending on the pathogen. Wild-type, npr1, NahG, sid2, dde2, jar1, ein2, and sid2/dde2 plants were pretreated with 0.2 mg mL−1 of RLs or water (control) and infected with B. cinerea (A), Pst (B), and H. arabidopsidis (C). Protection assays were performed as described in Figures 2–4. Asterisks indicate significant differences between the RL-treated samples and controls according to Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.005). The figures represent means + /− sd (n = 24) from one representative experiment. Each experiment was repeated three times.
Consistent with previous reports, we observed that the profile of Pst growth was similar in npr1, jar1, and wild-type plants (Laurie-Berry et al., 2006; Niu et al., 2011) and that NahG plants were more susceptible to Pst (Delaney et al., 1994; Fig. 7B). Surprisingly, we did not observe any differences in susceptibility to the bacterium between sid2, dde2, and wild-type plants as it was previously described (Nawrath and Métraux, 1999; Raacke et al., 2006). These differences may be explained by the method of bacterial inoculation (infiltration versus dipping in our study). Npr1, jar1, and dde2 mutations did not compromise RL-mediated reduction of bacterial growth. However, susceptibility of NahG, sid2, ein2 (and to a lesser extent, sid2/dde2) plants to Pst were unchanged after treatment with RLs. These results suggest that SA and ET play a role in RL-induced resistance to the hemibiotroph bacterium, but that this resistance is NPR1 independent and does not involve JA.
NahG plants were more susceptible to H. arabidopsidis in absence of treatment, which is in accordance with previous work (Donofrio and Delaney, 2001; Fig. 7C). Moreover, we observed a strong susceptibility of sid2 mutant to H. arabidopsidis as described previously with Peronospora parasitica (Nawrath and Métraux, 1999). We did not observe any change in the protective effect of RLs toward H. arabidopsidis in npr1, jar1, and dde2 mutants compared with wild-type plants (Fig. 7C), suggesting that resistance against the oomycete does not go through JA alone and is NPR1 independent. Resistance induced by RLs was compromised in NahG and ein2, suggesting that both SA and ET may be involved in RL-mediated resistance to H. arabidopsidis. The mixed results obtained with sid2 plants could be due to the high level of susceptibility of the mutant to H. arabidopsidis. It is interesting that resistance induced by RLs to the oomycete was completely compromised in sid2/dde2 plants, suggesting that SA and JA may act in synergy to account for the protection.
DISCUSSION
RLs are glycolipids produced by various bacterial species including some Pseudomonas spp. and Burkholderia spp. RLs have several potential functions in bacteria. They are involved in the uptake and biodegradation of poorly soluble substrates and are essential for surface motility and biofilm development (Abdel-Mawgoud et al., 2010). Recently, they have been highlighted as potential molecules recognized by animal cells that stimulate innate immunity (Andrä et al., 2006; Bauer et al., 2006; Howe et al., 2006; Vatsa et al., 2010). RLs have also been shown to induce defense responses in grapevine, wheat (Triticum durum), and tobacco cells (Varnier et al., 2009; Vatsa et al., 2010). We demonstrated here that RLs induce the typical Arabidopsis defense marker genes PR1, PDF1.2, and PR4, suggesting that Arabidopsis cells perceive these glycolipids as elicitors. Therefore, RLs display a nonspecific perception profile affecting a broad range of plant genera. RLs behaviors are very similar to those of cyclic lipopeptides that are involved in bacterial motility and biofilm development and that have also been recently described as inducers of plant innate immunity (Ongena et al., 2007; Raaijmakers et al., 2010). Surfactin, the most studied cyclic lipopeptide from Bacillus subtilis, has been shown to trigger early signaling events and late defense responses in tobacco cell suspensions (Jourdan et al., 2009). Other cyclic lipopeptides including massetolide A and fengycin were also identified as elicitors inducing a systemic resistance in tomato (Solanum lycopersicum) and bean (Phaseolus vulgaris; Ongena et al., 2007; Tran et al., 2007). Owing to their physical and chemical properties (i.e. amphiphilic molecules) and their potential mode of perception, RLs and lipopeptides can be considered as a new class of MAMPs produced by either pathogenic or nonpathogenic bacteria (Raaijmakers et al., 2010; Vatsa et al., 2010). Recently, some data indicated that surfactin perception relies on a lipid-driven process at the plasma membrane level (Henry et al., 2011). Such a sensor role of the lipid bilayer is quite uncommon considering that plant basal immunity is usually triggered upon recognition of microbial molecular patterns by high-affinity proteic receptors. It is yet unclear whether the induction of the defense response by RLs requires a specific pattern recognition receptor in the plant plasma membrane, as it is the case for flg22 and oligogalacturonides (Gómez-Gómez and Boller, 2000; Brutus et al., 2010) or whether they interfere directly with the plant plasma membrane as it has been postulated for surfactin or Nep1-like proteins (Qutob et al., 2006; Ottmann et al., 2009; Raaijmakers et al., 2010; Henry et al., 2011). However, the similarities in physical and chemical properties of lipopeptides and RLs suggest that RLs could be perceived in the same manner. Further experiments will be needed to clarify this point.
