Abstract
Background
We investigated whether Makorin ring finger protein 1 (MKRN1), an E3 ligase, affects p14ARF-associated cellular senescence and tumorigenesis by posttranslational modification in gastric tumorigenesis.
Methods
A link between MKRN1 and ARF was examined in MKRN1 null mouse embryonic fibroblasts (MEFs) and in human fibroblasts and gastric cancer cells by silencing MKRN1 using small interfering RNA (siRNA) and short hairpin RNA (shRNA). Ubiquitination and proteasomal degradation assays were used to assess p14ARF degradation associated with MKRN1. MKRN1 and p14ARF expression levels were analyzed with immunohistochemistry in malignant and normal tissues from gastric cancer patients and with χ2 tests. The tumor growth of gastric cancer cells stably expressing MKRN1 shRNA, p14ARF shRNA, or both was examined in mouse xenograft models (n = 4–6) and analyzed with unpaired t tests. All statistical tests were two-sided.
Results
MKRN1 knockout MEFs exhibited premature senescence and growth retardation with increased p19ARF protein expression. Similar results were obtained for human fibroblasts or gastric cancer cell lines by MKRN1 knockdown. Biochemical analyses confirmed that MKRN1 targets p14ARF for ubiquitination and subsequent proteasome-dependent degradation. A statistically significant association was shown between MKRN1 overexpression and p14ARF underexpression (P = .016). Xenograft analyses using p53-functional AGS or -dysfunctional SNU601 cells displayed statistically significant tumor growth retardation by silencing MKRN1, which was reversed under depletion of p14ARF (AGS cells, MKRN1 knockdown tumors vs MKRN1 and p14ARF knockdown tumors: 164.6 vs 464.8mm3, difference = 300.2mm3, 95% CI = 189.1 to 411.3mm3, P < .001).
Conclusions
We demonstrated that MKRN1 functions as a novel E3 ligase of p14ARF and that it potentially regulates cellular senescence and tumorigenesis in gastric cancer.
ARF (Alternative reading frame; p14ARF in humans and p19ARF in mice) is a product of the CDKN2/INK4a/ARF locus, which also encodes p16INK4a (1,2). Upon activation, ARF breaks the p53/Mdm2 feedback loop by sequestering Mdm2 into the nucleolus, freeing p53 from its negative regulatory circuit, and therefore promotes oncogene-induced senescence (3–5). ARF also functions as a tumor-suppressive regulator separate from the ARF/p53 axis through interactions with a variety of proteins including NPM and HIF1, allowing versatile roles as a tumor suppressor (6–9). These were previously confirmed in ARF knockout mice, which display a predisposition toward cancer development at an early age (10,11). Until recently, the loss of p14ARF function in various cancers, including gastric tumors, was believed to be caused by INK4a/ARF locus deletion or silencing triggered by promoter hypermethylation (12–16). The possibility that the posttranslational modification of p14ARF regulated its protein stability was also addressed, but only through a handful of reports implicating degradation of p14ARF through N-terminal ubiquitination (17,18). Moreover, the direct effects of p14ARF modification in tumorigenesis have not been defined.
Makorin ring finger protein 1 (MKRN1) is the source gene of the intronless MKRN gene family, and it shows high sequence conservation from invertebrates to vertebrates (19). It is a zinc-finger protein that was first identified as an E3 ligase for hTERT (20). In a previous report, we showed that MKRN1 simultaneously induces p53 and p21 ubiquitination and proteasome-dependent degradation. This suggests that the presence of MKRN1 in cancer cells might affect p53- and p21-dependent apoptosis and cell growth (21).
In this study, we found that MKRN1 could function as an E3 ligase of p14ARF. MKRN1-knockout mouse embryonic fibroblasts (MEFs) or ablation of MKRN1 in gastric cancer cells induced stabilization of p14ARF (p19ARF in mouse), resulting in cellular senescence and growth retardation. Ablation of p14ARF rescued the cell growth that was inhibited by MKRN1 depletion. The same observations were accomplished via xenografted mouse models. Moreover, inverse associations between expression levels of MKRN1 and p14ARF were determined in the tissues of human gastric cancer patients. Based on these results, we suggest that MKRN1 could affect gastric tumorigenesis by repressing cellular senescence and tumor-suppressive effects through downregulation of p14ARF in a p53-dependent or -independent manner.
Materials and Methods
Plasmids, Small Interfering RNA (siRNA), and Antibodies
pEGFP-N1 containing p14ARF was obtained from FHGB (21C Frontier Human Gene Bank, Seoul, South Korea). p14ARF WT-3XFLAG/CMV and p14ARF deletion mutants (1–64 and 65–132)-3XFLAG/CMV were prepared by PCR. MKRN1 cDNA and its mutants were as reported previously (21,22). pEGFP-C2 used as a transfection control was from Clontech (San Diego, CA). MKRN1 siRNA #5 (5′-CAGGCGAAGCTGAGTCAAGAA-3′), #6 (5′-CGGGATCCTCTCCAACTGCAA-3′), #7 (5′-caggcgaa gctgagtcaag-3′), p14ARF siRNA #1 (5′-GAACAUGGUGCGCA GGUUCTT-3′), and control siRNA were made by Qiagen-Xeragon (Valencia, CA). MKRN1 short hairpin RNA (shRNA) #2 (TRCN0000033796), #5 (TRCN0000296491), and p14ARF shRNA (TRCN00000010482) were purchased from Sigma-Aldrich (St Louis, MO). Polyclonal rabbit anti-HA (Y-11) (1:1000 dilution), monoclonal mouse anti-HA (F-7) (1:1000 dilution), and polyclonal rabbit anti-GFP (FL) (1:5000 dilution) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal mouse anti-FLAG (M2) (1:2000 dilution), polyclonal rabbit anti-FLAG (1:2000 dilution), and monoclonal mouse anti-actin (1:5000 dilution) antibodies were obtained from Sigma-Aldrich. Polyclonal rabbit anti-MKRN1 antibody (1:1000 dilution) and polyclonal rabbit anti-p14ARF antibodies (A300-340A and A300-342A) (1:1000 dilution) were purchased from Bethyl Laboratories (Montgomery, TX). Monoclonal mouse anti-p14ARF antibody (ab-3) (1:1000 dilution) was from Labvision (Fremont, CA). Polyclonal rabbit anti-p19ARF antibody (ab80) (1:1000 dilution) was from Abcam (UK). Monoclonal mouse anti-p14ARF antibody (Cell Signaling) and polyclonal rabbit anti-MKRN1 antibody (Bethyl Laboratories) were used for immunohistochemistry.
Cell Culture
Human non–small cell lung carcinoma (H1299), embryonic kidney cell line expressing SV40 large T antigen (293T), cervical cancer cell line (HeLa), lung fibroblast (IMR90), and human foreskin fibroblast (HFF) were obtained from American Type Culture Collection (ATCC, Manassas, VA) and grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. IMR90 and HFF cells of passage number 8 or less were used. All human gastric cancer cell lines including human stomach adenocarcinoma cell line (AGS) and gastric carcinoma cell lines (SNU-5, SNU-216, SNU638, SNU-668, SNU601, SNU-719, MKN28, YCC-2, and KATO-III) were purchased from the Korea Cell Line Bank (KCLB, Seoul, Korea) and were grown in RPMI medium supplemented with 10% fetal bovine serum. The ATCC and KCLB authenticate the phenotypes of these cell lines on a regular basis.
