Abstract
Image analysis of Arabidopsis (Arabidopsis thaliana) rosettes is an important nondestructive method for studying plant growth. Some work on automatic rosette measurement using image analysis has been proposed in the past but is generally restricted to be used only in combination with specific high-throughput monitoring systems. We introduce Rosette Tracker, a new open source image analysis tool for evaluation of plant-shoot phenotypes. This tool is not constrained by one specific monitoring system, can be adapted to different low-budget imaging setups, and requires minimal user input. In contrast with previously described monitoring tools, Rosette Tracker allows us to simultaneously quantify plant growth, photosynthesis, and leaf temperature-related parameters through the analysis of visual, chlorophyll fluorescence, and/or thermal infrared time-lapse sequences. Freely available, Rosette Tracker facilitates the rapid understanding of Arabidopsis genotype effects.
Integration of tools for simultaneous measurement of plant growth and physiological parameters is a promising way to rapidly screen for specific traits. Remote analysis with minimal handling is essential to avoid growth disturbance. For instance, fluorescence imaging can provide information on chlorophyll content without the need for pigment extraction, and at the same time, can be used for size estimations of green plants (Jansen et al., 2009). Also, thermal images can be used to evaluate relative transpiration differences without having to measure stomatal conductance.
Analysis of plant size or growth is frequently performed by destructive techniques that involve the harvesting of whole plants or plant parts at regular time points. This often requires extensive growth room or greenhouse space. In recent years, a number of methods for systematically tracking plant growth have been developed. For growth analysis of the primary root or hypocotyl, Root Trace (French et al., 2009) and HypoTrace (Wang et al., 2009) were developed primarily for the Arabidopsis (Arabidopsis thaliana) community. For analysis of rosette growth, a number of tools have been developed (Leister et al., 1999; Jansen et al., 2009; Arvidsson et al., 2011). However, the threshold for implementation is high because the software is tuned for specific hardware, restricting its accessibility for smaller labs. Hardware for large-scale screening is usually based on cameras steered into position with a robot (Walter et al., 2007; Jansen et al., 2009; Arvidsson et al., 2011). Therefore, because many labs do not have the means to invest in such a system, phenotypic analysis of plant growth is still frequently done manually. Plants are either weighed (destructive), scanned (destructive), or photographed, followed by manual analysis using image annotation software (nondestructive). These methods are time consuming and call for low-budget, user-friendly alternatives.
Several computer vision-based methods have been proposed to measure and analyze leaf growth in a nondestructive way. We summarize some of these methods below:
“Semiautomatic image analysis” (Jaffe et al., 1985; Guyer et al., 1986; Leister et al., 1999) has been proposed to automatically analyze plant growth. This method requires the user to select either a set of training pixels or to manually tweak an intensity threshold to get robust measurements. The methods are able to detect plant rosettes on a clear background, but are hampered by a nonuniform background, e.g. soil (Guyer et al., 1986). This restriction can be solved by illuminating the plants with IR (infrared) light.
“Motion-based methods” (Barron and Liptay, 1994; Barron and Liptay, 1997; Schmundt et al., 1998; Aboelela et al., 2005) exploit information from multiple time frames. These methods show accurate results for high quality images of isolated leaves, but are of course restricted to time-lapse sequences. A major drawback of these “optical flow” approaches is that they have difficulties with regions, which are partially occluded during a specific time frame.
“Vegetation segmentation” (Shimizu and Heins, 1995; Onyango and Marchant, 2001, 2003; Clément and Vigouroux, 2003) represents a group of algorithms specifically developed for crop segmentation, i.e. automatically delineating a crop from the soil in an image. These methods only require a visual (VIS) image of a plant in its natural growing environment. Thus, these tools can be used to measure plant growth over time. The vegetation segmentation methods are invariant to light conditions and independent of the camera system and crop size. Unfortunately, they rely on the assumption that the images are bimodal, i.e. the images consist of two types of pixels: pixels belonging to a plant or pixels corresponding to soil. The methods will fail if the image contains other types of pixels, e.g. corresponding to a tray, moss, cloth, etc. Although these methods are interesting for specific applications, they are too restrictive for a generic growth analysis system. Moreover, none of these methods have an implementation publicly available.
“Growscreen , Growscreen Fluoro, and Lemnagrid” (Walter et al., 2007, 2009; Arvidsson et al., 2011) combines image analysis with specific plant monitoring systems, e.g. the imaging-pulse amplitude-modulated fluorometer or the ScanalyzerHTS from Lemnatec. All three systems provide a wide range of measurements such as area, relative growth rate, and compactness. With Growscreen Fluoro, it is also possible to analyze chlorophyll fluorescence images. The downside of these frameworks is that they only work in combination with specific monitoring systems. A change of tray, camera, focus, lighting conditions, etc. needs a complete resetting of parameters, e.g. thresholds used for the segmentation of rosettes in the work of Arvidsson et al. (2011) are hard-coded, i.e. they are not dynamically calculated, instead they have to be manually set, which is tedious and error-prone because there are no guidelines on how to set specific parameters. Another important disadvantage is that neither Growscreen, Growscreen Fluoro, nor Lemnagrid is publicly available.
“Montpellier RIO Imaging Cell Image Analyzer” (Baecker, 2007) is a general purpose image analysis ImageJ plug-in. It is controlled by a visual scripting system, which is easier to use than regular scripting or programming languages, but less user-friendly than a specialized program with a graphical user interface. However, it is only able to analyze images with a single rosette in a VIS image, which is obviously a great disadvantage when large numbers of plants are to be analyzed without an automated/robotized image capturing. MRI Cell Image Analyzer can measure the rosette area over time. All parameters are trained for a specific dataset and should be adjusted for time-lapse sequences captured with a different camera system, or with different lighting conditions. The system is open source, and thus can be adapted to the needs of the user. Both ImageJ and the plug-in are freely available.
“Virtual Leaf” (Merks et al., 2011) is a cell-based computer modeling framework for plant tissue morphogenesis, providing a means to analyze the function of developmental genes in the context of the biophysics of growth and patterning. This framework builds a model by alternating between making experimental observations and refining the model. The necessary processing of experiments into quantitative observations, however, is lacking in this work.
“Leaf GUI (for graphical user interface) and Limani” (Dhondt et al., 2012; Price et al., 2011) both provide a framework for extensive analysis of leaf vein and areole measurements. This image analysis framework requires a high-resolution, high-magnification image of an isolated, cleared leaf. Thus, this method is invasive and does not allow the measuring of the same leaf over time. The framework provides a detailed analysis of the fine structures of a leaf but does not give any information about the global leaf or plant growth.
This list is not exhaustive. For a more in-depth overview of image analysis algorithms, we refer to Shamir et al. (2010) and Russ (2011).
