CONSPECTUS
The potential of gene therapy to benefit human health is tremendous because almost all human diseases have a genetic component, from untreatable monogenic disorders to cancer and heart disease. Unfortunately, a method for gene therapy that is both effective and safe has remained elusive. It has been said that “there are only three problems in gene therapy - delivery, delivery, and delivery.” (quote from I. M. Verma in Jaroff, L. TIME, 1999; Jan 11).
This Account describes an alternative strategy to viral gene delivery: the design of biodegradable polymers that are able to deliver DNA like a synthetic virus. Using high-throughput synthesis and screening techniques, we have created libraries of over 2000 structurally unique poly(β-amino esters) (PBAEs). PBAEs are formed by the conjugate addition of amines to diacrylates. These biomaterials are promising for nonviral gene delivery due to their ability to condense plasmid DNA into small and stable nanoparticles and their ability to promote cellular uptake and endosomal escape. Our laboratory has iteratively improved PBAE nanoparticles through polymer end modifications and nanoparticle coatings. Lead PBAEs have high gene delivery efficacy and low cytotoxicity both in vitro and in vivo.
Certain polymer structural characteristics are important for effective gene delivery. The best PBAEs are linear polymers of ~10 kDa that contain hydroxyl side chains and primary amine end groups. These polymers bind DNA to form nanoparticles that are small (<200 nm) and stable and have near-neutral ζpotential in the presence of serum-containing media. Lead PBAEs also contain tertiary amines that can buffer the low pH environment of endosomes and facilitate escape of polymer/DNA particles into the cytoplasm.
Diamine end-modified 1,4-butanediol diacrylate-co-5-amino-1-pentanol polymers (C32) bind DNA more tightly and form smaller nanoparticles than other PBAEs. These nanoparticles also have higher cellular uptake and the best gene expression of all gene delivery polymers in the library. These polymers are more effective for gene delivery than top commercially available nonviral vectors including jet-PEI and Lipofectamine 2000 and are comparable to adenovirus for in vitro gene delivery to human primary cells. In vivo, these PBAE/ DNA particles are promising as cancer therapeutics. This Account summarizes the results of our laboratory in using a combinatorial polymer library approach to elucidate polymer structure/function relationships and enable the development of polymeric gene delivery nanoparticles with viral-like efficacy.
Introduction
There are two approaches to gene delivery: viral and nonviral. Viral delivery is the more conventional approach because viruses have evolved to infect cells with high efficacy. As of July 2007, there have been over 1300 gene therapy clinical trials and 70% of these trials use a viral vector.1 While there have been some successes using viral gene therapy, there are still no FDA-approved products. Clinical trials have underscored the safety risks of viral gene therapy as viral approaches have caused cancer and death.2,3 For these reasons, new attention has been focused on nonviral approaches for gene therapy as these have the potential to overcome many of the inherent challenges of viral vectors.
Nonviral gene delivery systems make use of physical methods or a synthetic chemical vector or both to deliver the gene of interest. Limitations with viral approaches, including small cargo capacity, resistance to repeated infection, difficulty in production and quality control, and low safety, can be potentially overcome with a nonviral approach. Originally, nonviral gene delivery was simply delivery of naked plasmid DNA. To improve transfection, physical strategies such as hydrodynamic delivery, electroporation, a gene gun, ultrasound, jet injection, magnetofection, and photochemical internalization have been used.4 Though these techniques can aid in delivery of nucleic acids, there are problems due to inefficient uptake and poor biostability.
Numerous biomaterials have been studied as potential nonviral gene delivery vectors to enable improved DNA stability and uptake including inorganic surfaces, cationic lipids, polysaccharides, cationic polymers, and dendrimers.5,6 These biomaterials either bind to, complex with, or encapsulate DNA into systems that are comparatively easier to manufacture and scale-up than viral systems, although they have orders of magnitude lower efficacy. To rationally design a synthetic virus to deliver therapeutic genes, researchers have attempted to deconstruct the overall process into specific mechanistic steps of transport to identify delivery bottlenecks. The putative mechanism of nonviral gene delivery is shown in Figure 1. First, a biomaterial must bind to and condense or encapsulate DNA (1) to protect it from degradation and facilitate cellular uptake of the DNA-containing particle. Following endocytosis (2), the DNA-containing particle is inside the cell but is in the endosomal compartment instead of the cytoplasm. As the particle trafficks through the cell (3), it must also have a mechanism for escape to the cytoplasm (4). Finally, the DNA must unpack from the particle (5) and be imported into the nucleus (6) for gene expression to occur (7).
FIGURE 1.
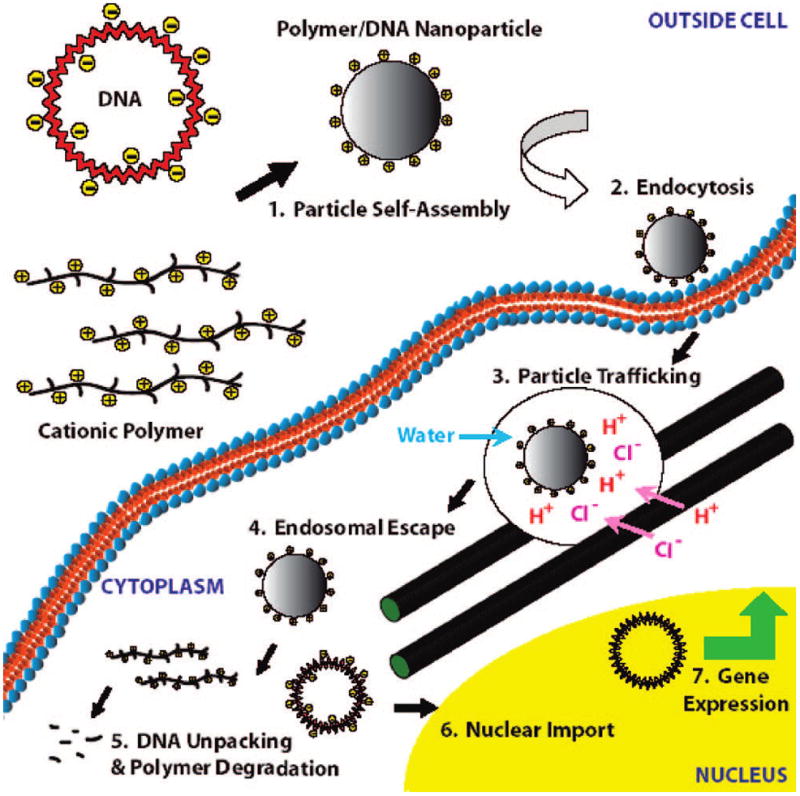
Mechanism of nonviral gene delivery.