Cellular and Biochemical Analyses
For the crystal violet staining assay, cells were transfected with 30nM siRNA using Lipofectamine RNAiMAX three times, followed by one wash with phosphate buffered saline (PBS) and fixation with 1% glutaraldehyde for 10min. After washing with PBS, cells were stained with 0.5% crystal violet for 10min at room temperature (RT). For immunoprecipitation assays, cells were plated in 100-mm dishes and transfected with plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After 24h of transfection, cells were lysed with a lysis buffer (50mM Tris-HCl [pH 7.5], 150mM NaCl, 0.5% NP-40, 0.5% Triton X-100, 0.1% Na-deoxycholate, 1mM EDTA, and protease inhibitors). Lysates were immunoprecipitated with antibodies for 2h at 4°C and incubated with protein G-sepharose beads (GE healthcare, Buckinghamshire, UK) for 2h at 4°C. Beads were centrifuged and eluted with 2× sample buffer. Immunofluorescence assays were performed as follows: Cells were plated in 12-well plates with cover slips. Transfection of plasmids was conducted using Lipofectamine 2000. After 24-h transfection, cells were fixed in 10% paraformaldehyde for 10min and then washed three times with PBS. The fixed cells were permeabilized by 0.5% Triton X-100 in PBS for 10min and washed three times with PBS. The cells were blocked with 5% bovine serum albumin in PBS for 30min and incubated with primary antibodies in 2.5% bovine serum albumin overnight. The cells were then washed three times for 5min each prior to incubation with Alexa Fluor 488- conjugated anti-mouse or Alexa 594-conjugated anti-rabbit antibodies (Molecular Probes, Eugene, OR) for 1h. The cells were washed, stained with 4′,6-diamidino-2-phenylindole for 10min, washed again, mounted on slides, and analyzed using N-SIM superresolution microscopy. For the cell viability assay, H1299, HeLa, AGS, and SNU 601 cells were plated and cultured for 24, 48, 72, and 96h. The cells were incubated with CellTiter-Glo Reagent (Promega, G7571) for 15min at 37°C. Luminescence was recorded using a luminometer.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was subjected to RT-PCR using p14ARF primers, 5′-cgg aat tca tgg tgc gca ggt tct tg-3′ (forward) and 5′-cg gaa ttc ttt ggt ctt cta gga agc gg-3′ (reverse). MKRN1 primers and PCR method were as reported previously (21).
Preparation of MKRN-1 shRNA–Loaded Lentivirus and Stable AGS Cell Lines
We prepared stable AGS cell lines with reduced MKRN1 and/or p14ARF expression by interference with specific shRNAs. Lentiviral vectors containing MKRN1 or p14ARF shRNA sequences were purchased from Sigma-Aldrich. The lentivirus particles were generated by cotransfection of 293FT cells with lentiviral vectors and three plasmids, VSVG, RSV-REV, and PMDLg/pPRE, using Lipofectamine 2000 transfection reagent following the manufacturer’s instructions (Invitrogen). Two days after transfection, the cell culture media were filtered using a 0.45-µm filter. AGS cells were transfected with these lentiviral particles and sorted using puromycin. MKRN1 and p14ARF protein and mRNA expression levels were determined by western blot and RT-PCR analysis, respectively.
Ubiquitination Assay
Ubiquitination assays were performed under denaturation conditions using Ni2+-nitrilotriacetic acid beads, as reported previously (22) as well as by immunoprecipitation of HA/Ub. After the denaturation of proteins in cells by sodium dodecyl sulfate (SDS) and lysis by lysis buffer (50mM Tris-HCl [pH 7.5], 150mM NaCl, 0.5% NP-40, 0.5% Triton X-100, 0.1% Na-deoxycholate, 1mM EDTA, and protease inhibitors), cell lysates were immunoprecipitated with anti-HA antibody.
SA-β-Galactosidase Assay
The β-galactosidase assay for senescence was performed using a senescence detection kit (#K320-250, BioVision, Mountain View, CA). Briefly, cells were plated in 60-mm dishes and cultured for 2–3 days and then washed once with PBS and fixed with fixation solution for 15min at RT. Cells were washed two times with PBS and incubated with staining solution overnight at 37°C, followed by microscopic analyses.
Preparation of MKRN1 Null MEFs
FVB/NJxC57BL/6J F2 mice with a MKRN1-knockout allele (heterozygous mice, +/−) were kindly provided by Drs Deyu Feng and TA Gray (Northwestern University and David Axelrod Institute) (23). MKRN1+/− mice were further backcrossed until N6 in the B6 background. Female and male N6 MKRN1+/− mice were crossed to produce MKRN1+/+, MKRN1−/− MEFs. To obtain MEFs from 13.5-day old embryos, embryos were washed using PBS (Welgene) and minced. After mincing, MEFs were incubated with 3-ml trypsin/EDTA (Gibco) at 37°C for 15min. Trypsinized MEFs were transferred to 150-mm culture dishes, and 20mL DMEM containing 10% fetal bovine serum (GIBCO) was added. The dishes were incubated at 37°C for 4–8h. After 4–8h, media were exchanged, MEFs were retained in DMEM containing 10% fetal bovine serum, and they were subcultured at 1:3 ratio upon reaching confluence.
In Vivo Gastric Cancer Xenograft Mouse Models
All animal experiments were approved by the Institutional Review Board of National Cancer Center (NCC, Korea) and performed under specific pathogen-free facilities and conditions in accordance with the Guidelines for the Care and Use of Laboratory Animals of NCC (NCC-11-034D). Six-week-old female specific pathogen-free Balb/c nude mice were purchased from Central Lab (Animal Inc, Korea). Mice were inoculated subcutaneously into both flanks with 106 stable AGS cells into each flank and with 106 SNU601 cells in each flank under 20 µL of ketamine/rompun (9:1) anesthesia. From palpable tumor formation until termination, tumor sizes were measured every 2 or 3 days using calipers, and tumor volume was calculated according to the formula, length × width2 × 0.5236. Mice were killed in 7.5% CO2 chamber, and tumors were harvested for immunohistochemical and other analyses.
Human Gastric Cancer Tissue Microarray Analyses
For immunohistochemical analysis, core tissue biopsy specimens (2-mm diameter) were obtained from individual paraffin-embedded gastric carcinomas (donor blocks) and arranged in new recipient paraffin blocks (tissue array blocks) using a trephine apparatus (Superbiochips Laboratories, Seoul, South Korea). Immunohistochemical analysis of MKRN1 and p14ARF was performed as described previously (24).
Statistical Analyses
The Pearson correlation coefficient was calculated to estimate the colocalization of p14ARF and MKRN1 within cells. We employed chi-square test to evaluate the relationship between p14ARF and MKRN1 levels in gastric tumor patient tissues. Two tumors per mouse were obtained and analyzed as mean tumor volume per mouse. Unpaired t tests were used to analyze the tumor volume means of xenograft mice. All statistical tests were two-sided, and the values were expressed as means with 95% confidence intervals. P values of less than .05 were considered to be statistically significant. Statistical analyses were performed using Graphpad Prism software (version 6; GraphPad Software Inc, La Jolla, CA).
Results
MKRN1 Ablation Inhibits Cellular Growth by Increasing p14ARF Protein Levels
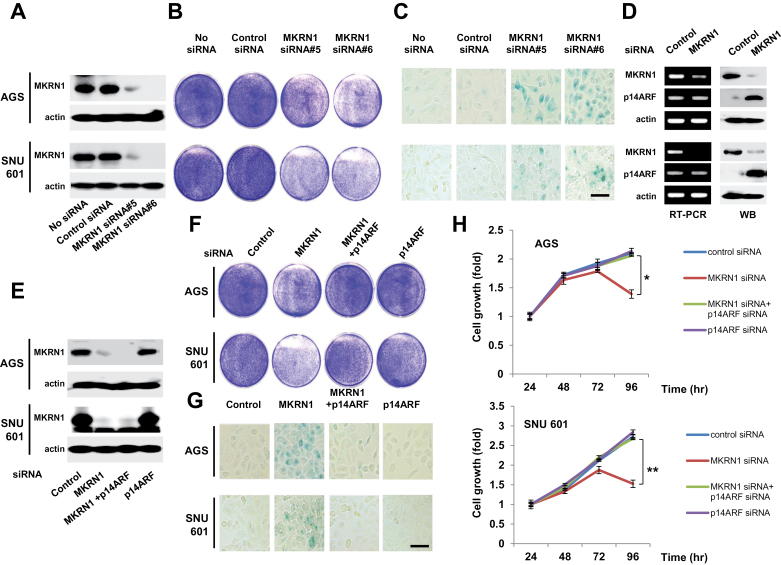
To understand the possible role of MKRN1 in tumor formation, we employed two gastric tumor cancer cell lines, AGS and SNU601. The former express wild-type p53, whereas the latter had a point mutation of p53 at R273L that renders p53 nonfunctional. First, knockdown of MKRN1 was carried out using MKRN1 siRNA#5 or #6 in both cell lines (Figure 1, A). Because two of the MKRN1 siRNAs (#5 and #6) worked equally well, we used #6 in the following experiments. Upon ablation of MKRN1, both cells exhibited growth retardation and senescence acceleration detected using SA-β-galactosidase staining (Figure 1, B and C). Because p14ARF is a major tumor suppressor inducing senescence in a p53-dependent or -independent manner, we tested whether p14ARF was affected in the absence of MKRN1. Interestingly, depletion of MKRN1 induced stabilization of p14ARF without any effects on its mRNA levels (Figure 1, D). When both gene expressions were simultaneously depleted (Figure 1, E), the effects of MKRN1 depletion on cells were annulled by p14ARF ablation, indicating an inverse association between MKRN1 and p14ARF (cell growth at 96h relative to 24 h: control siRNA vs MKRN1 siRNA: in AGS, mean fold change = 2.10 vs 1.39, difference = 0.71, 95% CI = 0.56 to 0.86, P = .004; in SNU601, mean fold change = 2.72 vs 1.53, difference = 1.19, 95% CI = 0.9 to 1.48, P = .005) (Figure1, F–H). We further tested two cancer cell lines with no p53 or nonfunctional p53, H1299 (p53 null human lung cancer cells), and HeLa (human cervical cancer cells) as described above. In line with the previous observations, we observed similar outcomes (Supplementary Figure 1, available online). These indicate that ablation of MKRN1 stabilizes and stimulates the activities of p14ARF, which in turn impedes cellular growth with or without functional p53.