We demonstrated that RLs induced a resistance to B. cinerea, Pst, and H. arabidopsidis, three pathogens that are members of different lifestyle categories. This is to our knowledge the first report that describes a MAMP-induced resistance, at the local level, to a necrotrophic fungus, a hemibiotrophic Gram-negative bacterium, and a biotrophic oomycete. RLs are known to have antimicrobial properties (Varnier et al., 2009; Vatsa et al., 2010), but the contribution of both the direct effect of RLs, and the induced defense responses in the resistance process is not known. Our results confirm that at high concentrations, RLs have strong inhibitory effects on B. cinerea, and this inhibition is not restricted to a specific strain of the fungus. This direct effect of RLs is thought to participate in the protection that we observed at high concentration. However, resistance to B. cinerea is impaired in NahG, sid2, jar1, and dde2 plants pretreated with a low concentration of RLs, demonstrating that activation of defense responses participate in RL-mediated resistance to the fungus. Similarly, although RLs are known to induce the direct lysis of oomycete zoospores (Stanghellini and Miller, 1997; Vatsa et al., 2010), our results show that RL-mediated resistance to H. arabidopsidis requires functional signaling pathways in the plant. RLs do not directly affect Pst growth or swarming motility, so bacterial resistance induced by RLs is essentially due to activation of plant defense responses.
In our study, levels of resistance to the virulent strain Pst observed in plants treated with RLs at 1 mg mL−1 were strong and very similar to those observed in control plants infected with the avirulent strain Pst-avrRPM1. At this concentration, RLs can induce micronecroses reminiscent to micro-HR in Arabidopsis (Supplemental Fig. S2) and in grapevine leaves (Varnier et al., 2009). HR induction and robust defense responses are characteristic of the ETI (Tsuda and Katagiri, 2010), and our results strengthen the similarities between RLs and some general elicitors/MAMPs or toxins including Nep-1-like proteins (Qutob et al., 2006), fungal toxin fumonisin B1 (Asai et al., 2000), lipopolysaccharides, (Desaki et al., 2006), or elicitins (Baillieul et al., 2003) that display ETI-like defense responses associated with a HR. Our data further reinforce the new concept that there is a continuum between MTI and ETI (Thomma et al., 2011) and that distinction between MAMPs and effectors is not completely relevant, at least in terms of physiological responses.