Polymers for Gene Delivery
Synthetic polymers are promising because they allow a high level of design flexibility for biomaterial construction. Typically, cationic polymers are used because they can electrostatically bind to and condense anionic DNA into nanometersized particles. Two “off-the-shelf” cationic polymers that have been investigated for gene delivery ability include poly(L-lysine) (PLL) and polyethylenimine (PEI) and are shown in Figure 2. While polymeric gene delivery is promising due its large design flexibility, overall efficacy is typically orders of magnitude lower than that of viral gene delivery.
FIGURE 2.
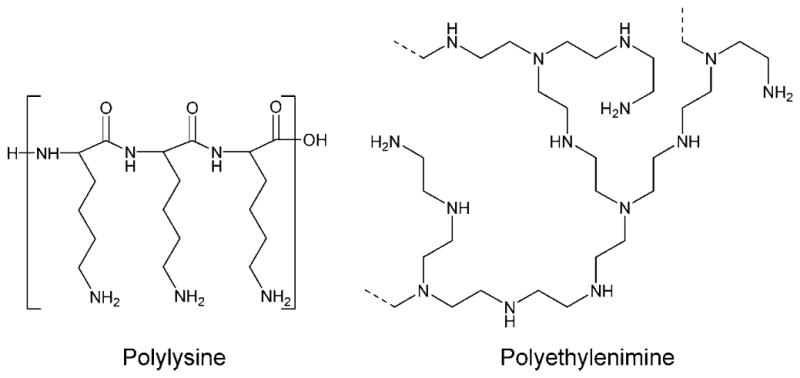
Structures of “off-the-shelf” gene delivery polymers.
A major advance in the field of nonviral gene delivery was the demonstration by Boussif et al. that polyethylenimine (PEI) could successful deliver DNA in vitro and in vivo.7 PEI was chosen as a vector because it has the highest cationic-charge-density potential of any organic macromolecule. PEI is more effective than PLL due to a high density of near-neutral pKa groups that can buffer the acidic environment of the endosome and facilitate endosomal escape through the “proton sponge” mechanism.8 PEI is not ideal, however, because its efficacy is still considerably lower than that of a virus, it is not biodegradable, and it causes considerable cytotoxicity via necrosis and apoptosis.9
Poly(β-amino esters)
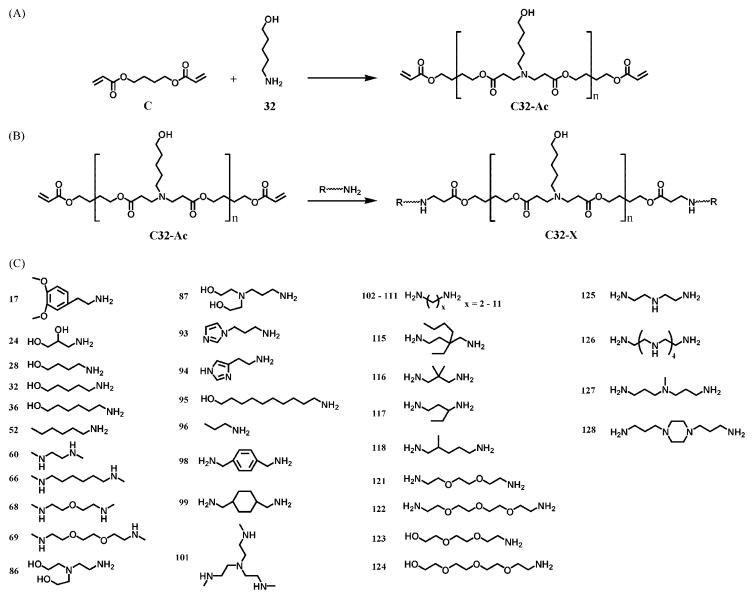
One particularly interesting class of polymers for gene delivery is poly(β-amino esters), which were first developed by David Lynn and co-workers in our laboratory in 2000.10 Poly(β-amino esters) (PBAEs) are promising compared with PEI due to their biodegradability via hydrolytically degradable ester groups, their reduced cytotoxicity, their ability for triggered DNA release within the cell, and their potential for structural diversity.10,11 They are easily synthesized by the conjugate addition of amine monomers to diacrylates, a one step reaction without the production of any byproducts (Figure 3).
FIGURE 3.
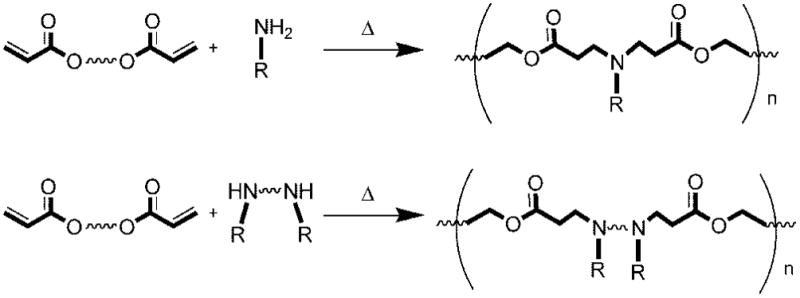
Synthesis of PBAEs by conjugate addition of amines to diacrylates.
