The use of small molecules to transiently modulate protein function circumvents the limitations of classical genetic approaches (Dobson, 2004; Hicks and Raikhel, 2012). These approaches are limited by functional gene redundancy because mutation of a single gene can result in a lack of phenotype due to compensation (Borevitz and Ecker, 2004). Alternatively, when an essential gene is mutated, embryo lethality can result (Candela et al., 2011). These obstacles can be circumvented by a chemical genomics approach that uses small molecules to temporarily modulate protein function. Small molecules often bind to proteins, transiently inhibiting proper function. This marriage of synthetic chemistry and biology, coupled with the grand scale of genomics, is a powerful approach that produces large amounts of useful phenotypic data prior to genetics (Hicks and Raikhel, 2012). Linking this to metabolomics studies will facilitate the detection of metabolic modulators, the dissection of the cross talk in the metabolic network, and the development of hypotheses on how changes in metabolism affect developmental or cellular responses.
Forward chemical genomic screens can uncover new specific reagents that can target transitory compartments to dissect their functions in a systematic manner. Such screens were performed in plants and resulted in the dissection of pathways including cellulose biosynthesis (DeBolt et al., 2007; Yoneda et al., 2007), callose activation (Zabotina et al., 2008), pathogen defense (Serrano et al., 2007), and the elusive identification of the abscisic acid receptor (Park et al., 2009). Furthermore, chemical screens probing the endomembrane system led to the identification of Sortins (Zouhar et al., 2004; Chanda et al., 2009), Gravacin (Surpin et al., 2005; Rojas-Pierce et al., 2007), and Endosidins (Robert et al., 2008; Drakakaki et al., 2011), which were used to elucidate endomembrane trafficking within the cell. Hitherto, chemical genomics has only rarely led to new insights or to the identification of target proteins involved in basic cellular metabolism as in a few case studies of the plant cell wall (Desprez et al., 2002), herbicide targets (Grossmann et al., 2012), and phenolics (Rosado et al., 2011). Nevertheless, there are great prospects to identify and dissect novel aspects of metabolism, particularly in multicellular organisms. Certainly, a large portion of the metabolome awaits discovery as our knowledge regarding the complex metabolic network and intracellular trafficking of metabolites is considerably limited in most organisms.
Metabolomics, a recently established “omics” approach, allows a broad detection of hundreds, if not thousands, of small molecules in a single analytical run of a given sample (Saito and Matsuda, 2010). Despite the frequently ambiguous identification of metabolites due to the lack of reference compounds and intrinsic limitations of mass spectrometry data interpretation, this technology facilitates new insights into currently unexplored metabolic pathways (Mintz-Oron et al., 2008; Hall and Hardy, 2012). The powerful feature of metabolomics is in its ability to perform nontargeted analysis, which permits the detection and to a certain extent identification of “unexpected” metabolites that were not initially “under the spotlight.”
The development of metabolomics approaches was largely driven by quantum leaps in analytical chemistry tools that allow for the separation and detection of metabolites (Allwood et al., 2011). The most widespread technology used currently in metabolomics assays is liquid chromatography-mass spectrometry (Lei et al., 2011; Moing et al., 2011). Instruments with rapid and high-resolution separation capacity through ultra-pressure chromatography are typically coupled with mass spectrometry detectors with extreme resolution and accurate mass determination capabilities. Complementary technologies include gas chromatography-mass spectrometry (Almstetter et al., 2012) and NMR (Kim et al., 2011; Ward et al., 2011). Another key feature of these cutting-edge analytical tools is their versatility in examining any given extract from bacteria to fungi, plants, and mammalian cells. Nevertheless, to date, no extraction method nor any technological platform exists that enables the measurement of all endogenous metabolites in a single extract.
Plants are especially appropriate targets for metabolomics approaches because they produce a wide range of metabolites (Clardy and Walsh, 2004). Several of these metabolites belong to unique chemical classes and are of great importance in our diet and daily life. The most widespread use of metabolomics technologies is currently in functional genomics, which links genetic and metabolic changes (Fiehn et al., 2000). This can be observed in the influence of abiotic/biotic stresses on metabolism (Obata and Fernie, 2012), or as an analysis tool employed in metabolic engineering approaches (Wu and Chappell, 2008). Furthermore, metabolomics becomes an integral part of systems biology where changes at the metabolome, transcriptome, and proteome levels are analyzed in an integrated fashion, as exemplified by Liberman et al. (2012). Recent advancements in the isolation of individual cell populations (e.g. through laser capture microdissection and fluorescence-activated cell sorting) can now be coupled to metabolomics analysis for a comprehensive investigation of the time- and space-resolved distribution of analytes (Rogers et al., 2012). Finally, given that metabolism is highly compartmentalized, more and more attention is also paid to the study of metabolism at a subcellular level (Krueger et al., 2011).