With the image analysis software proposed in this paper, we attempt to combine the strong points of the previous methods, while resolving major drawbacks. The software tool is able to detect multiple rosettes in an image without assuming bimodal images, i.e. the images can contain parts that are neither soil nor plants, and it does not require the use of a specific monitoring system with fixed lighting conditions, trays, resolution, etc. Specific parameters (e.g. the scale or number of plants in the field of view) might have to be tuned to cope with time-lapse sequences captured with different systems, but these parameters are straightforward and can easily be adjusted with a few mouse clicks using a simple graphical user interface. The software tool provides a wide range of rosette parameter measurements, i.e. area, compactness, average intensity, maximum diameter, stockiness, and relative growth rate. Apart from analyzing regular color images (VIS images) and/or chlorophyll fluorescence images, the proposed software tool is also able to measure average rosette intensity in thermal IR images. To the best of our knowledge, this is the first image analysis tool proposed in literature offering all of these features.
Although several image analysis tools have been proposed in the past, a robust measurement tool independent of the monitoring system has not been reported yet. We have developed such a freeware tool to analyze time-lapse sequences of Arabidopsis rosettes. In this work, we elaborate on the most important components of the proposed tool, called Rosette Tracker. The first part of the next section will explain how rosettes are detected in different image modalities. This will provide some technical insight in how Rosette Tracker works. Although this technical knowledge is not necessary to use the software tool, it helps to obtain a view on how the tool functions and to identify the cause of occasional failure. Next, practical issues, such as which measurements the tool can handle and what the requirements are for good time-lapse sequences, will be discussed. Finally, as an example, two plant-growth experiments are analyzed using Rosette Tracker to illustrate its versatility and usefulness.
RESULTS AND DISCUSSION
In the following section, we elaborate on the five different components of our image analysis system. These key components consist of (1) calibration of the system, (2) segmentation methods for VIS and chlorophyll fluorescence images, (3) rosette detection, (4) registration of VIS images with IR images, and (5) the set of measurements provided for plant growth analysis. Our proposed image analysis tool, Rosette Tracker, is implemented in the programming language Java 1.6.2 and can be used as a plug-in for ImageJ. The combination with ImageJ allows us to extend the proposed analysis tools with extra functionality and measurements available in ImageJ (Abràmoff et al., 2004). Both a compiled plug-in and the source code are freely available (http://telin.ugent.be/~jdvylder/RosetteTracker/).
Calibration
The conversion of pixels to physical measurement units, such as millimeters, is necessary to analyze measurements like rosette diameter or area. Rosette Tracker has two main options to calculate the actual scale in millimeters. The first option assumes that the actual scale is known and allows the user to enter the ratio of pixels to millimeters in a textbox. If the actual scale is not known in advance, as is often the case, Rosette Tracker provides an easy-to-use graphical tool to set the scale. The user can click on two points in the image between which the real distance is known, for example the two corners of a tray. Alternatively, the user can capture an image that includes a ruler. Based on the distance in the image (in pixels) and the distance in reality (in millimeters), the software tool can approximate the resolution. To get a good estimation, however, it is important that both control points and all rosettes lie nearly in the same focal plane, such that they are approximately at the same distance from the camera.
If the user does not set a scale, all distance and area measurements will be expressed in pixels. Although this does not result in absolute measurements, it allows for relative comparison of plants, provided that they are monitored with the same imaging system.
Segmentation of VIS Images
Color is probably one of the most distinct features for the detection of rosettes in VIS images (Clément and Vigouroux, 2003; Walter et al., 2007; Arvidsson et al., 2011). Many imaging systems represent color using three components: red, green, and blue (RGB). All possible colors that such a system can represent can be organized as a three-dimensional (3-D) cube. Many image analysis tools try to find a region in this RGB cube that corresponds to the green of rosettes in an image. This is not always an easy task because this region can have any shape and might vary between different monitoring systems. A different approach is to first transform the RGB cube to a different representation that is more intuitive for image analysis and processing. An interesting color transformation reorders the RGB cube into the HSV (for hue, saturation, and value) cylinder (Agoston, 2005; Walter et al., 2007; Supplemental Fig. S1, A and B). In this cylinder, color is represented by the three measurements in the name. Hue represents color tone, while saturation corresponds to the colors’ distance from white and value to the luminosity (Supplemental Fig. S1). It is important to note that the hue value is sufficiently discriminative to define pixels corresponding to chlorophyll (from leaf, algae, or moss), whereas the other two parameters, saturation and value, are less important. Thus, finding a color region corresponding to the green of plants is now reduced to finding an interval in one dimension, i.e. the hue dimension, instead of finding an arbitrary shape in three dimensions. An example of the hue channel of a color image is shown in Figure 1, A and B. Note that all pixels corresponding to rosettes show similar hue values. A detailed description and definition of different color space, such as RGB and HSV, has been reported previously (Agoston, 2005).
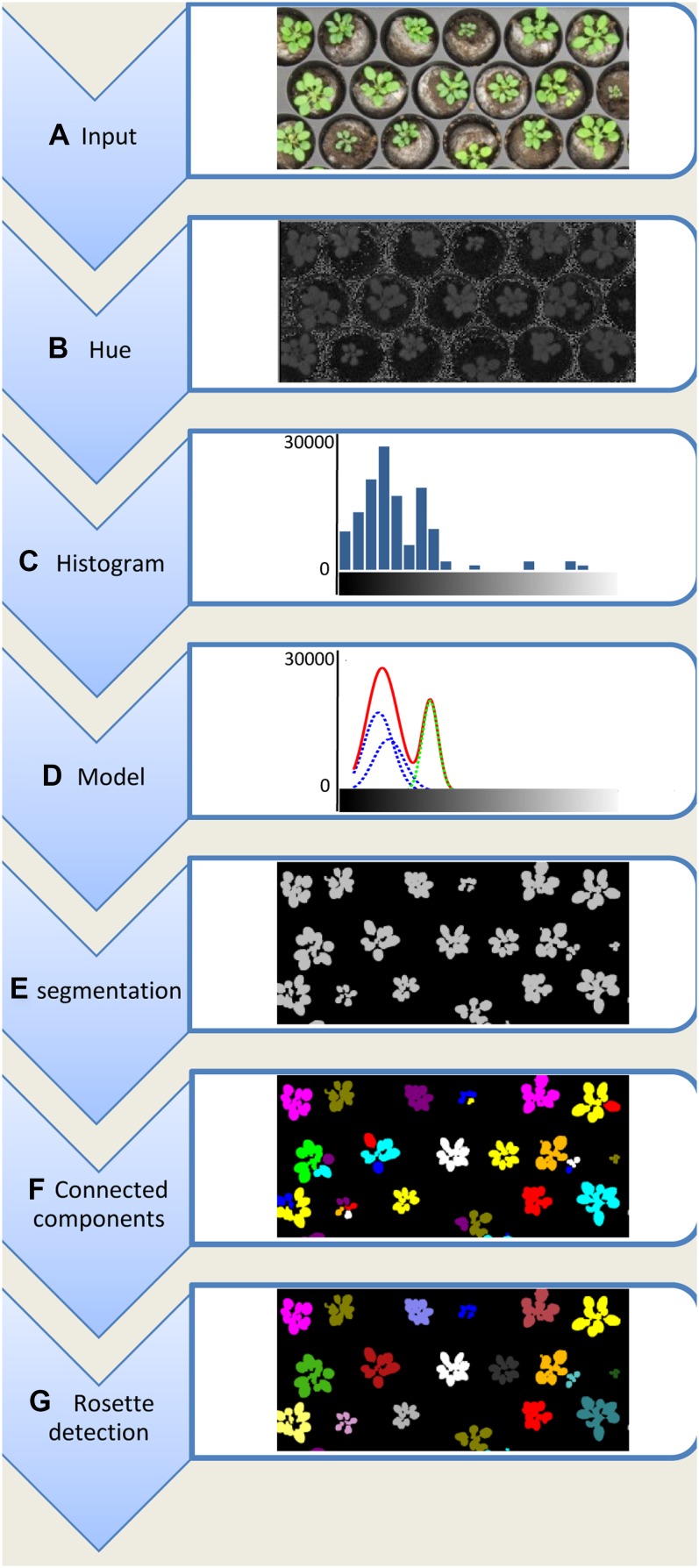
Figure 1.
