Abstract
This study quantified the antibiotic release kinetics and subsequent bactericidal efficacy of rifampicin (RIF) against Gram-positive and Gram-negative bacteria under in vitro static conditions. Antibiotic-loaded scaffolds were fabricated by electrospinning poly(caprolactone) (PCL) with 10% or 20% (w/w) RIF. Scaffold fiber diameter and RIF loading were characterized, and RIF release kinetics were measured. RIF-releasing and RIF-free scaffolds were inoculated with Pseudomonas aeruginosa and Staphylococcus epidermidis, and the suspended concentration live and dead bacteria were determined by fluorescent microscopy. Adherent bacteria and biofilm formation were examined using scanning electron microscopy. Mean fiber diameters were 557 ± 399 nm for RIF-free, 402 ± 225 nm for 10% RIF, and 665 ± 402 nm for 20% RIF scaffolds. RIF release kinetics exhibited a short-burst release during the first hour, followed by a 7 h, zero-order release during which both RIF scaffolds released ~50% of their initial RIF mass loading. P. aeruginosa and S. epidermidis suspended cell populations proliferated in accordance with logarithmic growth models when exposed to control scaffolds; however both RIF-containing scaffolds completely inhibited bacterial growth in suspension and, subsequently, prevented biofilm formation within the scaffolds through the first 6 h.
1 Introduction
The incidence of peri-prosthetic infections after major orthopedic surgeries such as total joint arthroplasty (i.e., hip and knee) and internal fracture fixation has increased over the past decade [1, 2]. The current standard of care to treat a chronic infection is a two-stage re-implantation procedure beginning with the removal of infected tissue and all prosthetic components, administration of intravenous antibiotics, placement of an antibiotic-loaded cement spacer [3, 4], and then a revision surgery after the infection has cleared (approximately 6 weeks). Thus, a patient may undergo multiple surgeries and prolonged antibiotic treatment in order to treat a periprosthetic infection [5]. In severe infections, arthrodesis, resection arthroplasty and amputation may be necessary [6], and can result in increased mortality of elderly patients [7]. In addition, these infections represent an enormous economic burden due to medical costs and lost wages and production.
Peri-prosthetic infections are thought to be caused by invading bacteria at the time of surgery. Staphylococcus epidermidis and Pseudomonas aeruginosa are Gram-positive and Gram-negative bacterial species involved with approximately 30 and 8% of bacterial colonies in orthopedic implants [8, 9]. Treating infected implant materials is exceedingly difficult, especially due to the inherent difficulties of treating an established biofilm [10]. Preventing the incidence of infection provides a viable therapeutic strategy [11–13]. Prophylactic antibiotic administration has been shown to significantly reduce the incidence of hospital-acquired infections [14], which cost an average of $68,000 per patient [15]. Antibiotics including gentamicin, rifampicin, vancomycin and tobramycin have been investigated either alone or in combination [16, 17] to prevent the formation of Gram-positive and Gram-negative microbial communities on prosthesis surfaces [18]. Prophylactic measures such as antibiotic-loaded bone cement or antibiotic implant coatings [19] offer a means to reduce the occurrence and severity of these infections, and diminish the need for subsequent surgeries.
Preventative strategies against orthopedic peri-prosthetic infections have often favored using antibiotics loaded into bone cements and spacers [10]. Synthetic bone graft scaffolds, such as electrospun polymeric nanofibers, are currently under development as effective approaches for augmenting and enhancing the regenerative functionality of total joint arthroplasties to enhance wound healing [20]. In particular, poly(caprolactone) (PCL) nanofiber scaffolds have been shown to support osteogenic differentiation of bone marrow stromal cells for orthopedic tissue engineering [21, 22]. Fibers may form a surface coating on an implant or a stand-alone nanofiber matrix, from which a desired molecule can be released while supporting new tissue formation [23]. Porous nanofiber scaffolds have been effective in delivering biologically active molecules such as growth factors [24, 25], short peptides [26], and antibiotics [27]. By releasing drugs locally, highly effective concentrations may be achieved while avoiding potentially toxic systemic concentrations. In the case of prophylactic designs against peri-prosthetic infections, localized antibiotic release is particularly appealing because microbes at the site of the implant will be exposed to antibiotic concentrations well above the mean inhibition concentration. This may avoid the development of antibiotic resistant bacterial strains in the microbe-rich gut [28].
In this work, nanofiber PCL scaffolds were loaded with two concentrations of rifampicin (RIF), and the RIF release kinetics from PCL scaffolds were measured. Then the bactericidal efficacy of PCL-RIF scaffolds was evaluated using Gram-positive and Gram-negative strains of bacteria—S. epidermidis and P. aeruginosa—in static in vitro conditions.
2 Materials and methods
2.1 Fabrication and characterization of nanofiber scaffolds
Control nanofiber scaffolds were fabricated by electrospinning a solution of PCL (12% w/v) and oleic acid (OLA) (0.36% w/v) in 3:1 (volume ratio) chloroform:methanol. RIF-encapsulating scaffolds were fabricated from a similar solution that contained RIF so that the final scaffold composition was 10 or 20% (w/w) RIF. The polymer solution was loaded into a glass syringe and fed to a 20-gauge blunt-tip needle by a syringe pump at a rate of 2.3–2.6 mL/h. A high-voltage power supply was used to apply voltage in the range of 18–21 kV to the blunt-tip needle that was positioned 10–11.5 cm from the grounded collector plate.
Scaffold morphology and fiber diameter were examined using a field-emission scanning electron microscope (SEM, JEOL JSM-6500F). Scaffolds were sputter-coated with 10 nm of gold and fiber diameters were measured using SEM image analysis software. At least 30 measurements were made on each scaffold (nmin = 30) and the fiber diameter distribution was plotted. Polymer crystallinity was determined using differential scanning calorimetry (DSC, TA Instruments DSC 2920). The scaffolds were heated from 5 to 120°C at 5°C/min and the crystallinity was calculated by the following equation:
| (1) |
where ΔHm,sample is the enthalpy of melting of the nanofiber scaffold and ΔHm,std is the enthalpy of melting of 100% crystalline PCL (ΔHm,std = 139 J/g) [29]. To control for the effect of electrospinning on crystallinity, polymer pellets that had not been subjected to electrospinning were used as controls and noted as “Source PCL”.