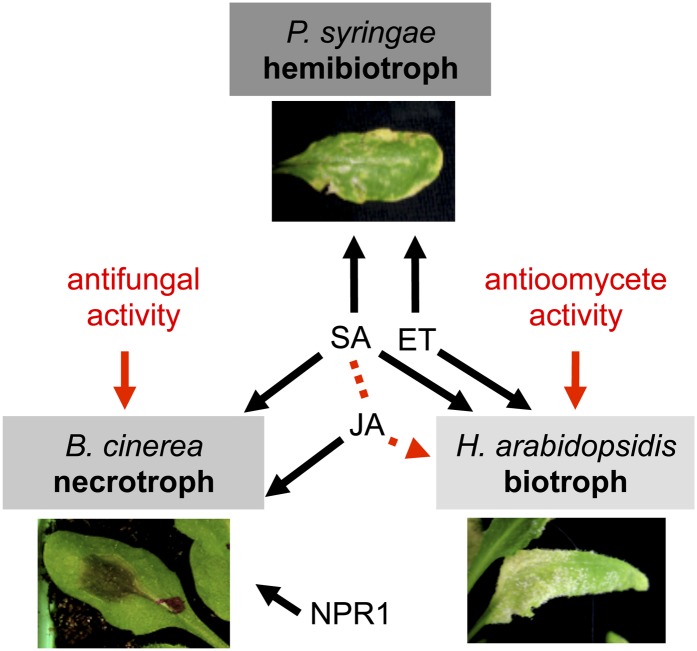
Our results with defense signaling in deficient mutants showed that RL-induced resistance to B. cinerea, H. arabidopsidis, and Pst requires some common signaling pathways but also differs for others. SA is essential for the resistance to the three pathogens, whereas ET is involved in the resistance to the hemibiotrophic bacterium and the biotrophic oomycete, and JA participates in the protection against the necrotrophic fungus (Fig. 8). It is interesting that RL-induced resistance to B. cinerea differs from resistance induced by typical MAMPs and danger-associated molecular patterns in terms of signaling pathways. Indeed, it has been shown that protection against B. cinerea in flg22- and oligogalacturonide-treated Arabidopsis plants is independent of SA, ET, and JA signaling (Ferrari et al., 2003; Ferrari et al., 2007). We found that RL-mediated PDF1.2 expression is overinduced in NahG and sid2 plants. This result is in agreement with a compensation of JA/ET signaling in SA-depleted plants, but this compensation effect does not allow for better protection in RL-treated NahG and sid2 plants. Moreover, RLs potentiate the expression of the SA marker PR-1 after B. cinerea challenge, reinforcing the potential role of SA-dependent responses for the resistance to the fungus. Signaling pathways involved in MAMP-mediated resistance against biotrophic bacteria seem to be more conserved because flg22 (like RLs) triggers resistance against Pst that is compromised in SA-deficient sid2 mutants (Kunze et al., 2004; Zipfel et al., 2004; Mishina and Zeier, 2007). Before our study, few data were available concerning elicitor-induced resistance against H. arabidopsidis. Only recently, Massoud et al. (2012) presented evidence that phosphite could prime Arabidopsis defenses against this oomycete. SA-dependent defenses are thought to play a role in limiting oomycete growth as demonstrated by experiments carried out in eds5 and sid2 plants (Nawrath and Métraux, 1999). Moreover, no enhanced susceptibility to oomycete was observed in npr1-1 (Col-0 background; Bowling et al., 1997). SA-dependent and NPR1-independent resistance responses that limit growth of oomycetes seem to be conserved in RL-induced resistance but not in phosphite-induced protection, which is SA and NPR1 dependent (Massoud et al., 2012). Until now, there was no evidence that ET-dependent responses were normally active in limiting H. arabidopsidis (Glazebrook, 2005), but our data suggest that ET is involved in RL-mediated resistance to the oomycete.
Figure 8.
Proposed model showing how RLs protect Arabidopsis against biotrophic hemibiotrophic and necrotrophic pathogens. RL-mediated resistance to Pst (hemibiotroph) and H. arabidopsidis (biotroph) involves SA and ET signaling (JA synergy with SA is illustrated by the dotted arrow). RL-mediated resistance to B. cinerea (necrotroph) involves SA and JA signaling and is affected by NPR1 mutation. Direct antimicrobial activities of RLs participate to protection against fungus and oomycete pathogens (far left and far right arrows). [See online article for color version of this figure.]
The similarity between RLs and lipopeptides could suggest that common signaling pathways may be involved in induced resistance to different pathogens by these molecules. However, the sole available study suggests that this seems not to be the case for resistance to oomycetes because tomato protection mediated by massetolide A against Phytophthora infestans is independent of SA signaling (Tran et al., 2007). Unfortunately, there is no data available on the most studied lipopeptide, surfactin, regarding the signaling sectors involved in induced resistance to pathogens.
In conclusion, we propose a model in which RLs mediate a MTI that efficiently restricts Arabidopsis colonization of biotrophic, hemibiotrophic, and necrotrophic pathogens (Fig. 8). SA is a central signaling sector in overall RL-induced resistance, whereas ET and JA are differentially required depending on the pathogen lifestyle. In addition to activation of plant defense responses, RLs possess antimicrobial properties that reinforce their efficiency in restricting fungi and oomycete spread.