The work flow of the proposed method: a top view of a rosette tray is captured using a color reflex camera (A); the color image is converted to the HSV color space, of which mainly the hue channel is used (B); the histogram of the hue values of the image is calculated (C); the histogram is modeled using a mixture of Gaussians (D). E, Pixels that have a hue value where the Gaussian distribution corresponding to green is higher than any other Gaussian distribution are classified as foreground pixels; all other pixels are classified as background. F, All connected components are labeled. G, All connected components corresponding to the same rosette are merged and relabeled.
Instead of defining fixed thresholds on a color component, such as is done in Leister et al., (1999) and Arvidsson et al. (2011), a more dynamic approach is necessary for a software tool that is not coupled with any specific monitoring system. To dynamically detect plants, Clément and Vigouroux (2003) proposed the detection of peaks in a histogram of color components. Whereas their method works in RGB space and constrains the image to include only soil or plants, we use the same concept but omit these constraints. First, the histogram of the hue values of the image is calculated (Fig. 1C). The peaks or modes in this histogram generally correspond to specific object types in the image, e.g. rosettes will correspond with a peak near the hue values corresponding with green, whereas soil, trays, labels, etc., will result in different peaks. To detect these peaks we model the histogram, h(.), with a mixture of Gaussians, i.e. we approximate the histogram as a weighted sum of Gaussian probability density functions (Fig. 1D):
where αi represents a weighting parameter and N(μi,σi2) stands for a Gaussian probability density function with mean μi and variance σi2. An example of a description using a mixture of Gaussians can be seen in Figure 1D, where the hue histogram is approximated by the red curve. This red curve is the sum of a set of Gaussian distributions, shown by the green and blue dashed lines. The optimal parameters, i.e. weights, means, and variances that result in the best approximation of the histogram, can be calculated using expectation maximization optimization (Bilmes, 1997). The expectation maximization method is an iterative optimization technique that alternates between two steps: an expectation step, where the expectation of the logarithm of the likelihood (log-likelihood) of Equation 1 is evaluated using the current estimates for the parameters, and a maximization step, where the parameters are optimized to maximize the expected log-likelihood found in the first step. For the exact equations used in each step, we refer to Bilmes (1997).
Each Gaussian probability density function generally corresponds with the physical appearance of different objects, e.g. a Gaussian probability density function for plant pixels, a Gaussian probability density function corresponding with tray pixels, etc. Although there can be numerous objects, and thus Gaussians, only the distinction between plant and other pixels is relevant. The Gaussian probability density function corresponding with rosette pixels is defined as the Gaussian whose mean, μj, is closest to the expected green hue (the green dashed line in Figure 1D). This expected green hue has a default value of 60 but can easily be changed in the software tool by clicking on a plant pixel. The hue value of this selected pixel will then be considered as the expected green hue. Given the mixture of Gaussians, each pixel can be classified based on its hue value; if the probability that the hue value belongs to a rosette is higher than the probability that it belongs to a different class, and hence a different Gaussian, the pixel is classified as a rosette pixel. In all other cases, the pixel is discarded as background.
Note that the expectation maximization algorithm is able to find the optimal parameters for the Gaussian mixture model for a fixed number of Gaussian probability density functions. Predefining this number is not an easy task because it limits the possible setups; two Gaussians only allow plant and soil pixels where the soil is more or less homogenous (Onyango and Marchant, 2003). To avoid predefining, Rosette Tracker starts the segmentation method using two Gaussian probability distribution functions and iteratively increases the number of Gaussians until correct segmentation is achieved. A segmentation result is considered correct if a Gaussian is found with a mean sufficiently close to the expected green hue. If the absolute difference between the expected hue and the mean of each Gaussian is higher than a specific threshold, the number of Gaussians is increased. This threshold is set to 10, assuming that the hue values of the image range from 0° to 360°. Note that the hue values have a cyclic nature, i.e. a hue of 0° is equal to a hue of 360° (Supplemental Fig. S1B). This wraparound, however, does not cause problems because a hue equal to 0° corresponds to red (light with a wavelength of approximately 650 nm), which rarely occurs in images of vegetative plant tissues.
The method proposed above detects all green pixels; however, this does not exclude green pixels that do not belong to a rosette, e.g. pixels corresponding to moss or algae growing on the surface of the plant substrate. These pixels have the same hue as rosettes but are generally darker than rosettes. Therefore, we can exclude them by applying our proposed segmentation algorithm not only in the hue channel but also in the value channel. A true rosette pixel is considered to be a pixel that is detected as a rosette pixel both by the segmentation of the hue and the saturation channels.
Segmentation of Chlorophyll Fluorescence Images
Although the previous segmentation method was developed for the analysis of VIS images, it can easily be adapted to work for chlorophyll fluorescence images as well. Because these are grayscale images instead of color images, the method is simplified in the following way. Instead of working in the HSV color space, a histogram is directly calculated for the chlorophyll fluorescence intensities. Subsequently, this histogram is modeled using a mixture of Gaussians in the same way as for VIS images, i.e. by iteratively increasing the number of Gaussians. However, the criteria to cease iteration are different because there is no default expected chlorophyll fluorescence intensity as is the case for a green hue. Therefore, Rosette Tracker estimates the expected intensity for each image independently, using the following steps:
-
1.
Calculate a rough initial segmentation. This segmentation will only be used to calculate an estimate of the expected intensity, and is thus less sensitive to small errors in the segmentation as long as the overall statistics of the detected foreground resemble the real statistics. Such an initial segmentation can be calculated using different algorithms such as:
-
a.