Figure 1.
MKRN1 knockdown induces cell growth retardation and cellular senescence through p14ARF regulation in gastric cancer cells. A) Treatment of MKRN1 siRNAs suppressed the expression of MKRN1. After 96-h transfection with control, MKRN1 #5, or MKRN1 #6 siRNAs, cell lysates were subjected to immunoblotting using anti-MKRN1 and actin antibodies. B) The suppression of cell proliferation in p53 wt AGS cells and p53 mutant SNU601 cells under MKRN1 knockdown. After treatment with siRNAs as described above, the cells were stained with crystal violet. C) Treatment of MKRN1 siRNAs induced cellular senescence. Cells transfected with MKRN1 siRNAs as above were stained for β-galactosidase activities. The scale bar indicates 80.26 µm. D) Knockdown of MKRN1 induces stabilization of p14ARF without changes in mRNA levels of p14ARF. Cells transfected with control or MKRN1 #6 were analyzed by RT-PCR using primers specific for p14ARF, MKRN1, and GAPDH. Cell lysates were analyzed using the antibodies indicated. E) MKRN1 siRNA #6 and p14ARF siRNAs treated in AGS and SNU601 cells suppressed the expression of MKRN1, p14ARF, or both. (F) p14ARF depletion blocks the inhibition of gastric cancer cell growth induced by MKRN1 ablation. G) Cellular senescence induced by MKRN1 depletion is reversed by p14ARF ablation. The scale bar indicates 80.26 µm. All experiments from E) to G) were carried out as described above. H) Ablation of p14ARF rescued the cell growth retardation induced by MKRN1 depletion. AGS and SNU601 cells were transfected with siRNA as indicated. After 24, 48, 72, and 96h, cell viability was assessed using CellTiter-Glo Reagent. The error bars indicate 95% confidence intervals. *, P = .004; **, P = .005 using a two-sided t test. Three independent experiments were performed. MKRN1 = Makorin ring finger protein 1; p14ARF = p14 alternative reading frame.
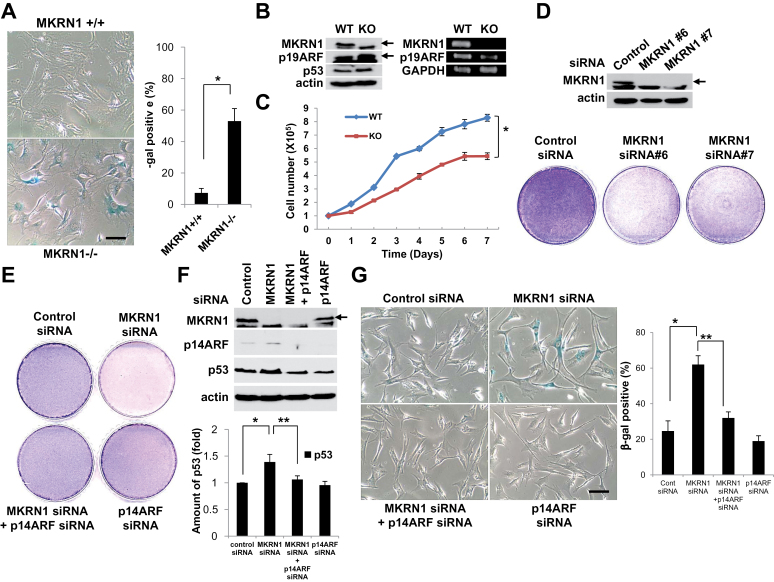
Next, the growth and senescence of MKRN1 null and wild-type MEFs were further analyzed (23). The results indicated that 53% of MKRN1−/− MEFs that were counted displayed premature senescence, whereas only 7.3% of wild-type MEFs went through senescence at passage 6 (MKRN1+/+ vs MKRN1−/−, mean percentage = 7.3% vs 53%, difference = 45.7%, 95% CI = 39.07% to 52.33%, P = .005) (Figure 2, A). The protein data comparing both cells exhibited an increase of p19ARF protein (Figure 2, B, left panels), whereas there was no detectable change of p19ARF mRNA levels (Figure 2, B, right panels). Moreover, MKRN1−/− MEFs showed growth retardation compared with MKRN1+/+ MEFs (MKRN1+/+ and MKRN1−/− at day 7, mean cell number = 8.29 × 105 vs 5.43 × 105, difference = 2.86 × 105, 95% CI = 2.53 × 105 to 3.19 × 105, P = .003) (Figure 2, C). Based on these observations, we investigated the cellular growth and senescence of two different human fibroblast cell lines, HFF and IMR90 (human fetal lung fibroblasts), in the absence of MKRN1, p14ARF, or both. Two MKRN1 siRNAs tested almost completely ablated the expression of endogenous MKRN1 (Figure 2, D and Supplementary Figure 2, A, available online). Under these conditions, each fibroblast cell line displayed retarded cellular growth compared with the control. Ablation of p14ARF, with or without concurrent MKRN1 knockdown, was able to recover cell growth to a level comparable to that of the control (Figure 2, E and Supplementary Figure 2, B, available online). Consistently, expression of p14ARF in the fibroblasts was increased by MKRN1 depletion and decreased following treatment with p14ARF siRNA, as shown in Figure 2, F and Supplementary Figure 2, C, available online (density of p53 relative to control: siRNA vs MKRN1 siRNA, mean fold change = 1 vs 1.39, difference = 0.39, 95% CI = 0.27 to 0.51, P = .01; MKRN1 siRNA vs MKRN1 siRNA + p14ARF siRNA, mean fold change = 1.39 vs 1.06, difference = 0.33, 95% CI = 0.19 to 0.47, P = .03, Figure 2, F). In both cell lines, the levels of p53 were associated with levels of p14ARF, underscoring the regulatory role of p14ARF on p53 (25, 26). SA-β-galactosidase staining assays indicated that depletion of MKRN1 accelerated the progression of senescence in both fibroblast cell lines and that this was reversed by depletion of p14ARF as shown in Figure 2, G and Supplementary Figure 2, D, available online (% β-gal-positive cells: control siRNA vs MKRN1 siRNA, mean percent = 24.7% vs 62%, difference = 37.3%, 95% CI = 30.67% to 43.93%, P = .008; MKRN1 siRNA vs MKRN1 siRNA + p14ARF siRNA, mean percent = 62% vs 32%, difference = 30%, 95% CI = 27.01% to 32.99%, P = .002, Figure 2, G). Although ablation of MKRN1 in both fibroblast cell lines induced stabilization of p14ARF and p53, a concurrent depletion of MKRN1 and p14ARF did not have a positive effect on p53 stability, implying that MKRN1 acts mainly on p14ARF, rather than on p53 in these cells.