RIF encapsulation within PCL scaffolds was determined colorimetrically using UV–visible spectroscopy. 10 mm diameter PCL scaffolds, containing either 10 or 20% RIF, were weighed then placed in 1 mL chloroform. Next, 15 mL of methanol was added to precipitate the PCL and the solution was centrifuged at 4,300 rpm for 2 min. Absorbance values of the RIF solutions were measured at 357 nm (UV-1601, Shimadzu Scientific Instruments). Standard solutions of RIF in 1:15 chloroform:methanol were used to calculate the weight percent of RIF in the PCL scaffolds.
2.2 Rifampicin release from nanofibers
The release kinetics of RIF from PCL nanofibers was evaluated from 0 to 8 h in sterile phosphate buffered saline (PBS). 10 mm diameter circular discs were placed in 1 mL sterile PBS at 37°C and 100% humidity. At 1, 4 and 8 h, a 200-μL aliquot of PBS was removed and replaced with 200 μL of fresh PBS. Each aliquot was immediately stored at −80°C until the completion of the release study. The concentration of antibiotic in each aliquot was measured colorimetrically by measuring the absorbance at 475 nm by UV–visible spectroscopy. Concentration data points were correlated to the Peppas equation for diffusion-mediated release including a burst effect, which takes the form:
| (2) |
where t is a time point, Mt, and Mtot are the mass released at time, t, and the total mass, which means that Mt/Mtot is the fraction released at t. The constant B offsets the curve for an initial burst effect, k is a constant accounting for the ground matrix (polymer nanofibers) and drug (RIF) characteristics, and n is the diffusional exponent that accounts for the transport mechanism [30, 31].
2.3 Static bacterial challenge
Parent bacterial cultures were started via loop inoculation from stock agar plates into 5 mL of medium [10 g/L lysogeny broth (LB) for P. aeruginosa (PA 01) growth, 10 g/L trypticase soy broth (TSB) for S. epidermidis (SE RP62A) growth] and incubated overnight in a rotary shaker incubator at 37°C. Scaffolds were placed in 3 mL of LB or TSB and inoculated with 10 μL of overnight culture, which resulted in final cell suspension concentration of P. aeruginosa, 7.6 × 106 cells, and S. epidermidis, 1.8 × 107 cells, respectively. Each hour for 0–6 h, 100 μL aliquots of the cell suspension were removed, diluted in PBS, and filtered through a 100 nm black polycarbonate membrane (VWR). Trapped bacteria were then stained with Live/Dead BacLight™ stains (Invitrogen) and counted microscopically. Cells were not directly visualized on the scaffolds since the polymer scaffolds absorbed the Live/Dead stain. Live and dead bacteria were visualized at identical locations and the images were used to count live and dead bacteria using ImageJ software (NIH); cell counts were averaged over five random areas on each filter specimen (n = 3). In order to count individual bacteria, different serial dilutions were used for samples collected from the RIF-free and PCL-RIF scaffolds, and those dilutions were accounted for when calculating bacterial suspended concentrations. After 6 h of bacterial culture, scaffolds were fixed for SEM analysis in 3% glutaraldehyde, dehydrated in an ethanol wash series, then sputter coated with 7 nm of gold prior to SEM imaging.
The growth of the suspended bacterial population exposed to the PCL scaffolds was modeled assuming classical exponential population growth. The exponential growth constant, μ, for the population change rate was determined by the microbial population equation:
| (3) |
where N = suspended cell concentration (cell count/L3); μ = growth constant (time−1); t = time (t). When this equation is integrated over time, t, and the terms arranged to solve for μ it takes the form:
| (4) |
where N0 or Nt is the initial number of bacteria and the number at time = t, respectively.
2.4 Statistical analysis
All nanofiber scaffolds (RIF-free, 10% RIF and 20% RIF) were cultured and assayed in triplicate at each time point specified. A one-way ANOVA was performed to determine the significance of RIF concentration within the polymer nanofibers and, where appropriate, the effect of time as well as the interaction of RIF by time (RIF × time). Significance was set at P value <0.05. All statistics are presented here as a mean ± standard deviation.