Initial studies of this polymer class showed that by varying synthesis conditions the polymer molecular weight can be varied from 2000 to 50000 Da.11 In physiological conditions, these polymers have a degradation half-life on the order of hours, but this is slowed considerably at a pH of 5 or when the polymers electrostatically condense DNA and form nanoparticles.10 Importantly, it was demonstrated that a wide range of PBAEs were able to self-assemble with DNA to form cationic nanoparticles.10,12 Investigative studies on the proton sponge mechanism and endosomal release by Akinc et al. also revealed that PBAEs can successfully buffer the endosomal compartment, similarly to PEI.12,13 A library of 140 PBAEs composed of 7 diacrylate monomers and 20 amine monomers was synthesized in parallel and screened for gene delivery efficacy.11,12 In general, the best performing complexes were found to have effective diameters smaller than 250 nm and positive ζpotentials in 10 mM HEPES buffer. While it was determined that the majority of PBAE/DNA particles tested were limited by poor cell uptake, two of the polymers had high uptake and mediated gene delivery 4–8-fold higher than PEI, comparable to the efficacy of Lipofectamine 2000. These studies demonstrated the utility of using parallel synthesis and screening of a polymer library to identify novel gene delivery polymers with efficacy greater than PEI. PBAEs that were poor transfection agents failed for a variety of reasons.12 Some polymers were simply not sufficiently water soluble, while others were unable to electrostatically bind DNA tightly enough to prevent its movement during gel electrophoresis. Of the polymers that could bind DNA, many formed particles with low cellular uptake. Of those that had sufficient cellular uptake, some particles also caused too high cytotoxicity. Two leading PBAEs were further optimized by varying polymer molecular weight, polymer end group, and polymer to DNA weight ratio.14 Interestingly, it was also discovered that polymers terminated with diacrylate monomers were unable to deliver DNA to cells, unlike the nearly identical polymers that were terminated with amine monomers instead.
High-Throughput Synthesis and Testing of a Polymer Library
Based on these preliminary studies with a small library of polymers, in 2003, we created a large library of 2350 structurally unique PBAEs using automation and high-throughput combinatorial chemistry.15 To achieve lower viscosity of the reagent solutions for high-throughput synthesis, DMSO was chosen as a solvent since it is well-tolerated by cells. All monomers (Figure 4) were diluted to 1.6 M in DMSO, and then a fluid-handling robot and a 12-channel micropipette were used to automate the mixing of monomers allowing us to set up all 2350 reactions in a single day. This large library of polymers was screened to determine which polymers could efficiently bind DNA and transfect COS-7 cells in serum-free conditions, an easy-to-transfect cell system useful for high-throughput biological assays. From this study, 46 PBAEs were found to transfect as well as or better than PEI.15 The best-performing polymers were next analyzed and refined to a library of 486 second-generation polymers for a more detailed investigation of structure/function relationships.16
FIGURE 4.
Diacrylate monomers (letters) and amine monomers (numbers) used to synthesize PBAEs.
Nanoparticle Biophysical Properties are Dependent on Polymer Structure
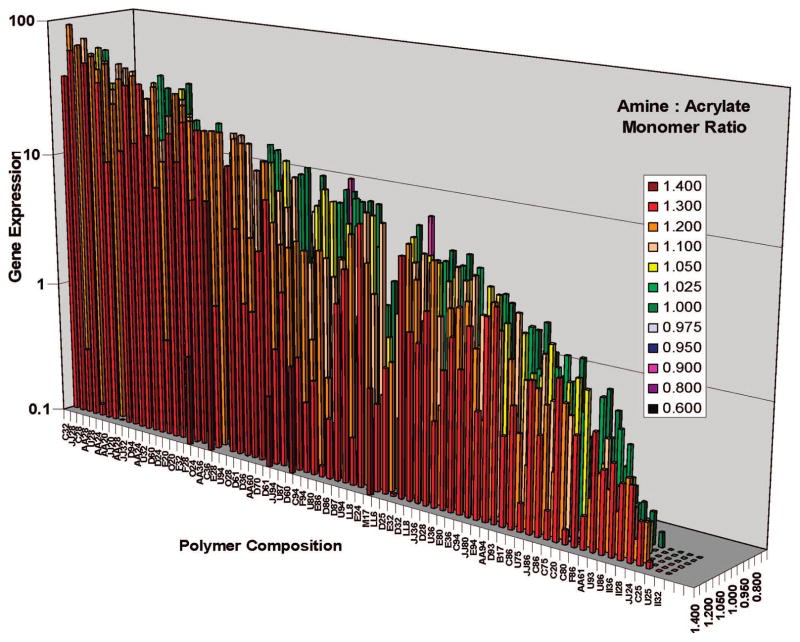
The first step of nonviral polymeric gene delivery is formation of a gene delivery nanoparticle. For efficient transfection, it is crucial that DNA is condensed into a particle to protect the DNA and allow for efficient uptake.17,18 Analysis of the second-generation poly(β-amino ester) library revealed that the top performing polymer complexes had sizes smaller than 150 nm and positive ζpotentials when biophysical properties were measured in buffer.16 Optimal formulations also tended to have polymer weight to DNA weight ratios greater than or equal to 40, molecular weights above 10000 Da, and other structural similarities. A comparison of the gene delivery efficacy of the top polymers in the PBAE library can be seen in Figure 5 as a function of monomer type and ratio. The top nine polymers were all formed from amino alcohols and hydrophobic diacrylate monomers, and the three best performing polymers (C32, JJ28, and C28) converged in structure (Figure 6).
FIGURE 5.
Gene delivery efficacy of the polymer library. COS-7 cells were transfected with polymer/DNA particles. Amine/acrylate monomer ratios below 1:1 generally produced polymers with much lower gene delivery and were not measured for all PBAEs. Reproduced with permission from ref 16.
FIGURE 6.
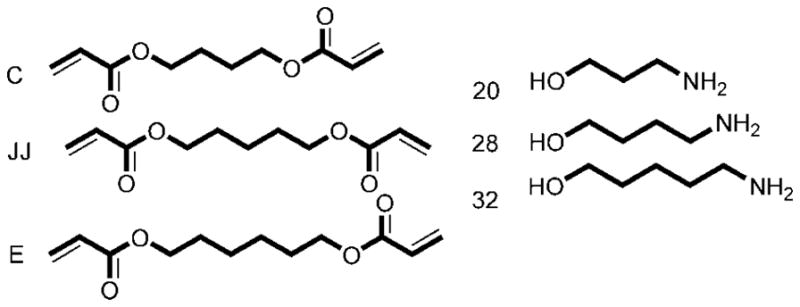
Subset of diacrylate and amine alcohol monomers especially promising for gene delivery. Interestingly, these monomers converge in structure to differ by only single carbons.