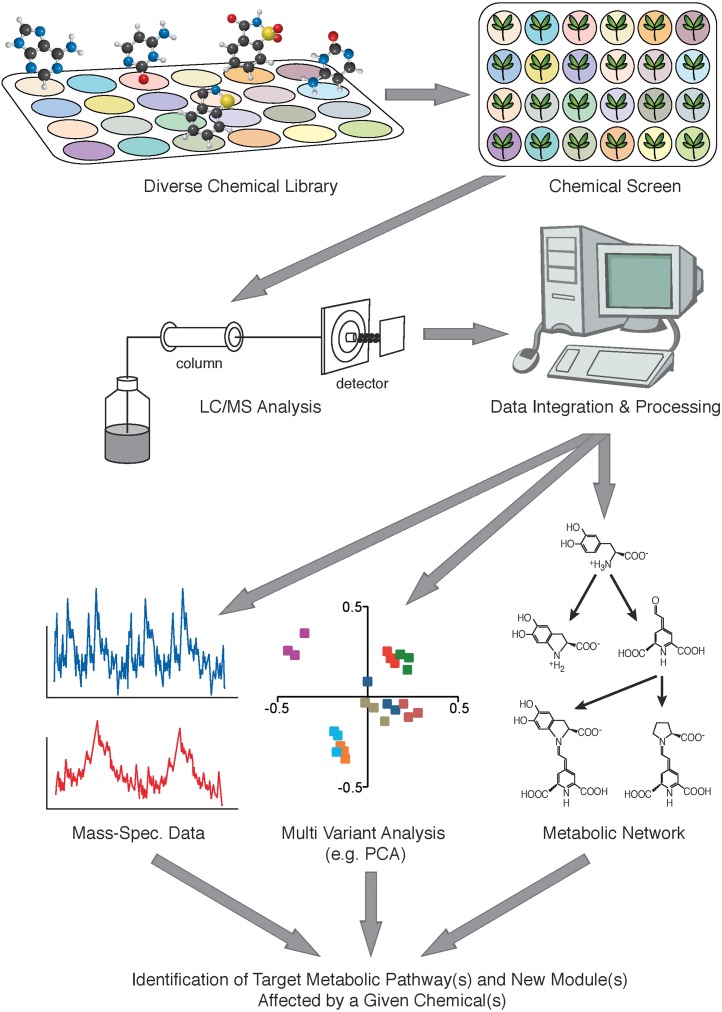
Combining metabolomics with chemical genomics, also referred to as “ChemoMetabolomics,” is a promising future application of both technologies. In such experiments, samples derived from cell cultures or whole organisms that were treated with a library of chemicals would be subjected to metabolomics analyses as, for example, those employing high-resolution mass spectrometry instruments. As compared with genetic approaches, ChemoMetabolomics, as well as chemical genomics, has the advantage of a time-resolved conditional disturbance, whereby the samples are exposed to a given chemical treatment over different time periods. This allows for the identification of perturbations, which when constitutively active (i.e. in a mutant) are lethal and thus not accessible for analysis. Examination of the metabolic response of the biological system to the chemical treatment can result in a number of potential outcomes (Fig. 1). A first obvious one is the identification of lead compounds that represent a starting point for the development of highly potent inhibitors, or modulators, of a metabolic pathway. Depending on the intermediate metabolites that can be identified in a given pathway, it might even be possible to predict the target protein/enzyme due to the specific change of metabolites in this putative pathway. Furthermore, whereas established target-oriented high-throughput setups that use in vitro systems monitor only one enzymatic reaction per experiment, the proposed approach facilitates screening products of many enzymatic reactions at a time. Moreover, a predicted change in the metabolic profile upon chemical treatment can be used to identify a chemical that targets a given enzyme. Leads identified in this way may be useful as inhibitors, blocking or enhancing certain specific biochemical pathways (e.g. resulting in the discovery of new herbicides, or compounds that boost a biosynthetic pathway of interest). A second, more complex outcome is the reconstruction of metabolic networks based on the perturbation data obtained using a large number of chemicals. Once a chemical affecting multiple metabolic pathways is identified, this will open the way to dissect the exceedingly intricate cross talk in the metabolic network. A third obvious outcome is based on the integrated analysis of metabolic changes and corresponding alterations in developmental or cellular processes, thus allowing for the development of a hypothesis regarding the effect of changes in metabolism on developmental or cellular responses. As a consequence, applying chemicals that could perturb vesicular-based metabolite trafficking will enable new insights into where and how metabolites are delivered to their site of action and storage.
Figure 1.
Work flow of a ChemoMetabolomics study. In a ChemoMetabolomics study, plants or cell cultures are treated with a chemical(s) prior to metabolomics analyses. Metabolic data outputs such as the one obtained by high-resolution liquid chromatography-mass spectrometry are then processed by multivariant analysis, for example, to identify target metabolic pathways and new modules in the metabolic network.