Figure 2.
Knockout or knockdown of MKRN1 induces senescence in MKRN1 KO MEF and HFF cells through the stabilization of ARF. A) Premature senescence was induced in MKRN1−/− MEF. MKRN1−/− and +/+ MEFs were stained for SA-β-galactosidase activities (×200 magnification). The graph shows the percentage of β-galactosidase-positive cells. The error bars indicate 95% confidence intervals. *, P = .005 using two-sided t test. Three independent experiments were performed. The scale bar indicates 160.51 µm. B) p19ARF was stabilized in MKRN1−/− MEFs. Cell lysates of MKRN1−/− and +/+ MEFs were detected with anti-p19ARF, p53, and MKRN1 antibodies. Cells were analyzed by RT-PCR using primers specific for p19ARF, MKRN1, and GAPDH. Arrows indicate MKRN1 and p19ARF proteins. C) MKRN1−/− MEFs showed growth retardation. The same amount of MKRN1 +/+ and −/− MEFs was plated and counted 1, 2, 3, 4, 5, 6, and 7 days later. The error bars indicate 95% confidence intervals. *, P = .003 using two-sided t test. Three independent experiments were performed. D) Knockdown of MKRN1 suppresses growth of HFF cells. After 96-h transfection with control, MKRN1 #6, or MKRN1 #7 siRNAs, the cells were stained with crystal violet. Cell lysates were subjected to immunoblotting using anti-MKRN1 and actin antibodies. The arrow indicates MKRN1. E) p14ARF depletion blocks the inhibition of cell growth induced by MKRN1 ablation. HFF cells were transfected with control or MKRN1 #6 siRNA, with or without p14ARF siRNA. After 96h, cells were stained with crystal violet. F) p14ARF protein is stabilized by MKRN1 depletion. HFF cells were treated as indicated, and their lysates were subjected to western blotting using antibodies against p14ARF, MKRN1, p53, and actin. The arrow indicates MKRN1. The p53 protein level was quantified using Image-J software. The error bars indicate 95% confidence intervals. *, P = .01; **, P = .03 using two-sided t test. Three independent experiments were performed. G) Cellular senescence induced by MKRN1 depletion is reversed by p14ARF ablation. HFF cells were transfected with control or MKRN1 #6 siRNA, with or without p14ARF siRNA, as indicated. After 96h, cells were stained for β-galactosidase activities (×200 magnification). The scale bar indicates 160.51 µm. The graph shows percentage of β-galactosidase-positive cells. The error bars indicate 95% confidence intervals. *, P = .008; **, P = .002 using two-sided t test. Three independent experiments were performed. MKRN1 = Makorin ring finger protein 1; p14ARF = p14 alternative reading frame; p19ARF = p19 alternative reading frame; MEFs = mouse embryonic fibroblasts; HFF = human foreskin fibroblast.
MKRN1 Induces p14ARF Destabilization
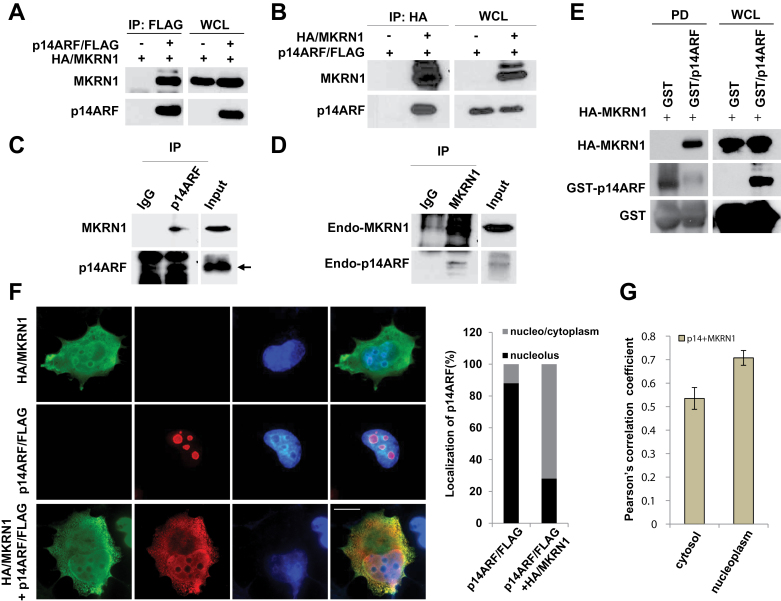
Next, we examined an interaction between MKRN1 and p14ARF. Both exogenous and endogenous p14ARF and MKRN1 were shown to interact by immunoprecipitation assays (Figure 3, A–D). We confirmed that MKRN1 directly binds to p14ARF through a GST pull-down assay (Figure 3, E). Immunofluorescent analyses showed that approximately 88% of p14ARF overexpressed in cells is localized to the nucleolus, whereas coexpression of MKRN1 resulted in relocalization of p14ARF to the nucleoplasm/cytoplasm in 72% of cells counted (Figure 3, F). Colocalization analysis using NIS-Elements AR software showed a Pearson correlation coefficient of 0.535 in cytoplasm and 0.708 in nucleoplasm (Figure 3, G). Pearson correlation coefficient values indicate that p14ARF and MKRN1 are moderately or strongly colocalized in the cytoplasm or nucleoplasm, respectively, suggesting that two proteins are generally found at the similar localization. Truncation mutation analyses indicated that p14ARF bound to the C-terminus of MKRN1, and MKRN1 bound to the N-terminus of p14ARF (Supplementary Figure 3, available online).
Figure 3.
MKRN1 interacts with p14ARF. A, B) MKRN1 and p14ARF proteins interact with each other. Plasmids expressing HA/MKRN1 alone or with p14ARF/FLAG were transfected into 293 cells. WCL were immunoprecipitated with anti-FLAG or anti-HA antibodies, and immunoprecipitates and WCLs were immunoblotted using the antibodies against MKRN1 and p14ARF. C, D) Endogenous MKRN1 and p14ARF bind to each other. HeLa cell lysates were immunoprecipitated with anti-p14ARF or MKRN1 antibodies, and immunoprecipitated proteins were analyzed using the same antibodies. Arrow indicates p14ARF. E) Recombinant p14ARF and MKRN1 proteins bind to each other directly. Purified GST/p14ARF and in vitro translated HA-MKRN1 were incubated at 37°C (input) followed by pull-down (PD) using glutathione-sepharose. The precipitated samples were detected using anti-MKRN1 and p14ARF antibodies. F) MKRN1 induces p14ARF translocation to nucleo/cytoplasm. H1299 cells were transfected with plasmids expressing HA/MKRN1 and p14ARF/FLAG. Fixed cells were detected by monoclonal anti-HA mouse and polyclonal anti-FLAG rabbit antibodies and stained by Alexa Fluor 594-conjugated anti-rabbit and Alexa Fluor 488-conjugated anti-mouse antibodies, analyzed using N-SIM superresolution microscopy (×150 magnification). The percentages on panels 6 and 10 and graph indicate the fractions of counted cells (200) that displayed such p14ARF/FLAG localization. G) The Pearson correlation coefficient for the colocalization of p14ARF and MKRN1. The error bars indicate 95% confidence intervals using two-sided t test. Thirty cells were tested for the analyses. The tan bars show the Pearson correlation for p14ARF and MKRN1 colocalization. MKRN1 = Makorin ring finger protein 1; p14ARF = p14 alternative reading frame.