MATERIALS AND METHODS
Plant Material and Elicitation Treatments
Arabidopsis (Arabidopsis thaliana) Col-0 plants were used in this work. The mutants npr1-1 (Cao et al., 1994), jar1.1 (Staswick et al., 2002), dde2-2 (von Malek et al., 2002), ein2-1 (Guzmán and Ecker, 1990), sid2-2 (Wildermuth et al., 2001), sid2-2/dde2-2 (Tsuda et al., 2009) or transgenic nahG plants (Delaney et al., 1994) were all in the Col-0 background. Plants were grown in soil (Gramoflor) at 21°C with 60% relative humidity and a 12-h-light/12-h-dark cycle (light intensity 150 µE m−2 s−1) for 5 weeks. RLs from Pseudomonas aeruginosa (mix of α-l-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate; RL-1,210: 40%) and 2-O-α-l-rhamnopyranosyl-α-l-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate (RL-2,210: 60%) used in this study were previously characterized in Varnier et al. (2009). Fully expanded leaves were sprayed with different concentrations of RLs or water (control). For gene expression, leaves (from at least five plants) were collected and mixed 6, 9, 24, and 48 h after elicitation and conserved in liquid nitrogen for RNA extraction.
Pathogen Assays in Planta
All of the protection experiments were repeated three times, unless otherwise indicated in the figure legends. Botrytis cinerea B05.10 cultures were initiated by transferring pieces of solid tomato (Solanum lycopersicum)/agar medium containing mycelium to fresh solid tomato/agar medium and incubated at 28°C. Conidia were collected from 3-week-old cultures in 2 mL of growth culture medium (KH2PO4 1.75 g L−1, MgSO4 0.75 g L−1, Glc 4 g L−1, peptone 4 g L−1, Tween 20 0.02% [v/v]). The suspension was adjusted at 105 conidia mL−1 of culture medium and agitated (130 rpm) during 9 h at 22°C to initiate spore germination. For each protection experiment, at least six plants and five leaves per plant were inoculated 4 d after elicitation with one droplet containing 1 × 105 germinative conidia mL−1. The diameter of each lesion was measured 48, 72, and 96 hpi.
Inoculation with the bacterial leaf pathogen Pseudomonas syringae pv tomato (Pst) strain DC3000 or Pst AvrRPM1 was realized by dipping. Briefly, bacteria were cultured overnight at 28°C in liquid King’s B medium, supplemented with rifampicin (50 µg mL−1) and kanamycin (50 µg mL−1). Subsequently, bacterial cells were collected by centrifugation and resuspended in 10 mm MgCl2 Silwet L77 0.02% to a final density of 108 colony forming units (cfu) mL−1 (optical density = 0.1). Plants were dipped in a suspension of Pst at 108 cfu mL−1 or in 10 mm MgCl2 Silwet L77 0.02% as control. At 3, 24, and 72 hpi, 10 foliar discs from five leaves were excised using a cork borer, weighed, and ground in 1 mL MgCl2 (10 mm) with a plastic pestle. Appropriate dilutions were plated on King’s B medium containing rifampicin (50 µg mL−1) and kanamycin (50 µg mL−1), and bacterial colonies were counted. Data are reported as means and sd of the log (cfu 0.1 g−1 fresh weight) of three replicates. Growth assays were performed three times with similar results.
Hyaloperonospora arabidopsidis isolate Noco2 was propagated at 7-d intervals in the Arabidopsis Col-0 wild type. Inoculum was prepared by placing heavily sporulating leaves into water, gently vortexing, and centrifuging the liquid to collect the conidiospores, which were resuspended in water (5 × 104 conidiospores mL−1). Infections were performed by spray-inoculation with asexual inoculum suspension (5 × 104 mL−1) on 5-week-old plants. The inoculated plants were maintained in a box for 7 d at 16°C with 8 h of light/day (100 μE m−2 s−1) and high humidity (80%–100%), which is optimal for H. arabidopsidis germination and growth. For the resistance test, each infected leaf was collected 7 d after infection and photographed to determine foliar surface (using ImageJ software) and placed in 400 µL of distilled water. Spores were then washed from infected leaves by vortexing, and an aliquot of spore suspension was counted under a microscope.