Supervised pixel classification methods (Cristianini and Shawe-Taylor, 2000; Rabunal and Dorado, 2006), which have shown good results but require large amounts of training data. This training is in contrast to the goal of Rosette Tracker, which attempts to be a dynamic analysis system with minimal user input.
-
b.
Pixel Clustering-based methods (Lloyd, 1982; Ester et al., 1996; Hall et al., 2009) represent another group of generic intensity-based segmentation methods. These methods have been efficiently used for the segmentation of biological objects but are strongly dependent on initialization, i.e. an initial estimate of the location of clusters.
-
c.
Threshold methods separate foreground from background by comparing pixel intensities with a threshold. If the intensity is higher than the threshold, the pixel is classified as foreground, otherwise as background. Rosette Tracker uses a threshold-based method because it does not require training or initialization. The threshold used by Rosette Tracker is calculated using the AutoThreshold function provided in ImageJ. This method is based on Ridler iterative thresholding (Ridler and Calvard, 1978), where a threshold is iteratively established using the average of the foreground and background class means. For a more detailed description on automatic threshold methods, we refer to Otsu (1979), Abràmoff et al. (2004), and Sezgin and Sankur (2004).
-
a.
-
2.
Calculate the mean intensity of all plant pixels, i.e. all pixels above the threshold.
-
3.
Use this mean as expected chlorophyll fluorescence intensity.
It is important to note that the expected intensity will depend on the quality of the segmentation method in Step 1. Tests showed that the proposed initialization method resulted in a good approximation of the expected intensity. If this automatic initialization would fail, however, it is also possible to predefine the expected intensity in a similar way as for VIS images, i.e. by clicking a well-chosen pixel in the chlorophyll fluorescence image.
Rosette Detection
The proposed segmentation method only classifies pixels as corresponding with a rosette pixel or belonging to the background. However, it does not detect to which rosette a pixel belongs. In several high-throughput systems, this is achieved using prior knowledge about the plant’s location and the pot in which the rosette is grown (Walter et al., 2007; Arvidsson et al., 2011). This knowledge has to be reprogrammed for each different setup, e.g. for different trays, different locations of pots, etc. Rosette Tracker proposes a method without this prior knowledge, thus allowing a dynamic setup while requiring a minimal user input.
The output of the segmentation algorithm is represented as a binary image where pixels corresponding to plants are represented by 1, and all other pixels have value 0 (Fig. 1E). Using a connected-component algorithm, this binary image can be transformed to a grayscale image where all foreground pixels, which are connected to each other, bear the same label (ideally corresponding to a single rosette) whereas pixels that are not connected bear a different label. Two pixels are considered to be connected whenever a path of consecutive neighboring foreground pixels exists between them (Fig. 1F).
Because of shadows, it might be that a single rosette is not recognized as such, but corresponds to multiple connected components, each with different labels. To overcome this, components are merged, i.e. relabeled until a predefined number of components remain. The number of final components corresponds to the number of plants visible in the image, which the user has to enter in the settings of Rosette Tracker. This relabeling is done using a clustering approach. Although several clustering techniques will show good results (Lloyd, 1982; Ester et al.; 1996; Hall et al., 2009), we propose a nearest-neighbor approach for its simplicity and efficiency. The connected components are relabeled by iteratively applying the following steps:
-
1.
Calculate the centroid for each connected component.
-
2.
Calculate the distance between all centroids.
-
3.
Merge the two components that are closest to each other.
An example of this relabeling is shown in Figure 1G. It is important that rosettes do not touch each other in the image because this would result in one connected component corresponding to two rosettes. For a hands-on comparison of different clustering techniques, we refer to the WEKA Data Mining Software (Hall et al., 2009).
To aid the user, Rosette Tracker relabels all rosettes such that they are ordered row by row. Vertical rows are automatically detected in the image, based on prior input of the number of rows. This is done using nearest-neighbor clustering, i.e. the same clustering method used for the relabeling of connected components in the rosette detection algorithm. The distance between rosettes, i.e. the basis for clustering rosettes into vertical rows, is defined as the absolute difference in y coordinates of the centroid of each rosette.
Analyzing Thermal IR Images
Although the proposed segmentation method is relatively generic, e.g. it works on color and grayscale images without major modifications, it will not work for all possible image modalities. Near-IR images generally have too low of a contrast and are too noisy to obtain proper segmentation results. Instead, Rosette Tracker uses the segmentation result from a VIS or chlorophyll fluorescence image to measure intensity in a thermal IR image, i.e. the segmentation result is projected as a mask onto the IR image.
To ensure proper correspondence between the segment and the rosette in the IR image, the segment first has to be warped, i.e. deformed in a specific way. This has to be done to cope with difference in scale, focus, small differences in location of the VIS and IR camera, lens distortion, etc. This is often done based on a set of landmarks, i.e. corresponding points that are annotated by the user in both VIS and IR images (Bookstein, 1989; Beatson et al., 2007). The result of this warping strongly depends on the accuracy with which a user can detect corresponding points in IR and VIS images. Low contrast and blur, however, hamper the accurate detection of landmarks in IR images. Therefore, Rosette Tracker requires users to click on corresponding regions in both images instead of accurate landmarks. The location of these regions does not have to be as accurate because Rosette Tracker will calculate the location that results in optimal warping. The technical details about this warping method can be found in our previous work (De Vylder et al., 2012).
Postprocessing
Because of noise, shadows, or clutter, some pixels might be erroneously classified. To correct for these small errors, two postprocessing steps are available: removing small holes and removing clutter. Both methods work on the same principle; objects/holes, which can be enclosed by a small disk, are removed. The size of this disk can be easily tuned using the settings menu of Rosette Tracker. These postprocessing steps are implemented using morphological opening and closing, i.e. a succession of growing and shrinking the objects, or vice versa (Russ, 2011).