Initial high-throughput screening experiments took place in a relatively easy to transfect COS-7 cell line in serum-free conditions. Because it has been reported that nonviral vector formulations can have lower efficacy in the presence of serum,19,20 we wanted to investigate this relationship for leading PBAEs from the polymer library. Whereas all leading PBAE polymers condensed DNA into small nanoparticles below 150 nm while in buffer, particle size dramatically changed when these particles were sized in serum.21 Some polymers (formed from the monomers shown in Figure 6) formed small, stable particles in the 200 nm range, whereas others with differential polymer structure aggregated in serum to micrometer-sized particles. ζ-Potential was found to change from positive in PBS to negative in serum, likely due to negatively charged serum proteins adsorbing to the positively charged nanoparticles for an overall net negative surface charge. Interestingly, the difference in ζpotential and size between similarly structured polymers in serum correlated to single atom changes to constituent monomer structure (Figure 7). Biophysical particle properties in serum-containing media but not in buffer were found to be predictive of gene delivery efficacy to hard-to-transfect primary human umbilical vein endothelial cells (HUVECs) in serum-containing media.21 This analysis highlights the use of a large polymeric library in elucidating structure/function relationships.
FIGURE 7.
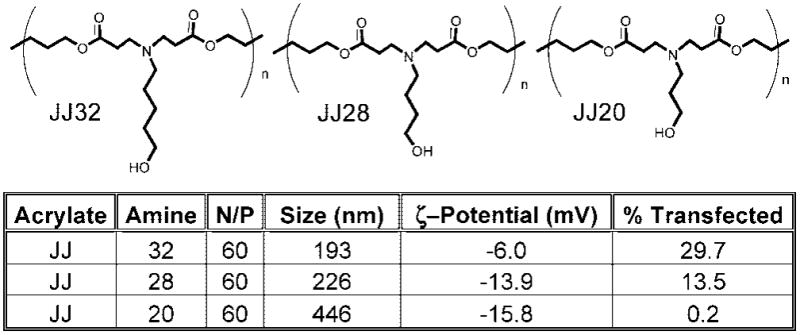
Single carbon differences to polymer structure change nanoparticle biophysical properties in serum and can lead to dramatic differences in transfection efficacy of HUVECs in serum. N/P is the ratio of polymer amines (N) to DNA phosphates (P).
Designing Polymeric Nanoparticles for Increased Uptake and Ligand-Specific Delivery
Once the particles are formed, the next key step in delivery is cellular uptake. Uptake can be nonspecific or targeted to specific receptors on cell types of interest. For efficient nonspecific uptake, it is important for the complexes to have an overall positive charge so that they will be electrostatically attracted to the negatively charged proteoglycan cell surface.22 It has been shown that the size of the gene delivery particles influences uptake and small particles, ~100 nm, are taken up the best.18 However, there are conflicting reports over whether smaller or larger particles exhibit higher overall transfection.18,19 This demonstrates the potential difficulty of analysis of these complexes because biophysical parameters such as particle size are coupled together with other parameters such as polymer molecular weight and charge ratio. This example also highlights the differences between an in vitro experiment, where larger particles could sediment and increase transfection, and a systemic in vivo application, where larger particles would be problematic for physical delivery to distant sites. An exception to this is delivery to the lung, where it has been shown that large aggregating PEI/DNA particles passively target the lung following tail-vein injection.23,24 For receptor-specific endocytosis, ligands such as epidermal growth factor (EGF)25 and RGD peptide26 have been utilized to increase cell specificity and overall uptake in vitro and in vivo. Though targeting often increases transfection, some experiments show that uptake that is both specific and efficient only occurs at a narrow window of charge ratios or not at all.25,26
To generate cell-specific targeting, we have looked at two complementary approaches for attaching targeting ligands to our polymeric gene delivery nanoparticles. The first approach is the covalent modification of the polymer side chain to allow for facile ligand incorporation. To accomplish this, Gregory Zugates and co-workers created a novel PBAE with a new amine monomer, 2-(pyridyldithio)-ethylamine (PDA).27 These cationic, degradable polymers contain pyridyldithio functionalities in the side chains that react with high specificity toward thiol ligands.
One potential issue with covalently coupling targeting ligands to a polymer is that it can alter the biophysical properties and gene delivery efficacy of the corresponding polymeric nanoparticle. For example, researchers have shown that as ligand substitution increases, overall gene delivery can decrease. This is presumably due to alteration of the original polymer’s functionality for DNA condensation, endosomal buffering, or both.28,29 To add targeting to gene delivery nanoparticles without changing the polymer’s functionality, we have developed an electrostatic coating approach where the positively charged nanoparticles self-assemble with anionic peptide coats that contain targeting ligands (Figure 8). These particles are small (100–200 nm) and stable in the presence of serum proteins as determined by TEM (Figure 8).30 This method is advantageous because in addition to incorporating ligand-specific delivery into the nanoparticle, the overall charge ratio of the nanoparticle can be tuned from cationic to neutral, which can lower nonspecific gene delivery as well as reducing potential serum protein interactions.31 When the overall charge ratio nears neutrality, poly(glutamic acid)–polyglycine–RGD coated nanoparticles transfect human endothelial cells an order of magnitude better than our negative control, the same nanoparticles coated with the near identical poly(glutamic acid)–polyglycine–RDG scrambled sequence.30
FIGURE 8.