3 Results
3.1 Fabrication and characterization of nanofiber scaffolds
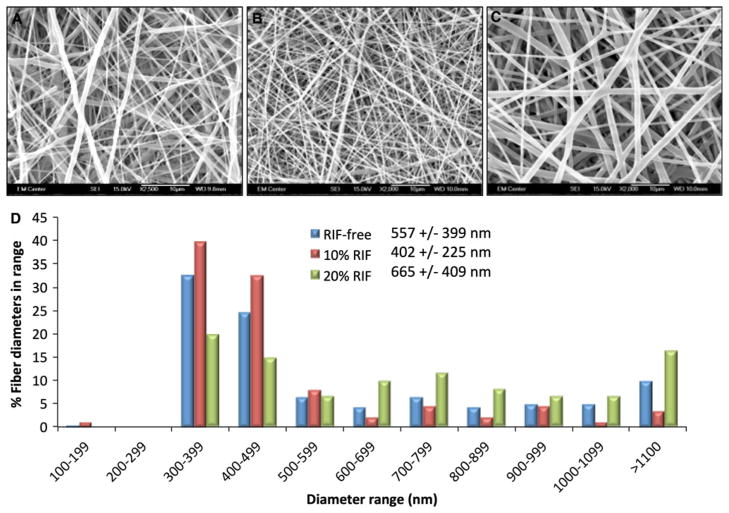
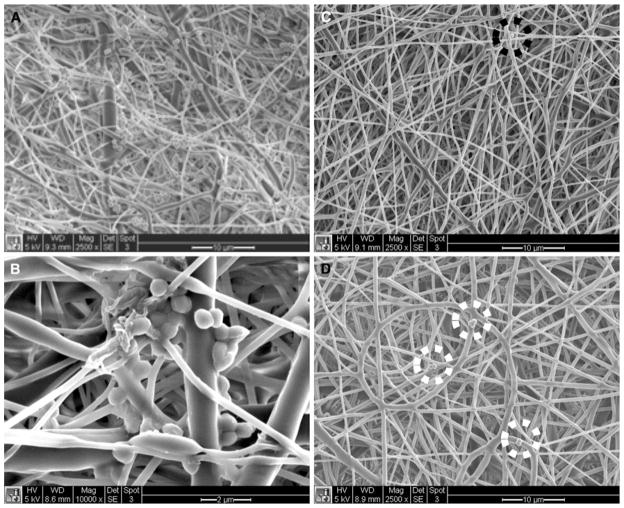
RIF-encapsulated PCL nanofiber scaffolds were successfully fabricated with two different RIF concentrations—10% or 20% (w/w). Control and RIF-encapsulated scaffolds were imaged and fiber diameters were measured under SEM. The 10% RIF scaffolds exhibited the narrowest distribution and smallest mean fiber diameter (Fig. 1). The 20% RIF scaffolds exhibited a broad distribution of fiber diameters, particularly with large fibers (>1 μm diameter). Although the average fiber diameters for RIF-free (557 ± 399 nm) and 20% RIF scaffolds (665 ± 407 nm) were similar, their distributions were different. The average fiber diameters for RIF-free and 10% RIF (402 ± 225 nm) scaffolds were different; however, both scaffolds exhibited fibers (>60%) with diameters between 300 and 500 nm. DSC results showed that the PCL in RIF-free scaffolds had the highest value of crystallinity of 67% and a t-test revealed that this difference was significant compared to both RIF-loaded scaffolds of ~55% (data not shown). However, PCL crystallinity between 10 and 20% RIF scaffolds was not significantly different.
Fig. 1.
Scanning electron micrographs of RIF-free (a: 557 ± 399 nm), 10% RIF (b: 402 ± 225 nm), and 20% RIF (c: 665 ± 409 nm) PCL nanofiber scaffolds. d Scaffold fiber diameters for the control and RIF-encapsulated scaffolds are represented as mean ± standard deviation
3.2 Rifampicin encapsulation and release from nanofibers
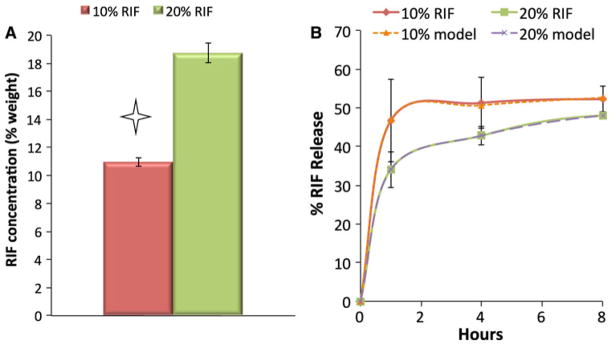
RIF encapsulation was quantified by precipitating PCL out of solution and then measuring the concentration of RIF colorimetrically against a standard curve. The 10% RIF scaffolds contained 11.0% (w/w) RIF and 20% RIF scaffolds 18.8% (w/w) (Fig. 2a). However, both were within ~1% (w/w) of their intended value, which is indicative of very efficient and accurate RIF loading. Loading data was used to calculate RIF release rates. The release kinetics of RIF from nanofiber scaffolds was quantified after 1, 4 and 8 h of incubation in PBS using UV–visible spectroscopy. RIF concentration was determined by measuring the absorbance at 475 nm then converting to concentration based a calibration curve. An adjustment accounting for the removed medium was used when calculating the mass released of RIF, and the percent released was fit to Eq. 2. Results in Fig. 2b show that both scaffolds initially released a burst of antibiotic, followed by slow, or minimal, drug release. The 10% RIF scaffolds released a greater percent of their theoretical loading, and the release occurred more rapidly compared to the 20% RIF scaffolds. The 10% RIF scaffolds released 51% of their total RIF mass content within the first 4 h, while the 20% RIF scaffolds released 48% of their RIF by 8 h. Cessation of RIF release was verified by sampling 1 day later (elapsed time 24 h), and neither scaffold released any additional RIF after the 8 h mark (data not shown). Models for both release profiles agreed well with experimental values over the entire 8 h.
Fig. 2.