Measurements
Quantifying plant growth using image analysis can be done by measurement of many different parameters (Walter et al., 2007, 2009; Arvidsson et al., 2011). Rosette Tracker enables a wide range of these measurements, which we briefly enumerate here:
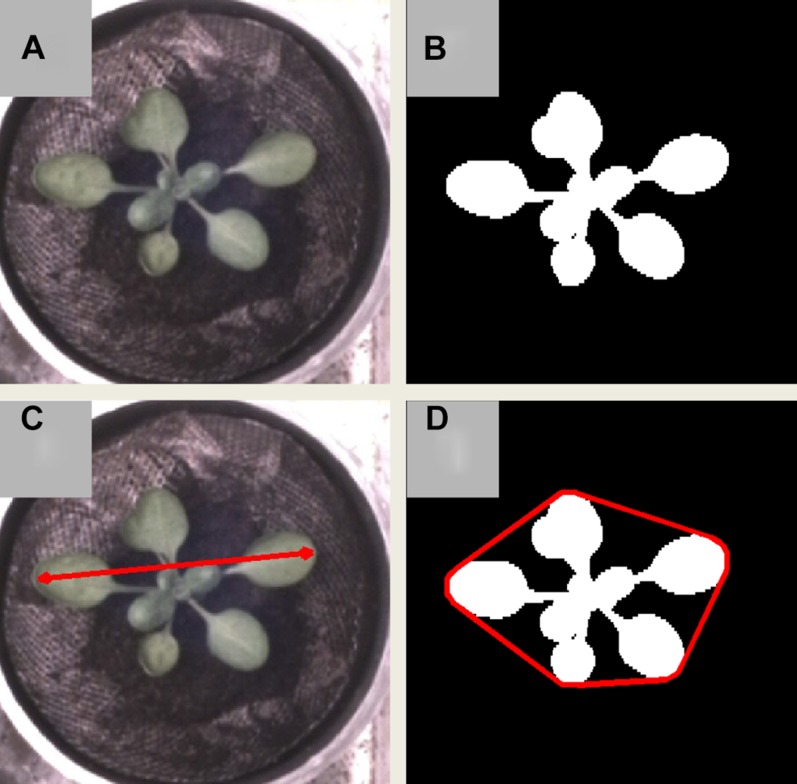
“Area” expresses the area the rosette occupies (Fig. 2B). This is expressed in square millimeters if Rosette Tracker is properly calibrated by setting a scale; otherwise, it is expressed in pixels. The area is measured in a two-dimensional image, i.e. an overhead projection of a 3-D plant. However, because rosettes of Arabidopsis remain relatively flat, this is a very reasonable approximation of the 3-D area (Leister et al., 1999; Walter et al., 2007; Arvidsson et al., 2011).
“Diameter” corresponds with the maximal distance between two pixels belonging to the rosette (Fig. 2C). Used in combination with area, this parameter can hint to changes in petiole length.
“Stockiness” is based on the ratio between the area of a rosette and its perimeter. It is a useful measurement to detect serration of leaves (Jansen et al., 2009) Stockiness is defined as 4 × π × Area/Perimeter2. This can be seen as a measure of circularity, i.e. a circular object’s stockiness is 1. Values vary between 0 and 1.
“Relative Growth Rate” (RGR) expresses the growth of a rosette between two consecutive frames (Blackman, 1919; Walter et al., 2007; Jansen et al., 2009). RGR is defined as 1/t × ln(Area2/Area1), where Area2 and Area1 correspond to the area of the rosette in the current and previous frames, respectively, whereas t represents the time between the two frames.
Average rosette “intensity” is a relevant measure for chlorophyll fluorescence and IR images, which relate to photosynthesis and temperature, respectively (Wang et al., 2003; Chaerle et al., 2004, 2007).
“Compactness” expresses the ratio between the area of the rosette and the area enclosed by the convex hull of the rosette (Jansen et al., 2009; Arvidsson et al., 2011). The convex hull of a rosette corresponds to the contour with the shortest perimeter possible that envelopes the rosette. This is represented in Figure 2D, where the compactness corresponds to the white area over the area enclosed by the red contour.
Figure 2.
An example of different rosette measurements on a VIS image: the actual VIS image of a rosette (A), the area detected by Rosette Tracker (B), the diameter of the rosette corresponds to the length of the red line (C), and depict the compactness of the rosette (D), i.e. the ratio of the area corresponding to the actual rosette over the area enclosed by the convex hull, shown as a red line. [See online article for color version of this figure.]
Image Requirements
Rosette Tracker has been developed to allow robust image analysis for a wide variety of images, with as few assumptions of the monitoring system as possible. There are, however, some limitations a user should be aware of to obtain reliable and accurate measurements. The most notable constraint is that rosettes should not touch one another in the image, otherwise they will be detected as a single rosette, and as a result, another rosette might be detected as two. This can be avoided by leaving enough space between the plants when performing an experiment.
Rosette labels are ordered by vertical rows, where label 1 is the top rosette of the utmost left row and the last label corresponds with the bottom rosette of the utmost right row (Supplemental Document S1). If the trays in the images are oriented such that the rosettes do not correspond with vertical rows, we suggest to first rotate the images using image processing software to get a logical order in rosette labels. To correlate the output of the analysis with the exact rosettes, it is useful to verify the labeling by looking at the segmentation images, which can be saved during analysis.
Although the program does not require any specific scaling, we advise careful development of experiments. High-resolution images result in accurate measurements but require more computational resources, e.g. memory or computational time. However, having a scale (millimeter/pixel) that is too high (i.e. too low of a resolution) will result in a loss of accuracy and, in extreme cases, might even result in an occasional corrupt measurement, where a leaf is classified as belonging to a wrong rosette (Supplemental Fig. S2). Relative errors induced by increased scaling will depend on the size of the plant, with smaller plants being more prone to error. For good results, we suggest using a maximum scale of 0.33/mm (resolution of 3 pixels/mm) for standard wild-type rosettes. In this way, a camera resolution of 12 megapixels could yield 500 rosettes per shot.
Rosette Tracker puts no constraints on the monitoring system used. This was tested by imaging 35 Arabidopsis rosettes using three different cameras. The measurements showed, on average, a relative sd of 5.2% (this is the sd of the measurements normalized by the average measurement itself).
Example 1: Quantification of Rosette Growth Using VIS Images
It has been demonstrated that the rosette area is directly proportional to its weight (Leister et al., 1999). Although the relation is not linear for older rosettes as older leaves can become occluded by newly formed leaves, tracking the rosette area can be a useful tool for monitoring growth.