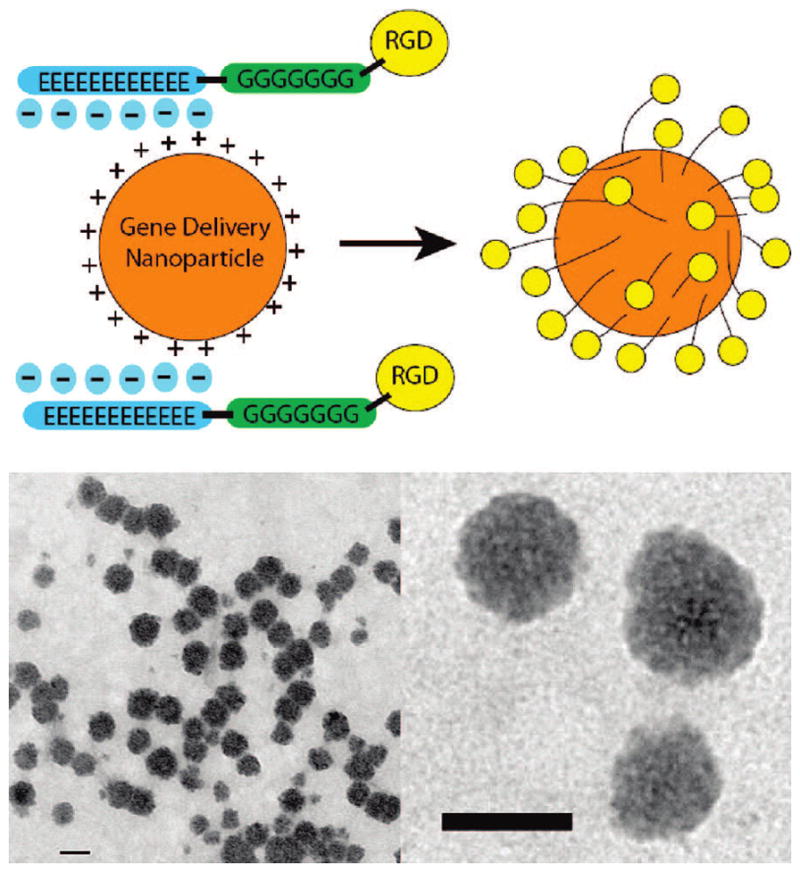
Electrostatic coating of cationic nanoparticles (top) with anionic ligand-containing peptides and TEM images (bottom) of C32 particles coated with poly(glutamic acid)–polyglycine–RGD peptides in serum media. Scale Bar is 100 nm in both figures. Reproduced with permission from ref 30. Copyright 2007 American Chemical Society.
Polymer Structure Determines Endosomal Release to Cytoplasm
During uptake and internalization, particles are packaged into endosomes and become part of the endosomal sorting pathway. Without any action by the vector, DNA would typically be degraded by the acidic environment of late endosomes, enzymatically degraded by lysosomes, or recycled out of the cell. In contrast, PEI vectors have enhanced gene delivery due to their ability to directly enable endosomal escape through the “proton sponge” mechanism.7,13 We have found that when PBAEs are designed for gene delivery, polymer structure can be tuned to similarly promote endosomal escape.
Akin Akinc and co-workers in our laboratory were able to probe the endosomal compartment and the proton sponge mechanism by double labeling plasmid DNA with both pH-insensitive and pH-sensitive fluorophores.13 We found that the ratio of pH-sensitive to pH-insensitive fluorescence determined the average pH environment of the delivered DNA and was indicative of the compartment of the DNA: whether the DNA was trafficked primarily to acidic lysosomes (polylysine vectors had an average pH of 4.5) or whether the DNA was able to escape this pathway (PEI vectors had an average pH of 5.9). When using this assay to investigate PBAEs from the library, we find the average pH environment of the delivered DNA can range from 4.0 to greater than 7.0, depending on polymer structure. Interestingly, the PBAEs with the highest gene delivery efficacy were all able to maintain a near-neutral pH environment (pH 6.5–7.0) for the delivered DNA.12 Polymers containing imidazole groups (pKa ≈ 6.2) are particularly good at facilitating endosomal escape.12,32 This “proton sponge” mechanism was recently proven by Sonawane et al. who showed that endosomes containing vectors with physiologically titratable amine groups like those formed from PEI but not vectors lacking these groups like vectors formed from polylysine buffered H+ ions, increased the counterion Cl− concentration within the endosome, swelled the endosome in size, and led to lysis of the endosome.8 This process is shown in Figure 1. The studies mentioned above highlight the importance of polymer structure including physiologically titratable amine groups in polymeric vector construction.
Polymer Structure Determines DNA Binding and Unpacking
Polymer/DNA binding affinity is important for self-assembly of small and stable polymeric gene delivery nanoparticles. To initially screen the PBAE library for polymers that could efficiently bind and condense DNA into a particle, a high-throughput electrophoretic DNA-binding assay was utilized.11 Investigation showed that molecular weight and structure can be correlated with DNA binding. For example, some lower molecular weight PBAEs (Mw < 11 kDa) were unable to bind and complex DNA even at a high 150:1 polymer to DNA weight ratio, whereas the same PBAEs at higher molecular weight (Mw > 13 kDa) were able to sufficiently bind and complex DNA even at a low 10:1 polymer to DNA weight ratio.14
Polymer/DNA binding helps determine particle stability, which is important to ensure that the DNA stays protected from degradation and has high cellular uptake efficiency. However, it is also important to consider that the particle should not bind or encapsulate the DNA so tightly as to prevent the timely release of the DNA once in the cytoplasm. It has been shown that polymeric vector/DNA complexation significantly inhibits RNA synthesis and that above an optimal length, overall transfection decreases as polylysine length increases.33 Similarly, 25 kDa PEI vectors are known to have higher transfection efficacy than those formed with higher molecular weight PEI.34 However, as previously mentioned, it is also known that PEI and PBAEs with molecular weights below 8 kDa typically exhibit poor transfection compared with higher molecular weight versions.14,16,34 Thus, it appears that there may be a biphasic relationship between vector/DNA binding and transfection efficacy among a broad range of polymers.
Terminal Groups of a Polymer Are Key for Gene Delivery Efficacy
We have determined that the terminal monomer unit of the polymer can be crucial to the efficacy of the overall polymer.14,16 The structural diversity of the PBAEs mentioned above is limited by chemical requirements of conjugate addition.10,11,14–16 We hypothesized that an investigation into larger PBAE chemical space could improve performance. To accomplish this, we synthesized a library of end-modified PBAEs using three diacrylate-terminated base polymers and 12 amine monomers as end-capping reagents.35 A one-step reaction was used to conjugate the new terminal groups to the base polymers and the combined effects of internal structure and terminal structure on PBAE efficacy were systematically assessed (Figure 9A, B). Lead polymer C32 was end-capped with an expanded library of 36 different amines by Zugates et al. to show wider structure/function relationships (Figure 9C).36,37
FIGURE 9.