Spore Germination Assay
B. cinerea spore germination assay was realized as described by Prost et al. (2005). Briefly, B. cinerea strain B05.10 was grown in sterile, flat-bottom, 96-well microplates in a final volume of 100 µL growth culture medium (KH2PO4 1.75 g L−1, MgSO4 0.75 g L−1, Glc 4 g L−1, peptone 4 g L−1, Tween 20 0.02% [v/v]). Cultures were started with 5,000 spores and RLs were added after 16 h of growth. Growth was monitored by measuring the absorbance of the microcultures at 595 nm with a microplate reader (Bio-Rad or DYNEX technologies) at 0 h and after 5, 8, and 24 h of incubation in the presence or absence of RLs. Germ tube growth was observed using inverted light microscopy (Leica) 5, 8, and 24 h after RLs addition.
Swarming Motility Assays
Swarm motility plates (0.5% agar) consisted of TSB/10 medium supplemented or not with RLs (0.2 and 1 mg mL−1). Once poured, swarm plates were allowed to dry at room temperature for 16 to 18 h prior to inoculation. Five microliters of overnight culture of bacteria (Pst or Pst-avrRPM1) grown in King’s B medium with appropriate antibiotics, was spot inoculated into the middle of the plate, allowed to dry, and incubated at 37°C for 16 to 18 h. The assay was performed at least three times for each condition.
RNA Extraction and Real-Time qRT-PCR
For each sample, 100 mg of leaves were ground in liquid nitrogen. Total RNA was isolated using Extract’All (Eurobio), and 1 µg was used for reverse transcription using the ABsolute MAX 2-Step qRT-PCR SYBR Green Kit (ThermoElectron) according to the manufacturer’s instructions. The transcript levels were determined by real-time qRT-PCR using the Chromo4 system (BIO-RAD) and the SYBR Green Master Mix PCR kit as recommended by the manufacturer (Applied Biosystems).
PCR reactions were carried out in duplicates in 96-well plates (15 µL per well) in a buffer containing 1× SYBR Green I mix (including Taq polymerase, deoxyribonucleotide triphosphates, SYBR green dye), 280 nm forward and reverse primers, and 1:10 dilution of reverse transcript RNA. After denaturation at 95°C for 15 min, amplification occurred in a two-step procedure: 15 s of denaturation at 95°C and 1 min of annealing/extension at 60°C, with a total of 40 cycles. Identical thermal cycling conditions were used for all targets. Specific primers were designed using the Primer Express software (Applied Biosystems) and are presented in Supplemental Table S1. PCR efficiency of the primer sets was calculated by performing real-time PCR on serial dilutions. For each experiment, PCR reactions were performed in duplicate, and three independent experiments were analyzed. Results correspond to means ± sd of duplicate reactions of one representative experiment out of three. Relative gene expression was determined with the formula fold induction: 2–ΔΔCt, where ΔΔCt = (Ct GI [unknown sample] – Ct GI [reference sample]) – (Ct actin [unknown sample] – Ct actin [reference sample]). GI is the gene of interest. Actin is used as internal control. The reference sample is the nontreated sample chosen to represent 1× expression of the gene of interest.
Seedling Assay and Histochemical GUS Detection
Seedling assay and GUS detection was performed according to Denoux et al. (2008) with minor modifications. For aseptic growth of seedlings, seeds were sterilized by treating them for 1 min in a mix of 95% ethanol/2% commercial bleach (9:1), supplemented with Tween 20 (final concentration 0.01% [v/v]), followed by three quick washes with 99% ethanol and placement to dry under the hood. Ten to 15 seeds were dispensed into each well of a 12-well tissue culture plate with 1 mL of Murashige and Skoog Basal medium with vitamins (Duchefa) supplemented with 0.5% Suc and 0.5 g L−1 2-(N-morpholino)ethanesulfonic acid, pH 5.7. Plates were sealed with Parafilm to prevent evaporation of the medium. Seedlings were grown at 22°C with a 16-h photoperiod at a light intensity of 100 µm−2 s−1 for 10 d before treatment. On the eighth day, the media were replaced with 1 mL of fresh media. Seedlings were treated with elicitors by adding directly to the medium either mono-RLs, di-RLs, or a mix of 40% mono- and 60% di-RLs. RLs were purified according to Varnier et al. (2009). flg22, a synthetic peptide of 22 amino acids (Boller and Felix, 2009), was used as positive control to a final concentration of 1 µm. GUS enzyme activity of PR-1::GUS Arabidopsis seedlings was determined histochemically. Seedling medium in each well was removed and after a quick wash with sodium phosphate buffer, was replaced by 2 mL of 50 mm sodium phosphate, pH 7, 0.1% Triton X-100 and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Duchefa). Seedlings were incubated for 8 h at 37°C. The samples were then fixed with acetic acid/ethanol 1:3 (v/v); the chlorophyll was entirely removed by several washes in 70% ethanol, and the seedlings were mounted in 100% lactic acid.