a RIF loading, measured by UV–VIS spectroscopy. Star denotes statistically significant difference (P ≤ 0.05). b RIF release profiles from 0 to 8 h for 10 and 20% w/w RIF scaffolds. Percent of RIF mass released is presented
3.3 Static bacterial challenge
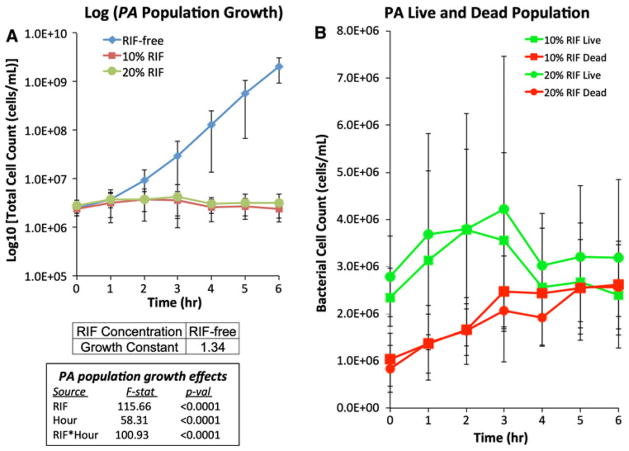
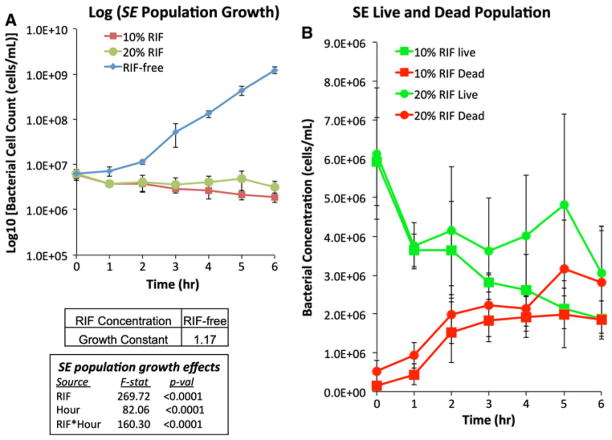
To determine the inhibitory effect of RIF release on suspended bacterial growth, scaffolds were inoculated with one of two species of bacteria, Gram-negative P. aeruginosa and Gram-positive S. epidermidis. Both bacterial species displayed uninhibited suspended growth in medium exposed to RIF-free scaffolds; the calculated growth constants, μ, were 1.17 and 1.34 for P. aeruginosa (Fig. 3a) and S. epidermidis (Fig. 4a), respectively. Over the course of the 6-h experiment, the number of dead P. aeruginosa suspended cells steadily increased while the number of suspended live P. aeruginosa cells only slightly increased for the first 3 h and then decreased (Fig. 3b). The total suspended cell concentration (both live and dead cells) of S. epidermidis exposed to either RIF-containing scaffold steadily decreased over the 6 h period, dropping to ~60% of the initial value (Fig. 4a). Dead S. epidermidis suspended cell number concentrations also increased steadily when exposed to either RIF-containing scaffold, while suspended live S. epidermidis cells steadily decreased (Fig. 4b).
Fig. 3.
Time-course measurements of suspended cell concentrations of Pseudomonas aeruginosa cultures (22.8 × 106 cells/mL) exposed to various scaffolds for 6 h at 37°C. a Total suspended cell concentrations exposed to RIF-free (diamond), 10%-RIF (square) and 20%-RIF (circle) scaffolds. b Live (green) and dead (red) suspended cell concentrations for cultures exposed to 10%-RIF (square) and 20%-RIF (circle) scaffolds
Fig. 4.
Time-course measurements of suspended cell concentrations of Staphylococcus epidermidis cultures (5.4 × 107 cells/mL) exposed to various scaffolds for 6 h at 37°C. a Total suspended cell concentrations exposed to RIF-free (diamond), 10%-RIF (square) and 20%-RIF (circle) scaffolds. (b) Live (green) and dead (red) suspended cell concentrations for cultures exposed to 10%-RIF (square) and 20%-RIF (circle) scaffolds
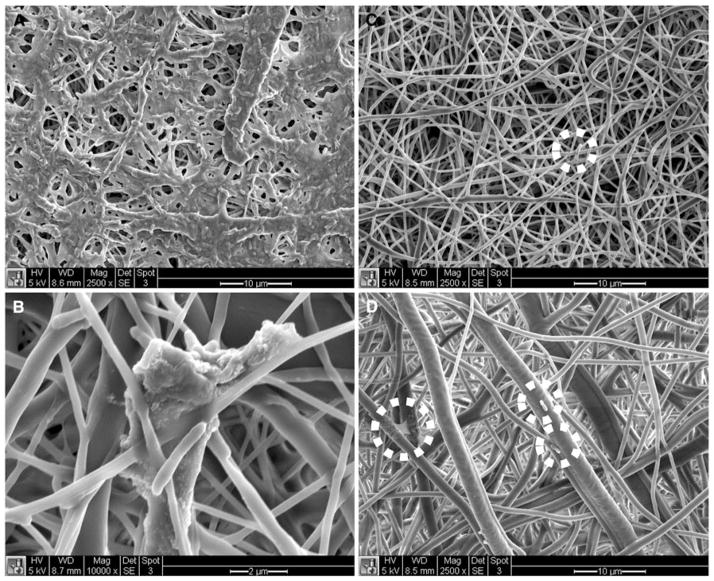
After 6 h, RIF-free and RIF-encapsulating scaffolds were fixed for SEM in order to analyze adherent cell accumulation for both P. aeruginosa and S. epidermidis.. On RIF-free scaffolds, both bacterial species multiplied prolifically and populated scaffold surfaces within dense colonies (Figs. 5a, b, 6a, b). The two bacterial strains behaved differently on RIF-free scaffolds as P. aeruginosa secreted abundant extracellular polysaccharides (EPS) on scaffold surfaces around bacterial colonies (Fig. 5a, b) and on the scaffold surface; the EPS produced by P. aeruginosa was so thick that it obstructed the view into the interior of the scaffolds over large portions of the surface (Fig. 5a). S. epidermidis produced EPS sparingly on the surface of the RIF-free scaffolds (Fig. 6a, b). On the 10 and 20% RIF scaffolds, no visible differences in bacterial populations were seen in the SEM images for either bacterial species. P. aeruginosa cells appeared as an occasional cell pair on the RIF-loaded scaffold surfaces, with minimal EPS secretion (Fig. 5c, d). Likewise, only occasional pairs of S. epidermidis cells were observed on either RIF-loaded scaffold with minimal EPS secretion (Fig. 6c, d).
Fig. 5.
SEM images of adherent Pseudomonas aeruginosa on RIF-free (a, b), 10% RIF (c), and 20% RIF (d) scaffolds 6 h after inoculation. Magnification for a, c, and d is 2,500× and scale bar is 10 μm. Magnification for b is 10,000× and scale bar is 2 μm
Fig. 6.