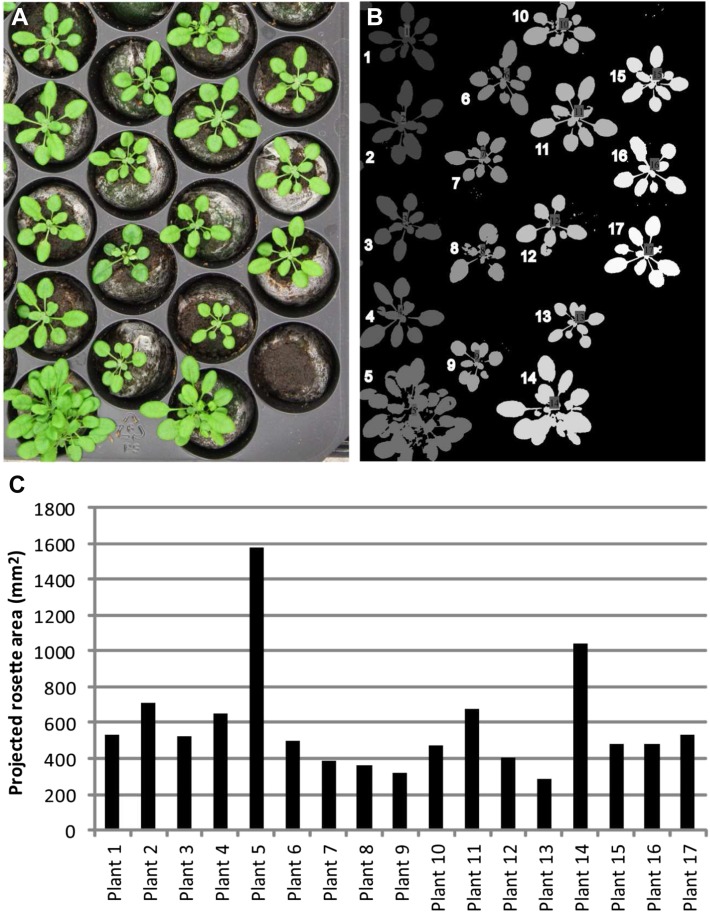
For images in the visual range, current single-lens reflex cameras offer a good value solution. A 12-megapixel reflex camera is suitable for the monitoring of 150 (55-mm lens) to 600 rosettes (18-mm lens) in one shot, from a distance of 2 m. Images captured with wide-angle lenses, such as 18 mm, should be treated for lens distortion prior to analysis. This can be done using camera calibration software such as ROS or CAMcal (Shu et al., 2003; Bader, 2012). Figure 3, A and B show a picture taken with a handheld reflex camera with a 55-mm lens and the corresponding segmentation picture as generated with Rosette Tracker, respectively. Figure 3C shows the eventual output as a bar graph in a spreadsheet, using values generated and saved by Rosette Tracker. The work flow that produced these data are shown in Supplemental Document S2 and illustrated by the supplemental video online (http://telin.ugent.be/~jdvylder/RosetteTracker/).
Figure 3.
Example 1: determination of projected rosette area with Rosette Tracker. A, Original color image showing Columbia-0 plants. B, Segmentation image produced from the color image by Rosette Tracker. Centered numbers in the rosettes from the Rosette Tracker image were repeated on the picture, next to the plant, for clarity. C, Graph showing the projected rosette area per plant from A, as calculated by Rosette Tracker. [See online article for color version of this figure.]
As stated above, time series can be analyzed in a straightforward way as well. Rosette Tracker analyzes all images in a specific file system folder and displays a graph of the measurements of different rosettes as a function of time (see Supplemental Document S1).
These measurements were compared with manually annotated measurements. The ratio between the Rosette Tracker and the manual-based measurements was over 0.97 on average, which is acceptable for the purpose of plant phenotyping based on image analysis.
Example 2: Analyzing a Composite Time-Lapse Sequence of Fluorescence and Thermal (IR) Images
Because of its ubiquitous accumulation in the shoot, fluorescence of chlorophyll can be used to determine the projected leaf area of green rosette plants. Fluorescence images of rosettes, therefore, are a valuable alternative for VIS images. IR images can be used to estimate leaf temperature. Temperature is dependent on evaporation and transpiration of the leaf, which is partly determined by stomatal opening. Hence, IR imaging can be used to monitor transpiration differences (Wang et al., 2003).
Using Rosette Tracker, we followed Arabidopsis ecotype (Burren-0 [Bur-0], Martuba-0 [Mt-0], Rschew-4 [Rsch-4], Lipowiec-0 [Lip-0], and Catania-1 [Ct-1]) and abscisic acid mutant rosettes in time, using a robotized time-lapse imaging system containing fluorescence and thermal cameras (Chaerle et al., 2007). It should be noted that similar analysis based on VIS images (see Example 1) instead of fluorescence images is also possible. The original image set and segmentation files, as generated by Rosette Tracker, are available online (http://telin.ugent.be/~jdvylder/RosetteTracker/). Afterward, these files were loaded into a spreadsheet and average values were calculated (Supplemental Document S3).
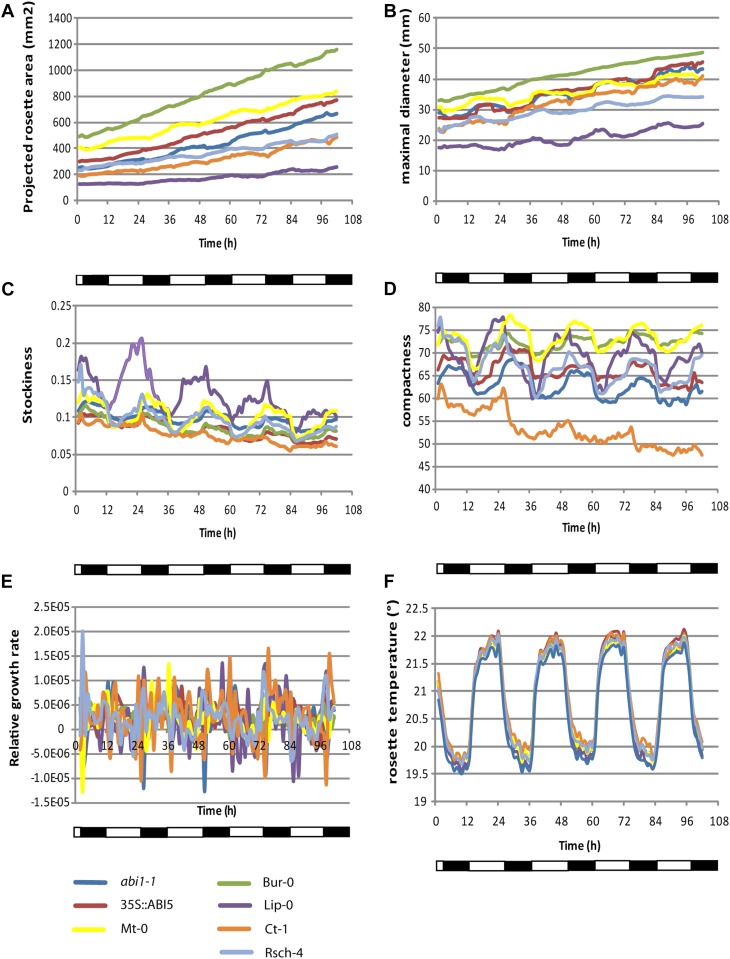
The projected leaf area data for ecotypes Bur-0, Mt-0, and Ct-1 confirm the previously observed large rosette size of Bur-0, the intermediate size of Mt-0, and the small size of Ct-1 (Fig. 4A; Granier et al., 2006). The diurnal difference in area, maximal diameter, stockiness, and compactness, visible in all lines, reflects up- and downward leaf movements (Fig. 4, B–D); at midday, rosettes are flatter than at midnight (Mullen et al., 2006). Figure 4E shows the level of small variations in RGR when measured in 1-h intervals. It can be noticed that in some lines, RGR is the lowest at the beginning of the night, contrasting what was found for circadian hypocotyl extension (Dowson-Day and Millar, 1999). This readout may be influenced by the relatively more compact appearance of the projected rosette image, which is related to the diurnal leaf movements (more hyponasty at night onset). The effect becomes even clearer when 4-h intervals are used for determining RGR (Supplemental Fig. S3). In the 4-h interval measurements, there is a difference between Lip-0, with lower RGR in the morning, and the other lines, most of which have a lower RGR at dusk. This can be directly related to the leaf movements and consequent phase differences between Lip-0 and the other lines regarding projected leaf area, maximal diameter, stockiness (very clear), and compactness (compare Fig. 4 with Supplemental Fig. S3). Optimal use of the RGR measurement tool in relation to growth, therefore, is best suited for image sequences with a 1-d interval (Supplemental Fig. S3). There it becomes apparent that during the experiment, there was an increase in RGR for the small rosettes of Lip-0, for the large rosettes of Bur-0, Mt-0, and 35S::ABI5, the RGR decreased, whereas the RGR remained, albeit with some fluctuation, at the same level in the other lines. Note that the first 24 h show “0” for RGR, because there was no available data for 24 h before the point of measurement for comparison (Supplemental Fig. S3).