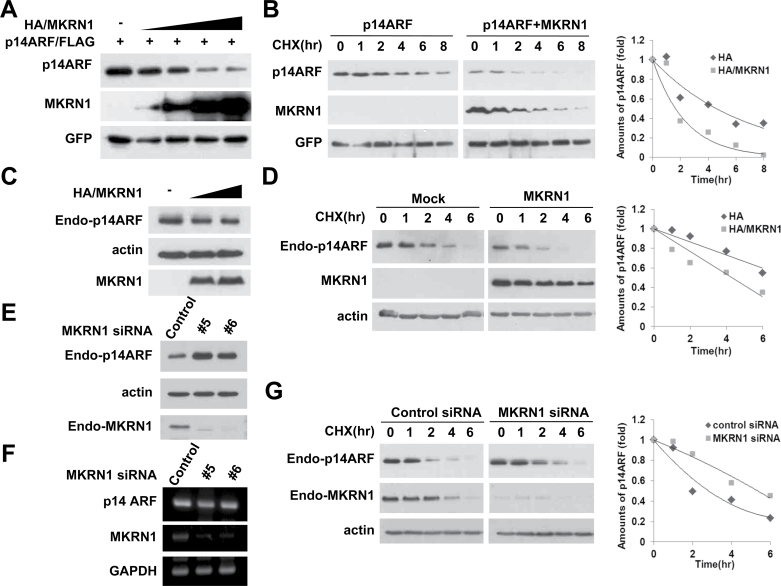
Because MKRN1 is known to be an E3 ligase, its ability to promote p14ARF degradation was evaluated (20–22). MKRN1 induced degradation of exogenous p14ARF in H1299 cells in a concentration-dependent manner (Figure 4, A). Experiments employing cycloheximide (CHX) treatment to measure the protein half-life showed that approximately half of the exogenous p14ARF protein disappeared in less than 2h in the presence of MKRN1, compared with almost 5h without MKRN1 (Figure 4, B). We observed similar regulatory effects of MKRN1 on endogenous p14ARF (Figure 4, C and D). The fact that MKRN1 depletion increased p14ARF protein levels without any noticeable effects on its mRNA levels indicates that MKRN1-mediated p14ARF regulation occurs at the posttranslational level (Figure 4, E and F). Lastly, MKRN1 ablation prolonged the half-life of endogenous p14ARF, corroborating the previous data (Figure 4, G). In summary, MKRN1 appears to function as a p14ARF repressor by inducing its destabilization through posttranslational modification.
Figure 4.
MKRN1 induces the degradation of p14ARF. A) MKRN1 induces the degradation of p14ARF in a dose-dependent manner. H1299 cells were transfected with mixtures of plasmids expressing p14ARF/FLAG and HA/MKRN1 with increasing concentrations, or with GFP. The cell lysates were detected with anti-FLAG, HA, and GFP antibodies. B) MKRN1 decreases the half-life of p14ARF. H1299 cells were transfected with mixtures of plasmids expressing p14ARF/FLAG, HA/MKRN1, or GFP followed by cycloheximide (CHX) treatment before harvest. The lysates were immunoblotted using antibodies against FLAG, HA, and GFP. C) Exogenous MKRN1 induces degradation of endogenous p14ARF. HeLa cells were transfected with increasing amounts of HA/MKRN1 plasmids. The cell lysates were immunoblotted with anti-p14ARF, HA, and actin antibodies. D) Exogenous MKRN1 reduces the stability of endogenous p14ARF. HeLa cells were transfected with mock vector or plasmid expressing HA/MKRN1 followed by CHX treatment before harvest. The cell lysates were immunoblotted with anti-p14ARF, HA, and actin antibodies. E) MKRN1 depletion stabilizes endogenous p14ARF. HeLa cells were transfected with control, MKRN1 #5, or MKRN1 #6 siRNAs and lysed after 48h. WCL were immunoblotted with anti-p14ARF, MKRN1, and actin antibodies. F) MKRN1 ablation does not affect p14ARF mRNA levels. HeLa cells were treated as above and analyzed by RT-PCR using primers specific for p14ARF, MKRN1, and GAPDH. G) MKRN1 depletion increases stability of endogenous p14ARF. HeLa cells were transfected with control and MKRN1 siRNAs followed by treatment with CHX and were then lysed and immunoblotted with anti-p14ARF, MKRN1, and actin antibodies. The graphs indicate the relative amounts of p14ARF protein compared with the levels of actin or GFP in the western blot. The expression of each protein was analyzed using the Image J program. MKRN1 = Makorin ring finger protein 1; p14ARF = p14 alternative reading frame; CHX = cycloheximide; WCL = whole cell lysates.
MKRN1 Induces p14ARF Ubiquitination and Proteasomal Degradation
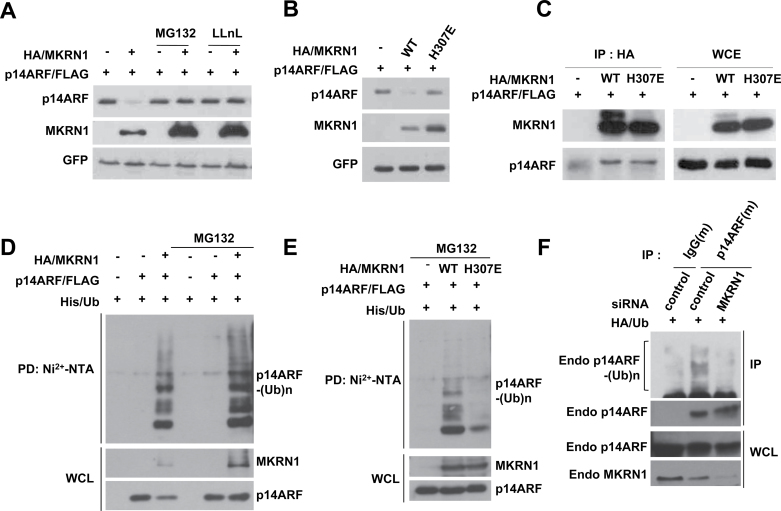
The effect of proteasome inhibitors, MG132 or LLnL, on the MKRN1-mediated p14ARF degradation process indicated that MKRN1-mediated degradation of p14ARF is proteasome-dependent (Figure 5, A). Using H307E, an E3 ligase defective form of MKRN1 that retains its ability to bind to p14ARF, we further demonstrated that the ligase activities of MKRN1 are required for p14ARF degradation (Figure 5, B and C) (20). In accordance with these data, MKRN1, but not the H307E mutant, was able to mediate ubiquitination of p14ARF (Figure 5, D and E). Furthermore, MKRN1 depletion led to decreased levels of endogenous ubiquitinated p14ARF (Figure 5, F). These data demonstrate that MKRN1 functions as an E3 ligase to mediate ubiquitination and degradation of p14ARF.
Figure 5.
MKRN1 functions as an E3 ligase by inducing ubiquitination and subsequent proteasomal degradation of p14ARF. A) MKRN1 induces proteasomal degradation of p14ARF. H1299 cells were transfected with mixtures of plasmids expressing p14ARF/FLAG, HA/MKRN1, or GFP followed by 3-h treatment with 20 µM MG132 or LLnL. The lysates were immunoblotted with anti-FLAG, HA, and GFP antibodies. B) E3 ligase activity of MKRN1 is required for p14ARF degradation. H1299 cells were transfected with mixtures of plasmids expressing p14ARF/FLAG, HA/MKRN1, HA/H307E (E3 ligase activity–deficient mutant), or GFP. The cell lysates were immunoblotted using anti-FLAG, HA, and GFP antibodies. C) Both MKRN1 WT and H307E interact with p14ARF. Mixtures of plasmids expressing p14ARF/FLAG, HA/MKRN1, or HA/H307E were transfected into 293T cells followed by immunoprecipitation using anti-HA antibodies and detection of the immunoprecipitated proteins with anti-FLAG and HA antibodies. D) MKRN1 ubiquitinates p14ARF. H1299 cells were transfected with mixtures of plasmids expressing His/Ub, p14ARF/FLAG, HA/MKRN1, or mock as indicated, with or without MG132. The cell lysates were pulled down with Ni2+-NTA resin and detected using anti-HA and FLAG antibodies. E) MKRN1 H307E is defective in inducing p14ARF ubiquitination. H1299 cells were transfected with mixtures of plasmids expressing His/Ub, p14ARF/FLAG, HA/MKRN1, or mock as indicated. The cell lysates were pulled down with Ni2+-NTA resin and detected with anti-FLAG and HA antibodies. F) MKRN1 depletion suppresses ubiquitination of endogenous p14ARF. HeLa cells were transfected with control or MKRN1 siRNAs followed by MG132 treatment. The cell lysates were immunoprecipitated with anti-p14ARF antibodies, and the immunoprecipitates were detected with anti-p14ARF and HA ubiquitin antibodies. All of the ubiquitination analyses were performed under conditions of denaturation. MKRN1 = Makorin ring finger protein 1; p14ARF = p14 alternative reading frame; Ni2+-NTA = Ni2+-nitrilotriacetic acid.