Whereas carrying out metabolomics assays undoubtedly has multiple advantages, largely due to the quantity and diversity of metabolite coverage and the “untargeted approach,” it might not always be the preferred method for metabolite analysis. As in other technologies, increasing metabolite coverage decreases capabilities in terms of sensitivity and quantification (Sweetlove et al., 2003). Moreover, the capacity to accurately and unambiguously identify metabolites is reduced to a great extent (Fernie et al., 2011). In some cases, a particular section of the metabolic network consisting of several metabolites that are structurally or functionally/biologically related (e.g. carotenoids, vitamins, hormones, and amino acids) are of interest. In this view, one might choose to utilize the “metabolite profiling” approach in ChemoMetabolomics, in which defined, well-established procedures are used to extract, separate, detect, quantify, and identify the metabolites in a targeted manner (Gu et al., 2012).
An important part of data handling and quality control procedures includes attempts to identify the metabolic “fate” of the chemical used for treatment, and subsequently filtering out its derivatives, which in most cases form through endogenous metabolism and can be the primary cause of the metabolic phenotype observed. A next step in ChemoMetabolomics would consist of genetic screens to isolate mutants that are hypersensitive or resistant to the chemical of interest, which will facilitate the location of the component(s)/target(s). Another approach for identifying targets is analyzing the similarity between metabolic profiles obtained from genetic mutants to those obtained from ChemoMetabolomics, which will also allow for the identification of the target protein and pathway of a given chemical. The output of such experiments could also be integrated and mutually queried with additional data derived from various chemical screens at the developmental and cell biological levels. The generation and mining of such a compendium of data sets could be an excellent instrument to probe as yet undiscovered links between metabolic and developmental programs and to obtain a more detailed view of biological processes at the system level. Our labs have just recently initiated ChemoMetabolomics experiments with the most encouraging preliminary data. It is expected that once established, this concept will open new frontiers in metabolic studies by leveraging on the gradually vanishing boundary between chemistry and biology.
References
- Allwood JW, De Vos RC, Moing A, Deborde C, Erban A, Kopka J, Goodacre R, Hall RD. (2011) Plant metabolomics and its potential for systems biology research background concepts, technology, and methodology. Methods Enzymol 500: 299–336 [DOI] [PubMed] [Google Scholar]
- Almstetter MF, Oefner PJ, Dettmer K. (2012) Comprehensive two-dimensional gas chromatography in metabolomics. Anal Bioanal Chem 402: 1993–2013 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Ecker JR. (2004) Plant genomics: the third wave. Annu Rev Genomics Hum Genet 5: 443–477 [DOI] [PubMed] [Google Scholar]
- Candela H, Pérez-Pérez JM, Micol JL. (2011) Uncovering the post-embryonic functions of gametophytic- and embryonic-lethal genes. Trends Plant Sci 16: 336–345 [DOI] [PubMed] [Google Scholar]
- Chanda A, Roze LV, Kang S, Artymovich KA, Hicks GR, Raikhel NV, Calvo AM, Linz JE. (2009) A key role for vesicles in fungal secondary metabolism. Proc Natl Acad Sci USA 106: 19533–19538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clardy J, Walsh C. (2004) Lessons from natural molecules. Nature 432: 829–837 [DOI] [PubMed] [Google Scholar]
- DeBolt S, Gutierrez R, Ehrhardt DW, Melo CV, Ross L, Cutler SR, Somerville C, Bonetta D. (2007) Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc Natl Acad Sci USA 104: 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H. (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. (2004) Chemical space and biology. Nature 432: 824–828 [DOI] [PubMed] [Google Scholar]
- Drakakaki G, Robert S, Szatmari AM, Brown MQ, Nagawa S, Van Damme D, Leonard M, Yang Z, Girke T, Schmid SL, et al. (2011) Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc Natl Acad Sci USA 108: 17850–17855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Aharoni A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V. (2011) Recommendations for reporting metabolite data. Plant Cell 23: 2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Grossmann K, Hutzler J, Tresch S, Christiansen N, Looser R, Ehrhardt T. (2012) On the mode of action of the herbicides cinmethylin and 5-benzyloxymethyl-1, 2-isoxazolines: putative inhibitors of plant tyrosine aminotransferase. Pest Manag Sci 68: 482–492 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. (2012) Rapid LC-MS/MS profiling of protein amino acids and metabolically related compounds for large-scale assessment of metabolic phenotypes. Methods Mol Biol 828: 1–11 [DOI] [PubMed] [Google Scholar]