SEM images of adherent Staphylococcus epidermidis on RIF-free (a, b), 10% RIF (c), and 20% RIF (d) scaffolds 6 h after inoculation. Magnification for a, c, and d is 2,500× and scale bar is 10 μm. Magnification for b is 10,000× and scale bar is 2 μm
4 Discussion
The overall goal of this project was to develop a poly(caprolactone) (PCL) nanofiber tissue scaffold that can release a range of antibiotic molecules and provide supportive architecture for tissue regrowth once the threat of infection has been mitigated. In this study, rifampicin (RIF), an antibiotic with bactericidal action primarily against Gram-positive bacteria [32, 33], was incorporated at two weight concentrations, 10 and 20%, into the PCL electrospinning solution so that it could freely and quickly diffuse out of the polymer nanofibers. Successful RIF encapsulation was confirmed by UV–visible measurements (Fig. 2a). RIF encapsulation measurements (% weight) correlated very well with estimated loading, suggesting no loss of RIF during electrospinning. RIF encapsulation did have an effect on fiber crystallinity. PCL degradation occurs relatively slowly in the absence of hydrolyzing enzymes [34, 35], and the decrease in crystallinity due to RIF encapsulation may increase the degradation rate; hydrolysis of ester bonds in PCL occurs preferentially in amorphous regions [36, 37]. DSC analysis was applied to measure any differences in PCL crystallinity between the RIF-free and RIF-encapsulating PCL scaffolds. RIF-free scaffolds exhibited 67% crystallinity while 10% RIF and 20% RIF loaded scaffolds dropped to 54 and 58%, respectively, with no statistical differences between the scaffolds (data not shown). Approximating scaffold mass loss with new tissue deposition has been identified as an ideal characteristic of biodegradable scaffolds [38] and will be examined in the future along with mammalian cell culture.
Using SEM for high-magnification imaging, the fiber diameters and scaffold morphology were characterized (see Fig. 1). Fiber diameter can be important not only for changing the small-molecule release rate, but also for applications in treating or preventing infections related to orthopedic trauma or surgery because changes in fiber diameter affect some orthopedic cell phenotypes. Osteoblast and fibroblast phenotypes have demonstrated relative insensitivity to changes in fiber diameters over the ranges observed in all three of the scaffolds in this study (~200–800 nm) [39]. Thus, any of the three scaffolds would have morphologies supportive of tissue regrowth, as has been demonstrated previously [21].
Release profiles for both 10 and 20% RIF scaffolds exhibited a burst release for 1–2 h (Fig. 2b), followed by a period of nominal drug release up to 8 h, after which release ceased for both RIF scaffolds. This is a common observation in studies of small-molecule release from nano-scale substrates [40], and it affected the model parameters. RIF release from 10 and 20% RIF encapsulated scaffolds were fit to a diffusion-mediated release model (Eq. 2) [30]. The exponential variable, n, accounts for the diffusional characteristics of a soluble molecule in particular ground substrate geometries, and it is theoretically 0.45 for cylindrical substrates such as nanofibers. Multiple iterations of model fitting analyses were performed varying the burst release offset variable, B, between 0 and 50% of the t = 1 h release values for both RIF scaffolds. However, none of the iterations led to a fit of n close to 0.45, which is the theoretical value for cylindrical geometries of non-swellable matrices [30], and the best model fits were achieved with B set to a value of zero. The model parameters were accurate in fitting a predicted release profile lasting approximately 8 h so long as they were allowed to differ between the 10 and 20% RIF scaffolds and the exponential variable was not constrained to a value of 0.45. The values calculated by the statistical analysis software were 0.165 and 0.055 for 20 and 10% RIF scaffolds, respectively. This suggests that there may be a deviation from the assumption of diffusion-mediated release once 45–50% of the initial RIF mass has been released into the surrounding medium. Both PCL and RIF are hydrophobic, and therefore desorption may limit additional RIF release in these static conditions. Thus, future experiments examining the bactericidal efficacy of these scaffolds in dynamic conditions will also examine the RIF release profile.
RIF released from PCL nanofiber scaffolds was examined by inoculating the scaffold solution with either S. epidermidis or P. aeruginosa. These are two of the three most common strains present in peri-prosthetic infection cultures, appearing in 30 and 8% of such infections respectively. S. aureus infections occur at approximately the same rate as S. epidermis, and S. aureus differs physiologically from S. epidermis by expressing two coagulase enzymes [41]. Coagulases facilitate the fibrinogen conversion to fibrin, which is instrumental in quickly forming a pseudocapsule that protects S. aureus from immune cells [42]. The lysogeny broth used in these experiments lacks coagulase targets (i.e., Fibrinogen and prothrombin), and thus this key difference between S. aureus and S. epidermis would not have its protective effect for S. aureus.
The population growth on RIF-encapsulated scaffolds was compared against RIF-free control scaffolds. RIF released from PCL scaffolds clearly inhibited suspended bacterial growth through 6 h (Figs. 3, 4). A close examination of scaffolds under SEM showed that neither S. epidermidis nor P. aeruginosa were able to form biofilms on either scaffold containing RIF, while both strains were able to form biofilms on RIF-free scaffolds (Figs. 5a, b, 6a, b). In particular, P. aeruginosa formed thick biofilms on large portions of the scaffold surfaces (Fig. 5a). Interestingly, there did not appear to be any additional benefit with 20% RIF scaffolds over the 10% scaffolds. The closed batch nature of the experimental conditions may explain this observation.
The static, batch conditions permit a cumulative buildup of RIF in the liquid phase without any clearance except for small aliquots removed each hour. This is a key detail for interpreting the translation of these scaffolds into clinical settings. The static nature of this ex vivo model is appropriate for healing scenarios in which extracellular fluid is slowly cleared from the site of implantation. For example, space adjacent to cauterized tissues where blood flow is temporarily obstructed would have a slow RIF clearance, and this slow clearance would lead to sustained high RIF concentrations until blood flow was restored. However, highly dynamic environments would diffuse the RIF quickly into the blood stream where it would be metabolized by the liver and gall bladder [43]. Thus, further evaluation in dynamic systems should be pursued to determine the efficacy of scaffolds with 10 or 20% RIF against biofilm formation.