Cell Death Assay
Cell death was measured in the leaves by staining with Evans blue according to the method described by Kato et al. (2007), with some modifications. Excised leaflets from Arabidopsis plants were vacuum infiltrated with 0.2% (w/v) Evans blue (Sigma-Aldrich, France) for 10 min in eppendorfs to maintain plant tissues in the dying solution. After staining, the leaves were washed three times with distilled water until they were fully decolorized. All experiments were repeated at least three times, and at least 10 leaves collected from multiple seedlings (5 weeks old) were inspected in each experiment. Pictures of representative leaves were taken with a Canon Powershot G12 digital camera.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RLs induce SA and JA accumulation in Arabidopsis.
Supplemental Figure S2. RL-induced cell death in Arabidopsis.
Supplemental Figure S3. Purified mono- and di-RLs induce Arabidopsis defense response.
Supplemental Figure S4. RL-mediated potentiation of gene expression after pathogen challenge.
Supplemental Figure S5. Effect of RLs on B. cinerea spore germination.
Supplemental Table S1. Primers sequences used in qRT-PCR
Supplementary Material
Acknowledgments
We thank Fanja Rabenoelina (Université de Reims Champagne-Ardenne, Reims, France) for technical support, Patrick Saindrenan (Institut de Biologie des Plantes, Orsay, France) for providing H. arabidopsidis isolate Noco2, and Dimitri Heintz (Institut de Biologie Moléculaire des Plantes, Strasbourg, France) for SA and JA analysis. Sid2-2, dde2-2, and sid2-2/dde2-2 plants were kindly provided by J.P. Metraux (University of Fribourg, Fribourg, Switzerland) and F. Katagari (University of Minnesota, St. Paul, MN).
Glossary
- SA
salicylic acid
- RL
rhamnolipid
- PAMP
pathogen-associated molecular pattern
- MAMP
microbe-associated molecular pattern
- MTI
MAMP-triggered immunity
- ETI
effector-triggered immunity
- JA
jasmonic acid
- ET
ethylene
- HR
hypersensitive response
- hpt
h post treatment
- Col-0
Columbia-0
- hpi
h post inoculation
- cfu
colony forming units
- qRT-PCR
quantitative reverse transcription-PCR
- Pst
Pseudomonas syringae pv tomato
References
- Abdel-Mawgoud AM, Lépine F, Déziel E. (2010) Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86: 1323–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrä J, Rademann J, Howe J, Koch MH, Heine H, Zähringer U, Brandenburg K. (2006) Endotoxin-like properties of a rhamnolipid exotoxin from Burkholderia (Pseudomonas) plantarii: immune cell stimulation and biophysical characterization. Biol Chem 387: 301–310 [DOI] [PubMed] [Google Scholar]
- Asai T, Stone JM, Heard JE, Kovtun Y, Yorgey P, Sheen J, Ausubel FM. (2000) Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12: 1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillieul F, de Ruffray P, Kauffmann S. (2003) Molecular cloning and biological activity of α-, β-, and γ-megaspermin, three elicitins secreted by Phytophthora megasperma H20. Plant Physiol 131: 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Brandenburg K, Zähringer U, Rademann J. (2006) Chemical synthesis of a glycolipid library by a solid-phase strategy allows elucidation of the structural specificity of immunostimulation by rhamnolipids. Chemistry 12: 7116–7124 [DOI] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Pöschl Y, Gust AA, Scheel D, Lee J. (2009) Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bostock RM. (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43: 545–580 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, de Lorenzo G. (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, Shanks RM, O’Toole GA. (2005) Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187: 7351–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EJ, Schenk PM, Kazan K, Penninckx IA, Anderson JP, Maclean DJ, Cammue BP, Ebert PR, Manners JM. (2003) Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol 133: 1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowski L, Lawniczak L, Czaczyk K. (2012) Why do microorganisms produce rhamnolipids? World J Microbiol Biotechnol 28: 401–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates ME, Beynon JL. (2010) Hyaloperonospora Arabidopsidis as a pathogen model. Annu Rev Phytopathol 48: 329–345 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, de Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant 1: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaki Y, Miya A, Venkatesh B, Tsuyumu S, Yamane H, Kaku H, Minami E, Shibuya N. (2006) Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol 47: 1530–1540 [DOI] [PubMed] [Google Scholar]