Synthesis of end-modified poly(β-amino ester)s by (A) reaction of acrylate-terminated C32 polymer with (B) primary diamine molecules in DMSO and (C) structures of amine-capping molecules.
Lead modified polymers C32-103 and C32-117 were found to bind DNA with higher affinity and form nanoparticles that were ~30% smaller than those formed with regular amino alcohol terminated C32.35 We also found that these modified PBAE nanoparticles enable an up to 5-fold increase in cellular DNA uptake compared with regular C32. It is important to note that without end-modification, there are no primary amines present in the leading PBAEs including C32. We reason that the modification of end group from alcohol to primary amine increases the polymers’ cationic charge, improving DNA binding affinity and condensation of DNA into nanoparticles. This leads to smaller nanoparticles with increased cellular uptake. We find that end-modified PBAEs have gene delivery efficacy comparable to lentivirus38 and adenovirus35 (Figure 10) for transfection of HUVECs in vitro. In comparison to the previous “gold standard” for polymeric transfection, 25 kDa polyethylenimine, the PBAE nanoparticles presented here have 2 orders of magnitude higher efficacy while simultaneously having 2 orders of magnitude lower toxicity.21,35 We believe that these polymeric vectors highlight the strength of a combinatorial polymer library approach, and they could set a new benchmark in nonviral transfection capability.
FIGURE 10.
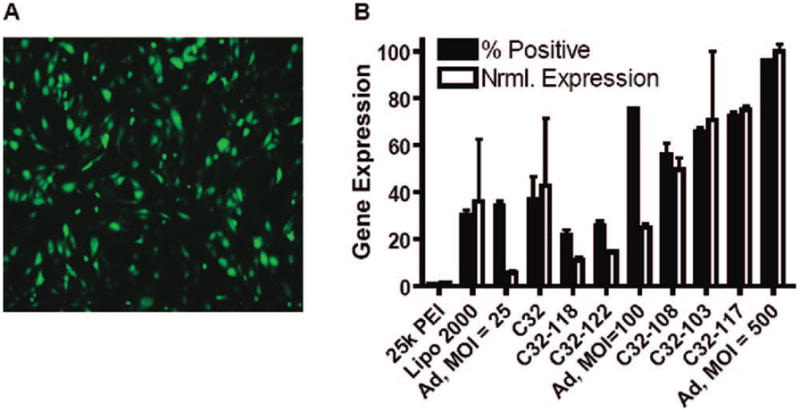
Gene expression of leading nonviral polymeric vectors compared with adenovirus vectors: (A) fluorescent micrograph of GFP gene delivery mediated by polymer C32-103 at 24 h post-transfection; (B) gene expression compared on the basis of both the percentage of cells positively transfected and the normalized total gene expression per cell at 48 h post-transfection. Reproduced with permission from ref 35. Copyright 2007 Wiley-VCH Verlag GmbH & Co. KGaA.
In Vivo Gene Delivery
Specific challenges for in vivo nonviral gene delivery include preventing serum protein interactions that inhibit transfection, minimizing activation of the complement system, minimizing toxicity, and ensuring targeted delivery to only the tissues and cell-types of interest. One difficulty is that positively charged complexes can also bind to blood constituents such as albumin, immunoglobulins, and fibrinogen, interact with macrophages and neutrophils, and nonspecifically bind to many other untargeted tissues and cell types.6,31 It is also known that these interactions increase clearance and lead to a shorter plasma half-life of the vectors.6 To more effectively screen PBAEs for in vivo gene delivery, leading polymers were screened for small size, stability, neutral surface charge, and high transfection efficacy in the presence of serum proteins.
Following intratumoral injection, lead PBAE C32 was found to have 4-fold higher gene delivery efficacy than jet-PEI. We also used polymer C32 to deliver a gene encoding a chain of diptheria toxin i.t. to prostate cancer xenografts. Treatment prevented tumor growth and also caused 40% of the tumors to regress in size.39 In contrast, healthy muscle was transfected poorly by C32. In a similar prostate cancer mouse model, intraprostate injection of C32/diptheria toxin nanoparticles resulted in apoptosis of 80% of the tumor cells at the site of injection.40
Following intraperitoneal (i.p.) injection of PBAE/DNA nano-particles, whole-body gene expression levels for end-modified C32 polymers ranged from 175-fold (C32-116) to 500-fold (C32-103) higher than those in buffer-injected control mice.36 In comparison to other gene delivery polymers, these end-modified PBAEs were 4–12-fold more effective than regular C32 and 15–42-fold more effective than jet-PEI. We tested the duration of luciferase expression after DNA delivery with C32 and C32-117 and found sustained expression past 1 week both with end-modified C32 and with regular C32, but end-modified C32 was expressed at significantly higher levels. For C32-117 nanoparticles, low levels of luciferase expression were still detected 2 months after administration. End-modification also altered the biodistribution of gene expression, with small polymer modifications leading to greater than 100-fold differences in certain tissues. For example, in an ovarian cancer mouse model, i.p. injected end-modified polymer C32-117 had greater than 100-fold higher tumor gene expression than unmodified C32.35 Intravenous injection of C32 particles led to lower overall gene expression that was primarily localized to the spleen and the liver. End-modified C32-117 particles had similar expression to unmodified C32 except that delivery to the lung was greater than 10-fold.36 Following all routes of administration, structural differences to the end of C32 polymers significantly altered gene expression in vivo, motivating a more detailed inquiry into the structure/function relationships of these polymers.
Steven Little and co-workers in our laboratory have also explored the use of PBAEs as copolymers in the fabrication of insoluble microspheres for genetic vaccines.41 Combining a pH-sensitive PBAE with conventional poly(lactic-co-glycolic acid) increases in vitro gene delivery to macrophages by 3–5 orders of magnitude as well as increases activation of dendritic cells in vitro. When these PBAE-containing microparticles are used in vivo as genetic vaccines, they are able to cause antigen-specific rejection of transplanted syngenic tumor cells.41
Conclusions
We have used high-throughput synthesis and screening techniques to create large libraries of structurally unique PBAEs for gene delivery. These have been iteratively improved, as well as end-modified, to further explore polymer structure. PBAEs are promising for nonviral gene delivery due to their (1) large potential for structural diversity, (2) ability to condense DNA into small and stable nanoparticles, (3) ability for ligand-specific uptake, (4) ability to buffer the endosome and facilitate endosomal escape, (5) biodegradability via hydrolytic cleavage of ester groups, (6) low cytotoxicity compared with other cationic polymers, and (7) high efficacy in vitro and in vivo.