RIF is appealing because it can act against bacteria in any state of growth (i.e., dormant, actively growing, or intracellular) [44], and MIC50 are on the order of 0.004–0.008 μg/mL for S. epidermidis [45]. The mass released by 10% RIF and 20% RIF scaffolds after 4 h were 256 ± 77 μg and 445 ± 17 μg respectively. In 3 mL of broth, the mean concentrations of RIF were 85.33 μg/mL and 148 μg/mL, which are ~5 orders of magnitude greater than the MIC and minimum bactericidal concentration (MBC) for S. epidermidis. Thus, although the % RIF release did not increase past 50% for either scaffold, the masses released led to highly effective RIF concentrations in the static, closed batch experiments.
Clinically, oral RIF therapy is viewed with some skepticism primarily because bacteria can develop resistance with a relatively simple mutation in the β subunit of RNA poly-merase [46]. RIF-resistant strains of P. aeruginosa have demonstrated 50% minimum inhibitory concentrations (MIC50) of up to 1,600 μg/mL in vitro [47]. Additionally, oral dosage regiments need to increase in frequency because the serum half-life decreases due to increased metabolism rates as liver enzymes are up-regulated in response to the RIF administration. Consequently, oral RIF monotherapy is strongly discouraged, although multi-drug antibiotic regimens frequently involve RIF [48]. Taking these complications into consideration, it is conceivable that ‘old’ antibiotics such as RIF could find new use as locally delivered adjuvant therapy for prophylactic infection prevention.
Recently, several publications have employed polymer nanofibers as a platform for silver nanoparticle release as an antimicrobial design, showing that silver release takes place over the course of several weeks and is effective at inhibiting bacterial growth for S. epidermidis [49, 50]. The result that P. aeruginosa proliferates rapidly while also secreting EPS within 6 h in broth exposed to RIF-free scaffolds indicates that the initial hours after inoculation are critical for inhibiting biofilm formation. Should bacteria be permitted to form a biofilm before silver concentrations reached inhibitory levels, the bacteria would be partially protected from the bactericidal action of silver. However, both the 10 and 20% RIF scaffolds proved effective at inhibiting proliferation and EPS secretion through 8 h. Together, this suggests that an antimicrobial tissue scaffold could be effective by delivering two phases of antibiotic molecules—one that releases very quickly over a short time interval, and a second that releases more slowly over a long time interval.
5 Conclusions
PCL nanofiber scaffolds were fabricated to include either 10 or 20% (w/w) RIF, and the RIF release kinetics and bactericidal efficacies of the scaffolds were evaluated compared to RIF-free control scaffolds. There were significant differences between the RIF release profiles, though both scaffolds showed an initial burst release, and RIF release was completed after 8 h. Approximately, 50% of the loaded RIF remained entrapped within the scaffolds. Bacterial growth and EPS secretion was examined over 6 h of batch suspended bacterial growth. Bacterial growth in free suspension exposed to RIF-releasing scaffolds was significantly hindered compared to control materials. SEM images showed clear differences between bacterial growth on RIF-free and PCL-RIF scaffolds. Both bacterial species formed dense populations and secreted EPS on the RIF-free scaffolds. P. aeruginosa or S. epidermidis exhibited minimal colonization on both RIF scaffolds. Future work will build on these results by investigating the efficacy in dynamic culture conditions, and to incorporate a second phase of drug delivery to combat persistent bacteria.
Contributor Information
Timothy T. Ruckh, School of Biomedical Engineering, Colorado State University, Fort Collins, CO 80523-1376, USA
Rachael A. Floreani, Department of Bioengineering, University of Washington, Seattle, WA 98195-5061, USA
Derek A. Carroll, Department of Mechanical Engineering, Colorado State University, Campus Delivery, Fort Collins, CO 80523-1374, USA
Krasimira Mikhova, Department of Bioengineering, University of Washington, Seattle, WA 98195-5061, USA.
James D. Bryers, Department of Bioengineering, University of Washington, Seattle, WA 98195-5061, USA
Ketul C. Popat, Email: Ketul.Popat@colostate.edu, School of Biomedical Engineering, Colorado State University, Fort Collins, CO 80523-1376, USA. Department of Mechanical Engineering, Colorado State University, Campus Delivery, Fort Collins, CO 80523-1374, USA.