There is a restricted functional homology between human p14ARF and mouse p19ARF, and these two proteins are known to have different binding partners in cells (2, 27, 28). We were able to observe an increase of p19ARF in MKRN1 null MEFs (Figure 2, B), suggesting a relationship between p19ARF and mouse MKRN1 (mMKRN1) that is comparable to that between p14ARF and human MKRN1. To examine these relationships, we first tested the interaction between mMKRN1 and p19ARF. As shown in supplementary Figure 4, A and B, exogenous mMKRN1 and p19ARF do bind to each other. Moreover, mMKRN1 also degrades p19ARF in a dose-dependent manner (Supplementary Figure 4, C, available online) through ubiquitination of p19ARF (Supplementary Figure 4, D, available online). These showed that human MKRN1 and mouse MKRN1 display similar suppressive regulation of p14ARF and p19ARF, respectively. In conclusion, our data strongly indicate that MKRN1 is an E3 ligase that targets ARF as a substrate.
MKRN1 and p14ARF Levels Are Inversely Associated in Human Gastric Cancer
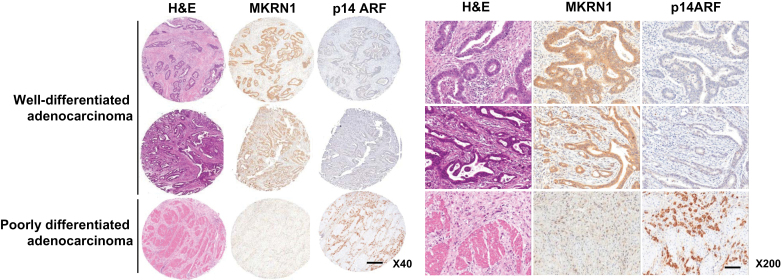
To further examine the physiological connection between MKRN1 and p14ARF, several gastric cancer cell lines were tested for their expression levels of MKRN1 and p14ARF. Among 12 cell lines tested, 10 lines (indicated by asterisks) display an inverse association between protein levels of MKRN1 and p14ARF (Supplementary Figure 5, available online). To investigate the clinical implication of this relationship, immunohistochemical studies were further performed to evaluate in situ expression of MKRN1 and p14ARF in 59 gastric cancer tissues. MKRN1 and p14ARF were mainly expressed in the cytoplasm and nucleoplasm of cancer cells, although nuclear localization of MKRN1 was occasionally detected (Figure 6). Of 57 gastric carcinoma patients, MKRN1 was expressed in 22 (38.6%) and p14ARF in 15 (28.3%). MKRN1 overexpression was statistically significantly associated with well-differentiated status of gastric carcinoma (P = .0083, two-sided χ 2 test). p14ARF expression tended to be associated with poor differentiation and advanced staging of gastric carcinoma, although this was not statistically significant. Among patients with MKRN1 overexpression, 16 (72.7%) showed p14ARF underexpression; these cases were statistically significantly associated with a well-differentiated tumor (P = .016, two-sided χ 2 test) and tended to show association with an earlier stage. Overall, these results indicate that the protein levels of MKRN1 and p14ARF display a relatively high inverse association, supporting the possible role of MKRN1 as a negative regulator of p14ARF in vivo as well as in vitro.
Figure 6.
Protein levels of MKRN1 and p14ARF in malignant tissues of gastric cancer patients. In situ expression of MKRN1 and p14ARF in gastric cancer tissues was detected by immunohistochemical analysis and hematoxylin and eosin (H&E) staining as described in “Materials and Methods” (magnification: left, ×40; right, ×200). The upper two cases are well-differentiated adenocarcinomas, and the lower one is a poorly differentiated adenocarcinoma. The scale bars indicate 200 µm (×40) and 40 µm (×200). MKRN1 = Makorin ring finger protein 1; p14ARF = p14 alternative reading frame.
MKRN1 Depletion Suppresses the Formation of Tumors in Nude Mice
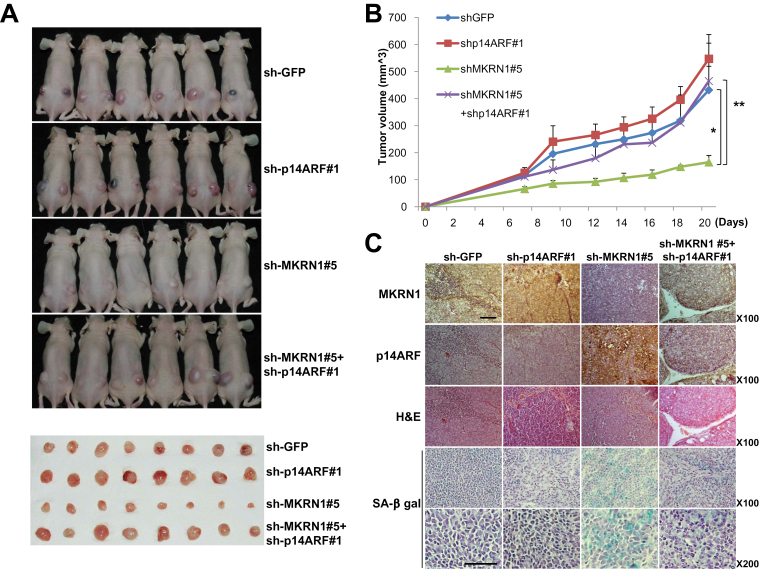
The biochemical analyses of the effect of MKRN1 on p14ARF function led to the hypothesis that MKRN1 plays a critical regulatory role in tumor formation. We tested this by constructing human gastric cancer AGS cell lines encompassing wild-type p53 that stably express MKRN1 shRNA #1 or #5, and comparing these with control cells (sh-GFP or AGS). Comparison of MKRN1 expression among these cell lines showed a 50% and 90% decrease in MKRN1 protein in lines #1 and #5, respectively (Supplementary Figure 6, A, available online). These cell lines were subcutaneously injected into the backs of nude mice (n = 4), and each tumor was followed for approximately a month. As anticipated, the tumor sizes were inversely associated with the levels of MKRN1 expression in the stable cell lines (Supplementary Figure 6, B and C, available online). The average size of tumors originating from cell line #5 with the lowest levels of MKRN1 was one-third of that originating from cell line #1 with medium levels of MKRN1. The size of tumors originating from cell line #5 was a sixth of that of the controls, whereas tumors originating from cell line #1 were half the size of controls. The difference in average size between tumors derived from stable cell lines #1 and #5 suggests that the protein level of MKRN1 could produce a graded regulatory effect on tumor formation. The similarity of tumor sizes between AGS and sh-GFP cell lines indicates that lentiviral infection had no major effects on tumor growth in xenograft mice inoculated with AGS cells. The relationship between MKRN1 and p14ARF protein levels in the tumors originating from these cell lines was similar to that those previously described for gastric cancers (Supplementary Figure 6, D, available online). To test whether the MKRN1 depletion-mediated growth retardation of xenografted tumor cells was due to an increase in p14ARF levels, we next made cell lines that were stably transfected with MKRN1 shRNA#5, p14ARF shRNA#1, or both (Supplementary Figure 7, A, available online). MKRN1 knockdown induced a reduction in tumor volume compared with the wild type, as shown above. This decrease in tumor volume was reversed by a concomitant ablation of p14ARF, suggesting that MKRN1 and p14ARF do indeed show an inverse relation. For example, mice inoculated with AGS cells displayed mean tumor volume of 164.6mm3 when MKRN1 was ablated (mean tumor volume: sh-GFP vs sh-MKRN1#5, 431.8mm3 vs 164.6mm3, difference = 267.2mm3, 95% CI = 189.7mm3 to 344.7mm3, P = .001), whereas tumors with MKRN1 and p14ARF knockdown had mean tumor volume of 464.8mm3 (sh-MKRN1#5 vs sh-p14ARF#1 + sh-MKRN1#5, 164.6mm3 vs 464.8mm3, difference = 300.2mm3, 95% CI = 158.3mm3 to 442.1mm3, P = .009) (Figure 7, A and B). Next, the tumors from the xenografts were analyzed using immunohistochemical studies (Figure 7, C). The data showed that depletion of MKRN1 was associated with increased expression of p14ARF, with most cells displaying senescence by β-gal assay (Figure 7, C, sh-MKRN1#5 panels). On the other hand, concomitant depletion of p14ARF and MKRN1 annihilated the effects described above (Figure 7, C, panels 4).