Figure 4.
Example 2: edited output of results obtained by Rosette Tracker, using a combined analysis of fluorescence and thermal images. 4-week-old rosettes of abi1-1, 35S::ABI5, Bur-0, Ct-1, Mt-0, Lip-0, and Rsch-4 were photographed each hour, and the following average rosette parameters were calculated: projected rosette area (A), maximal diameter (B), stockiness (C), compactness (D), RGR (E), and temperature (F).
Analysis of IR images demonstrated that temperatures are lowest in abi1-1 mutants, indicating strong transpiration (Fig. 4F). The Arabidopsis ecotypes are all warmer than abi1-1. Earlier observations have shown similar transpiration rates for the ecotypes Bur-0, Mt-0, and Ct-1 (Granier et al., 2006). The abscisic acid-insensitive mutant abi1-1 is known to have reduced stomatal closure and should indeed appear colder in thermal images. By contrast, ABI5 does not influence water loss from plants (Finkelstein, 1994); in this image sequence, the values of the 35S::ABI5 are similar to those of the wild-type ecotypes. Additionally, diurnal temperature variation is observed, with peaks during the day, due to irradiation heat (Fig. 4F).
CONCLUSION
Rosette Tracker offers a user-friendly ImageJ plug-in for rapid analysis of rosette parameters. It is designed for Arabidopsis but can work for any other rosette plants species. The use is not limited to any specific indoor image acquisition hardware, lowering the threshold for implementation in field experiments. It can be used to analyze size and temperature of single snapshots, including multiple rosettes and more complex time series with a high number of frames. In combination with a standard to high-end camera system and modern PC, it is a powerful and affordable tool for plant growth evaluation.
MATERIALS AND METHODS
For Testing the Robustness against Different Cameras
A photograph of 34 Arabidopsis (Arabidopsis thaliana) rosettes was taken using three different cameras: an Olympus C5050 camera, a Canon Powershot SX110IS, and a built-in camera in an LG5000 mobile phone. The images were captured at a distance of 50 cm.
For Example 1
Arabidopsis Columbia-0 wild-type seeds were acquired from the Nottingham Arabidopsis Stock Center. Plants were grown for 4 weeks in a growth room with 22°C and 16-h/8-h light/dark cycles. Light intensity was 60 µmol m−2 s−1 from cool-white light tubes (Philips). A photograph was taken using a Canon 500D reflex camera with an 18- to 55-mm lens (www.Canon.com), at 55 mm, from a distance of 50 cm. Image 1 used for this analysis can be found at http://telin.ugent.be/~jdvylder/RosetteTracker.
For Example 2
Arabidopsis accessions and abi1-1 were acquired from the Nottingham Arabidopsis Stock Center. 35S::ABI5 was kindly provided by J. Smalle. Plants were grown in trays for 3 weeks in 13-h/11-h light/dark cycles in a growth room at 21°C. Daytime photosynthetic photon flux density was 150 µmol m−2 s−1. Rosettes were photographed every hour by a robotized camera system (Chaerle et al., 2006). Fluorescence images were acquired with an in-house-developed fluorescence imaging system (Chaerle et al., 2004), whereas thermal images were acquired with a FLIR-AGEMA Thermovision THV900SWTE (Flir Systems). The output files (supplemental data/online) were rearranged, and average values were calculated from up to eight replicate plants, depending on the genotype (Supplemental Document S3). A compilation of all unarranged measurements, photographs, and configuration files used for this example can be downloaded from http://telin.ugent.be/~jdvylder/RosetteTracker.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Two possible representations of color and examples of the representation of a VIS image in HSV color space.
Supplemental Figure S2. The relation between scale and error on area measurements.
Supplemental Figure S3. Comparison of RGR results using different time intervals.
Supplemental Document S1. Quick-start guide for new users.
Supplemental Document S2. Work flow for Example 1.
Supplemental Document S3. Measurements for Example 2.
Supplementary Material
Acknowledgments
The authors wish to thank Pieter Callebert, Laury Chaerle, and Xavier Vanrobaeys for setup and image acquisition of the time-lapse sequence in Example 2. Supplemental Figure S1, A and B, is modified from the works of Michael Horvath.