- Déziel E, Lépine F, Milot S, Villemur R. (2003) rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149: 2005–2013 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Dong X. (2004) NPR1, all things considered. Curr Opin Plant Biol 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Donofrio NM, Delaney TP. (2001) Abnormal callose response phenotype and hypersusceptibility to Peronospoara parasitica in defence-compromised arabidopsis nim1-1 and salicylate hydroxylase-expressing plants. Mol Plant Microbe Interact 14: 439–450 [DOI] [PubMed] [Google Scholar]
- El Oirdi M, Trapani A, Bouarab K. (2010) The nature of tobacco resistance against Botrytis cinerea depends on the infection structures of the pathogen. Environ Microbiol 12: 239–253 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, de Lorenzo G, Ausubel FM, Dewdney J. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, de Lorenzo G, Ausubel FM. (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J 35: 193–205 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Govrin EM, Levine A. (2002) Infection of Arabidopsis with a necrotrophic pathogen, Botrytis cinerea, elicits various defense responses but does not induce systemic acquired resistance (SAR). Plant Mol Biol 48: 267–276 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim VA, Altmann S, Ellinger D, Eschen-Lippold L, Miersch O, Scheel D, Rosahl S. (2009) PAMP-induced defense responses in potato require both salicylic acid and jasmonic acid. Plant J 57: 230–242 [DOI] [PubMed] [Google Scholar]
- Heck S, Grau T, Buchala A, Métraux JP, Nawrath C. (2003) Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J 36: 342–352 [DOI] [PubMed] [Google Scholar]
- Henry G, Deleu M, Jourdan E, Thonart P, Ongena M. (2011) The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cell Microbiol 13: 1824–1837 [DOI] [PubMed] [Google Scholar]
- Howe J, Bauer J, Andrä J, Schromm AB, Ernst M, Rössle M, Zähringer U, Rademann J, Brandenburg K. (2006) Biophysical characterization of synthetic rhamnolipids. FEBS J 273: 5101–5112 [DOI] [PubMed] [Google Scholar]
- Jourdan E, Henry G, Duby F, Dommes J, Barthélemy JP, Thonart P, Ongena M. (2009) Insights into the defense-related events occurring in plant cells following perception of surfactin-type lipopeptide from Bacillus subtilis. Mol Plant Microbe Interact 22: 456–468 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Tsuda K. (2010) Understanding the plant immune system. Mol Plant Microbe Interact 23: 1531–1536 [DOI] [PubMed] [Google Scholar]
- Kato Y, Miura E, Matsushima R, Sakamoto W. (2007) White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol 144: 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T, Curty LK, Barja F, van Delden C, Pechère JC. (2000) Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182: 5990–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, van Loon LC, Pieterse CM. (2008) Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol 147: 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CM. (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Berry N, Joardar V, Street IH, Kunkel BN. (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact 19: 789–800 [DOI] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. (1995) Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact 8: 863–870 [DOI] [PubMed] [Google Scholar]
- Lawton KA, Potter SL, Uknes S, Ryals J. (1994) Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell 6: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 16: 223–233 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Du Y, Koornneef A, Proietti S, Körbes AP, Memelink J, Pieterse CM, Ritsema T. (2010a) Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic Acid. Mol Plant Microbe Interact 23: 187–197 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Spoel SH, de Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RA, Ritsema T, Pieterse CM. (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, van der Does D, de Lange ES, Delker C, Wasternack C, van Wees SC, Ritsema T, Pieterse CM. (2010b) Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232: 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makandar R, Nalam V, Chaturvedi R, Jeannotte R, Sparks AA, Shah J. (2010) Involvement of salicylate and jasmonate signaling pathways in Arabidopsis interaction with Fusarium graminearum. Mol Plant Microbe Interact 23: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoud K, Barchietto T, Le Rudulier T, Pallandre L, Didierlaurent L, Garmier M, Ambard-Bretteville F, Seng JM, Saindrenan P. (2012) Dissecting phosphite-induced priming in Arabidopsis infected with Hyaloperonospora arabidopsidis. Plant Physiol 159: 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R, Alban S, de Ruffray P, Jamois F, Franz G, Fritig B, Yvin JC, Kauffmann S. (2004) β-1,3 Glucan sulfate, but not β-1,3 glucan, induces the salicylic acid signaling pathway in tobacco and Arabidopsis. Plant Cell 16: 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S. (2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Atzorn R, Miersch O, Wasternack C. (2006) The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP. (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP. (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu DD, Liu HX, Jiang CH, Wang YP, Wang QY, Jin HL, Guo JH. (2011) The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol Plant Microbe Interact 24: 533–542 [DOI] [PubMed] [Google Scholar]