Figure 7.
The suppressive effect of MKRN1 on tumor growth was rescued by p14ARF depletion. A–C) Mice (N = 6) were inoculated subcutaneously into both flanks with 106 cells of each AGS cell line. Lentiviruses expressing sh-RNA targeting p14ARF, MKRN1, or both were employed to produce stable AGS cell lines. Lentivirus expressing sh-RNA targeting GFP was used as a control. A) At 23 days after inoculation, mice were killed in 7.5% CO2 chamber, and mice and excised tumors from mice were photographed. B) Quantification of tumor formation was performed by measurement of tumor size 7, 9, 12, 14, 16, 18, and 20 days after inoculation (N = 6). The error bars indicate 95% confidence intervals. *, P = .001; **, P = .009, two-sided t test. All statistical tests were two-sided. C) Immunohistochemical analysis and hematoxylin and eosin (H&E) staining were performed for each group of tumors as described in “Materials and Methods” (magnification ×100). SA-β-galactosidase-stained slides were viewed under ×100 and ×200 magnification. The scale bar indicates 100 µm. MKRN1 = Makorin ring finger protein 1; p14ARF = p14 alternative reading frame.
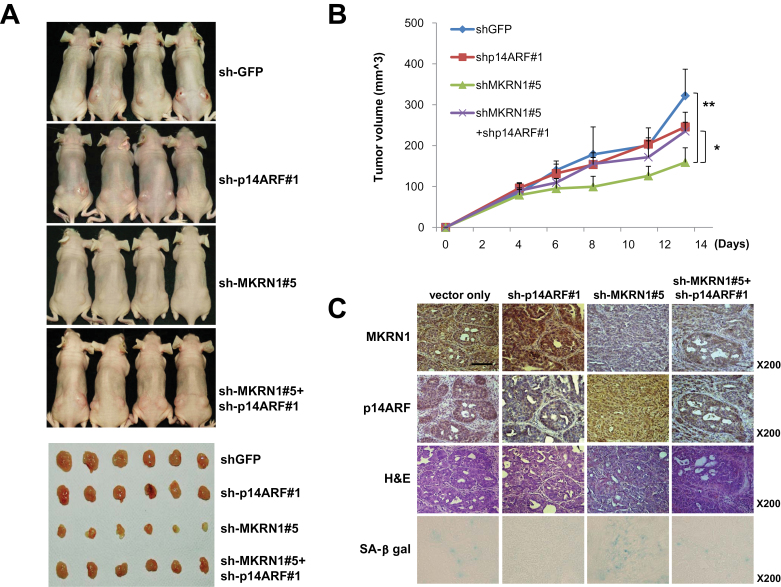
Using SNU601 gastric cancer cells containing dysfunctional p53 mutant R273L, similar xenograft assays were performed as described above to establish MKRN1 effects on p14ARF tumorigenesis independent of p53. As with AGS cell lines, stable transfections of MKRN1 shRNA#5, p14ARF shRNA#1, or both into SNU601 were carried out (Supplementary Figure 7, B, available online). The results indicated that depletion of MKRN1 was able to suppress cell growth, and that this effect was reversed by a simultaneous depletion of p14ARF. For example, mice inoculated with SNU601 cells with MKRN1 knockdown showed mean tumor volume of 158.9mm3 (mean tumor volume: sh-GFP vs sh-MKRN1#5, 321.8mm3 vs 158.9mm3, difference = 162.9mm3, 95% CI = 104.24mm3 to 221.56mm3, *: P =.01), whereas tumors with MKRN1 and p14ARF knockdown displayed 262.6mm3 (sh-MKRN1#5 vs sh-p14ARF#1 + sh-MKRN1#5, 158.9mm3 vs 234.9mm3, difference = 76mm3, 95% CI = 38.71mm3 to 107.29mm3, P = .02) (Figure 8, A and B). The depletion of MKRN1, p14ARF, or both proteins was detected using immunohistochemical studies, and the cell senescence was further confirmed using β-gal analyses (Figure 8, C), which exhibited comparable results as shown in Figure 7. Overall, the data suggest that depletion of MKRN1 could enhance p14ARF function, which led to the hampering of cell growth both in p53 functional and nonfunctional cell lines.
Figure 8.
The tumor-suppressive effects of MKRN1 knockdown via stabilization of p14ARF were observed in the p53 mutated gastric cancer cell line SNU601. A–C) Mice (N = 4) were inoculated subcutaneously in both flanks with 106 cells of each SNU601 cell line. The stable SNU601 cell lines were prepared in the same ways as mentioned in Figure 7. A) At 23 days after inoculation, mice were killed in 7.5% CO2 chamber, and mice and excised tumors from mice were photographed. B) Quantification of tumor formation was performed by measurement of tumor size 4, 6, 8, 11, and 13 days after inoculation (N = 4 tumors for four mice). The error bars indicate 95% confidence intervals. *, P = .02; **, P = .01, two-sided t test. All statistical tests were two-sided. C) Immunohistochemical analysis and hematoxylin and eosin (H&E) staining were performed for each group of tumors as described in Materials and Methods (magnification ×200). SA-β-galactosidase-stained slides were viewed under ×200 magnification. The scale bar indicates 100 µm. MKRN1 = Makorin ring finger protein 1; p14ARF = p14 alternative reading frame.
Discussion
Genetic studies of mice demonstrated that the absence of p19ARF results in the development of tumors such as sarcoma, lymphoma, and carcinoma at early ages (10, 11). Similar to these observations, p14ARF expression is suppressed in numerous cancers, including gastric tumors, due to silencing of its expression by promoter hypermethylation or allelic loss (13, 29–32). The recent discovery of the ULF, which induces p14ARF degradation, highlighted posttranslational regulatory mechanisms, but the direct effects on turmorigenesis remain unknown (33).
In this study, we introduce MKRN1 as a novel E3 ligase of p14ARF that affects gastric cancer development. Premature senescence with p19ARF stabilization in MKRN1−/− MEF suggests that MKRN1 might regulate ARF-induced cellular senescence. The importance of the regulatory role of MKRN1 in p14ARF stability is emphasized by the observation that p14ARF becomes stabilized upon MKRN1 ablation, resulting in growth retardation of both human fibroblasts and gastric cancer cells. Moreover, MKRN1 ablation suppressed gastric tumor formation in xenografted mice, which was reversed under depletion of p14ARF with independent p53 function.
Upon these results, one crucial issue of MKRN1 regulatory pathways is how MKRN1 determines a major target among its multiple substrates that include p14ARF, p53, and p21. Because we previously reported that MKRN1 mediates p53 and p21 degradation through the ubiquitination and proteasome-dependent degradation pathways (21), interestingly, it appears that ablation of MKRN1 induced p14ARF and p53 at the same time, as expected, whereas a simultaneous knockdown of p14ARF and MKRN1 was not able to induce p53. These observations indicate that the MKRN1-dependent p53 degradation might be overruled by a more dominant MKRN1-dependent p14ARF regulation, at least in the cell lines used here. Further experiments using p53 dysfunctional or null gastric cancer cells indicated that MKRN1 knockdown was capable of inducing p14ARF stabilization with concurrent cell growth retardation. These data suggest possible physiological roles of MKRN1 and p14ARF in regulating gastric tumorigenesis without p53 functions. The central importance of the interaction between p14ARF and MKRN1 was more clearly demonstrated by the xenografted mice data, showing an eruption of tumor growth upon the ablation of p14ARF in AGS gastric cancer cells expressing wild-type p53 in which growth was otherwise retarded by the absence of MKRN1. Because the system employed here cannot exclude the possible role of p53 in coordination with p14ARF in reversing MKRN1-dependent retardation of tumor growth, SNU601 gastric cancer cells with dysfunctional p53 were tested in xenografted mice analyses. This cell line has mutation of p53 on residue 273 from arginine to leucine, which is known as a hot spot in various cancer cells (34,35). MKRN1 depletion–mediated tumor-suppressive function was rescued by p14ARF ablation in this cell line. It indicates that MKRN1 could mediate tumorigenesis via p14ARF regulation without functional p53 and suggests that MKRN1 inhibition might be a meaningful target for cancer therapy through p14ARF activation in tumors with nonfunctional p53.