References
- 1.Kurtz SM, Lau E, Schmier J, Ong KL, Zhao K, Parvizi J. Infection burden for hip and knee arthroplasty in the United States. J Arthroplasty. 2008;23(7):984–91. doi: 10.1016/j.arth.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Bozic KJ, Ries MD. The impact of infection after total hip arthroplasty on hospital and surgeon resource utilization. J Bone Joint Surg Am. 2005;87(8):1746–51. doi: 10.2106/JBJS.D.02937. [DOI] [PubMed] [Google Scholar]
- 3.Mont M, Waldman B, Hungerford DS. Evaluation of preoperative cultures before second-stage reimplantation of a total knee prosthesis complicated by infection: a comparison-group study. J Bone Joint Surg. 2000;82:1552–7. doi: 10.2106/00004623-200011000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Shi Z, Neoh KG, Kang ET, Wang W. Antibacterial and mechanical properties of bone cement impregnated with chitosan nanoparticles. Biomaterials. 2006;27(11):2440–9. doi: 10.1016/j.biomaterials.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 5.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298(15):1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 6.Moyad TF, Thornhill T, Estok D. Evaluation and management of the infected total hip and knee. Orthopedics. 2008;31(6):581–8. doi: 10.3928/01477447-20080601-22. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Singletary R, Schmader K, Anderson DJ, Bolognesi M, Kaye KS. Surgical site infection in the elderly following orthopaedic surgery. J Bone Joint Surg. 2006;88:1705–12. doi: 10.2106/JBJS.E.01156. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Wang H, Kaplan JB, Lee WY. Microfluidic approach to create 3D tissue models for biofilm-related infection of orthopaedic implants. Tissue Eng Part C Methods. 2010 doi: 10.1089/ten.TEC.2010.0285. [DOI] [PubMed] [Google Scholar]
- 9.Campoccia D, Montanaro L, Arciola CR. The significance of infection related to orthopedic devices and issues of antibiotic resistance. Biomaterials. 2006;27(11):2331–9. doi: 10.1016/j.biomaterials.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 10.Francolini I, Donelli G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol Med Microbiol. 2010;59:227–38. doi: 10.1111/j.1574-695X.2010.00665.x. [DOI] [PubMed] [Google Scholar]
- 11.Hetrick EM, Schoenfisch MH. Reducing implant-related infections: active release strategies. Chem Soc Rev. 2006;35:780–9. doi: 10.1039/b515219b. [DOI] [PubMed] [Google Scholar]
- 12.Campoccia D, Montanaro L, Arciola CR. The significance of infection related to orthopedic devices and issues of antibiotic resistance. Biomaterials. 2006;27:2331–9. doi: 10.1016/j.biomaterials.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 13.Zhao L, Chu PK, Zhang Y, Wu Z. Antibacterial coatings on titanium implants. J Biomed Mater Res. 2009;91B:470–80. doi: 10.1002/jbm.b.31463. [DOI] [PubMed] [Google Scholar]
- 14.Gillespie WJ, Walenkamp GH. Antibiotic prophylaxis for surgery for proximal femoral and other closed long bone fractures. Cochrane Database Syst Rev. 2010;3:CD000244. doi: 10.1002/14651858.CD000244.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu VH, Crosslin DR, Friedman JY, Reed SD, Cabell CH, Griffiths RI, et al. Staphylococcus aureus bacteremia in patients with prosthetic devices: costs and outcomes. Am J Med. 2005;118(12):1416. doi: 10.1016/j.amjmed.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Bliziotis IA, Ntziora F, Lawrence KR, Falagas ME. Rifampin as adjuvant treatment of Gram-positive bacterial infections: a systematic review of comparative clinical trials. Eur J Clin Microbiol Infect Dis. 2007;26(12):849–56. doi: 10.1007/s10096-007-0378-1. [DOI] [PubMed] [Google Scholar]
- 17.Trampuz A, Widmer AF. Infections associated with orthopedic implants. Curr Opin Infect Dis. 2006;19(4):349–56. doi: 10.1097/01.qco.0000235161.85925.e8. [DOI] [PubMed] [Google Scholar]
- 18.van de Belt H, Neut D, Schenk W, van Horn JR, van der Mei HC, Busscher HJ. Infection of orthopedic implants and the use of antibiotic-loaded bone cements. Acta Orthop Scand. 2001;72:557–71. doi: 10.1080/000164701317268978. [DOI] [PubMed] [Google Scholar]
- 19.Nablo BJ, Prichard HL, Butler RD, Klitzman B, Schoenfisch MH. Inhibition of implant-associated infections via nitric oxide release. Biomaterials. 2005;26(34):6984–90. doi: 10.1016/j.biomaterials.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Ko EK, Jeong SI, Rim NG, Lee YM, Shin H, Lee BK. In vitro osteogenic differentiation of human mesenchymal stem cells and in vivo bone formation in composite nanofiber meshes. Tissue Eng Part A. 2008;14(12):2105–19. doi: 10.1089/ten.tea.2008.0057. [DOI] [PubMed] [Google Scholar]
- 21.Ruckh TT, Kumar K, Kipper MJ, Popat KC. Osteogenic differentiation of bone marrow stromal cells on poly(epsilon-caprolactone) nanofiber scaffolds. Acta Biomater. 2010;6(8):2949–59. doi: 10.1016/j.actbio.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimoto H, Shin YM, Terai H, Vacanti JP. A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials. 2003;24(12):2077–82. doi: 10.1016/s0142-9612(02)00635-x. [DOI] [PubMed] [Google Scholar]
- 23.Sill TJ, von Recum HA. Electrospinning: applications in drug delivery and tissue engineering. Biomaterials. 2008;29(13):1989–2006. doi: 10.1016/j.biomaterials.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Choi JS, Leong KW, Yoo HS. In vivo wound healing of diabetic ulcers using electrospun nanofibers immobilized with human epidermal growth factor (EGF) Biomaterials. 2008;29(5):587–96. doi: 10.1016/j.biomaterials.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Schneider A, Wang XY, Kaplan DL, Garlick JA, Egles C. Biofunctionalized electrospun silk mats as a topical bioactive dressing for accelerated wound healing. Acta Biomater. 2009;5(7):2570–8. doi: 10.1016/j.actbio.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartman O, Zhang C, Adams EL, Farach-Carson MC, Petrelli NJ, Chase BD, et al. Biofunctionalization of electrospun PCL-based scaffolds with perlecan domain IV peptide to create a 3-D pharmacokinetic cancer model. Biomaterials. 2010;31(21):5700–18. doi: 10.1016/j.biomaterials.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim K, Luu YK, Chang C, Fang D, Hsiao BS, Chu B, et al. Incorporation and controlled release of a hydrophilic antibiotic using poly(lactide-co-glycolide)-based electrospun nanofibrous scaffolds. J Control Release. 2004;98(1):47–56. doi: 10.1016/j.jconrel.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 28.de Vries LE, Vallès Y, Agersø Y, Vaishampayan PA, García-Montaner A, Kuehl JV, et al. The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PLoS ONE. 2011;6(6):e21644. doi: 10.1371/journal.pone.0021644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guarino V, Taddei P, Di Foggia M, Fagnano C, Ciapetti G, Ambrosio L. The influence of hydroxyapatite particles on in vitro degradation behaviour of Pcl based composite scaffolds. Tissue Eng Part A. 2009;15:3655–68. doi: 10.1089/ten.TEA.2008.0543. [DOI] [PubMed] [Google Scholar]