- Ongena M, Jourdan E, Adam A, Paquot M, Brans A, Joris B, Arpigny JL, Thonart P. (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol 9: 1084–1090 [DOI] [PubMed] [Google Scholar]
- Ottmann C, Luberacki B, Küfner I, Koch W, Brunner F, Weyand M, Mattinen L, Pirhonen M, Anderluh G, Seitz HU, et al. (2009) A common toxin fold mediates microbial attack and plant defense. Proc Natl Acad Sci USA 106: 10359–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, de Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF. (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF. (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perneel M, D’hondt L, de Maeyer K, Adiobo A, Rabaey K, Höfte M. (2008) Phenazines and biosurfactants interact in the biological control of soil-borne diseases caused by Pythium spp. Environ Microbiol 10: 778–788 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, van der Ent S, van Wees SC. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, Carbonne F, Griffiths G, Esquerré-Tugayé MT, Rosahl S, et al. (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 139: 1902–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob D, Kemmerling B, Brunner F, Küfner I, Engelhardt S, Gust AA, Luberacki B, Seitz HU, Stahl D, Rauhut T, et al. (2006) Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18: 3721–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raacke IC, Mueller MJ, Berger S. (2006) Defects in allene oxide synthase and 12-oxa-phytodienoic acid reductase alter the resistance to Pseudomonas syringae and Botrytis cinerea. J Phytopathol 154: 740–744 [Google Scholar]
- Raaijmakers JM, de Bruijn I, Nybroe O, Ongena M. (2010) Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 34: 1037–1062 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF. (1997) Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant Microbe Interact 10: 69–78 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanghellini ME, Miller RM. (1997) Biosurfactants: their identity and potential efficacy in the biological control of zoosporic plant pathogens. Plant Dis 81: 4–12 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML. (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Humphrey PT, Whiteman NK. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17: 260–270 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Tierens KF, Broekaert WF. (1999) Requirement of functional ethylene-insensitive2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH. (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Penninckx IA, Broekaert WF, Cammue BP. (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63–68 [DOI] [PubMed] [Google Scholar]
- Tran H, Ficke A, Asiimwe T, Höfte M, Raaijmakers JM. (2007) Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol 175: 731–742 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F. (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. (2008) Interplay between MAMP-triggered and SA-mediated defense responses. Plant J 53: 763–775 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SC, de Swart EA, van Pelt JA, van Loon LC, Pieterse CM. (2000) Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 8711–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnier AL, Sanchez L, Vatsa P, Boudesocque L, Garcia-Brugger A, Rabenoelina F, Sorokin A, Renault JH, Kauffmann S, Pugin A, et al. (2009) Bacterial rhamnolipids are novel MAMPs conferring resistance to Botrytis cinerea in grapevine. Plant Cell Environ 32: 178–193 [DOI] [PubMed] [Google Scholar]
- Vatsa P, Sanchez L, Clément C, Baillieul F, Dorey S. (2010) Rhamnolipid biosurfactants as new players in animal and plant defense against microbes. Int J Mol Sci 11: 5095–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic Acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- von Malek B, van der Graaff E, Schneitz K, Keller B. (2002) The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Williamson B, Tudzynski B, Tudzynski P, van Kan JA. (2007) Botrytis cinerea: the cause of grey mould disease. Mol Plant Pathol 8: 561–580 [DOI] [PubMed] [Google Scholar]
- Yoo DS, Lee BS, Kim EK. (2005) Characteristics of microbial biosurfactant as an antifungal agent against plant pathogenic fungus. J Microbiol Biotechnol 15: 1164–1169 [Google Scholar]
- Zimmerli L, Métraux JP, Mauch-Mani B. (2001) β-Aminobutyric acid-induced protection of Arabidopsis against the necrotrophic fungus Botrytis cinerea. Plant Physiol 126: 517–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.