Several polymer structural characteristics were important for high gene delivery. Specifically, out of the more than 2000 polymers tested, polymers synthesized by the conjugate addition of amino alcohols containing 3–5 linear carbons to diacrylates containing 4–6 interior carbons produced the top-performing polymers. The best PBAEs were linear, were synthesized at an amine/acrylate ratio of 1.2:1, and had a molecular weight of ~10 kDa. Acrylate-terminated polymers that contained neither primary nor secondary amines at the polymer ends led to polymer/DNA particles with low cellular uptake and low gene delivery. In contrast, amine monomer-terminated polymers (containing secondary amines near the polymer ends) formed particles with much higher uptake and gene delivery. By conjugation of a diamine instead of an amino alcohol to the ends of the same base polymer, cellular uptake and final gene expression were further increased. This analysis shows that having both primary and secondary amines near the ends of the gene delivery polymer significantly improves delivery. We have also shown that the tertiary amines that are a major component of the PBAEs are important to the endosomal buffering capacity of these polymers. These titratable amines provide PBAE-containing particles with a mechanism of endosomal escape to deliver cargo to the cytoplasm. The hydrophilic, biodegradable structure of lead PBAEs may also be important in minimizing their cytotoxicity.
The molecular characteristics of the lead PBAEs mentioned above are also important in forming polymer/DNA nanoparticles with favorable biophysical properties. Lead PBAEs all formed nanoparticles that were small (<200 nm) and stable and had near neutral ζpotential in the presence of serum-containing media. Small molecular changes to the polymer backbone, polymer side chains, and polymer terminal groups were found to lead to dramatic changes in gene delivery via known mechanistic steps including DNA binding, nanoparticle biophysical properties, particle uptake, endosomal escape, and final protein expression. Diamine end-modified C32 polymers bound DNA more tightly and formed smaller nanoparticles than other PBAEs. These nanoparticles also had higher cellular uptake that resulted in the best gene expression of all gene delivery polymers in the library. Lead PBAEs are more effective for gene delivery than top commercially available nonviral vectors including jet-PEI and Lipofectamine 2000 and are comparable to adenovirus for in vitro gene delivery to human primary cells. The polymers have also been demonstrated to facilitate gene delivery in vivo following local administration. This polymer library approach helped elucidate polymer structures for effective gene delivery and motivates a similar approach to other areas of biomaterial design.
Acknowledgments
This work was supported by a grant from the National Institutes of Health, EB 00244.
Biographies
Jordan J. Green was born in Ottawa, Canada. He received his B.S. in biomedical engineering and chemical engineering from Carnegie Mellon University in 2003 and studied at Imperial College London from 2001–2002. He received his Ph.D. in biological engineering from the Massachusetts Institute of Technology in 2007. Dr. Green is currently a postdoctoral associate in Institute Professor Robert Langer’s lab with research interests in bio-materials and drug delivery.
Robert Langer is one of 13 Institute Professors (MIT’s highest honor). He has written over 1000 articles, has over 600 issued or pending patents, and holds 11 honorary doctorates. He has received over 150 awards including the Charles Stark Draper Prize, considered the engineering equivalent of the Nobel Prize; the Albany Medical Prize, the nation’s largest medical prize; the National Medal of Science; and the American Chemical Society’s (ACS) Polymer Chemistry, Applied Polymer Science, and Chemistry of Materials Awards. He has been elected to the National Academy of Science, the Institute of Medicine of the National Academy of Science, and the National Academy of Engineering.
Daniel G. Anderson is a research associate at the David H. Koch Institute for Integrative Cancer Research at the Massachusetts Institute of Technology. He received his Ph.D. in molecular genetics from the University of California at Davis. At MIT, he pioneered the development of high-throughput methods for the synthesis, analysis, and formulation of biomaterials for drug delivery and tissue engineering. The delivery systems he has developed have provided new methods for nanoparticulate and microparticulate drug delivery, nonviral gene therapy, siRNA delivery, stem cell manipulation, and vaccines. His work has resulted in the publication of over 70 papers and patents.