This study also had some limitations. Because there are diverse combinations of p53 mutations or deletion with epigenetic suppression of p14ARF proteins in human tumors (12–16), targeting of MKRN1 for tumor therapy could be limited to tumors displaying similar cellular context as the gastric cancer cells investigated above.
Taken together, we can draw noteworthy conclusion from these observations that MKRN1 E3 ligase remains unique in its ability to negatively regulate the major tumor suppressors including p14ARF and p53 (21). This indicates a possible role of MKRN1 as a potentially meaningful oncogene. The human gastric tumor tissue array data corroborate this notion, showing an inverse association between MKRN1 and p14ARF in approximately half of the gastric cancer tissues tested. However, we were not able to determine the associations between p53, MKRN1, and p14ARF because up to 60% of patient samples examined displayed p53 mutant expression, consistent with a previous report of p53 mutation frequency in gastric cancer (35).
Our results verified MKRN1 as an ubiquitin E3 ligase of p14ARF. The data presented strongly indicate that MKRN1 could instigate tumorigenesis by regulating the p14ARF-associated pathways and thus potentially represents an important therapeutic target in gastric tumors.
Funding
This work was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korean government (MEST) (2010-0017787) and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0087610).
Supplementary Material
Notes
The contributions by authors were as follows: A Ko—study concept, design and acquisition of data; J-Y. Shin, J Seo, K-D. Lee, E-W. Lee, and M-S. Lee—acquisition of data; I-J. Choi—provided gastric cancer patient samples and information; J. S. Jeong—pathological analysis; H-W. Lee and K. H. Chun—analysis and interpretation of data; J. W. Song—study concept, design, funding, and supervisor.
References
- 1. Stone S, Jiang P, Dayananth P, et al. Complex structure and regulation of the P16 (MTS1) locus. Cancer Res 1995. 55(14):2988–2994 [PubMed] [Google Scholar]
- 2. Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 1995. 83(6):993–1000 [DOI] [PubMed] [Google Scholar]
- 3. Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 1998. 12(15):2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bates S, Phillips AC, Clark PA, et al. p14ARF links the tumour suppressors RB and p53. Nature 1998. 395(6698):124–125 [DOI] [PubMed] [Google Scholar]
- 5. de Stanchina E, McCurrach ME, Zindy F, et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev 1998. 12(15):2434–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Itahana K, Bhat KP, Jin A, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell 2003. 12(5):1151–1164 [DOI] [PubMed] [Google Scholar]
- 7. Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol 2004. 24(3):985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brady SN, Yu Y, Maggi LB, Jr, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol 2004. 24(21):9327–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fatyol K, Szalay AA. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1alpha (HIF-1alpha) and inhibits HIF-1-mediated transcription. J Biol Chem 2001. 276(30):28421–28429 [DOI] [PubMed] [Google Scholar]
- 10. Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res 1999. 59(9):2217–2222 [PubMed] [Google Scholar]
- 11. Kamijo T, Zindy F, Roussel MF, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 1997. 91(5):649–659 [DOI] [PubMed] [Google Scholar]
- 12. Sharpless E, Chin L. The INK4a/ARF locus and melanoma. Oncogene 2003. 22(20):3092–3098 [DOI] [PubMed] [Google Scholar]
- 13. Gardie B, Cayuela JM, Martini S, Sigaux F. Genomic alterations of the p19ARF encoding exons in T-cell acute lymphoblastic leukemia. Blood 1998. 91(3):1016–1020 [PubMed] [Google Scholar]
- 14. Kasahara T, Bilim V, Hara N, Takahashi K, Tomita Y. Homozygous deletions of the INK4a/ARF locus in renal cell cancer. Anticancer Res 2006. 26(6B):4299–4305 [PubMed] [Google Scholar]
- 15. Iwato M, Tachibana O, Tohma Y, et al. Alterations of the INK4a/ARF locus in human intracranial germ cell tumors. Cancer Res 2000. 60(8): 2113–2115 [PubMed] [Google Scholar]
- 16. Esteller M, Tortola S, Toyota M, et al. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res 2000. 60(1):129–133 [PubMed] [Google Scholar]
- 17. Roberti A, Rizzolio F, Lucchetti C, de Leval L, Giordano A. Ubiquitin-mediated protein degradation and methylation-induced gene silencing cooperate in the inactivation of the INK4/ARF locus in Burkitt lymphoma cell lines. Cell Cycle 2011. 10(1):127–134 [DOI] [PubMed] [Google Scholar]
- 18. Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev 2004. 18(15):1862–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gray TA, Hernandez L, Carey AH, et al. The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics 2000. 66(1):76–86 [DOI] [PubMed] [Google Scholar]
- 20. Kim JH, Park SM, Kang MR, et al. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev 2005. 19(7):776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee EW, Lee MS, Camus S, et al. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J 2009. 28(14):2100–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ko A, Lee EW, Yeh JY, et al. MKRN1 induces degradation of West Nile virus capsid protein by functioning as an E3 ligase. J Virol 2010. 84(1):426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gray TA, Wilson A, Fortin PJ, Nicholls RD. The putatively functional Mkrn1-p1 pseudogene is neither expressed nor imprinted, nor does it regulate its source gene in trans. Proc Natl Acad Sci USA 2006. 103(32): 12039–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee SA, Choi SR, Jang JS, et al. Expression of VEGF, EGFR, and IL-6 in gastric adenomas and adenocarcinomas by endoscopic submucosal dissection. Dig Dis Sci 2010. 55(7):1955–1963 [DOI] [PubMed] [Google Scholar]
- 25. Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1999. 1(1):20–26 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998. 92(6):725–734 [DOI] [PubMed] [Google Scholar]
- 27. Guan KL, Jenkins CW, Li Y, et al. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev 1994. 8(24):2939–2952 [DOI] [PubMed] [Google Scholar]
- 28. Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell 1996. 85(1):27–37 [DOI] [PubMed] [Google Scholar]
- 29. Nakamura M, Watanabe T, Klangby U, et al. p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol 2001. 11(2):159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Domínguez G, Carballido J, Silva J, et al. p14ARF promoter hypermethylation in plasma DNA as an indicator of disease recurrence in bladder cancer patients. Clin Cancer Res 2002. 8(4):980–985 [PubMed] [Google Scholar]
- 31. Shintani S, Nakahara Y, Mihara M, Ueyama Y, Matsumura T. Inactivation of the p14(ARF), p15(INK4B) and p16(INK4A) genes is a frequent event in human oral squamous cell carcinomas. Oral Oncol 2001. 37(6):498–504 [DOI] [PubMed] [Google Scholar]
- 32. Randerson-Moor JA, Harland M, Williams S, et al. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet 2001. 10(1):55–62 [DOI] [PubMed] [Google Scholar]
- 33. Chen D, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature 2010. 464(7288):624–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaneuchi M, Yamashita T, Shindoh M, et al. Induction of apoptosis by the p53-273L (Arg –> Leu) mutant in HSC3 cells without transactivation of p21Waf1/Cip1/Sdi1 and bax. Mol Carcinog 1999. 26(1):44–52 [PubMed] [Google Scholar]
- 35. Chen PH, Lin SY, Wang CK, Chen YJ, Chen TC, Chang JG. “Hot spots” mutation analysis of p53 gene in gastrointestinal cancers by amplification of naturally occurring and artificially created restriction sites. Clin Chem 1993. 39(10):2186–2191 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.