Glossary
- VIS
visual
- IR
infrared
- 3-D
three-dimensional
- RGB
red, green, and blue
- HSV
hue, saturation, and value
- RGR
relative growth rate
- Bur-0
Burren-0
- Ct-1
Catania-1
- Mt-0
Martuba-0
- Lip-0
Lipowiec-0
- Rsch-4
Rschew-4
References
- Aboelela A, Liptay A, Barron JL. (2005) Plant growth measurement techniques using near-infrared imagery. Int J Robot Autom 20: 42–49 [Google Scholar]
- Abràmoff MD, Magalhães PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Agoston MK. (2005) Computer Graphics and Geometric Modeling: Implementation and Algorithms. Springer, London
- Arvidsson S, Pérez-Rodríguez P, Mueller-Roeber B. (2011) A growth phenotyping pipeline for Arabidopsis thaliana integrating image analysis and rosette area modeling for robust quantification of genotype effects. New Phytol 191: 895–907 [DOI] [PubMed] [Google Scholar]
- Bader M. (2012) How to calibrate a monocular camera. ROS Tutorials. http://www.ros.org/wiki/camera_calibration/Tutorials/MonocularCalibration (September 24, 2012)
- Baecker V. (2007) Automatic measurement of plant features using ImageJ and MRI Cell Image Analyzer. In Workshop on Growth Phenotyping and Imaging in Plants. AGRON-OMICS, Montpellier, France, p 17
- Barron JL, Liptay A. (1994) Optic flow to measure minute increments in plant growth. Bioimaging 2: 57–61 [Google Scholar]
- Barron JL, Liptay A. (1997) Measuring 3D plant growth using optical flow. Bioimaging 5: 82–86 [Google Scholar]
- Beatson RK, Powell MJD, Tan AM. (2007) Fast evaluation of polyharmonic splines in three dimensions. IMA J Numer Anal 27: 427–450 [Google Scholar]
- Bilmes JA. (1997) A gentle tutorial on the EM algorithm and its application to parameter estimation for gaussian mixture and hidden markov models. ICSI-TR-97-021 Technical Report. International Computer Science Institute, Berkeley, CA
- Blackman VH. (1919) The compound interest law and plant growth. Ann Bot (Lond) 33: 353–360 [Google Scholar]
- Bookstein FL. (1989) Principal warps: thin-plate splines and the decomposition of deformations. IEEE Trans Pattern Anal Mach Intell 11: 567–585 [Google Scholar]
- Chaerle L, Hagenbeek D, De Bruyne E, Valcke R, Van Der Straeten D. (2004) Thermal and chlorophyll-fluorescence imaging distinguish plant-pathogen interactions at an early stage. Plant Cell Physiol 45: 887–896 [DOI] [PubMed] [Google Scholar]
- Chaerle L, Hagenbeek D, Van der Straeten D, Leinonen I, Jones H. (2006) Monitoring and screening plant populations with thermal and chlorophyll fluorescence imaging. Comp Biochem Physiol A Mol Integr Physiol 143: S143–S144 [Google Scholar]
- Chaerle L, Leinonen I, Jones HG, Van Der Straeten D. (2007) Monitoring and screening plant populations with combined thermal and chlorophyll fluorescence imaging. J Exp Bot 58: 773–784 [DOI] [PubMed] [Google Scholar]
- Clément A, Vigouroux B. (2003) Unsupervised segmentation of scenes containing vegetation (Forsythia) and soil by hierarchical analysis of bi-dimensional histograms. Pattern Recognit Lett 24: 1951–1957 [Google Scholar]
- Cristianini N, Shawe-Taylor J. (2000) An Introduction to Support Vector Machines: and Other Kernel-Based Learning Methods. Cambridge University Press, Cambridge, UK
- De Vylder J, Douterloigne K, Vandenbussche F, Van Der Straeten D, Philips W. (2012) A non-rigid registration method for multispectral imaging of plants. In Sensing for Agriculture and Food Quality and Safety: Proceedings of the Society of Photo-Optical Instrumentation Engineers (SPIE). SPIE, Baltimore, MD, pp 1–8
- Dhondt S, Van Haerenborgh D, Van Cauwenbergh C, Merks RMH, Philips W, Beemster GTS, Inzé D. (2012) Quantitative analysis of venation patterns of Arabidopsis leaves by supervised image analysis. Plant J 69: 553–563 [DOI] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ. (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17: 63–71 [DOI] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Sander J, Xu X. (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. In E Simoudis, J Han, UM Fayyad, eds, International Conference on Knowledge Discovery and Data Mining (KDD-96). The Association for the Advancement of Artificial Intelligence, Portland, OR, pp 226–231
- Finkelstein RR. (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- French A, Ubeda-Tomás S, Holman TJ, Bennett MJ, Pridmore T. (2009) High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol 150: 1784–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier-Combaud S, Lebaudy A, et al. (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169: 623–635 [DOI] [PubMed] [Google Scholar]
- Guyer DE, Miles GE, Schreiber MM, Mitchell OR, Vanderbilt VC. (1986) Machine vision and image processing for plant identification. Trans ASABE 29: 1500–1507 [Google Scholar]
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. (2009) The WEKA data mining software: an update. SIGKDD Explor 11: 10–18 [Google Scholar]
- Jaffe MJ, Wakefield AH, Telewski F, Gulley E, Biro R. (1985) Computer-assisted image analysis of plant growth, thigmomorphogenesis and gravitropism. Plant Physiol 77: 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Gilmer F, Biskup B, Nagel KA, Rascher U, Fischbach A, Briem S, Dreissen G, Tittmann S, Braun S, et al. (2009) Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol 36: 902–914 [DOI] [PubMed] [Google Scholar]
- Leister D, Varotto C, Pesaresi P, Niwergall A, Salamini F. (1999) Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol Biochem 37: 671–678 [Google Scholar]
- Lloyd SP. (1982) Least-squares quantization in PCM. IEEE Trans Inf Theory 28: 129–137 [Google Scholar]
- Merks RMH, Guravage M, Inzé D, Beemster GTS. (2011) VirtualLeaf: an open-source framework for cell-based modeling of plant tissue growth and development. Plant Physiol 155: 656–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JL, Weinig C, Hangarter RP. (2006) Shade avoidance and the regulation of leaf inclination in Arabidopsis. Plant Cell Environ 29: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Onyango CM, Marchant JA. (2001) Physics-based colour image segmentation for scenes containing vegetation and soil. Image Vis Comput 19: 523–538 [Google Scholar]
- Onyango CM, Marchant JA. (2003) Segmentation of row crop plants from weeds using colour and morphology. Comput Electron Agric 39: 141–155 [Google Scholar]
- Otsu N. (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9:62–66 [Google Scholar]
- Price CA, Symonova O, Mileyko Y, Hilley T, Weitz JS. (2011) Leaf extraction and analysis framework graphical user interface: segmenting and analyzing the structure of leaf veins and areoles. Plant Physiol 155: 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabunal JR, Dorado J. (2006) Artificial Neural Networks in Real-Life Applications. Idea Group, Hershey, PA
- Ridler TW, Calvard S. (1978) Picture thresholding using an iterative selection method. IEE Trans Syst Man Cybern 8: 630–632 [Google Scholar]
- Russ JC. (2011) The Image Processing Handbook, Ed 6. CRC Press, Boca Raton, FL
- Schmundt D, Stitt M, Jähne B, Schurr U. (1998) Quantitative analysis of the local rates of growth of dicot leaves at a high temporal and spatial resolution, using image sequence analysis. Plant J 16: 505–514 [Google Scholar]
- Sezgin M, Sankur B. (2004) Survey over image thresholding techniques and quantitative performance evaluation. J Electron Imaging 13: 146–165 [Google Scholar]
- Shamir L, Delaney JD, Orlov N, Eckley DM, Goldberg IG. (2010) Pattern recognition software and techniques for biological image analysis. PLoS Comput Biol 6: e1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Heins RD. (1995) Computer-vision-based system for plant growth analysis. Trans ASABE 38: 959–964 [Google Scholar]
- Shu C, Brunton A, Fiala M. (2003) Automatic grid finding in calibration patterns using Delaunay triangulation. Technical Report. National Research Council, Ottawa, Canada
- Walter A, Jansen M, Gilmer F, Biskup B, Nagel KA, Rascher U, Fischbach A, Briem S, Dreissen G, Tittmann S, et al. (2009) Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol 36: 902–914 [DOI] [PubMed] [Google Scholar]
- Walter A, Scharr H, Gilmer F, Zierer R, Nagel KA, Ernst M, Wiese A, Virnich O, Christ MM, Uhlig B, et al. (2007) Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: a setup and procedure designed for rapid optical phenotyping of different plant species. New Phytol 174: 447–455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.