References
- 1.Gene Therapy Clinical Trials Worldwide provided by the Journal of Gene Medicine. 2007 doi: 10.1002/jgm.1100. www.wiley.co.uk/genmed/clinical/ [DOI] [PubMed]
- 2.Hollon T. Researchers and regulators reflect on first gene therapy death. Nat Med. 2000;6:6. doi: 10.1038/71545. [DOI] [PubMed] [Google Scholar]
- 3.Check E. Gene therapy put on hold as third child develops cancer. Nature. 2005;433:561. doi: 10.1038/433561a. [DOI] [PubMed] [Google Scholar]
- 4.Wagner E, Kircheis R, Walker GF. Targeted nucleic acid delivery into tumors: new avenues for cancer therapy. Biomed Pharmacother. 2004;58:152–161. doi: 10.1016/j.biopha.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Putnam D. Polymers for gene delivery across length scales. Nat Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 6.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Delivery Rev. 2002;54:715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 7.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci US A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 9.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Lynn DM, Langer R. Degradable poly(beta-amino esters): Synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 11.Lynn DM, Anderson DG, Putnam D, Langer R. Accelerated discovery of synthetic transfection vectors: Parallel synthesis and screening of a degradable polymer library. J Am Chem Soc. 2001;123:8155–8156. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- 12.Akinc A, Lynn DM, Anderson DG, Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J Am Chem Soc. 2003;125:5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 13.Akinc A, Langer R. Measuring the pH environment of DNA delivered using nonviral vectors: Implications for lysosomal trafficking. Biotechnol Bioeng. 2002;78:503–508. doi: 10.1002/bit.20215. [DOI] [PubMed] [Google Scholar]
- 14.Akinc A, Anderson DG, Lynn DM, Langer R. Synthesis of poly(beta-amino ester)s optimized for highly effective gene delivery. Bioconjugate Chem. 2003;14:979–988. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- 15.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem, Int Ed. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 16.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol Ther. 2005;11:426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Liu G, Molas M, Grossmann GA, Pasumarthy M, Perales JC, Cooper MJ, Hanson RW. Biological properties of poly-L-lysine-DNA complexes generated by cooperative binding of the polycation. J Biol Chem. 2001;276:34379–34387. doi: 10.1074/jbc.M105250200. [DOI] [PubMed] [Google Scholar]
- 18.Ogris M, Steinlein P, Carotta S, Brunner S, Wagner E. DNA/polyethylenimine transfection particles: Influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci. 2001;3:21. doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5:1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 20.Guo W, Lee RJ. Efficient gene delivery via non-covalent complexes of folic acid and polyethylenimine. J Controlled Release. 2001;77:131–138. doi: 10.1016/s0168-3659(01)00456-4. [DOI] [PubMed] [Google Scholar]
- 21.Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjugate Chem. 2006;17:1162–1169. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 22.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci US A. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircheis R, Wightman L, Schreiber A, Robitza B, Rossler V, Kursa M, Wagner E. Polyethylenimine/DNA complexes shielded by transferrin target gene expression to tumors after systemic application. Gene Ther. 2001;8:28–40. doi: 10.1038/sj.gt.3301351. [DOI] [PubMed] [Google Scholar]
- 24.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J Gene Med. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 25.Schaffer DV, Lauffenburger DA. Optimization of cell surface binding enhances efficiency and specificity of molecular conjugate gene delivery. J Biol Chem. 1998;273:28004–28009. doi: 10.1074/jbc.273.43.28004. [DOI] [PubMed] [Google Scholar]
- 26.Kunath K, Merdan T, Hegener O, Haberlein H, Kissel T. Integrin targeting using RGD-PEI conjugates for in vitro gene transfer. J Gene Med. 2003;5:588–599. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- 27.Zugates G, Anderson D, Little S, Lawhorn I, Langer R. Synthesis of poly(beta-amino ester)s with thiol-reactive side chains for DNA delivery. J Am Chem Soc. 2006;128:12726–12734. doi: 10.1021/ja061570n. [DOI] [PubMed] [Google Scholar]
- 28.Kursa M, Walker GF, Roessler V, Ogris M, Roedl W, Kircheis R, Wagner E. Novel shielded transferrin-polyethylene glycol-polyethylenimine/DNA complexes for systemic tumor-targeted gene transfer. Bioconjugate Chem. 2003;14:222–231. doi: 10.1021/bc0256087. [DOI] [PubMed] [Google Scholar]
- 29.Suh W, Han SO, Yu L, Kim SW. An angiogenic, endothelial-cell-targeted polymeric gene carrier. Mol Ther. 2002;6:664–672. [PubMed] [Google Scholar]
- 30.Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Electrostatic ligand coatings of nanoparticles enable ligand-specific gene delivery to human primary cells. Nano Lett. 2007;7:874–879. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 31.Trubetskoy VS, Wong SC, Subbotin V, Budker VG, Loomis A, Hagstrom JE, Wolff JA. Recharging cationic DNA complexes with highly charged polyanions for in vitro and in vivo gene delivery. Gene Ther. 2003;10:261–271. doi: 10.1038/sj.gt.3301888. [DOI] [PubMed] [Google Scholar]
- 32.Putnam D, Gentry CA, Pack DW, Langer R. Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc Natl Acad Sci US A. 2001;98:1200–1205. doi: 10.1073/pnas.031577698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 34.Choosakoonkriang S, Lobo BA, Koe GS, Koe JG, Middaugh CR. Biophysical characterization of PEI/DNA complexes. J Pharm Sci. 2003;92:1710–1722. doi: 10.1002/jps.10437. [DOI] [PubMed] [Google Scholar]
- 35.Green JJ, Zugates GT, Tedford NC, Huang Y, Griffith LG, Lauffenburger DA, Sawicki JA, Langer R, Anderson DG. Combinatorial modification of degradable polymers enables transfection of human cells comparable to adenovirus. Adv Mater. 2007;19:2836–2842. [Google Scholar]
- 36.Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, Sawicki JA, Anderson DG. Rapid optimization of gene delivery by parallel end-modification of poly(β-amino ester)s. Mol Ther. 2007;15:1306–1312. doi: 10.1038/sj.mt.6300132. [DOI] [PubMed] [Google Scholar]
- 37.Zugates GT, Tedford NC, Zumbuehl A, Jhunjhunwala S, Kang CS, Griffith LG, Lauffenburger DA, Langer R, Anderson DG. Gene delivery properties of end-modified poly(beta-amino ester)s. Bioconjugate Chem. 2007;18:1887–1896. doi: 10.1021/bc7002082. [DOI] [PubMed] [Google Scholar]
- 38.Totsugawa T, Kobayashi N, Maruyama M, Okitsu T, Noguchi H, Watanabe T, Matsumura T, Fujiwara T, Tanaka N. Successful lentivirus-based delivery of a LacZ gene into human endothelial cells. Transplant Proc. 2003;35:499–500. doi: 10.1016/s0041-1345(02)03799-5. [DOI] [PubMed] [Google Scholar]
- 39.Anderson DG, Peng W, Akinc A, Hossain N, Kohn A, Padera R, Langer R, Sawicki JA. A polymer library approach to suicide gene therapy for cancer. Proc Natl Acad Sci US A. 2004;101:16028–16033. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng W, Anderson DG, Bao Y, Padera RF, Jr, Langer R, Sawicki JA. Nanoparticulate delivery of suicide DNA to murine prostate prostate tumors. Prostate. 2007;67:855–862. doi: 10.1002/pros.20576. [DOI] [PubMed] [Google Scholar]
- 41.Little SR, Lynn DM, Ge Q, Anderson DG, Puram SV, Chen JZ, Eisen HN, Langer R. Poly-beta amino ester-containing microparticles enhance the activity of nonviral genetic vaccines. Proc Natl Acad Sci US A. 2004;101:9534–9539. doi: 10.1073/pnas.0403549101. [DOI] [PMC free article] [PubMed] [Google Scholar]