- Hall RD, Hardy NW. (2012) Practical applications of metabolomics in plant biology. Methods Mol Biol 860: 1–10 [DOI] [PubMed] [Google Scholar]
- Hicks GR, Raikhel NV. (2012) Small molecules present large opportunities in plant biology. Annu Rev Plant Biol 63: 261–282 [DOI] [PubMed] [Google Scholar]
- Kim HK, Choi YH, Verpoorte R. (2011) NMR-based plant metabolomics: where do we stand, where do we go? Trends Biotechnol 29: 267–275 [DOI] [PubMed] [Google Scholar]
- Krueger S, Giavalisco P, Krall L, Steinhauser MC, Büssis D, Usadel B, Flügge UI, Fernie AR, Willmitzer L, Steinhauser D. (2011) A topological map of the compartmentalized Arabidopsis thaliana leaf metabolome. PLoS ONE 6: e17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Huhman DV, Sumner LW. (2011) Mass spectrometry strategies in metabolomics. J Biol Chem 286: 25435–25442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LM, Sozzani R, Benfey PN. (2012) Integrative systems biology: an attempt to describe a simple weed. Curr Opin Plant Biol 15: 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, et al. (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moing A, Aharoni A, Biais B, Rogachev I, Meir S, Brodsky L, Allwood JW, Erban A, Dunn WB, Kay L, et al. (2011) Extensive metabolic cross-talk in melon fruit revealed by spatial and developmental combinatorial metabolomics. New Phytol 190: 683–696 [DOI] [PubMed] [Google Scholar]
- Obata T, Fernie AR. (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69: 3225–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Chary SN, Drakakaki G, Li S, Yang Z, Raikhel NV, Hicks GR. (2008) Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc Natl Acad Sci USA 105: 8464–8469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ED, Jackson T, Moussaieff A, Aharoni A, Benfey PN. (2012) Cell type-specific transcriptional profiling: implications for metabolite profiling. Plant J 70: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Pierce M, Titapiwatanakun B, Sohn EJ, Fang F, Larive CK, Blakeslee J, Cheng Y, Cutler SR, Peer WA, Murphy AS, et al. (2007) Arabidopsis P-glycoprotein19 participates in the inhibition of gravitropism by gravacin. Chem Biol 14: 1366–1376 [DOI] [PubMed] [Google Scholar]
- Rosado A, Hicks GR, Norambuena L, Rogachev I, Meir S, Pourcel L, Zouhar J, Brown MQ, Boirsdore MP, Puckrin RS, et al. (2011) Sortin1-hypersensitive mutants link vacuolar-trafficking defects and flavonoid metabolism in Arabidopsis vegetative tissues. Chem Biol 18: 187–197 [DOI] [PubMed] [Google Scholar]
- Saito K, Matsuda F. (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Serrano M, Robatzek S, Torres M, Kombrink E, Somssich IE, Robinson M, Schulze-Lefert P. (2007) Chemical interference of pathogen-associated molecular pattern-triggered immune responses in Arabidopsis reveals a potential role for fatty-acid synthase type II complex-derived lipid signals. J Biol Chem 282: 6803–6811 [DOI] [PubMed] [Google Scholar]
- Surpin M, Rojas-Pierce M, Carter C, Hicks GR, Vasquez J, Raikhel NV. (2005) The power of chemical genomics to study the link between endomembrane system components and the gravitropic response. Proc Natl Acad Sci USA 102: 4902–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Last RL, Fernie AR. (2003) Predictive metabolic engineering: a goal for systems biology. Plant Physiol 132: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JL, Baker JM, Llewellyn AM, Hawkins ND, Beale MH. (2011) Metabolomic analysis of Arabidopsis reveals hemiterpenoid glycosides as products of a nitrate ion-regulated, carbon flux overflow. Proc Natl Acad Sci USA 108: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Chappell J. (2008) Metabolic engineering of natural products in plants; tools of the trade and challenges for the future. Curr Opin Biotechnol 19: 145–152 [DOI] [PubMed] [Google Scholar]
- Yoneda A, Higaki T, Kutsuna N, Kondo Y, Osada H, Hasezawa S, Matsui M. (2007) Chemical genetic screening identifies a novel inhibitor of parallel alignment of cortical microtubules and cellulose microfibrils. Plant Cell Physiol 48: 1393–1403 [DOI] [PubMed] [Google Scholar]
- Zabotina O, Malm E, Drakakaki G, Bulone V, Raikhel N. (2008) Identification and preliminary characterization of a new chemical affecting glucosyltransferase activities involved in plant cell wall biosynthesis. Mol Plant 1: 977–989 [DOI] [PubMed] [Google Scholar]
- Zouhar J, Hicks GR, Raikhel NV. (2004) Sorting inhibitors (Sortins): Chemical compounds to study vacuolar sorting in Arabidopsis. Proc Natl Acad Sci USA 101: 9497–9501 [DOI] [PMC free article] [PubMed] [Google Scholar]