- 30.Ritger P, Peppas NA. A simple equation for description of solute release I. Fickian and non-Fickian release from non-swellable devices in the form of slabs, spheres, and cylinders or discs. J Control Release. 1987;5:23–36. [PubMed] [Google Scholar]
- 31.Sinclair G, Peppas N. Analysis of non-Fickian transport in polymers using simplified exponential expressions. J Membr Sci. 1984;17:329–31. [Google Scholar]
- 32.Lavicky J, Urbanova Z, Raskova H, Rotta J, Vanecek J. The effects of peptidoglycan, a pyrogenic constituent of gram-positive microorganisms, on the pharmacokinetics of rifampicin. Toxicon. 1988;26(3):293–300. doi: 10.1016/0041-0101(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 33.Pohlod DJ, Saravolatz LD, Somerville MM. In vitro susceptibility of gram-positive cocci to LY146032 teicoplanin, sodium fusidate, vancomycin, and rifampicin. J Antimicrob Chemother. 1987;20(2):197–202. doi: 10.1093/jac/20.2.197. [DOI] [PubMed] [Google Scholar]
- 34.Hoshino A, Isono Y. Degradation of aliphatic polyester films by commercially available lipases with special reference to rapid and complete degradation of poly(L-lactide) film by lipase PL derived from Alcaligenes sp. Biodegradation. 2002;13(2):141–7. doi: 10.1023/a:1020450326301. [DOI] [PubMed] [Google Scholar]
- 35.Zeng J, Chen X, Liang Q, Xu X, Jing X. Enzymatic degradation of poly(L-lactide) and poly(epsilon-caprolactone) electrospun fibers. Macromol Biosci. 2004;4(12):1118–25. doi: 10.1002/mabi.200400092. [DOI] [PubMed] [Google Scholar]
- 36.Pitt C, Gratzl M, Kimmel C, Surles J, Schindler A. Aliphatic polyesters II. The degradation of poly (DL-lactide), poly (epsilon-caprolactone), and their copolymers in vivo. Biomaterials. 1981;2(4):215–20. doi: 10.1016/0142-9612(81)90060-0. [DOI] [PubMed] [Google Scholar]
- 37.Pulkkinen M, Malin M, Tarvainen T, Saarimäki T, Seppälä J, Järvinen K. Effects of block length on the enzymatic degradation and erosion of oxazoline linked poly-ε-caprolactone. Eur J Pharm Sci. 2007;31(2):119–28. doi: 10.1016/j.ejps.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Ishaug SL, Crane GM, Miller MJ, Yasko AW, Yaszemski MJ, Mikos AG. Bone formation by three-dimensional stromal osteo-blast culture in biodegradable polymer scaffolds. J Biomed Mater Res. 1997;36(1):17–28. doi: 10.1002/(sici)1097-4636(199707)36:1<17::aid-jbm3>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 39.Bashur CA, Dahlgren LA, Goldstein AS. Effect of fiber diameter and orientation on fibroblast morphology and proliferation on electrospun poly(D, L-lactic-co-glycolic acid) meshes. Biomate-rials. 2006;27(33):5681–8. doi: 10.1016/j.biomaterials.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Venugopal J, Prabhakaran MP, Low S, Choon AT, Deepika G, Dev VR, et al. Continuous nanostructures for the controlled release of drugs. Curr Pharm Des. 2009;15(15):1799–808. doi: 10.2174/138161209788186344. [DOI] [PubMed] [Google Scholar]
- 41.Bjerketorp J, Jacobsson K, Frykberg L. The von Willebrand factor-binding protein (vWbp) of Staphylococcus aureus is a coagulase. FEMS Microbiol Lett. 2004;234(2):309–14. doi: 10.1016/j.femsle.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 42.Cheng AG, McAdow M, Kim HK, Bae T, Missiakas DM, Schneewind O. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog. 2010;6(8):e1001036. doi: 10.1371/journal.ppat.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L, Xu Y, Shea C, Fowler JS, Hooker JM, Tonge PJ. Radio-synthesis and bioimaging of the tuberculosis chemotherapeutics isoniazid, rifampicin and pyrazinamide in baboons. J Med Chem. 2010;53(7):2882–91. doi: 10.1021/jm901858n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frippiat F, Meunier F, Derue G. Place of newer quinolones and rifampicin in the treatment of Gram-positive bone and joint infections. J Antimicrob Chemother. 2004;54(6):1158. doi: 10.1093/jac/dkh451. [DOI] [PubMed] [Google Scholar]
- 45.Pohlod DJ, Saravolatz LD, Somerville MM. In vitro susceptibility of Gram-positive cocci to LY146032 teicoplanin, sodium fusi-date, vancomycin, and rifampicin. J Antimicrob Chemother. 1987;20:197–202. doi: 10.1093/jac/20.2.197. [DOI] [PubMed] [Google Scholar]
- 46.Tupin A, Gualtieri M, Roquet-Baneres F, Morichaud Z, Brodolin K, Leonetti JP. Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. Int J Anti-microb Agents. 2010;35(6):519–23. doi: 10.1016/j.ijantimicag.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 47.Jatsenko T, Tover A, Tegova R, Kivisaar M. Molecular characterization of Rifr mutations in Pseudomonas aeruginosa and Pseudomonas putida. Mutation Res. 2010;683:106–14. doi: 10.1016/j.mrfmmm.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 48.Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. N Engl J Med. 2004;351(16):1645–54. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]
- 49.Xing Z-C, Chae W-P, Baek J-Y, Choi M-J, Jung Y, Kang I-K. In vitro assessment of antibacterial activity and cytocompatibility of silver-containing PHBV nanofibrous scaffolds for tissue engineering. Biomacromolecules. 2010;11:1248–53. doi: 10.1021/bm1000372. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Lin T, Fang J, Yao G, Zhao H, Dodson M, et al. In vivo wound healing and antibacterial performances of electrospun nanofibre membranes. J Biomed Mater Res. 2010;94A:499–508. doi: 10.1002/jbm.a.32718. [DOI] [PubMed] [Google Scholar]